Abstract
The NF-E2 p45-related factor 2 (NRF2) and the aryl hydrocarbon receptor (AHR) are transcription factors controlling pathways modulating xenobiotic metabolism. AHR has recently been shown to affect Nrf2 expression. Conversely, this study demonstrates that NRF2 regulates expression of Ahr and subsequently modulates several downstream events of the AHR signaling cascade, including (i) transcriptional control of the xenobiotic metabolism genes Cyp1a1 and Cyp1b1 and (ii) inhibition of adipogenesis in mouse embryonic fibroblasts (MEFs). Constitutive expression of AHR was affected by Nrf2 genotype. Moreover, a pharmacological activator of NRF2 signaling, CDDO-IM {1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole}, induced Ahr, Cyp1a1, and Cyp1b1 transcription in Nrf2+/+ MEFs but not in Nrf2−/− MEFs. Reporter analysis and chromatin immunoprecipitation assay revealed that NRF2 directly binds to one antioxidant response element (ARE) found in the −230-bp region of the promoter of Ahr. Since AHR negatively controls adipocyte differentiation, we postulated that NRF2 would inhibit adipogenesis through the interaction with the AHR pathway. Nrf2−/− MEFs showed markedly accelerated adipogenesis upon stimulation, while Keap1−/− MEFs (which exhibit higher NRF2 signaling) differentiated slowly compared to their congenic wild-type MEFs. Ectopic expression of Ahr and dominant-positive Nrf2 in Nrf2−/− MEFs also substantially delayed differentiation. Thus, NRF2 directly modulates AHR signaling, highlighting bidirectional interactions of these pathways.
NRF2 is a cap'n'collar (CNC) basic leucine zipper (bZIP) transcription factor and an important regulator of the transcription of cytoprotective enzymes and antioxidant genes, such as glutathione S-transferases (GSTs) and NAD(P)H (quinone acceptor) oxidoreductase 1 (NQO1), through binding to antioxidant response elements (AREs) found in their promoters (20, 28). The NRF2 pathway can be modulated in a dynamic way by multiple pharmacological agents {NRF2 activators, including 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole (CDDO-Im)} through interactions with KEAP1, a cytoplasmic binding partner of NRF2 (12, 50, 51). Studies using Nrf2 and Keap1 knockout mice have provided key insights into the importance of this pathway in protecting cells from exposure to environmental toxins, inflammatory stresses, neurodegeneration, and carcinogenesis (18). Although more resistant to toxins (32), Keap1 null mice, which have higher constitutive levels of NRF2, showed hyperkeratosis, which implies that the pathway may also be involved in keratinocyte differentiation (46).
Information generated from analyses of genetic networks and large-scale two-hybrid screens has indicated that to understand the complexity of a phenotype or disease, one needs to investigate the interplay of signals and transcription factors that leads to the expression of downstream genes (3, 24). Although much is known about the role of the NRF2 pathway itself, little is known about its interaction with other signaling pathways. Recent evidence suggests that cross talk may exist between aryl hydrocarbon receptor (AHR) and NRF2 pathways. AHR is a ligand-activated transcription factor that belongs to the basic helix-loop-helix/PER-aryl hydrocarbon receptor nuclear translocator (ARNT)-SIM family. When activated by a ligand, AHR translocates to the nucleus and dimerizes with ARNT, another basic helix-loop-helix protein. AHR is activated when bound with ligand, usually a planar polyaromatic chemical. Activated AHR then binds to xenobiotic response elements (XREs) in the promoters of various cytochrome P450s (CYPs) (1, 15), resulting in increased expression of these enzymes. Because several AHR ligands are suspected carcinogens, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and benzo[a]pyrene, the majority of studies on AHR have focused on its role in mediating the toxicity of these chemicals. However, studies utilizing Ahr knockout mice and cell lines revealed that AHR signaling may be involved also in physiological processes, such as growth and differentiation, in a manner that is independent of exogenous ligand (2, 34). Moreover, it has been reported that the AHR pathway is involved in maintaining a balance between promoting and preventing oxidative stress as well as in preventing toxic redox cycles of catechol estrogens (21, 29). AHR also regulates gene expression involved in cytoskeletal organization, bioenergetics, and cell proliferation (9). Several studies suggest that one of the physiological roles of the AHR is the negative regulation of adipocyte differentiation. Shimba et al. (41) have demonstrated that TCDD treatment suppresses the conversion of 3T3-L1 cells into adipocytes. Using MEF cell lines derived from Ahr knockout mice, Alexander et al. (2) reported that the AHR is a constitutive inhibitor of triglyceride synthesis and an early regulator of adipocyte differentiation. In addition, one of the phenotypes of Ahr null mice is transient fatty liver (39), implying an in vivo regulatory role of AHR in the adipogenic process.
Studies on cross talk between AHR and NRF2 have focused on their roles in controlling expression of xenobiotic-metabolizing enzymes. For example, it has been reported that the inducible expression of NQO1 by TCDD depends on both AHR and NRF2 (25). Miao et al. (27) demonstrated that Nrf2 gene transcription is directly modulated by AHR activation. The present study demonstrates that signaling in the opposite direction also occurs, namely, that transcription of the Ahr gene is directly affected by NRF2. Our data clearly demonstrate that expression of Ahr, Cyp1a1, and Cyp1b1 is partially dependent on NRF2, implying that NRF2 modulates both transcription of Ahr and its downstream targets. In addition, our results indicate that the NRF2 pathway inhibits adipogenesis via activation of the AHR signaling cascade. Thus, the NRF2 pathway has a broader reach than heretofore described, i.e., indirect regulation of the expression of CYPs, inhibition of adipogenesis, and possibly other AHR-dependent processes.
MATERIALS AND METHODS
Cell culture.
Immortalized mouse embryonic fibroblasts (MEFs) were established from the embryos of C57BL/6J Nrf2−/− or Nrf2+/+ littermates. Primary MEFs were established from the embryos of Keap1−/− or Keap1+/+ littermates. The cells were cultured in Iscove's minimal essential medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and incubated at 37°C in a humidified atmosphere of 5% CO2.
Plasmids.
The mouse Ahr gene regulatory region was isolated from C57BL/6J mouse liver genomic DNA by PCR. The region was directly cloned between KpnI and NcoI sites in pGL3-Basic (Promega) and confirmed by sequencing analysis. In this construct, ATG of AHR was fused to that of luciferase. Serial promoter deletion fragments were prepared by the S1 exonuclease III nuclease reaction method followed by NcoI digestion. Then, the fragments were ligated to the site between KpnI and NcoI sites of pGL3 basic. The mutant ARE reporter was produced by ligating oligonucleotide containing the mutated ARE (GGTACCCACTACGTCCTCCGTCCAACCGTGCTGCGAAGAGGGTGGGGCC) to the site between KpnI and ApaI of p-967 construct. All constructs were confirmed by sequencing. Because pRLTK (Promega) bears an ARE in the thymidine kinase (TK) promoter region, this ARE was deleted in order to use this vector for normalizing transfection efficiency. This improved normalizing vector, pRLTK-ΔARE, was constructed by deleting the SmaI and PvuII fragment from pRLTK. Constructs used in transfection experiments were purified with QIAGEN plasmid kits. For establishment of stable transfectants, cDNAs for murine Ahr and Nrf2ΔNeh2 were inserted into pTracer-EF/Bsd (Invitrogen). Ahr cDNA was isolated from pSportAHR (ATCC 63125). Nrf2ΔNeh2 cDNA (17) was isolated from pcDNA3-Nrf2ΔNeh2 (generously provided by Ken Itoh, Hirosaki University).
Transient transfection and measurement of luciferase activity.
Cells were transfected at 60 to 70% confluence with GeneJuice transfection reagent (Novagen). Briefly, cells were seeded in 24-well plates at a density of 1 × 104 cells/well. Cells were grown overnight; the transfection complex containing 0.45 μg of plasmid DNA, 0.05 μg of the pRLTK-ΔARE plasmid, and transfection reagent was added to each well, and cells were incubated for 15 h. Cells were then incubated for another 24 h with or without drug treatment. Renilla and firefly luciferase activities in cell lysates were measured with the Dual Luciferase assay kit (Promega) with a luminometer (Turner Designs). For forced expression studies, pcDNA3-Nrf2ΔNeh2 was cotransfected with promoter plasmids.
Isolation and purification of total RNA and RT-PCR.
Cells were seeded at 60% confluence the day before treatment with vehicle or CDDO-Im. Total RNA was purified using the Versagene RNA purification system (Gentra Systems). Genes were analyzed by SYBR green real-time quantitative reverse transcription-PCR (RT-PCR). cDNA was synthesized using the iScript system (Bio-Rad). Real-time PCR was performed on a Bio-Rad My-IQ real-time PCR instrument using Applied Biosystems SYBR green PCR master mix in 20-μl reaction mixture volumes. The PCR efficiency was determined from a standard curve and used in the Pfaffl method for calculation of relative quantification (33). Tata-binding protein (Tbp) was used as a normalizing control. Primers are shown in Table S1 in the supplemental material.
Chromatin immunoprecipitation (ChIP) assay.
Formaldehyde cross-linking and chromatin fragmentation were carried out as described previously (22). Diluted chromatin solution was incubated with an anti-NRF2 antibody or nonspecific immunoglobulin G (Santa Cruz Biotechnology) for 18 h at 4°C with rotation. After the DNA was washed and eluted, precipitated DNA was resuspended with 60 μl of water, and 2 μl of DNA was used for PCR amplification with the following primers for Ahr ARE, 5′-TTTTGAGGCTGGAAAACAGGTACT-3′ and 5′-ACGTGATGACGCAGGACGTA-3′. The primer sequences for promoters of β-actin and Gsta1 ARE have been described previously (22).
Design and transfection of Ahr-targeted specific siRNA.
Small interfering RNA (siRNA) duplexes were prepared by Ambion. Targeted coding regions of the Ahr oligonucleotide sequences were as follows: iAhr (5′-AAGACTGGAGAAAGTGGCATG-3′) (13) and iAhr2 (5′-AACGAGGAGTTCTTCAGAACT-3′) (predesigned by Ambion). A National Center for Biotechnology Information standard nucleotide-nucleotide BLAST program was used to verify that this sequence did not match that of any other mouse gene. Quantitative RT-PCR results confirmed that transfection with iAhr reduced mRNA levels of Ahr more efficiently than transfection with iAhr2 did; therefore, iAhr was used for subsequent experiments. The negative-control siRNA (AM4611) used in this study was a nonsilencing siRNA designed by Ambion. MEFs were seeded in a six-well plate and transfected in the presence of 30 nM of either siRNA or negative-control RNA in a final volume of 1 ml OPTI-MEM (Invitrogen) with Lipofectamine 2000 (Invitrogen). After 24 h, cells were replenished with fresh culture medium containing 10% fetal bovine serum (FBS). Cells were incubated for an additional 40 h, and extracted RNA and proteins were analyzed by RT-PCR and immunoblotting.
Protein isolation and immunoblots.
MEFs were lysed in the Tris-HCl buffer (10 mM, pH 7.5), 1% NP-40, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM sodium chloride, and 1 mM EDTA for 30 min at 4°C. The samples were centrifuged at 10,000 × g for 10 min to pellet debris. Cell lysates were loaded and run on a 8% sodium dodecyl sulfate-polyacrylamide gel before being transferred electrophoretically onto a polyvinylidene difluoride membrane. The membrane was blocked with 5% fat-free milk solution and then sequentially incubated with primary antibody and enzyme-conjugated secondary antibody. The results were documented on X-ray film by using an ECL detection kit (Amersham Biosciences). Immunoblot analysis confirmed the expression of AHR and NRF2ΔΝeh2 in the stable clones (data not shown) using anti-AHR antibody (Biomol) and anti-NRF2 antibody (generously provided by Ken Itoh, Hirosaki University). Polyclonal anti-β-actin antibody and anti-lamin B antibody were obtained from Santa Cruz Biotechnology.
Induction of differentiation.
A protocol for adipocyte differentiation of MEFs was used as described previously (2). Briefly, MEFs were plated at a 100% confluence in a 12-well plate 24 to 48 h before stimulation of differentiation. For the first 3 days, cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS, 10 μg/ml insulin (Sigma), 5 μM dexamethasone (Sigma), 0.2 mM isobutylmethylxanthine (Sigma), and 1 μM rosiglitazone (Cayman Chemical) for the first 3 days. Subsequently, cells were maintained in DMEM supplemented with 10% FBS, 10 μg/ml insulin, and 1 μM rosiglitazone. Oil Red O staining was done per the manufacturer's instructions (Chemicon).
RESULTS
AHR expression and signaling are regulated by NRF2 in murine embryonic fibroblasts.
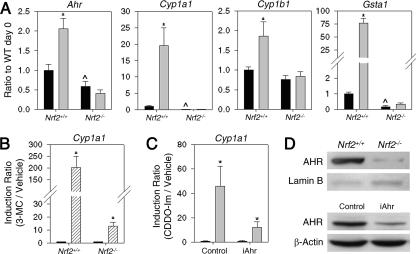
Microarray analyses of MEFs derived from Nrf2+/+ and Nrf2−/− mice have been conducted to compare the influence of the transcription factor genotype on global gene expression patterns (data not shown). These studies suggested that NRF2 may affect expression levels of Ahr and some of its downstream target genes. To follow up on this observation, a more direct analysis was undertaken in the present study. Both mRNA and protein levels of AHR were higher in Nrf2+/+ MEFs than in Nrf2−/− MEFs (Fig. 1A and D). Quantitative RT-PCR indicated that constitutive expression of AHR target gene Cyp1a1, but not Cyp1b1, was significantly lower in Nrf2−/− MEFs (Fig. 1A). The mRNA levels of Ahr were elevated 2.1-fold by CDDO-Im, a potent activator of NRF2 signaling, in Nrf2+/+ MEFs but not in Nrf2−/− MEFs. The mRNA levels of Arnt, which encodes a heterodimeric binding partner of AHR, were not affected by Nrf2 genotype or CDDO-Im (data not shown). CDDO-Im induced transcription of AHR target genes Cyp1a1 and Cyp1b1 in Nrf2+/+ MEFs but not in Nrf2−/− MEFs, implying that the NRF2 pathway influences the transcriptional function of AHR. Gsta1, a well-characterized NRF2-regulated gene, was induced over 75-fold in Nrf2+/+ MEFs but not at all in Nrf2−/− MEFs.
FIG. 1.
Differential expression analysis of Ahr and downstream genes in Nrf2+/+ and Nrf2−/− MEFs. (A) Transcript levels were measured following treatment with vehicle (black bars) and 25 nM CDDO-Im (gray bars) for 24 h. Ratios to Nrf2+/+ vehicle-treated controls on day 0 (Ratio to WT day 0) are shown on the y axes. Values are means plus standard errors (SEs) (error bars) (n = 4). Values that are significantly different (P < 0.05) from that of the respective vehicle (*) or that of the wild-type vehicle (WT) (∧) are indicated. (B) Transcript levels were measured following treatment with vehicle (black bar) and 2 μM 3-MC (hatched bar) for 24 h. Ratios to the respective vehicle-treated control (Induction Ratio) are shown. Values are means plus SEs (error bars) (n = 3). Values that are significantly different (P < 0.05) from that of the respective vehicle (*) are indicated. (C) MEF cells were transfected with 30 nM siRNA (iAhr) or control RNA. Cells were treated with 25 nM CDDO-Im for 24 h or vehicle. Ratios to the respective vehicle-treated control (Induction Ratio) are shown. Values are means plus SEs (error bars) (n = 4). Values that are significantly different (P < 0.05) from that of the respective vehicle (*) are indicated. (D) Protein levels of AHR were detected by immunoblotting. (Top blots) AHR protein levels in Nrf2+/+ and Nrf2−/− MEFs. (Bottom blots) AHR protein levels in control and iAhr-treated Nrf2+/+ MEFs.
Cells were treated with siRNA for Ahr (iAhr) to determine whether CDDO-Im induces Cyp1a1 transcription through the AHR pathway. Treatment with iAhr resulted in a 70 to 80% knockdown of AHR protein (Fig. 1D). After transfection with iAhr, induction of Cyp1a1 transcripts by CDDO-Im was only 25% of that observed in AHR-intact control cells (Fig. 1C). To test whether there is any off-target effect, mRNA levels of β-actin and hypoxanthine-guanine phosphoribosyltransferase (Hprt) were measured using quantitative RT-PCR. Transfection of iAhr had no effect on these genes (data not shown), confirming the specificity of the siRNA construct.
The fact that mRNA levels of Ahr are lower in Nrf2−/− MEFs than in Nrf2+/+ MEFs suggests that the response to AHR ligands may be impaired in Nrf2−/− MEFs. To determine whether the AHR-XRE pathway is intact in Nrf2−/− MEFs, 3-methylcholanthrene (3-MC), a prototypical ligand that directly binds to AHR, was used. In Nrf2+/+ MEFs, the mRNA levels of Cyp1a1 were increased by 200-fold by 2 μM 3-MC (Fig. 1B). In Nrf2−/− MEFs, the drug treatment resulted in 13-fold induction of Cyp1a1, suggesting that enhancement of AHR signaling by AHR agonists is suppressed in Nrf2−/− MEFs.
Transcription of the Ahr gene is regulated by an ARE located in its proximal promoter.
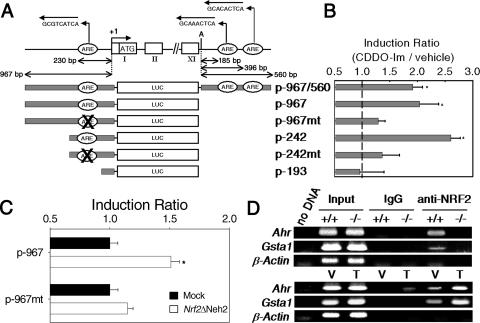
To analyze the regulation of Ahr by NRF2, the promoter region (−967 bp upstream of the transcription start site) was isolated from mouse liver genomic DNA by PCR amplification and ligated into the luciferase reporter pGL3 basic vector. Three potential AREs were identified by consensus sequence: one upstream of the transcription start site (−230 bp) and two downstream (185 bp and 396 bp) of the putative poly(A) addition signal (Fig. 2A).
FIG. 2.
Analysis of murine Ahr promoter. (A) Murine Ahr promoter constructs are shown. Three AREs in an inverted direction were identified, one upstream of the transcription start site (labeled “+1”) and two downstream of the putative poly(A) addition signal (labeled “A”) of the Ahr gene. Arrows indicate the orientations of these putative AREs. Boxes marked with roman numerals indicate exons of the murine Ahr promoter. LUC, luciferase. (B) Luciferase activities derived from full-length, truncated, or mutated promoters following treatment with 25 nM CDDO-Im are shown. Values that are significantly different (P < 0.05) from that of the respective vehicle-treated controls (*) are indicated. Values are means plus standard errors (SEs) (error bars) (n = 4). (C) Response of wild-type and mutated Ahr promoter to ectopic expression of Nrf2ΔNeh2 (white bar) and mock plasmid (black bar). Values that are significantly different (P < 0.05) from that of the mock plasmid-transfected control (*) are indicated. Values are means plus SEs (error bars) (n = 3). (D) Binding of NRF2 to Ahr promoter in intact cells. Water (no DNA), inputs, chromatin immunoprecipitants with immunoglobulin G (IgG), and anti-NRF2 antibody (anti-NRF2) were used for PCR amplification of each promoter. (Top gel) ARE-containing promoter regions from the Ahr promoter were detected in NRF2 immunoprecipitants obtained from Nrf2+/+ MEFs (+/+) but not from Nrf2−/− MEFs (−/−). (Bottom gel) Enhanced binding of NRF2 to the Ahr promoter following CDDO-Im (T) treatment compared to that in vehicle (V)-treated Nrf2+/+ MEFs.
The reporter construct with the promoter of the Ahr gene (p-967) showed higher basal luciferase activity (data not shown) and inducible luciferase activity than the construct with both promoter and downstream enhancer sequences (p-967/560) (Fig. 2B). These data suggest that the two AREs in the downstream region are not involved in NRF2-AHR cross regulation. To determine the location of the cis element(s) involved in this cross regulation, two truncated promoter constructs were made (p-242 and p-193). Wild-type MEFs were transfected with these constructs, and luciferase activities from transfected cells following CDDO-Im treatment or forced expression of Nrf2ΔNeh2 were measured. Neh2 is the negative regulatory domain of NRF2 that is involved with the interaction with KEAP1 (17, 19). The plasmid Nrf2ΔNeh2 expresses a dominant-positive form of NRF2 protein that is more resistant to proteosomal degradation than the wild-type protein is. The full-length promoter (p-967) was activated by CDDO-Im treatment (2.0-fold), as well as by forced expression of Nrf2ΔNeh2 (1.5-fold). This magnitude of response is consistent with the 2.1-fold increase in Ahr transcripts seen following treatment of wild-type MEFs with CDDO-Im (Fig. 1A). Responses to CDDO-Im were higher (2.6-fold) in proximal promoter constructs (p-242) than in the full-length promoter (Fig. 2B). These results suggest that a promoter containing 242 bp upstream of the transcription start site contains cis elements that can be activated by the NRF2-ARE. Further, when the upstream region containing the consensus ARE was deleted from the full-length promoter, the resulting construct (p-193) was not activated by CDDO-Im treatment (Fig. 2B). These results suggest that the −230 ARE may be necessary for the activation of the Ahr promoter by NRF2-ARE signaling. To confirm the results from promoter truncation, the −230 ARE was mutated in the p-967 and p-242 constructs (p-967mt and p-242mt [Fig. 2A]). This mutation in the ARE largely abolished promoter activation upon either CDDO-Im treatment (Fig. 2B) or Nrf2ΔNeh2 cotransfection in Nrf2+/+ MEFs (Fig. 2C). These results indicate that the ARE is required for the activation of the promoter by NRF2. ChIP assays were performed to confirm that NRF2 binds to the Ahr promoter in intact cells. The promoter region containing an ARE at −230 bp was detected by PCR amplification with NRF2-immunoprecipitated chromatin from Nrf2+/+ MEFs (Fig. 2D, top gel). As positive and negative controls, the ARE of the Gsta1 promoter (10) was detected in NRF2-immunoprecipitated samples, whereas the β-actin promoter was not amplified. Levels of binding of NRF2 to the Ahr promoter were higher in CDDO-Im-treated cells than in vehicle-treated cells (Fig. 2D, bottom gel). A similar pattern of binding was observed with the Gsta1 ARE. The Gsta1 ARE and Ahr promoters were not amplified from Nrf2−/− MEFs. Collectively, these data suggest that NRF2 regulates transcription of Ahr through direct binding to the ARE in the Ahr promoter in both uninduced and induced states.
Adipocyte differentiation was accelerated in Nrf2−/− MEFs.
As described above in the introduction, AHR inhibits conversion of 3T3-L1 preadipocytes and MEFs to mature lipid-containing adipocytes. Our findings show that immortalized Nrf2−/− MEFs have lower levels of AHR protein and mRNA, suggesting that NRF2 may influence adipogenesis via activation of AHR signaling. Therefore, the rate of adipogenesis of Nrf2−/− MEFs was compared to that of Nrf2+/+ MEFs.
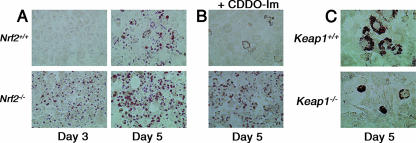
According to Alexander et al. (2), basal differentiation medium (containing insulin, dexamethasone, and isobutylmethylxanthine) used for preadipocyte differentiation was not sufficient to induce differentiation in immortalized MEF cell lines. Therefore, peroxisome proliferator-activated receptor gamma (PPARγ) agonist rosiglitazone was added to the basal medium. Neither Nrf2+/+ MEFs nor Nrf2−/− MEFs differentiate spontaneously to adipocytes (data not shown). However, lipid droplets were clearly detectable in immortalized Nrf2−/− MEFs 3 days after stimulation, whereas droplets were not visible until 5 days poststimulation in Nrf2+/+ MEFs (Fig. 3A). CDDO-Im treatment substantially inhibited adipogenesis only in Nrf2+/+ MEFs (Fig. 3B), further implying a negative role of NRF2 in adipogenesis.
FIG. 3.
Effects of Nrf2 and Keap1 genotypes on adipogenesis of MEFs. Cells were incubated with DMEM containing adipogenic reagents described in Materials and Methods. Representative photographs of the cells stained with Oil Red O are shown (400-fold magnified). (A) Nrf2+/+ and Nrf2−/− MEFs 3 days and 5 days after stimulation of differentiation. (B) Effects of CDDO-Im (25 nM) on differentiation of Nrf2+/+ and Nrf2−/− MEFs 5 days after stimulation of differentiation. (C) Keap1+/+ and Keap1−/− MEFs 5 days after stimulation of differentiation.
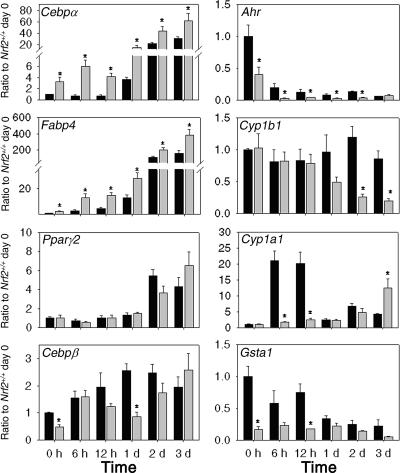
Adipocyte differentiation is a process marked by a set of gene expression changes. CCAAT/enhancer-binding protein alpha (CEBPα) and PPARγ are transcription factors that regulate adipocyte-specific genes, such as fatty acid-binding proteins (FABPs) (14, 37, 47). Transcript levels of CEBPα and PPARγ are increased during differentiation, leading to upregulation of their downstream genes. Therefore, expression levels of CEBPα, PPARγ, and FABP4 serve as markers of adipocyte differentiation. Transcript levels of Cebpα, Fabp4, and Pparγ2 were induced by stimulation with differentiation medium in both Nrf2+/+ and Nrf2−/− MEFs (Fig. 4). Basal transcript levels of Cebpα and Fabp4 were higher in Nrf2−/− MEFs than in Nrf2+/+ MEFs (3.3-fold and 2.4-fold, respectively), and these higher levels were sustained throughout the differentiation process. No differences in the mRNA levels of Pparγ2 between the two genotypes were detected. An increase in CEBPβ, which regulates transcription of PPARγ and CEBPα by binding to the CEBP response elements present in the promoter regions, is considered an early marker of adipogenesis (36). The basal transcript levels of this gene were lower in Nrf2−/− MEFs than in Nrf2+/+ MEFs at day 0 and day 1, but not at later time points. Our data suggest that during differentiation, Nrf2−/− MEFs accumulate larger amounts of the terminal effectors, such as CEBPα and FABP4, that play important roles in both differentiation and lipid accumulation (14).
FIG. 4.
Time-dependent changes of transcript levels throughout adipogenesis in Nrf2+/+ MEFs (black bars) and Nrf2−/− MEFs (gray bars). Ratios to Nrf2+/+ MEFs on day 0 (Ratio to Nrf2+/+ day 0) are shown on the y axes. Time (in hours [h] and days [d]) is shown on the x axes. Values are means plus standard errors (error bars) (n = 3). Values that are significantly different (P < 0.05) from that of the respective Nrf2+/+ MEFs (*) are indicated.
To determine whether the transcript levels and the actions (i.e., transactivation of downstream genes) of NRF2 and AHR are consistent with the adipogenesis pattern observed in Fig. 3, the mRNA levels of Ahr, Nrf2 and downstream genes were monitored throughout the differentiation process (Fig. 4). The mRNA levels of Nrf2 were not affected by differentiation (data not shown), but the mRNA levels of Gsta1, a marker of the action of NRF2, were lower in Nrf2+/+ MEFs throughout differentiation, indicating that the transcriptional action of NRF2 is diminished during differentiation. Shimba et al. (40) reported that AHR was undetectable following adipogenesis in preadipocytes, and Alexander et al. (2) have shown that AHR was downregulated following adipogenesis in MEFs. Our data also confirmed that Ahr mRNA levels decrease during differentiation. A ChIP assay confirmed that NRF2 binding to the ARE at the −230 bp of Ahr promoter was suppressed 24 h after stimulation of differentiation (see Fig. S1 in the supplemental material). The basal levels of Ahr were lower in Nrf2−/− MEFs than in Nrf2+/+ MEFs not only before stimulation (day 0) but also throughout the differentiation process. Cyp1a1 and Cyp1b1 mRNA levels were used as markers of AHR function. In Nrf2+/+ MEFs, Cyp1b1 mRNA levels were not affected by the treatment, whereas in Nrf2−/− MEFs, the levels started decreasing from day 1. Cyp1a1 levels were markedly induced in Nrf2+/+ MEFs at early time points but were relatively stable in Nrf2−/− MEFs. In summary, AHR expression and its downstream actions were decreased following adipogenesis and lower in Nrf2−/− MEFs than in Nrf2+/+ MEFs. These data support our hypothesis that NRF2 acts as a negative regulator of adipogenesis through the regulation of Ahr transcription and signaling.
Adipocyte differentiation was delayed in Keap1−/− MEFs.
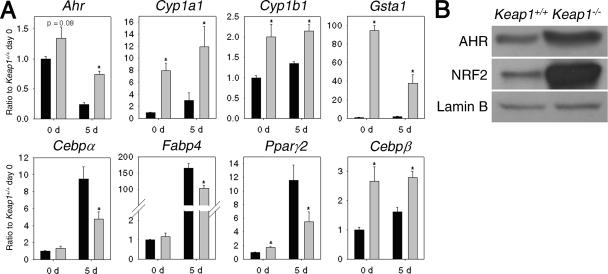
To determine whether enhanced accumulation of NRF2 influences the rate of adipocyte differentiation, primary MEFs were established from a Keap1-disrupted mouse and a congenic wild-type mouse (Keap1−/− MEFs and Keap1+/+ MEFs, respectively). After 5 days of adipocyte differentiation, lipid droplets were detectable both in primary Keap1+/+ MEFs and Keap1−/− MEFs (Fig. 3C). However, the number of differentiated cells and size of the lipid droplets were substantially larger in Keap1+/+ MEFs than in Keap1−/− MEFs. These results suggest that the enhanced NRF2 accumulation in Keap1−/− MEFs inhibits adipogenesis (after spontaneous immortalization, MEFs became smaller compared to primary cells; therefore, cells in Fig. 3C look bigger than cells in Fig. 3A).
The transcript levels of differentiation markers, Ahr and its downstream genes were detected at day 0 and day 5 after stimulation of adipogenesis in these cells (Fig. 5A). The mRNA levels of Cebpα, Fabp4, and Pparγ2 were induced by stimulation with differentiation medium. The basal transcript levels of Cebpα and Fabp4 were not affected by genotype at day 0 but were higher in Keap1+/+ MEFs at day 5. The mRNA levels of Pparγ2 were higher in Keap1−/− MEFs at day 0 but lower at day 5 compared to Keap1+/+ MEFs. The mRNA levels of Cebpβ were lower in Keap1+/+ MEFs than in Keap1−/− MEFs, both at day 0 and day 5. These results suggest that increased NRF2 accumulation affects steps between inductions of Cebpβ and Cebpα. These data are consistent with the pattern observed in Fig. 4. However, unlike the situation in immortalized Nrf2+/+ and Nrf2−/− MEFs, Pparγ2 may also play a role in primary MEFs.
FIG. 5.
Effects of Keap1 genotype on transcript and protein levels in MEFs. (A) Transcript levels of Ahr and downstream genes and markers at day 0 and day 5 after stimulation of adipogenesis in Keap1+/+ MEFs (black bars) and Keap1−/− MEFs (gray bars). Ratios to Keap1+/+ MEFs on day 0 (Ratio to Keap1+/+ day 0) are shown on the y axes. Time after stimulation of adipogenesis (in days [d]) is shown on the x axes. Values are means plus standard errors (error bars) (n = 3). Values that are significantly different (P < 0.05) from that of the respective Keap1+/+ MEFs (*) are indicated. (B) Protein levels of AHR and NRF2 were detected by immunoblotting.
Both transcript and protein levels of Ahr were increased by disruption of Keap1 at day 0 (Fig. 5A and B), and transcript levels of Ahr were 3.1-fold higher in Keap1−/− MEFs than in Keap1+/+ MEFs at day 5, suggesting that NRF2 regulates Ahr expression both before and during differentiation. The mRNA levels of Cyp1a1 and Cyp1b1 were not affected by adipogenesis but were higher in Keap1+/+ MEFs at day 0 and day 5. These data suggest that NRF2 may affect not only Ahr transcription but also its function. Gsta1 mRNA levels were decreased in Keap1−/− MEFs following adipogenesis but slightly increased in Keap1+/+ MEFs. Overall, an inverse association between NRF2 accumulation and adipogenesis was observed, further confirming the inhibitory role of NRF2 in this process.
Stable expression of Ahr and Nrf2ΔNeh2 rescued Nrf2−/− MEFs from adipocyte differentiation.
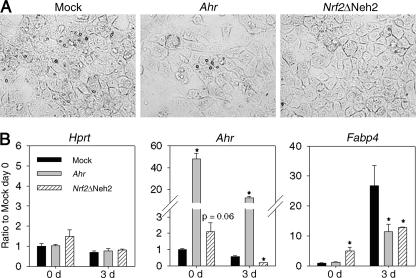
To determine whether ectopic expression of Ahr and Nrf2 can “rescue” Nrf2−/− MEFs from differentiation, Nrf2−/− MEFs were transfected with mock vector or with plasmids expressing mouse Ahr and Nrf2ΔNeh2, and stable cell lines were established from these cells. At poststimulation day 3, mock transfectants showed accelerated lipid accumulation and morphological changes compared to cells expressing Ahr and Nrf2ΔNeh2 (Fig. 6A). The mRNA levels of Fabp4 were substantially lower in Ahr- and Nrf2ΔNeh2-expressing cells than in control cells at day 3 (Fig. 6B). The fact that mRNA levels of Hprt were not affected by stable transfection and differentiation shows that stable transfection does not influence nonspecific targets. mRNA levels of Ahr were elevated in Ahr- and NrfΔNeh2-expressing cells than in mock-transfected cells. However, the levels were significantly decreased in differentiated cells, suggesting that there are additional mechanisms that control Ahr gene transcription beside NRF2. The fact that the Nrf2ΔNeh2-expressing cells are more resistant to differentiation stimuli compared to mock-transfected cells excludes the possibilities that Nrf2−/− MEFs have differences other than Nrf2 genotype that influence the phenotype. The finding that forced expression of Ahr rescued Nrf2−/− MEFs from adipogenesis provides direct support for our hypothesis that NRF2 inhibits adipogenesis through AHR signaling.
FIG. 6.
Effects of ectopic expression of Ahr and Nrf2ΔNeh2 on adipogenesis of Nrf2−/− MEFs. (A) Representative photographs of the cells stained with Oil Red O and magnified 400-fold (3 days after stimulation of differentiation). (B) Transcript levels of Hprt, Ahr, and Fabp4 at day 0 and day 3. The values for mock-transfected cells (black bars) and cells expressing Ahr (gray bars) and Nrf2ΔNeh2 (hatched bars) are shown. Ratios to mock transfectants at day 0 (Ratio to Mock day 0) are shown on the y axes. Time (in days [d]) is shown on the x axes. Values are means plus standard errors (error bars) (n = 3). Values that are significantly different (P ≤ 0.05) from that of the respective mock transfectant (*) are indicated.
DISCUSSION
Studying interactive networks between signaling pathways is indispensable for understanding the complexity of a phenotype. For example, several transcription factors have been implicated in the inflammatory process associated with asthma, including the glucocorticoid receptor, NF-κB, activator protein 1, nuclear factor of activated T-cells, cyclic AMP response element-binding protein, CEBP, PPAR, and NRF2 (38). Information on cross talk between signaling pathways may also provide insight into understanding the previously unrealized functions of a pathway and point to additional targets for interactions (11).
Nrf2 gene transcription is directly modulated by AHR activation of XRE-like elements in the Nrf2 promoter (27). NRF2 is also known to autoregulate its own expression through an ARE-like element in the proximal region of its promoter (22). However, this study is the first to show that NRF2 can affect Ahr transcription and downstream AHR signaling (Fig. 7). This bidirectional regulation of AHR and NRF2 pathways provides clues on the roles of NRF2 in complex diseases and in uncharacterized phenotypes.
FIG. 7.
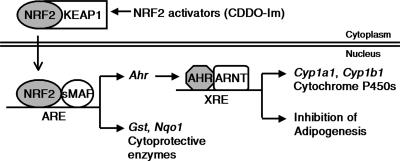
Schematic representation of regulation of the AHR-XRE pathway by NRF2. The figure illustrates that interaction of NRF2 with ARE in the promoter of Ahr induces transcription of Ahr and triggers downstream events, the subsequent upregulation of cytochrome P450s and inhibition of adipogenesis. sMAF, small MAF proteins.
The cross talk between NRF2 and AHR may help to explain the chemopreventive action of NRF2 activators. While studies on the AHR-CYP1A1 pathway often focus on production of toxic intermediates, the induction of both CYP1A1 and CYP1B1 resulted in an enhanced clearance of the procarcinogen benzo[a]pyrene (7). As suggested by Köhle and Bock (21), tightened coupling between the NRF2 and AHR pathways may result in attenuation of health risks caused by xenobiotics. The mode of action of bifunctional inducers, agents that enhance expression and activity of CYPs and cytoprotective enzymes (26), can be explained partially by our findings that NRF2 upregulates not only the expression of cytoprotective enzymes but also the expression of CYPs via AHR signaling. It is possible that drugs classified as bifunctional inducers increase expression of CYPs through NRF2-AHR-XRE interactions (Fig. 7).
Studies among various inbred strains of mice and F1 hybrids have shown that inducibility of AHR-regulated enzymes is a function of the number of AHR molecules per cell (8). However, our studies do not indicate complete concordance between NRF2 levels, AHR levels, and AHR-regulated gene expression. Clearly, transcriptional upregulation of Ahr is one of the mechanisms by which NRF2 modulates AHR signaling. However, the 41% reduction of Ahr transcript levels in Nrf2−/− MEFs compared to Nrf2+/+ MEFs may not be substantial enough to explain the complete abolishment of Cyp1a1 and Cyp1b1 induction by CDDO-Im in Nrf2−/− MEFs (Fig. 1A). Moreover, the transcript levels of Ahr were 1.3-fold higher in Keap1−/− MEFs than in Keap1+/+ MEFs, while Cyp1a1 and Cyp1b1 mRNA levels were 7.9-fold and 2.0-fold higher in Keap1−/− MEFs, respectively (Fig. 5A). Immunoblots comparing Keap1−/− and Nrf2−/− MEFs to wild-type MEFs (Fig. 1D and 5B) imply that Keap1 and Nrf2 genotypes may also affect AHR expression in a posttranscriptional manner. Thus, it is possible that a minimal change in the level of Ahr mRNA is amplified during or after its translation.
Although the mRNA levels of Cyp1a1 and Cyp1b1 were analyzed as markers of transactivation by AHR and Gsta1 as a marker of transactivation by NRF2, careful interpretation of the data is required. Isobutylmethylxanthine, which induces accumulation of cyclic AMP by inhibiting adenosine 3′,5′-cyclic monophosphate phosphodiesterase, was included in the differentiation medium. It is known that cyclic AMP activates AHR and induces its nuclear translocation (31). Dexamathasone, another component of the differentiation medium, potentiates induction of Cyp1a1 transcription by TCDD (23). Our data suggest that the differentiation medium induced transcription of Cyp1a1 in Nrf2+/+ MEFs (Fig. 4). These data confirm that the activation of AHR-Cyp1a1 signaling is dependent on Nrf2 genotype because Cyp1a1 transcription was not induced in Nrf2−/− MEFs. Transcript levels of Cyp1b1 also did not correlate perfectly with the mRNA levels of Ahr (Fig. 4 and 5). This can be explained perhaps by the fact that Cyp1b1 mRNA levels are regulated in an AHR-independent manner during adipogenesis (6). Our data show that Gsta1 mRNA levels were decreased in Nrf2+/+ MEFs and Keap1−/− MEFs throughout adipogenesis, suggesting that the action of NRF2 decreased during differentiation. However, it is possible that Gsta1 transcripts also reflect diminished expression of AHR because it is well-known that GSTs are also induced by AHR agonists, such as TCDD (21, 48).
Data shown in Fig. S1 in the supplemental material suggest that NRF2 binding to the promoter of Ahr was decreased during adipocyte differentiation. However, RT-PCR data confirmed that Ahr transcription was suppressed from 6 h after stimulation of differentiation (Fig. 4), while the change in NRF2 binding to ARE was detectable only after 24 h. Furthermore, Ahr mRNA levels were affected in Nrf2−/− MEFs and Nrf2+/+ MEFs. These results suggest that NRF2 is not the only factor that controls Ahr gene transcription during adipocyte differentiation. Indeed, our results are consistent with the report of Shimba et al. (40) demonstrating that the sequence of the Ahr promoter region responsible for differentiation-dependent suppression of Ahr transcription does not contain an ARE. Our findings also suggest that the level of expression of Ahr before stimulation of differentiation might be more important for determination of cell fate than transcriptional regulation during adipogenesis, since suppression of Ahr transcription was observed in Nrf2−/− MEFs and Nrf2+/+ MEFs (Fig. 4) and in mock-transfected cells and Nrf2ΔNeh2-transfected cells (Fig. 6).
In addition to regulation of expression of CYPs, AHR plays a role in a number of processes, such as development, apoptosis, growth, and adipogenesis (34, 49, 52). Among these AHR-dependent phenotypes, our work evaluated the role of NRF2 on regulation of adipogenesis because of the central role of adipogenesis in multiple diseases, such as obesity (a major risk factor for diabetes, cancer, and cardiovascular diseases) and lipodystrophy (5, 16, 35). Various in vitro models for adipogenesis, such as embryonic stem cells, preadipocytes, and MEFs, are available (4). Among these models, adipogenesis using MEFs is particularly useful to study the effects of gene knockouts. The fact that disruption of Nrf2 accelerated differentiation to adipocytes, while disruption of Keap1 delayed the process (Fig. 3A and C) implies a negative role of NRF2 in adipogenesis. Pharmacological targeting of the NRF2 pathway with CDDO-Im, which inhibited differentiation (Fig. 3B), confirmed that NRF2 modulates components of adipocyte differentiation.
To evaluate how NRF2 regulates adipogenesis, differentiation markers involved at multiple stages of adipogenesis were analyzed. CEBPs and PPARs are the two families of transcription factors that play critical roles in adipogenesis (47). CEBPβ and CEBPδ function at an early phase of the differentiation process by sensing adipogenic stimuli and initiating expression of CEBPα and PPARγ (37, 47). CEBPα and PPARγ play roles at a later stage by inducing and maintaining expression of adipocyte-specific genes, such as Fabp4 (14). The elevation of CEBPβ upon adipogenic stimulation is transient, while CEBPα and PPARγ remain upregulated for the duration of adipogenesis (43).
Although the mechanism by which AHR regulates adipogenesis has not been fully characterized, recent work suggests that AHR affects differentiation stages that follow CEBPβ activation, i.e., CEBPα or PPARγ upregulation. In 3T3-L1 preadipocytes, forced expression of AHR resulted in lower induction of Fabp4 and Cebpα upon differentiation than in control cells, while the induction of Pparγ was not affected (41). Pparγ expression could be induced by differentiation in Ahr−/− MEFs, but levels were lower than in Ahr+/+ MEFs (2). In addition, Vogel and Matsumura (45) demonstrated that although TCDD suppressed adipogenesis of MEFs, it induced expression of Cebpβ rather than inhibiting it.
In the present study, mRNA levels of Cebpα and Fabp4 were higher in Nrf2−/− MEFs than in Nrf2+/+ MEFs both before and after differentiation, while induction of Pparγ2 was not affected by Nrf2 genotype (Fig. 4). mRNA levels of Cebpβ were lower in Nrf2−/− MEFs than in Nrf2+/+ MEFs. In primary Keap1−/− MEFs (Fig. 5A), disruption of Keap1 (which leads to accumulation of NRF2 in the nucleus) resulted in minimal induction of Cebpα, Fabp4, and Pparγ2 upon differentiation. Cebpβ mRNA levels remained higher in Keap1−/− MEFs than in Keap1+/+ MEFs, both before and after induction of adipogenesis. These data suggest that NRF2 negatively modulates expression of Cebpα and Pparγ2 but not Cebpβ during the course of adipogenesis. The stages of differentiation that are affected by NRF2 directly overlap with those affected by AHR, thereby supporting the hypothesis that NRF2 inhibits adipogenesis through cross talk with AHR signaling.
Although how AHR controls expression of XRE genes has been extensively studied, the molecular events that regulate Ahr expression itself have been less well investigated (40). Our findings represent a new perspective for control of Ahr expression by demonstrating that NRF2 directly binds to a functional ARE found in the proximal promoter of Ahr (Fig. 2). Our data also suggest that NRF2 affects AHR function (i.e., transcription of Cyp1a1 and Cyp1b1 and adipogenesis) in an exogenous ligand-independent manner. Tijet et al. (44) identified a number of genes for which expression is AHR dependent but TCDD independent, suggesting that the constitutive level of AHR may affect gene expression. Candidates for endogenous AHR ligand, such as indigo (42), may also play a role in the interaction with the NRF2 pathway.
In summary, the observation that NRF2 controls AHR signaling expands the function of NRF2 to include influencing the metabolism of xenobiotics and carcinogens via CYPs as well as controlling adipogenesis. Recent studies of Ahr Nrf2 double knockout mice in comparisons with wild-type and single transcription factor knockout mice offer additional evidence for interactions between these pathways (30). Although not addressed experimentally in this report, our findings also imply that the modifying influence of the NRF2 pathway can be expanded to other AHR-dependent processes, such as programmed cell death and development. Moreover, our findings may allow a broader clinical application of NRF2 activators, such as in prevention and treatment of obesity.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA94076, HL081205, and ES03819. S. Shin is the recipient of a Samsung Scholarship (Samsung Foundation of Culture).
We thank K. N. Anuzis (Johns Hopkins University) for help with adipocyte differentiation, A. Singh (Johns Hopkins University) and M. K. Kwak (Yeungnam University) for help with the ChIP assay. We also thank K. Itoh (Hirosaki University), Y. Fujii-Kuriyama, and J. Mimura (University of Tsukuba) for providing anti-NRF2 antibody and Nrf2ΔNeh2-expression plasmid and M. B. Sporn (Dartmouth Medical School) for providing CDDO-Im.
Footnotes
Published ahead of print on 20 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alexander, D. L., S. E. Eltom, and C. R. Jefcoate. 1997. Ah receptor regulation of CYP1B1 expression in primary mouse embryo-derived cells. Cancer Res. 57:4498-4506. [PubMed] [Google Scholar]
- 2.Alexander, D. L., L. G. Ganem, P. Fernandez-Salguero, F. Gonzalez, and C. R. Jefcoate. 1998. Aryl-hydrocarbon receptor is an inhibitory regulator of lipid synthesis and of commitment to adipogenesis. J. Cell Sci. 111:3311-3322. [DOI] [PubMed] [Google Scholar]
- 3.Boone, C., H. Bussey, and B. J. Andrews. 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8:437-449. [DOI] [PubMed] [Google Scholar]
- 4.Bost, F., M. Aouadi, L. Caron, and B. Binetruy. 2005. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 87:51-56. [DOI] [PubMed] [Google Scholar]
- 5.Camp, H. S., D. Ren, and T. Leff. 2002. Adipogenesis and fat-cell function in obesity and diabetes. Trends. Mol. Med. 8:442-447. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y. C., W. Zheng, M. Yamamoto, X. Liu, P. R. Hanlon, and C. R. Jefcoate. 2005. Differentiation of pluripotent C3H10T1/2 cells rapidly elevates CYP1B1 through a novel process that overcomes a loss of Ah receptor. Arch. Biochem. Biophys. 439:139-153. [DOI] [PubMed] [Google Scholar]
- 7.Ebert, B., A. Seidel, and A. Lampen. 2005. Induction of phase-1 metabolizing enzymes by oltipraz, flavone and indole-3-carbinol enhance the formation and transport of benzo[a]pyrene sulfate conjugates in intestinal Caco-2 cells. Toxicol. Lett. 158:140-151. [DOI] [PubMed] [Google Scholar]
- 8.Eisen, H. J., R. R. Hannah, C. L. Legraverend, A. B. Okey, and D. W. Nebert. 1983. The Ah receptor: controlling factor in the induction of drug-metabolizing enzymes by certain chemical carcinogens and other environmental pollutants, p. 227-258. In G. Litwack (ed.), Biochemical actions of hormones, vol. 10. Academic Press, New York, NY. [Google Scholar]
- 9.Fong, C. J., L. D. Burgoon, and T. R. Zacharewski. 2005. Comparative microarray analysis of basal gene expression in mouse Hepa-1c1c7 wild-type and mutant cell lines. Toxicol. Sci. 6:342-353. [DOI] [PubMed] [Google Scholar]
- 10.Friling, R. S., A. Bensimon, Y. Tichauer, and V. Daniel. 1990. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA 87:6258-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianchandani, E. P., D. L. Brautigan, and J. A. Papin. 2006. Systems analyses characterize integrated functions of biochemical networks. Trends Biochem. Sci. 31:284-291. [DOI] [PubMed] [Google Scholar]
- 12.Goldring, C., N. Kitteringham, R. Jenkins, I. Copple, J. F. Jeannin, and B. K. Park. 2006. Plasticity in cell defence: access to and reactivity of critical protein residues and DNA response elements. J. Exp. Biol. 209:2337-2343. [DOI] [PubMed] [Google Scholar]
- 13.Gouédard, C., R. Barouki, and Y. Morel. 2004. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol. Cell. Biol. 24:5209-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregoire, F. M., C. M. Smas, and H. S. Sul. 1998. Understanding adipocyte differentiation. Physiol. Rev. 78:783-809. [DOI] [PubMed] [Google Scholar]
- 15.Hankinson, O. 2005. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 433:379-386. [DOI] [PubMed] [Google Scholar]
- 16.Harp, J. B. 2004. New insights into inhibitors of adipogenesis. Curr. Opin. Lipidol. 15:303-307. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kensler, T. W., N. Wakabayashi, and S. Biswal. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47:89-116. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi, A., M. I. Kang, Y. Watai, K. I. Tong, T. Shibata, K. Uchida, and M. Yamamoto. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhle, C., and K. W. Bock. 2006. Activation of coupled Ah receptor and Nrf2 gene batteries by dietary phytochemicals in relation to chemoprevention. Biochem. Pharmacol. 72:795-805. [DOI] [PubMed] [Google Scholar]
- 21.Köhle, C., and K. W. Bock. 2007. Coordinate regulation of phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem. Pharmacol. 73:1853-1862. [DOI] [PubMed] [Google Scholar]
- 22.Kwak, M. K., K. Itoh, M. Yamamoto, and T. W. Kensler. 2002. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 22:2883-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, K. P., M. H. Wong, and C. K. Wong. 2004. Modulation of AhR-mediated CYP1A1 mRNA and EROD activities by 17beta-estradiol and dexamethasone in TCDD-induced H411E cells. Toxicol. Sci. 78:41-49. [DOI] [PubMed] [Google Scholar]
- 24.Loose, M., and R. Patient. 2006. Global genetic regulatory networks controlling hematopoietic cell fates. Curr. Opin. Hematol. 13:229-236. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Q., K. Kinneer, Y. Bi, J. Y. Chan, and Y. W. Kan. 2004. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem. J. 377:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao, W., L. Hu, M. Kandouz, D. Hamilton, and G. Batist. 2004. A cell-based system to identify and characterize the molecular mechanism of drug-metabolizing enzyme (DME) modulators. Biochem. Pharmacol. 67:1897-1905. [DOI] [PubMed] [Google Scholar]
- 27.Miao, W., L. Hu, P. J. Scrivens, and G. Batist. 2005. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J. Biol. Chem. 280:20340-20348. [DOI] [PubMed] [Google Scholar]
- 28.Nakata, K., Y. Tanaka, T. Nakano, T. Adachi, H. Tanaka, T. Kaminuma, and T. Ishikawa. 2006. Nuclear receptor-mediated transcriptional regulation in phase I, II, and III xenobiotic metabolizing systems. Drug Metab. Pharmacokinet. 21:437-547. [DOI] [PubMed] [Google Scholar]
- 29.Nebert, D. W., A. L. Roe, M. Z. Dieter, W. A. Solis, Y. Yang, and T. P. Dalton. 2000. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 59:65-85. [DOI] [PubMed] [Google Scholar]
- 30.Noda, S., N. Harada, A. Hida, Y. Fujii-Kuriyama, H. Motohashi, and M. Yamamoto. 2003. Gene expression of detoxifying enzymes in AhR and Nrf2 compound null mutant mouse. Biochem. Biophys. Res. Commun. 303:105-111. [DOI] [PubMed] [Google Scholar]
- 31.Oesch-Bartlomowicz, B., A. Huelster, O. Wiss, P. Antoniou-Lipfert, C. Dietrich, M. Arand, C. Weiss, E. Bockamp, and F. Oesch. 2005. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proc. Natl. Acad. Sci. USA 102:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okawa, H., H. Motohashi, A. Kobayashi, H. Aburatani, T. W. Kensler, and M. Yamamoto. 2006. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 339:79-88. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puga, A., C. R. Tomlinson, and Y. Xia. 2005. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol. 69:199-207. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, A., A. Duran, M. Selloum, M. F. Champy, F. J. Diez-Guerra, J. M. Flores, M. Serrano, J. Auwerx, M. T. Diaz-Meco, and J. Moscat. 2006. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3:211-222. [DOI] [PubMed] [Google Scholar]
- 36.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 37.Rosen, E. D. 2005. The transcriptional basis of adipocyte development. Prostaglandins Leukot. Essent. Fatty Acids 73:31-34. [DOI] [PubMed] [Google Scholar]
- 38.Roth, M., and J. L. Black. 2006. Transcription factors in asthma: are transcription factors a new target for asthma therapy? Curr. Drug Targets 7:589-595. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, J. V., G. H. Su, J. K. Reddy, M. C. Simon, and C. A. Bradfield. 1996. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA 93:6731-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimba, S., M. Hayashi, T. Ohno, and M. Tezuka. 2003. Transcriptional regulation of the AhR gene during adipose differentiation. Biol. Pharm. Bull. 26:1266-1271. [DOI] [PubMed] [Google Scholar]
- 41.Shimba, S., T. Wada, and M. Tezuka. 2001. Arylhydrocarbon receptor (AhR) is involved in negative regulation of adipose differentiation in 3T3-L1 cells: AhR inhibits adipose differentiation independently of dioxin. J. Cell Sci. 114:2809-2817. [DOI] [PubMed] [Google Scholar]
- 42.Sugihara, K., S. Kitamura, T. Yamada, T. Okayama, S. Ohta, K. Yamashita, M. Yasuda, Y. Fujii-Kuriyama, K. Saeki, S. Matsui, and T. Matsuda. 2004. Aryl hydrocarbon receptor-mediated induction of microsomal drug-metabolizing enzyme activity by indirubin and indigo. Biochem. Biophys. Res. Commun. 318:571-578. [DOI] [PubMed] [Google Scholar]
- 43.Tang, Q. Q., J. W. Zhang, and M. D. Lane. 2004. Sequential gene promoter interactions of C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem. Biophys. Res. Commun. 319:235-239. [DOI] [PubMed] [Google Scholar]
- 44.Tijet, N., P. C. Boutros, I. D. Moffat, A. B. Okey, J. Tuomisto, and R. Pohjanvirta. 2006. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 69:140-153. [DOI] [PubMed] [Google Scholar]
- 45.Vogel, C. F., and F. Matsumura. 2003. Interaction of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) with induced adipocyte differentiation in mouse embryonic fibroblasts (MEFs) involves tyrosine kinase c-Src. Biochem. Pharmacol. 66:1231-1244. [DOI] [PubMed] [Google Scholar]
- 46.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]
- 47.Wu, Z., N. L. Bucher, and S. R. Farmer. 1996. Induction of peroxisome proliferator-activated receptor γ during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol. Cell. Biol. 16:4128-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao, G. H., J. A. Pinaire, A. D. Rodrigues, and R. A. Prough. 1995. Regulation of the Ah gene battery via Ah receptor-dependent and independent processes in cultured adult rat hepatocytes. Drug Metab. Dispos. 23:642-650. [PubMed] [Google Scholar]
- 49.Yang, X., D. Liu, T. J. Murray, G. C. Mitchell, E. V. Hesterman, S. I. Karchner, R. R. Merson, M. E. Hahn, and D. H. Sherr. 2005. The aryl hydrocarbon receptor constitutively represses c-myc transcription in human mammary tumor cells. Oncogene 24:7869-7881. [DOI] [PubMed] [Google Scholar]
- 50.Yates, M. S., and T. W. Kensler. 2007. Chemopreventive promise of targeting the Nrf2 pathway. Drug News Perspect. 20:109-117. [DOI] [PubMed] [Google Scholar]
- 51.Yates, M. S., M. Tauchi, F. Katsuoka, K. C. Flanders, K. T. Liby, T. Honda, G. W. Gribble, D. A. Johnson, J. A. Johnson, N. C. Burton, T. R. Guilarte, M. Yamamoto, M. B. Sporn, and T. W. Kensler. 2007. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol. Cancer Ther. 6:154-162. [DOI] [PubMed] [Google Scholar]
- 52.Zaher, H., P. M. Fernandez-Salguero, J. Letterio, M. S. Sheikh, A. J. Fornace, Jr., A. B. Roberts, and F. J. Gonzalez. 1998. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 54:313-321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.