Abstract
Histone deacetylase (HDAC) inhibitors such as trichostatin A and valproic acid modulate transcription of many genes by inhibiting the activities of HDACs, resulting in the remodeling of chromatin. Yet this effect is not universal for all genes. Here we show that HDAC inhibitors suppressed the expression of steroidogenic gene CYP11A1 and decreased steroid secretion by increasing the ubiquitination and degradation of SF-1, a factor important for the transcription of all steroidogenic genes. This was accompanied by increased expression of Ube2D1 and SKP1A, an E2 ubiquitin conjugase and a subunit of the E3 ubiquitin ligase in the Skp1/Cul1/F-box protein (SCF) family, respectively. Reducing SKP1A expression with small interfering RNA resulted in recovery of SF-1 levels, demonstrating that the activity of SCF E3 ubiquitin ligase is required for the SF-1 degradation induced by HDAC inhibitors. Overexpression of exogenous SF-1 restored steroidogenic activities even in the presence of HDAC inhibitors. Thus, increased SF-1 degradation is the cause of the reduction in steroidogenesis caused by HDAC inhibitors. The increased SKP1A expression and SCF-mediated protein degradation could be the mechanism underlying the mode of action of HDAC inhibitors.
Histone deacetylase (HDAC) inhibitors like sodium butyrate (NaB), trichostatin A (TSA), valproic acid (VPA), and suberoylanilide hydroxamic acid are potent chemicals that modulate chromatin structure and alter transcription. These molecules inhibit HDAC activities, leading to histone hyperacetylation, change of chromatin structure, and removal of transcriptional corepressors and thus induce transcription of many genes (7, 9). Some of these HDAC inhibitors are currently being developed as anticancer drugs in clinical trials (7), and VPA has been successful in treating epilepsy (20). Despite the therapeutic potential of HDAC inhibitors, their wide effects on transcription, DNA repair, DNA replication, and mitosis result in several side effects, including disruption of the endocrine system and reduction of steroid secretion, which severely limit their usefulness (14, 32). Despite their general property of inducing transcription, HDAC inhibitors also repress the expression of some genes (2, 8, 23, 44). Therefore, the mechanism underlying the action of HDAC inhibitors has become an interesting issue.
Steroids are synthesized by steroidogenic enzymes regulated by steroidogenic factor 1 (SF-1), also known as Ad4BP or NR5A1 (26, 34). SF-1 is a member of the nuclear receptor superfamily that controls the expression of genes involved in steroidogenesis, including those encoding various steroidogenic enzymes (CYP11A1, HSD-3B, CYP21, CYP11, CYP19, and CYP17), peptide hormones (α- and β-subunits of gonadotropins), membrane-bound hormone receptor (MC-2R), and intracellular cholesterol carrier (StAR) (12, 25, 27); these genes are important in the function and development of steroidogenic tissues, including the adrenals and gonads (39).
Steroid receptors are usually activated through the binding of their cognate ligand in the cytoplasm. Although phospholipids were recently proposed to be the ligand for SF-1 based on cocrystallography data (24, 41), the ligand-binding domain of SF-1 can adopt an active conformation independently of any ligand (13), and thus the activation of SF-1 remains a topic of interest. Posttranslational modifications including phosphorylation (15), acetylation (10, 19), and conjugation by small ubiquitin modifier (SUMO) (11, 22, 29) can modulate SF-1 transcriptional activity. Phosphorylation mediated by mitogen-activated protein kinase and acetylation mediated by p300 and GCN5 (general control nonderepressed) enhanced SF-1 function. In contrast, SUMO conjugation represses its function. However, until now little was known about whether SF-1 was also modified by ubiquitination.
Protein ubiquitination is an important posttranslational modification that provides the signal for targeting proteins to the 26S proteasome for degradation. Ubiquitination is usually carried out by three enzymes, which include a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (40). The E3 ligases play an important role in substrate recognition, and their activities serve as a rate-limiting step of ubiquitination.
All known E3 ligases utilize one of two catalytic domains, a RING finger or a HECT domain, to interact with the E2-conjugating enzymes and facilitate ubiquitin chain formation (40). The SKP1/CUL1/F-box protein (SCF) complex is a multisubunit RING finger type E3 ligase that plays an important role in cell cycle regulation through proteolysis of many core components of the cell cycle, like cyclins, E2F1, p21, p27, and MYC proteins (3, 35). SCF E3 ligase consists of four components, including an adaptor protein (SKP1), a RING finger protein (RBX1), a scaffold protein (CUL1), and a variable F-box protein (36). The substrate specificity of SCF ligase depends on the associated F-box protein; thus far approximately 70 F-box proteins in humans have been identified (21, 36).
In this study, we found that HDAC inhibitors promoted the ubiquitination of SF-1 and led to proteasome-mediated SF-1 degradation. We also demonstrated that HDAC inhibitors enhanced the expression of SKP1, a subunit of SCF E3 ligase. RNA interference-mediated knockdown of SKP1 blunted degradation of SF-1 induced by HDAC inhibitors. Thus, our results provide further insight into SF-1 degradation and the mode of action of HDAC inhibitors.
MATERIALS AND METHODS
Plasmids and reagents.
SF1-712 Luc plasmid was a generous gift from J. Milbrandt (43). The mouse Cyp11a1 promoter-luciferase reporter was constructed by PCR amplification of the flanking fragment of the mouse Cyp11a1 gene (−2300 to +1) from mouse tail genomic DNA, followed by subcloning into XhoI and HindIII sites of pGL3-basic (Promega, Madison, WI). For the construction of pFLAG-CMV2 SF-1 and pFLAG-CMV2 PKAc, the coding sequences for mouse SF-1 and protein kinase A (PKA) catalytic subunit were amplified by PCR from pcDNA3.1 SF-1-HA (33) and pCMV-PKAc (Stratagene Inc., La Jolla, CA) and subcloned into the HindIII and XbaI sites of the pFLAG-CMV2 vector (Sigma, St. Louis, MO), respectively. All constructs were confirmed by direct DNA sequencing. Inhibitors of HDAC, TSA, NaB, and VPA, as well as 26S proteasome inhibitor MG132, were obtained from Sigma.
Cell culture and reporter assays.
Mouse Y1 and human NCI-H295 adrenocortical tumor cells were maintained in Dulbecco's modified Eagle medium (DMEM)-F12 medium supplemented with 10% fetal bovine serum. Stable Y1 cell clones 18 and 55 expressing SF-1-hemagglutinin (HA) have been described previously (10). Transient transfection was performed using Lipofectamine Plus (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For reporter assays, luciferase reporter plasmids (1 μg) were transfected into Y1 cells in 60-mm culture dishes. After 24 h, cells were subcultured into 24-well plates with or without 100 ng/ml TSA supplementation for another 24 h. Luciferase activities were determined and normalized to the total protein level.
Immunoblotting and immunoprecipitation.
The following antibodies were obtained commercially: anti-acetyl-histone H3 (Upstate, Lake Placid, NY), antiubiquitin (Serotec, Oxford, United Kingdom), anti-SKP1A (Santa Cruz Biotechnology Inc., Santa Cruz, CA), and anti-acetyl-tubulin and anti-FLAG tag (Sigma). The immune sera against SF-1 (11), CYP11A1 (17), and CYP21 (16) have been described previously. The anti-HSP70 antibody was a kind gift from C. Wang (IMB, Academia Sinica, Taiwan). For direct immunoblotting, cells were harvested and boiled in 1× gel loading buffer. Equal volumes of the whole-cell lysate were separated by 10%, 7.5% (see Fig. 5A), or 15% (see Fig. 6) polyacrylamide gel electrophoresis followed by immunoblotting with the antibodies indicated in the figures. All immunoblots were also probed with anti-HSP70 to ensure equal loading of samples. For immunoprecipitation, expression plasmids for FLAG-tagged SF-1 (FLAG-SF-1) and the FLAG-tagged catalytic subunit of PKA (FLAG-PKAc) were transfected into Y1 cells in 6-cm dishes. Twenty-four hours posttransfection, cells were treated with 100 ng/ml TSA or 2 mM VPA or left without treatment for another 6 h. Whole-cell extracts were prepared in IPH buffer (50 mM Tris-HCl [pH 8.0], 240 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 1× protease inhibitor cocktail; Sigma) and immunoprecipitated with anti-FLAG beads (Sigma). The immunoprecipitates were further processed for immunoblotting using antibodies against ubiquitin, SF-1, and the FLAG tag.
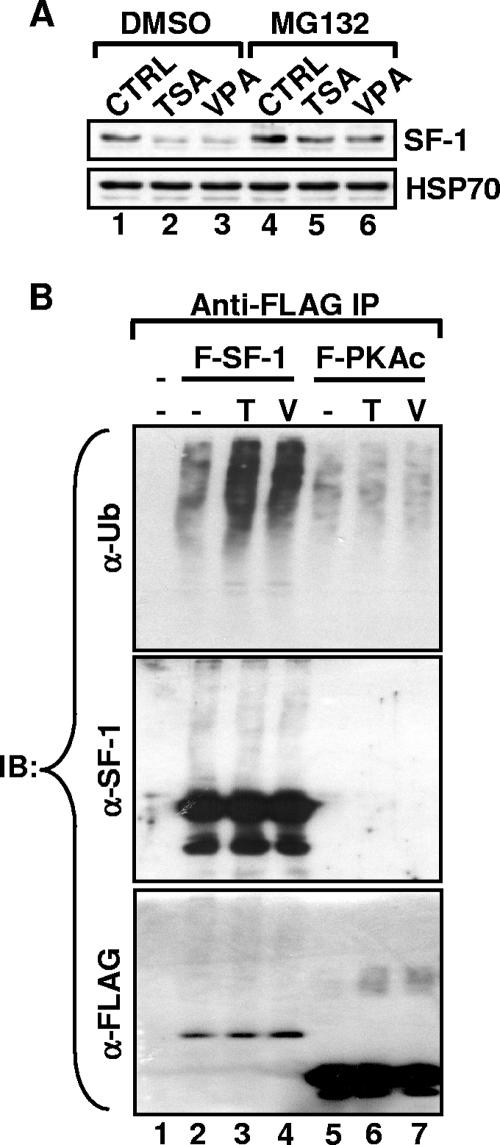
FIG. 5.
TSA and VPA induce polyubiquitination and proteasome-dependent degradation of SF-1. (A) Y1 cells were treated for 12 h without (CTRL) or with 100 ng/ml TSA or 2 mM VPA in the presence of 10 mM MG132 or vehicle control (dimethyl sulfoxide [DMSO]). Levels of SF-1 and HSP70, determined by immunoblotting analysis, are shown. (B) The FLAG-SF-1 and FLAG-PKAc subunits were expressed in Y1 cells. The cells were treated for 12 h with 100 ng/ml TSA (T) or 2 mM VPA (V) in the presence of 10 mM MG132. The anti-FLAG immunoprecipitates were analyzed by immunoblotting (IB) with antibodies against ubiquitin (α-Ub), SF-1 (α-SF-1), or FLAG (α-FLAG).
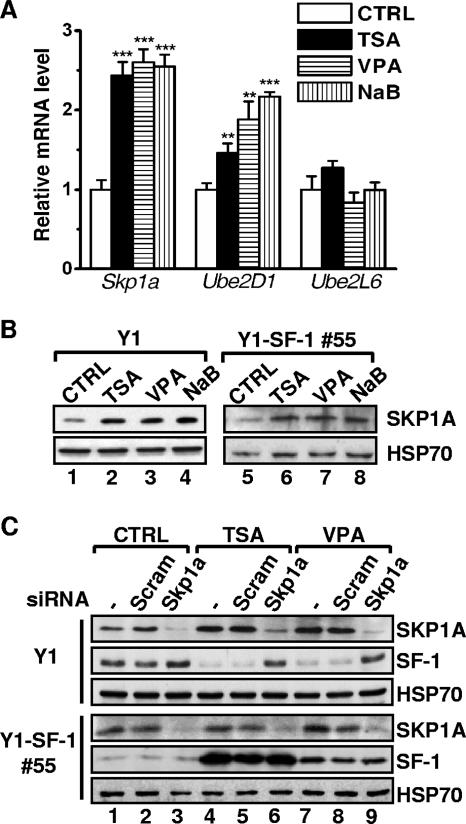
FIG. 6.
Silencing of Skp1a restores the HDAC inhibitor-reduced SF-1 level. (A) The mRNA levels of Skp1a, Ube2D1, and Ube2L6 from Y1 cells treated for 24 h without (CTRL) or with 100 ng/ml TSA, 2 mM VPA, or 2 mM NaB were determined by quantitative real-time PCR. Results were normalized to GAPDH expression and are expressed relative to the values for control cells. **, P < 0.01; ***, P < 0.001 (compared to control cells). (B) SKP1A levels were determined by immunoblotting analysis of whole-cell lysates of Y1 and Y1-SF-1 55 cells that had been treated for 24 h as in panel A. (C) Y1 and Y1-SF-1 55 cells were transfected with scrambled (Scram) or Skp1a-targeted siRNA oligonucleotides three times at 24-h intervals. In the third transfection, the cells were also treated without (CTRL) or with 100 ng/ml TSA or 2 mM VPA. The protein levels of SKP1A, SF-1, and HSP70 were determined by immunoblotting analysis.
Quantitative real-time reverse transcription-PCR (RT-PCR).
Total RNA was isolated from Y1 cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was performed with 1 μg total RNA and 200 U of Superscript II (Invitrogen) with random primers (Promega) for 50 min at 42°C. First-strand cDNA (5 ng) was used as the template together with 250 nM of each primer in a LightCycler quantitative PCR (Roche Diagnostics, Grenzacherstrasse, Switzerland) with QuantiTect SYBR green PCR master mixture (QIAGEN, Valencia, CA) to follow the progress of DNA synthesis. Primers used were GTCCAGTGTCCACCCTTATCC (forward) and GCACGTGAGCAGCCCGTAGTG (reverse) for Sf-1, CTGCCTCCAGACTTCTTTCG (forward) and TTCTTGAAGGGCAGCTTGTT (reverse) for Cyp11a1, CTCCTCCTCCTGAGGATGATG (forward) and GGCCACAGTCTTGCATGTGAC (reverse) for Skp1a, CTGACAGCGCCTATCAAGGTG (forward) and GGGACCTCAGGATGTCAAGAC (reverse) for Ube2D1, and CCCGAGATGATGGCCAGCAAG (forward) and GAAGGCTTTGAGGCCATAGGG (reverse) for Ube2L6.
Protein stability assay.
The expression plasmid for FLAG-SF-1 (1 μg) or FLAG-PKAc was transfected into Y1 cells in six-well plates. Twenty-four hours after transfection, the medium was replaced with methionine-free DMEM, and cells were pulsed for 1 h with [35S]methionine (100 μCi/ml), followed by chase in fresh DMEM-F12 medium with or without 100 ng/ml TSA. Whole-cell extracts were prepared in IPH buffer and immunoprecipitated with anti-FLAG beads. The immunoprecipitates were separated by 10% polyacrylamide gel electrophoresis followed by autoradiography at −70°C for 24 h. Quantitative analysis used Image Gauge, version 3.2, software with a FujiFilm LAS-1000plus image reader. Three independent experiments were performed.
DNA microarray.
DNA microarray analysis was performed through the service provided by the microarray core facility of the Institute of Molecular Biology, Academia Sinica, Taiwan (http://www.imb.sinica.edu.tw/mdarray/). Briefly, total RNA was isolated from control Y1 cells or Y1 cells treated with 100 ng/ml TSA using TRIzol reagent. Fluorescence-labeled cDNA probes (Alexa 555 for control cells; Alexa 647 for TSA-treated cells) were generated from 20 μg DNase-treated RNAs using 400 U of Superscript III (Invitrogen) with oligo(dT) primers (PerkinElmer Inc., Wellesley, MA), hydrolyzed in EDTA-NaOH mixture at 70°C for 15 min, and cleaned up using QIAquick columns (QIAGEN). Fluorescent probes from control and TSA-treated cells were cohybridized to oligonucleotides from a mouse 32K oligonucleotide array (QIAGEN; Array-Ready mouse oligonucleotide set, version 3.0). After extensive washing, the microarrays were scanned for the Alexa 555 and Alexa 647 fluorescent signals using a GenePix 4000B microarray scanner (Molecular Devices Co., Sunnyvale, CA). The images were analyzed using GeneSpring GX software (Agilent Technologies Inc., Santa Clara, CA).
RNA interference.
The nonsilencing control (scrambled) and mouse Skp1a-targeted (accession no. NM_001543 in GenBank) small interfering RNAs (siRNA) were obtained from Dharmacon Inc. (Chicago, IL). The siRNA (100 nM) was transfected into Y1 cells three times at 24-h intervals using Lipofectamine 2000 according to the manufacturer's recommendations. In the third transfection, the cells were treated with the TSA (100 ng/ml) or VPA (2 mM) for another 24 h and harvested in 1× gel loading buffer for immunoblotting analysis.
RESULTS
HDAC inhibitors reduce SF-1 and CYP11A1 levels in adrenocortical cell lines.
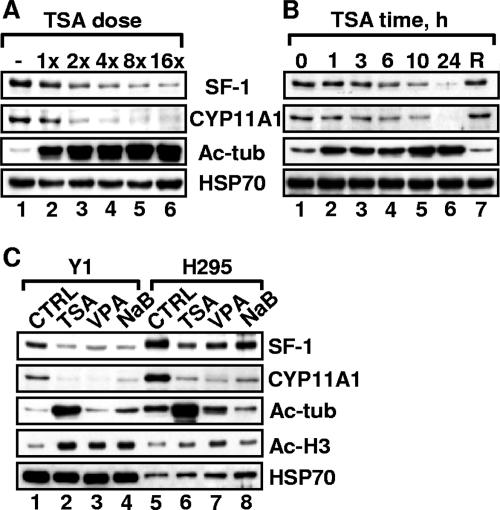
We have shown previously that p300-mediated acetylation of SF-1 correlates with its transcriptional activity (10). To further investigate the regulation of SF-1 by acetylation, we treated mouse adrenocortical Y1 cells with TSA, a potent inhibitor of both class I and class II HDACs. This treatment resulted in a general increase of acetylation, as exemplified by the increased levels of tubulin acetylation (Fig. 1A and B). Unexpectedly, SF-1 protein levels in Y1 cells were reduced by TSA treatment in a dose- and time-dependent manner, while the SF-1 target gene product CYP11A1 was also reduced by TSA treatment. SF-1 protein level was restored by the removal of TSA (Fig. 1B, lane 7), suggesting that this TSA effect is reversible and not due to cell death.
FIG. 1.
HDAC inhibitors reduce SF-1 and CYP11A1 levels in both mouse Y1 and human H295 adrenocortical tumor cell lines. (A and B) TSA reduces SF-1 and CYP11A1 levels in a dose- (A) and time (B)-dependent manner. Y1 cells were treated with 12.5 (1×), 25 (2×), 50 (4×), 100 (8×), and 200 (16×) ng/ml of TSA for 24 h (A) or with 100 ng/ml TSA for the indicated times (B). In the recovery (R) experiment, after a 24-h exposure to TSA, cells were replaced in fresh medium for another 24 h. The levels of SF-1, CYP11A1, acetyl-tubulin (Ac-tub), and HSP70 (loading control), determined by immunoblotting, are shown. (C) Y1 and H295 adrenocortical cell lines were treated for 24 h without (control [CTRL]) or with 100 ng/ml TSA, 2 mM VPA, or 2 mM NaB. The levels of SF-1, CYP11A1, acetyl-tubulin, acetyl-histone H3 (Ac-H3), and HSP70, determined by immunoblotting, are shown.
When Y1 cells were treated with class I-specific HDAC inhibitors NaB and VPA, we also observed increased acetylation of histone H3 as well as decreased levels of SF-1 and CYP11A1. Tubulin acetylation was not increased (Fig. 1C), because the deacetylation of tubulin is mediated by HDAC6, a class II HDAC unaffected by NaB and VPA. Similar effects were also observed in human adrenocortical H295 cells. Thus, the ability to reduce SF-1 and CYP11A1 levels is common for many HDAC inhibitors and probably occurs through inhibiting the activities of class I HDACs.
HDAC inhibitors diminish steroidogenesis through modulating SF-1 level.
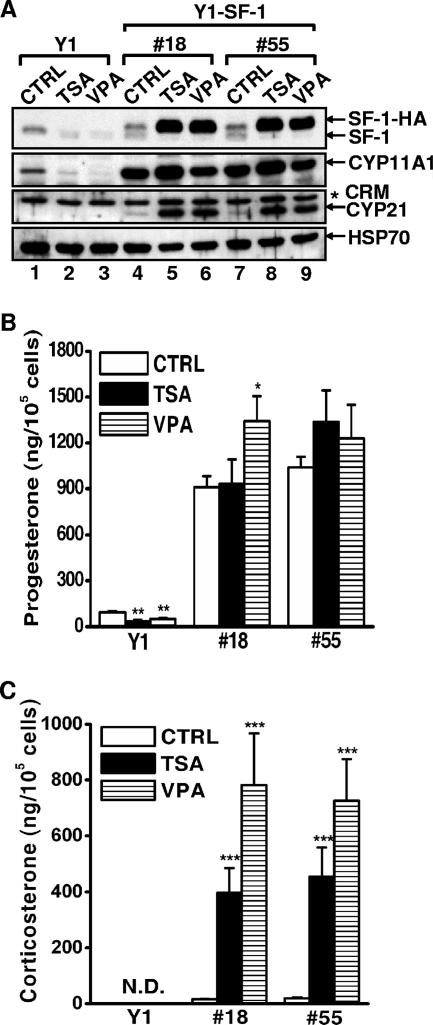
Since CYP11A1 expression is regulated by SF-1, we wished to know whether the reduction of CYP11A1 was a consequence of decreased SF-1 and whether SF-1 overexpression could restore the level of CYP11A1. We generated two stable Y1 clones (18 and 55) that overexpressed SF-1-HA from a cytomegalovirus promoter, which is known to be induced by butyrate and TSA (9). Although not robustly overexpressed, SF-1-HA was dramatically increased in stable clones 18 and 55 upon TSA or VPA treatment (Fig. 2A). The endogenous SF-1, on the other hand, was diminished after treatment. The levels of CYP11A1 in clones 18 and 55 were higher than that in the CTRL cells and were further increased after TSA or VPA treatment. Thus, the decreased expression of CYP11A1 is due to diminished SF-1 and could be restored when SF-1 was supplied exogenously.
FIG. 2.
HDAC inhibitors diminish steroidogenesis through modulating SF-1 level. (A) Normal Y1 cells and stable clones (18 and 55) were treated for 24 h without (CTRL) or with 100 ng/ml TSA or 2 mM VPA. Levels of SF-1, SF-1-HA, CYP11A1, CYP21, and HSP70, as determined by immunoblotting analysis, are shown. The asterisk denotes nonspecific cross-reacting material (CRM). (B and C) The progesterone (B) and corticosterone (C) levels in the medium of panel A were determined by enzyme immunoassay or radioimmunoassay and normalized to the corresponding cell numbers. All values represent the results of at least three separate experiments, with error bars representing standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared to control cells in each Y1 clone). N.D., not detected.
We also examined the level of another SF-1 target gene product, CYP21, which is usually not expressed in Y1 cells. Intriguingly, CYP21 was also expressed in clones 18 and 55, and the expression was further increased after TSA or VPA treatment. This indicates that a large amount of SF-1 can reactivate CYP21 expression.
Since both CYP11A1 and CYP21 are steroidogenic enzymes, we tested whether modulation of gene expression by HDAC inhibitors affected steroid production. Indeed, the progesterone level in Y1 cells was low and was further reduced by HDAC inhibitors (Fig. 2B). The progesterone levels in clones 18 and 55 were high and were not decreased after TSA or VPA treatment. Corticosterone, the reaction product of CYP21, was not detected in normal Y1 cells due to the absence of CYP21; it was produced at detectable levels in stable clones 18 and 55 and accumulated further after TSA or VPA treatment (Fig. 2C). Taken together, these results suggest the HDAC inhibitors reduce steroidogenesis mainly through modulating the level of SF-1.
HDAC inhibitors downregulate the expression of Cyp11a1 but not Sf-1.
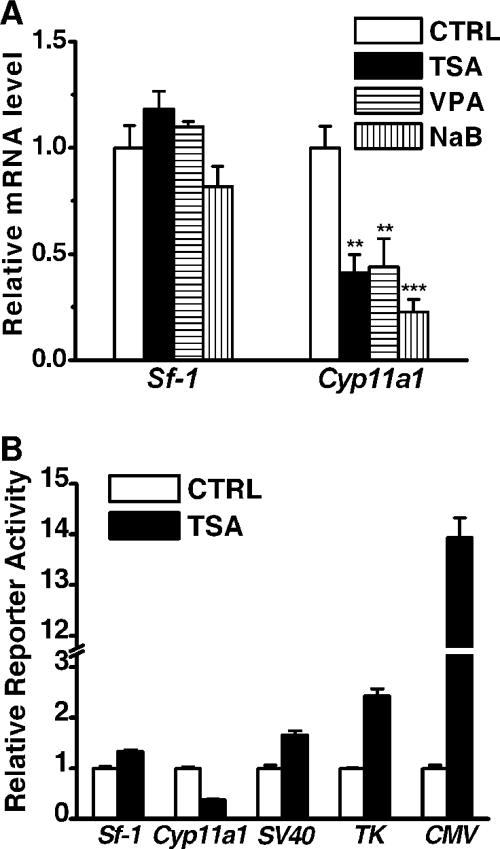
Given that class I HDACs are important regulators of gene expression, we next examined whether the expression of Sf-1 and Cyp11a1 was affected by HDAC inhibitors. As shown in Fig. 3A, Cyp11a1 mRNA levels were significantly reduced upon treatment with HDAC inhibitors, whereas Sf-1 mRNA levels were not affected by such treatments. Thus, the mechanisms of the reduction of SF-1 and CYP11A1 proteins are different.
FIG. 3.
HDAC inhibitors downregulate the expression of Cyp11a1 but not Sf-1. (A) Quantitative real-time RT-PCR analysis of mRNA levels for Cyp11a1 and Sf-1 from Y1 cells treated for 24 h without (CTRL) or with 100 ng/ml TSA, 2 mM VPA, or 2 mM NaB. Results were normalized to GAPDH expression and are expressed as values relative to ethanol-treated Y1 cells. **, P < 0.01; ***, P < 0.001 (compared to the control cells). (B) Expression of luciferase reporters driven by promoters of Cyp11a1 (2.3 kb), Sf-1 (0.7 kb), SV40, TK, and CMV in Y1 cells. After treatment without (CTRL) or with 100 ng/ml TSA for 24 h, luciferase activities were determined and normalized to total protein levels of each transfectant and are expressed as activity relative to the value for the control. All values show the results from at least three separate experiments, with error bars representing standard deviations.
Since SF-1 is a major transcription factor for Cyp11a1 expression, it is reasonable to suggest that reduced SF-1 levels will lead to decreased Cyp11a1 transcription, thus leading to decreased mRNA levels. To confirm this, a reporter assay was performed to analyze promoter strengths (Fig. 3B). Expression of a reporter gene driven by a 2.3-kb Cyp11a1 promoter was significantly repressed by TSA treatment. While the promoter activities of Sf-1, SV40, and TK were slightly induced by TSA treatment, the reporter driven by CMV was dramatically enhanced (∼14 times greater). These results indicate that the effects of HDAC inhibitors in modulating gene expression are promoter dependent.
Induction of proteasome-mediated degradation of SF-1.
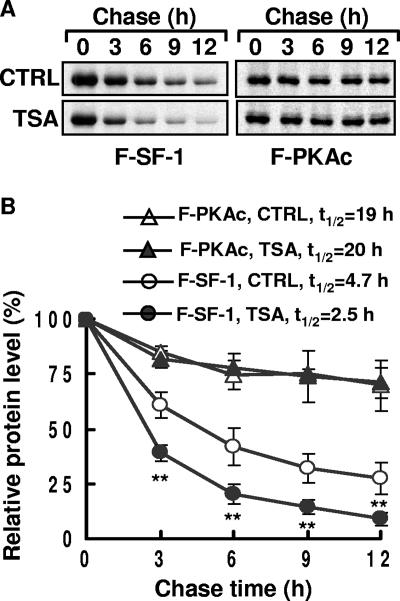
As described above, HDAC inhibitors reduce SF-1 protein levels but not its mRNA levels, indicating that these chemicals probably affect SF-1 protein stability. A pulse-chase protocol was employed to compare the stabilities of SF-1 in the presence and absence of TSA treatment. As shown in Fig. 4A, less SF-1 was detected in the presence of TSA. The relative half-life of SF-1 was about 4.7 h in the absence of TSA, but it was reduced to about 2.5 h in the presence of TSA (Fig. 4B). The stability of an unrelated protein, FLAG-PKAc, however, was not affected by TSA. Thus, TSA decreases SF-1 stability.
FIG. 4.
TSA increases SF-1 turnover. (A) Levels of FLAG-SF-1 (F-SF-1) and FLAG-PKAc (F-PKAc) at different time points in a pulse-chase experiment. The expression plasmid for F-SF-1 or F-PKAc was transfected into Y1 cells. After 24 h, cells were pulse-labeled with [35S]methionine for 1 h and chased in the absence (CTRL) or presence of 100 ng/ml TSA for the indicated times. 35S-F-SF-1 and 35S-F-PKAc were immunoprecipitated by anti-FLAG, separated by gel electrophoresis, and detected by autoradiography. (B) The relative amounts of F-SF-1 and F-PKAc in cells treated without (CTRL) or with 100 ng/ml TSA are shown. The graphs show the quantitative results of three independent experiments. t1/2, half-life of SF-1. **, P < 0.01 compared to the control cells.
To further investigate the mechanism of SF-1 degradation, the inhibitor for 26S proteasome MG132 was applied in combination with TSA or VPA. Treatment of Y1 cells with MG132 resulted in increased SF-1 in the control cells and also maintained reasonable levels of SF-1 even after TSA or VPA treatment (Fig. 5A). This indicates that SF-1 degradation is eased by MG132 treatment; thus, TSA and VPA decreased SF-1 levels, possibly through proteasome-mediated degradation.
As the most common mechanism of targeting proteins for 26S proteasome-mediated degradation depends on polyubiquitination, we examined HDAC inhibitor-induced ubiquitination of SF-1. FLAG-SF-1 was expressed in Y1 cells in the presence or absence of HDAC inhibitors. FLAG-SF-1 was precipitated by anti-FLAG antibody in the presence of MG132 to prevent its degradation and was analyzed by immunoblotting against ubiquitin, SF-1, or FLAG (Fig. 5B). In the absence of HDAC inhibitors, only a small amount of ubiquitinated SF-1 was observed (Fig. 5B, lane 2). In the presence of TSA or VPA the amounts of ubiquitinated SF-1 increased significantly, although the total levels of FLAG-SF-1 detected by anti-SF-1 or anti-FLAG antibodies were not much different. This degradation appears to be specific, as the total amounts and the levels of ubiquitin conjugation of an irrelevant control protein, FLAG-PKAc, were not affected. Thus, increased proteasomal degradation through polyubiquitination appears to be the most likely cause for the HDAC inhibitor-induced degradation of SF-1.
HDAC inhibitor-induced degradation of SF-1 is mediated by an SCF ligase complex.
To identify genes involved in polyubiquitination of SF-1 in response to HDAC inhibitors, a microarray analysis was employed to search for TSA-modulated genes in Y1 cells. While 471 genes were downregulated by TSA, 268 genes were upregulated (see http://www.ebi.ac.uk/arrayexpress/, accession numbers E-MEXP-1197 and A-MEXP-840). Special attention was given to genes involved in the ubiquitination pathway that were upregulated by TSA. Among them, the S-phase kinase-associated protein 1A gene (Skp1a), which encodes a subunit of the SCF ubiquitin E3 ligase complex, was selected for further study because of its role in protein degradation and its apparent upregulation by TSA. The effect of HDAC inhibitors on Skp1a expression was confirmed by real-time RT-PCR analysis: Skp1a expression increased after Y1 cells were treated with HDAC inhibitors (Fig. 6A). Expression of Ube2D1, which encodes an E2-conjugating enzyme in the SCF complex, also increased. The mRNA levels of another E2-conjugating enzyme gene, Ube2L6, which were increased by both TSA and VPA in murine F9 cells and human HeLa cells (23), were not changed by these treatments in Y1 cells. As with its mRNA, levels of SKP1A protein were also higher after treatment with HDAC inhibitors in both Y1 and Y1-SF-1 55 cells (Fig. 6B).
Since SKP1A and its associated SCF ligase complex are part of the protein degradation machinery, we tested whether the SCF complex mediated HDAC inhibitor-induced SF-1 degradation by removing SKP1A with an siRNA. Although SKP1A accumulated after treatment with TSA or VPA, it was efficiently eliminated by siRNA against Skp1a (Fig. 6C). In untreated Y1 cells, transfection of siRNA against Skp1a but not a scrambled sequence induced accumulation of SF-1 protein by 1.5-fold. In TSA- and VPA-treated cells, SF-1 level was significantly reduced, but it was restored after transfection with siRNA against Skp1a. Scrambled siRNA had no effect. Thus, the induction of Skp1a by HDAC inhibitors is a likely cause of the degradation of SF-1.
We also tested the relation of SF-1 and SKP1A1 in Y1-SF-1 55 cells that overexpress SF-1 (Fig. 6C). Although SKP1A level was induced by TSA or VPA, SF-1 in these cells was so strongly overexpressed that its level was not significantly affected by TSA or VPA. Similarly, reduction of SKP1A by its siRNA did not affect the level of SF-1 greatly in this overexpressed cell line. Thus, huge overexpression of SF-1 can offset its increased degradation, leading to accumulation of SF-1. This result is consistent with the finding in Fig. 2A.
DISCUSSION
SF-1 is an essential transcription factor controlling the function of steroidogenic tissues. In this report, we demonstrate that HDAC inhibitors reduced SF-1 stability by inducing its polyubiquitination for 26S proteasome-mediated degradation. Reduced SF-1 levels consequently resulted in decreased expression of SF-1 target genes and diminished steroid production. We also demonstrate that HDAC inhibitors induced the expression of the SCF E3 ligase subunit, SKP1A, and its associated E2 conjugase, Ube2D1. RNA interference-mediated gene silencing of Skp1a abolished HDAC inhibitor-induced degradation of SF-1. These results reveal a novel action of HDAC inhibitors: promotion of SF-1 degradation through SCF-mediated ubiquitination, consequently leading to reduced steroidogenesis.
Degradation of SF-1 through the ubiquitin-proteasome pathway.
Unlike what is found for most proteins, the cell cannot tolerate moderate changes in the amount of SF-1. Heterozygous SF-1+/− mice suffer from decreased adrenocortical volume and impaired corticosterone synthesis in response to stress (5, 6). In humans, two patients with adrenal insufficiency due to SF-1 haploid insufficiency have been described (1, 4). Thus, the regulation of SF-1 quantity appears to be very important, yet very little information is known about it.
SF-1 activity can be regulated by posttranslational modifications, such as phosphorylation (15), acetylation (10, 19), and SUMO conjugation (11, 22, 29), but these modifications did not affect SF-1 levels. Our current results suggest that SF-1 level is regulated by the ubiquitin-proteasome pathway, similar to the degradation of other steroid receptors like estrogen receptor (31), progesterone receptor (PR) (28), and glucocorticoid receptor (GR) (42).
We found that the half-life of SF-1 protein is approximately 5 h, which is short compared to the 18-h and 21-h half-lives of unliganded GR and PR, respectively. Upon treatment with cognate ligands, the half-life of GR fell to 9 h (42) and that of PR fell to 6 h (37), which are similar to the half-life of SF-1. Since transcriptionally active forms of steroid receptors are substrates for the ubiquitin-proteasome pathway (38), it is possible that the short half-life of SF-1 indicates that it is constitutively active. Indeed the ligand-binding domain of SF-1 can adopt an active transformation (13), and the proposed ligand for SF-1, phospholipid, is abundant in cells (24, 30, 41). However, mutations of SF-1 at sites of phospholipid interaction, sumoylation, acetylation, and phosphorylation did not change SF-1 levels (10, 11, 15, 24). It will be of interest to identify the motif that serves as the substrate for the ubiquitin-proteasome pathway.
We report here HDAC inhibitors destabilize SF-1 by increasing its degradation through the ubiquitin-proteasome pathway in adrenal tumor cells. In contrast to our result, Jacob and colleagues showed that TSA increased the half-life of SF-1 in transfected COS-1 cells (19). It is possible that the difference in stabilities seen here is a consequence of using different cell lines to determine the half-life of SF-1. In steroidogenic cells, our evidence showed that HDAC inhibitors indeed destabilized SF-1.
HDAC inhibitors induce SCF E3 ubiquitin-mediated protein degradation.
HDAC inhibitors have emerged as anticancer drugs because of their potential to kill transformed cells (7, 18). These chemicals exert their anticancer activities by blocking the catalytic activities of HDACs and consequently modulating the transcription of a subset of genes. However, other mechanisms have been reported. TSA promotes ubiquitin-proteasome-mediated destruction of cyclin D1 through the activity of the SCF-SKP2 E3 ligase complex in breast cancer MCF-7 cells (2). VPA induces degradation of HDAC2 by increasing the expression of Ubc8 E2 ubiquitin conjugase (23) and of the B56 regulatory subunit of protein phosphatase 2A, which triggers the degradation of p300 through dephosphorylation-dependent ubiquitination (8). Our results show that HDAC inhibitors TSA, VPA, and butyrate induced the expression of SKP1A and Ube2D1 and thus likely increase SCF E3 ubiquitin ligase-mediated ubiquitination. It has been proposed that SCF E3 ubiquitin ligase plays an important role in the proteolysis of core components that control cell cycles at G1/S and G2/M transitions (36). Interestingly, most of HDAC inhibitors also induce cell cycle arrest at G1/S and G2/M transitions (7). Therefore, activation of SCF E3 ligase activity might be a mechanism of the cell cycle dysregulation seen with HDAC inhibitors.
Long-term treatment with HDAC inhibitors causes decreased steroidogenesis.
We have shown here that HDAC inhibitors resulted in a decrease of steroid hormone secretion in Y1 cells. This result is consistent with the report that VPA caused a decrease in progesterone secretion in porcine follicular cells, even though we have not tested the effect of HDAC inhibitors in vivo (14). HDAC inhibitors decreased steroidogenesis by increasing the degradation of SF-1. This effect seemed to be contradictory to another effect of HDAC inhibitors, namely, the possible increase of SF-1 activity through enhancing its acetylation (10). In fact these two effects occur on different time scales. The SF-1 acetylation reaction can be accomplished in 30 min; thus, this stimulating effect is fast and short term. The effect of HDAC inhibitors on SF-1 degradation is secondary, involving the activation of other genes in the ubiquitin conjugation system, so this effect will gradually appear only after long-term exposure. Thus, the effects of HDAC inhibitors on SF-1 activity may be biphasic: they would increase SF-1 activity in the short term but decrease SF-1 amount on chronic exposure. Eventually steroidogenesis would be reduced after long-term exposure of cells to HDAC inhibitors.
Acknowledgments
We thank Shu-Yun Tung at the Microarray Core Facility of Institute of Molecular Biology, Academia Sinica, for her excellent technical support.
This work was funded by grant NSC95-2311-B-001-018 from the National Science Council and from Academia Sinica, Republic of China.
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Achermann, J. C., M. Ito, M. Ito, P. C. Hindmarsh, and J. L. Jameson. 1999. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat. Genet. 22:125-126. [DOI] [PubMed] [Google Scholar]
- 2.Alao, J. P., E. W. Lam, S. Ali, L. Buluwela, W. Bordogna, P. Lockey, R. Varshochi, A. V. Stavropoulou, R. C. Coombes, and D. M. Vigushin. 2004. Histone deacetylase inhibitor trichostatin A represses estrogen receptor alpha-dependent transcription and promotes proteasomal degradation of cyclin D1 in human breast carcinoma cell lines. Clin. Cancer Res. 10:8094-8104. [DOI] [PubMed] [Google Scholar]
- 3.Ang, X. L., and J. W. Harper. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24:2860-2870. [DOI] [PubMed] [Google Scholar]
- 4.Biason-Lauber, A., and E. J. Schoenle. 2000. Apparently normal ovarian differentiation in a prepubertal girl with transcriptionally inactive steroidogenic factor 1 (NR5A1/SF-1) and adrenocortical insufficiency. Am. J. Hum. Genet. 67:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bland, M. L., R. C. Fowkes, and H. A. Ingraham. 2004. Differential requirement for steroidogenic factor-1 gene dosage in adrenal development versus endocrine function. Mol. Endocrinol. 18:941-952. [DOI] [PubMed] [Google Scholar]
- 6.Bland, M. L., C. A. Jamieson, S. F. Akana, S. R. Bornstein, G. Eisenhofer, M. F. Dallman, and H. A. Ingraham. 2000. Haploinsufficiency of steroidogenic factor-1 in mice disrupts adrenal development leading to an impaired stress response. Proc. Natl. Acad. Sci. USA 97:14488-14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolden, J. E., M. J. Peart, and R. W. Johnstone. 2006. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5:769-784. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., J. R. St-Germain, and Q. Li. 2005. B56 regulatory subunit of protein phosphatase 2A mediates valproic acid-induced p300 degradation. Mol. Cell. Biol. 25:525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W. Y., E. C. Bailey, S. L. McCune, J. Y. Dong, and T. M. Townes. 1997. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl. Acad. Sci. USA 94:5798-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, W. Y., L. J. Juan, and B. C. Chung. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol. Cell. Biol. 25:10442-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, W. Y., W. C. Lee, N. C. Hsu, F. Huang, and B. C. Chung. 2004. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279:38730-38735. [DOI] [PubMed] [Google Scholar]
- 12.Clark, B. J., S. C. Soo, K. M. Caron, Y. Ikeda, K. L. Parker, and D. M. Stocco. 1995. Hormonal and developmental regulation of the steroidogenic acute regulatory protein. Mol. Endocrinol. 9:1346-1355. [DOI] [PubMed] [Google Scholar]
- 13.Desclozeaux, M., I. N. Krylova, F. Horn, R. J. Fletterick, and H. A. Ingraham. 2002. Phosphorylation and intramolecular stabilization of the ligand binding domain in the nuclear receptor steroidogenic factor 1. Mol. Cell. Biol. 22:7193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregoraszczuk, E., A. K. Wojtowicz, E. Tauboll, and E. Ropstad. 2000. Valproate-induced alterations in testosterone, estradiol and progesterone secretion from porcine follicular cells isolated from small- and medium-sized ovarian follicles. Seizure 9:480-485. [DOI] [PubMed] [Google Scholar]
- 15.Hammer, G. D., I. Krylova, Y. Zhang, B. D. Darimont, K. Simpson, N. L. Weigel, and H. A. Ingraham. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell 3:521-526. [DOI] [PubMed] [Google Scholar]
- 16.Hu, M. C., and B. C. Chung. 1990. Expression of human 21-hydroxylase (P450c21) in bacterial and mammalian cells: a system to characterize normal and mutant enzymes. Mol. Endocrinol. 4:893-898. [DOI] [PubMed] [Google Scholar]
- 17.Hu, M. C., I. C. Guo, J. H. Lin, and B. C. Chung. 1991. Regulated expression of cytochrome P-450scc (cholesterol-side-chain cleavage enzyme) in cultured cell lines detected by antibody against bacterially expressed human protein. Biochem. J. 274:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Insinga, A., S. Minucci, and P. G. Pelicci. 2005. Mechanisms of selective anticancer action of histone deacetylase inhibitors. Cell Cycle 4:741-743. [DOI] [PubMed] [Google Scholar]
- 19.Jacob, A. L., J. Lund, P. Martinez, and L. Hedin. 2001. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J. Biol. Chem. 276:37659-37664. [DOI] [PubMed] [Google Scholar]
- 20.Johannessen, C. U., and S. I. Johannessen. 2003. Valproate: past, present, and future. CNS Drug Rev. 9:199-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kipreos, E. T., and M. Pagano. 2000. The F-box protein family. Genome Biol. 1:REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu, T., H. Mizusaki, T. Mukai, H. Ogawa, D. Baba, M. Shirakawa, S. Hatakeyama, K. I. Nakayama, H. Yamamoto, A. Kikuchi, and K. Morohashi. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18:2451-2462. [DOI] [PubMed] [Google Scholar]
- 23.Kramer, O. H., P. Zhu, H. P. Ostendorff, M. Golebiewski, J. Tiefenbach, M. A. Peters, B. Brill, B. Groner, I. Bach, T. Heinzel, and M. Gottlicher. 2003. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22:3411-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343-355. [DOI] [PubMed] [Google Scholar]
- 25.Lala, D. S., Y. Ikeda, X. Luo, L. A. Baity, J. C. Meade, and K. L. Parker. 1995. A cell-specific nuclear receptor regulates the steroid hydroxylases. Steroids 60:10-14. [DOI] [PubMed] [Google Scholar]
- 26.Lala, D. S., D. A. Rice, and K. L. Parker. 1992. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 6:1249-1258. [DOI] [PubMed] [Google Scholar]
- 27.Lan, H. C., H. J. Li, G. Lin, P. Y. Lai, and B. C. Chung. 2007. Cyclic AMP stimulates SF-1-dependent CYP11A1 expression through homeodomain-interacting protein kinase 3-mediated Jun N-terminal kinase and c-Jun phosphorylation. Mol. Cell. Biol. 27:2027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange, C. A., T. Shen, and K. B. Horwitz. 2000. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. USA 97:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, M. B., L. A. Lebedeva, M. Suzawa, S. A. Wadekar, M. Desclozeaux, and H. A. Ingraham. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 25:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., M. Choi, G. Cavey, J. Daugherty, K. Suino, A. Kovach, N. C. Bingham, S. A. Kliewer, and H. E. Xu. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17:491-502. [DOI] [PubMed] [Google Scholar]
- 31.Masuyama, H., and Y. Hiramatsu. 2004. Involvement of suppressor for Gal 1 in the ubiquitin/proteasome-mediated degradation of estrogen receptors. J. Biol. Chem. 279:12020-12026. [DOI] [PubMed] [Google Scholar]
- 32.Mikkonen, K., L. K. Vainionpaa, A. J. Pakarinen, M. Knip, I. Y. Jarvela, J. S. Tapanainen, and J. I. Isojarvi. 2004. Long-term reproductive endocrine health in young women with epilepsy during puberty. Neurology 62:445-450. [DOI] [PubMed] [Google Scholar]
- 33.Monte, D., F. DeWitte, and D. W. Hum. 1998. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem. 273:4585-4591. [DOI] [PubMed] [Google Scholar]
- 34.Morohashi, K., S. Honda, Y. Inomata, H. Handa, and T. Omura. 1992. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J. Biol. Chem. 267:17913-17919. [PubMed] [Google Scholar]
- 35.Nakayama, K. I., and K. Nakayama. 2005. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin. Cell Dev. Biol. 16:323-333. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama, K. I., and K. Nakayama. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6:369-381. [DOI] [PubMed] [Google Scholar]
- 37.Nardulli, A. M., and B. S. Katzenellenbogen. 1988. Progesterone receptor regulation in T47D human breast cancer cells: analysis by density labeling of progesterone receptor synthesis and degradation and their modulation by progestin. Endocrinology 122:1532-1540. [DOI] [PubMed] [Google Scholar]
- 38.Nawaz, Z., and B. W. O'Malley. 2004. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol. Endocrinol 18:493-499. [DOI] [PubMed] [Google Scholar]
- 39.Parker, K. L., and B. P. Schimmer. 1997. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 18:361-377. [DOI] [PubMed] [Google Scholar]
- 40.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 41.Wang, W., C. Zhang, A. Marimuthu, H. I. Krupka, M. Tabrizizad, R. Shelloe, U. Mehra, K. Eng, H. Nguyen, C. Settachatgul, B. Powell, M. V. Milburn, and B. L. West. 2005. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc. Natl. Acad. Sci. USA 102:7505-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster, J. C., C. M. Jewell, J. E. Bodwell, A. Munck, M. Sar, and J. A. Cidlowski. 1997. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J. Biol. Chem. 272:9287-9293. [DOI] [PubMed] [Google Scholar]
- 43.Woodson, K. G., P. A. Crawford, Y. Sadovsky, and J. Milbrandt. 1997. Characterization of the promoter of SF-1, an orphan nuclear receptor required for adrenal and gonadal development. Mol. Endocrinol. 11:117-126. [DOI] [PubMed] [Google Scholar]
- 44.Xiong, Y., S. C. Dowdy, K. C. Podratz, F. Jin, J. R. Attewell, N. L. Eberhardt, and S. W. Jiang. 2005. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res. 65:2684-2689. [DOI] [PubMed] [Google Scholar]