Abstract
The conserved TOR (target of rapamycin) kinase is part of a TORC1 complex that regulates cellular responses to environmental stress, such as amino acid starvation and hypoxia. Dysregulation of Akt-TOR signaling has also been linked to the genesis of cancer, and thus, this pathway presents potential targets for cancer chemotherapeutics. Here we report that rapamycin-sensitive TORC1 signaling is required for the S-phase progression and viability of yeast cells in response to genotoxic stress. In the presence of the DNA-damaging agent methyl methanesulfonate (MMS), TOR-dependent cell survival required a functional S-phase checkpoint. Rapamycin inhibition of TORC1 signaling suppressed the Rad53 checkpoint-mediated induction of ribonucleotide reductase subunits Rnr1 and Rnr3, thereby abrogating MMS-induced mutagenesis and enhancing cell lethality. Moreover, cells deleted for RNR3 were hypersensitive to rapamycin plus MMS, providing the first demonstration that Rnr3 contributes to the survival of cells exposed to DNA damage. Our findings support a model whereby TORC1 acts as a survival pathway in response to genotoxic stress by maintaining the deoxynucleoside triphosphate pools necessary for error-prone translesion DNA polymerases. Thus, TOR-dependent cell survival in response to DNA-damaging agents coincides with increased mutation rates, which may contribute to the acquisition of chemotherapeutic drug resistance.
TOR (target of rapamycin) is a phosphatidylinositol 3-kinase-related kinase family member that regulates cellular responses to wide-ranging environmental stresses, including nutrient starvation, growth factor deprivation, and hypoxia (8, 20, 45). These diverse environmental cues are transmitted by multiprotein TOR complexes through a variety of downstream pathways to regulate cap-dependent mRNA translation, transcriptional stress responses, cell cycle progression from G1 to S phase, and cell survival. Rapamycin (RAP) is a macrocyclic lactone antibiotic that, in complex with FKBP12 (the Saccharomyces cerevisiae homolog is Fpr1), specifically targets TOR. Since dysregulation of Akt-TOR signaling has been associated with tumorigenesis (7, 8), this pathway provides potential targets for cancer chemotherapy, as evidenced by the development of RAP analogs in clinical trials. Yet despite intense investigation of RAP action and the phenotypic consequences of TOR inhibition, the mechanistic basis for the antitumor activity of RAP in preclinical and clinical studies remains unclear.
The TOR kinase was initially identified in the yeast S. cerevisiae, in a genetic screen for mutants conferring resistance to RAP. S. cerevisiae carries two closely related Tor1 and Tor2 kinases, while other eukaryotic genomes encode a single kinase, typified by mammalian mTOR (45). As in mammalian cells, TOR signaling in yeast regulates cell growth through the function of distinct multiprotein complexes (29, 39, 45, 46). In yeast, Tor1 or Tor2 participates in the formation of a RAP-sensitive TOR complex 1 (TORC1) that consists of Kog1, Lst8, and Tco89. Mammalian TORC1 consists of mTOR, raptor (an ortholog of Kog1), and mLst8. Under favorable environmental conditions, TORC1 regulates the accumulation of cell mass, in part by controlling the translation initiation of a limited subset of capped mRNAs, nutrient uptake, and ribosome biogenesis. RAP treatment or depriving cells of nutrients or essential growth factors induces a starvation response characterized by decreased protein synthesis, macroautophagy, and the induction of stress response transcription factors (45).
A second multiprotein complex, TORC2, is RAP “insensitive” and provides an essential function in regulating actin cytoskeletal organization during cell growth (45). Yeast TORC2 composition is restricted to Tor2 in association with other conserved proteins, including Avo3 (rictor in mammalian cells). TORC2 also functions in endocytosis and in regulating calcineurin and sphingolipid signaling (34, 42, 45), while recent studies suggest the functional interplay of TORC1 and membrane trafficking (3).
The phosphatidylinositol 3-kinase-related kinase family members ATM, ATR, and DNA-PK and the yeast Mec1 and Tel1 kinases play important roles in DNA repair and checkpoint responses to DNA damage (1). Although a direct role for TOR signaling in S phase has not been defined, several observations prompted us to investigate a requirement for TORC1 function in response to DNA replication stress. First, when p53−/− mouse embryo fibroblasts or mutant p53 cancer cells are cultured under serum-free conditions, RAP treatment induces apoptosis, which coincides with entry into S phase (22, 23). Second, the inhibition of mTOR signaling by RAP enhances the cytotoxic activity of the DNA-damaging agent cisplatin (5, 41). However, the underlying mechanisms affecting cell survival in S phase remain unclear. Third, we previously reported the isolation of conditional yeast mutants exhibiting enhanced sensitivity to the DNA topoisomerase I (Top1) poison camptothecin (17, 25, 37). Several of these mutants, including mutants in hypomorphic alleles of CDC45 and DPB11, exhibit alterations in DNA replication, which exacerbate the S-phase-dependent toxicity of camptothecin. Surprisingly, many of these mutants are also hypersensitive to RAP (see Table S1 in the supplemental material).
The TOR pathway regulates cellular responses to a variety of environmental stresses; however, these considerations further implicate TOR signaling as a determinant of cell survival in response to aberrations in DNA replication. To directly address this hypothesis, we examined the effects of RAP inhibition of TORC1 on S-phase progression and the viability of synchronized cells in response to genotoxic stress. Here we report that TORC1 signaling is required for replication fork progression and to maintain the elevated levels of ribonucleotide reductase (RNR) subunits Rnr1 and Rnr3 induced by Rad53 checkpoint activation, which contribute to cell survival in response to DNA damage. The RAP-induced decrease in Rnr1/3 suppressed the mutagenic activity of methyl methanesulfonate (MMS). These findings establish TORC1 as a survival pathway in response to genotoxic stress and provide a mechanistic basis for the antitumor activity of RAP analogs used in combination with cytotoxic agents in clinical studies.
MATERIALS AND METHODS
Chemicals, yeast strains, and media.
Hydroxyurea (HU) was purchased from U.S. Biological (Swampscott, MA). RAP, obtained from the NCI drug repository, was dissolved in dimethyl sulfoxide, and stock solutions of 1 mg/ml were stored at −20°C. The mating pheromone α-factor, from Diagnostic Chemicals Ltd. (Oxford, CT), was stored at −20°C at 1 mg/ml in methanol and used at a final concentration of 5 μg/ml. MMS, cycloheximide (CHX), and canavanine were purchased from Sigma (St. Louis, MO).
S. cerevisiae strains, cultured under standard conditions, were derived from strain FY251 (MATa ura3-52 his3Δ200 leu2Δ1 trp1Δ63) and carried TRP1, GAL1-RNR1-3HA, RNR1-3HA, RNR2-3HA, RNR3-3HA, RNR4-3HA, HTA2-3HA, TOR1RR (TOR1S1972R), TOR2RR (TOR2S1975R), mrc1Δ, rad9Δ, rnr3Δ, tof1Δ, rfx1Δ, tor1Δ, and sml1Δ or a combination thereof. Gene disruptions and hemagglutinin (HA) tagging were accomplished with PCR-amplified selectable markers and confirmed by PCR, using primers that flank the sites of integration. TOR1RR and TOR2RR mutant alleles were obtained by PCR-based mutagenesis and selection on RAP and were confirmed by DNA sequencing. In GAL1-RNR1-3HA cells, the GAL1 promoter was 100 bp upstream of the RNR1 start codon. YCp-T-RAD53-HA was kindly provided by K. Sugimoto (University of Medicine and Dentistry of New Jersey).
Cell cycle analysis and viability assays.
MATa cells, α-factor arrested in G1 phase of the cell cycle or arrested in early S phase with 10 mg/ml HU, were washed by filtration and released into medium alone containing 200 ng/ml RAP, 0.05% MMS, 0.05% MMS plus 200 ng/ml RAP (MMS+RAP), 100 μg/ml CHX, or 100 μg/ml CHX plus 0.05% MMS (CHX+MMS). In experiments with tor1Δ strains, a lower concentration of RAP (50 ng/ml) was also used. At the times indicated, aliquots of cells were washed by centrifugation to remove the drugs, serially 10-fold diluted, and plated on yeast-peptone-dextrose (YPD) agar plates to assess cell viability or on synthetic medium without Arg and with canavanine to assess the frequency of canavanine resistance among surviving colonies. Aliquots of cells were also fixed with 70% ethanol and stored at 4°C for subsequent fluorescence-activated cell sorting (FACS) analysis or microscopy. For DAPI (4′,6′-diamidino-2-phenylindole) staining of DNA, 3 to 5 μl of cells was spotted on polylysine-coated Teflon slides, followed by 5 μl of 1 μg/ml DAPI and 2 μl Prolong antifade solution (Molecular Probes, Inc.). Cells were viewed with a Zeiss Axioskop 2 microscope equipped with differential interference contrast (DIC) and epifluorescence, and images were acquired with a Micromax charge-coupled device camera (Princeton Instruments, Inc.) and IP lab software (Scanalytics).
Western and Northern blot analyses.
To assess epitope-tagged protein levels, NaOH-trichloroacetic acid (TCA) cell extracts were prepared following release from α-factor or HU as described previously (25). HA-tagged proteins were detected by immunoblotting with a monoclonal HA antibody (12CA5; Roche, Indianapolis, IN) and by chemiluminescence (Pierce, Rockford, IL). RNAs, purified using a Ribopure-yeast kit (Ambion, Austin, TX), were subjected to Northern blot analysis using a NorthernMax-Gly kit (Ambion) and gene-specific probes PCR amplified from yeast genomic DNA and radiolabeled by random priming using a DECAprime II kit (Ambion). Bands were visualized by phosphorimage analysis.
2-D gel analysis of replication intermediates.
Replication intermediates were purified 10 min, 30 min, 1 h, or 3 h following release of cells from α-factor into YPD or YPD plus RAP, MMS, or MMS+RAP, as described previously (31). To analyze ARS305 replication intermediates, purified DNAs, restricted with EcoRV and HindIII, were resolved by two-dimensional (2-D) gel electrophoresis, transferred to nylon membranes, hybridized with a 32P-labeled probe spanning ARS305, and visualized by phosphorimage analysis.
RESULTS
RAP-sensitive TORC1 signaling maintains cell viability and promotes S-phase progression in response to DNA damage.
RAP inhibition of TOR signaling induces yeast cell cycle arrest in early G1 phase, which precedes the G1 block induced by the α-factor mating pheromone (4). Thus, we could assess TOR signaling in S phase by releasing cells from α-factor arrest in late G1 phase into YPD medium containing RAP, in the presence or absence of the DNA-damaging agent MMS. As shown in Fig. 1A, wild-type cells released into medium (control) synchronously transited S phase and acquired a 2C (2N) DNA content by 40 min. When the cells were released into RAP, a subpopulation of α-factor-arrested cells failed to transit S phase. As previously reported (4), only 82% of the cells had entered S phase (as defined by cells forming buds), relative to the number of cells released into medium alone, 20 min following release from α-factor into RAP. Nevertheless, the kinetics of S-phase transit for these cells mirrored those of the untreated control cells, with RAP-treated cells accumulating in the next G1 phase. As expected, S-phase transit was decreased in the presence of MMS due to activation of the Rad53 checkpoint (30, 35) (Fig. 1A, MMS panel). Surprisingly, however, RAP treatment further delayed the slow S-phase transit induced by MMS (Fig. 1A, MMS+RAP panel). For instance, 220 min following α-factor release, the majority of MMS-treated cells had a DNA content approaching 2C, while cells released into MMS+RAP had a considerably reduced DNA content. The persistent accumulation of MMS+RAP-treated cells in early S phase relative to the late S-G2 DNA content of MMS-treated cells is highlighted by the superposition of the 220-min FACS profiles in Fig. S1 in the supplemental material. However, during this time course of drug exposure, the discrepancy in S-phase transit between MMS- versus MMS+RAP-treated cells became apparent from 100 min on, coinciding with a more pronounced reduction in cell viability in MMS+RAP-treated cells than that for MMS-treated cells (Fig. 1B). RAP treatment alone was growth inhibitory, not cytotoxic, with only a slight increase in the number of colonies from time zero to 220 min. In contrast, the cytotoxic activity of MMS or MMS+RAP was reflected in the decrease in colony formation over time following removal of the drugs and plating of cells on YPD agar.
FIG. 1.
RAP inhibition of TOR signaling decreases S-phase transit and cell viability in response to MMS treatment. (A) Wild-type cells released from α-factor into YPD containing no drug (control), MMS, RAP, or MMS+RAP were processed for flow cytometry at the times indicated. (B) Serial dilutions of cells treated as described for panel A were spotted onto YPD plates. Colony formation was assessed at 30°C. (C) Cells released from HU arrest into YPD containing no drug (control), MMS, RAP, or MMS+RAP were collected and serially diluted at the times indicated. The number of viable cells forming colonies on YPD plates following incubation at 30°C was plotted relative to that at time zero (release from HU) (n = 3).
To ensure that these effects were restricted to S phase and not due to RAP-induced alterations in cell cycle transit from late G1 to S phase, several independent experimental strategies were pursued. First, cells were arrested in early S phase with HU and then treated as described above. HU inhibition of RNR induces the activation of the Rad53 S-phase checkpoint as a consequence of alterations in replication fork progression. Consequently, the cell cycle arrest induced by HU occurs in early S phase. In these experiments, similar results to those for cells synchronized with α-factor were obtained: RAP alone was cytostatic, while cotreatment with MMS+RAP further slowed S-phase progression and increased cell killing induced by MMS (Fig. 1C; also see Fig. 3). Thus, independent of the mechanism of cell synchronization (α-factor in G1 phase or HU in early S phase), RAP induced the same effects on the S-phase transit and viability of cells exposed to MMS.
FIG. 3.
CHX and RAP induce distinct alterations in MMS-treated cell viability in S phase. (A) Cells released from HU arrest into YPD containing no drug (control), MMS, RAP, MMS+RAP, CHX, or MMS+CHX were collected at the indicated times and processed for flow cytometry. (B) Serial dilutions of cells treated as described for panel A were assayed for cell viability as described in the legend to Fig. 1B.
A second approach involved exposing cells that express high levels of Top1 to RAP and camptothecin. Since camptothecin cytotoxicity requires ongoing DNA replication to induce replication-dependent DNA lesions (6), any alteration in cell viability induced by RAP cotreatment would be attributable to S-phase-dependent events. Indeed, a similar increase in cytotoxicity was observed when α-factor-arrested cells were released into medium containing RAP and camptothecin (data not shown). In addition, in time course experiments where MMS and RAP were added at 10-minute intervals following release from G1 arrest, the effects of TOR inhibition appeared to be restricted to early S phase. RAP-induced phenotypes, evident in cells treated at 10 min, disappeared when cells where treated at 20 min, when the majority of cells had acquired a close-to-2C DNA content. Thus, in cells exposed to sufficient DNA damage to induce the intra-S-phase checkpoint, TOR signaling enhanced cell survival and S-phase transit.
We next asked if the alterations in S-phase transit suggested by FACS profiles and morphological examinations of MMS+RAP- versus MMS-treated cells (Fig. 1 and data not shown) might reflect diminished replication origin firing rather than a decrease in fork progression. To assess origin firing and fork stability in cells treated with MMS, with or without RAP, replication intermediates were purified, resolved in 2-D gels, and probed with sequences corresponding to an early origin of replication (ARS305) on chromosome III (Fig. 2). ARS305 efficiently fires early in S phase, while origins more proximal to the left telomere end of chromosome III, ARS301 to ARS304, are normally dormant. In these gels, a bubble arc reflects bidirectional origin firing and Y arcs result from the asymmetric movement of replication forks through the restriction fragment being probed. X spikes accompany origin firing and decrease in intensity as forks migrate (31).
FIG. 2.
TORC1 signaling maintains replication fork stability in the presence of MMS. At the indicated times following α-factor release into YPD, MMS, RAP, or MMS+RAP, replication intermediates were resolved in 2-D gels. The distribution of bubble arcs (circles), Y arcs (Y diagram), and X spikes (triple arrows), which indicate origin firing, passive DNA replication, and slow fork progression, respectively, was determined in Southern blots with a probe for ARS305, an early firing origin of replication on the left arm of chromosome III.
After release of cells into S phase, firing of ARS305 was unaffected by RAP, as evidenced by a robust bubble arc at 10 min (Fig. 2, RAP panel). Untreated cells continued to cycle: the bubble arc apparent after release from α-factor was not detected at 60 min and reappeared at 180 min as cells entered subsequent cell cycles. The decrease in replication intermediates after 60 min of RAP treatment coincided with accumulation of cells in G1 phase, as evidenced by FACS analysis and cell morphology (Fig. 1 and data not shown). With MMS, the accumulation of a strong Y arc and X spike indicates slow fork progression at 30 min. The decrease in intermediates at 60 min coincided with fork progression and the accumulation of cells with a 2C DNA content (as in Fig. 1A). A similar pattern of robust ARS305 firing was also obtained with MMS+RAP-treated cells at 10 min, although a slightly less intense pattern of replication intermediates was obtained at 30 min. However, the decrease in replication intermediates at 60 and 180 min (Fig. 2), relative to the levels with MMS alone, did not correspond with increased DNA content (Fig. 1A). A persistent cell cycle arrest in early S phase due to Rad53 checkpoint activation would yield stable replication intermediates over the course of these experiments. In contrast, these data suggest that MMS+RAP induces a decrease in fork stability that coincides with a failure to transit S phase and with a decrease in cell viability.
One downstream pathway regulated by RAP-sensitive TORC1 is the initiation of translation of a subset of mRNAs by eukaryotic initiation factor 4E and eukaryotic initiation factor 4G (8). To ensure that these observations were not simply an artifact of translation inhibition by RAP, we asked if the global inhibition of protein translation by CHX in the presence of MMS induced similar effects on cell cycle and viability to those induced by RAP. Because protein synthesis is required for cells to transit from G1 into S phase (16), these experiments were performed with cells synchronized in early S phase. When HU-arrested cells were released into YPD medium containing CHX, cells progressed through S phase, albeit at a lower rate than those of control and RAP-treated cells alone (Fig. 3A). However, these cells failed to enter the next cell cycle, as protein synthesis is required for nuclear division (9). A more pronounced delay in S-phase transit was induced by MMS+CHX (compare the 220-min profiles in Fig. 3A). Yet, in contrast to the increased lethality of MMS+RAP-treated cells, CHX treatment did not affect the viability of MMS-treated cells. Thus, these data refute the simple notion that global effects of RAP on protein translation decrease cell viability in response to DNA damage in S phase.
Several lines of investigation were next pursued to ensure that these phenotypes derived from RAP inhibition of TORC1 signaling. First, efficient inhibition of TOR signaling by RAP was demonstrated by the induction of a starvation response (Fig. 4A). Following the same experimental strategy as that described for Fig. 1A, RAP-induced autophagy (26) was apparent in the majority of cells 220 min after release from α-factor in either the presence or absence of MMS. Moreover, a persistent autophagic phenotype was evident in the terminal S-phase-arrested cells following a 24-h exposure to MMS+RAP (data not shown). In contrast, no autophagic bodies were observed in MMS-treated cells.
FIG. 4.
TORC1-dependent cell viability and S-phase transit in the presence of MMS requires a functional S-phase checkpoint. (A) DIC and DAPI images of cells treated with MMS, RAP, or MMS+RAP for 220 min following release from α-factor. Arrows indicate autophagic bodies, visible in DIC images as bumps in enlarged vacuoles. (B) Isogenic wild-type, tor1Δ, TOR1RR, and rad9Δ strains were released from α-factor into YPD alone (white bars), MMS (gray bars), or MMS+RAP (black bars). The number of colonies formed at 220 min was plotted relative to the number at time zero (n = 3). Values of >1 indicate cell proliferation, and values of <1 indicate cytotoxicity. RAP alone did not affect TORRR cell growth and allowed an ∼2-fold increase in cell number (relative to that at time zero) for other strains (data not shown). The number is the ratio of colonies obtained with MMS+RAP to that obtained with MMS. (C) Isogenic wild-type and tor1Δ cells were released from α-factor arrest into YPD alone (control), RAP, MMS, MMS+RAP (200 ng/ml), or MMS plus a low concentration of RAP (50 ng/ml). At 0 and 220 min, aliquots were serially diluted and spotted onto YPD agar. Similar numbers of colonies to those shown for wild-type cells were obtained for tor1Δ cells at time zero. (D) Isogenic wild-type and mrc1Δ cells released from α-factor into YPD alone (control), MMS, RAP, or MMS+RAP were serially diluted at the indicated times and plated onto YPD agar. The number of colonies was plotted relative to the number obtained at time zero (n = 3).
Second, a series of isogenic yeast strains were engineered to assess specific alterations in TOR signaling. In these experiments, the effects of RAP were evident from 100 min on (data not shown). However, to simplify the presentation, cell viabilities 220 min following release from α-factor arrest are presented, as these data distinguish the cytotoxic and protective effects of TOR inhibition in the presence of MMS. Relative to that of wild-type cells, deletion of TOR1 had no effect on MMS cytotoxicity (Fig. 4B). However, deletion of TOR1 increased the RAP sensitivity of MMS-treated cells >10-fold (Fig. 4B). In yeast, RAP inhibits the multiprotein complex TORC1, comprised of either Tor1 or Tor2 (29). Thus, tor1Δ would reduce the cellular complement of RAP-sensitive TORC1 to those complexes containing Tor2, thereby enhancing cell sensitivity to RAP. Indeed, consistent with this interpretation, higher concentrations of RAP were required to induce wild-type cell sensitivity to MMS than those needed for isogenic tor1Δ cells following release into S phase (compare MMS+50 ng/ml RAP patterns in Fig. 4C). We then asked if TORC1 comprised of Tor2 preferentially modulates cell sensitivity to DNA damage. A RAP-resistant TORRR strain, where the Ser1972 codon was mutated to an Arg codon in TOR1S1972R (10), showed no effect of RAP on MMS cytotoxicity (Fig. 4B). Similar results were obtained with a RAP-resistant TOR2S1975R mutant (data not shown). Since the RAP resistance conferred by these single amino acid substitutions in either Tor1 or Tor2 is dominant, these findings establish that RAP-sensitive signaling through TORC1 containing Tor1 or Tor2 modulates the survival of cells exposed to DNA damage in S phase.
S-phase checkpoint activation is required for the protective function of TOR.
In response to genotoxic stress, activation of the Rad53 checkpoint maintains cell survival by regulating events that enhance the stability and repair of stalled replication forks. The checkpoint also minimizes the potential for increased DNA damage by coordinating origin firing, DNA polymerization, and histone synthesis (13, 30, 32, 35, 43). We next asked if RAP inhibition of TORC1 affected S-phase checkpoint signaling in isogenic strains defective for Rad53 checkpoint activation or function. For instance, Mrc1 promotes replication fork progression and acts as a mediator to enhance Rad53 phosphorylation in response to replication stress (2, 27, 44). Indeed, the protective function of TORC1 in MMS-treated cells required Mrc1 (Fig. 4D). The kinetics of MMS-induced lethality of mrc1Δ cells were increased relative to those of wild-type cells and were unaffected by cotreatment with RAP. Moreover, this pattern of cell lethality coincided with a failure to slow S-phase progression (see Fig. S2 in the supplemental material). Because mrc1Δ cells exhibit checkpoint-independent defects in S phase, additional studies were carried out with strains deleted for Rad9, which promotes Rad53 phosphorylation in the DNA damage checkpoint (18); Tof1, which functions with Csm3 and Mrc1 to stabilize stalled forks (11); or the Rad53 checkpoint kinase. Similar results were obtained in each case, as follows: the enhanced sensitivities of rad9Δ, tof1Δ, and rad53Δ strains to MMS were not increased by cotreatment with RAP, and TORC1 signaling did not affect S-phase progression (Fig. 4B and data not shown). Thus, diminished activation or abrogation of the S-phase checkpoint eliminates the ability of TOR signaling to maintain cell survival and promote S-phase transit.
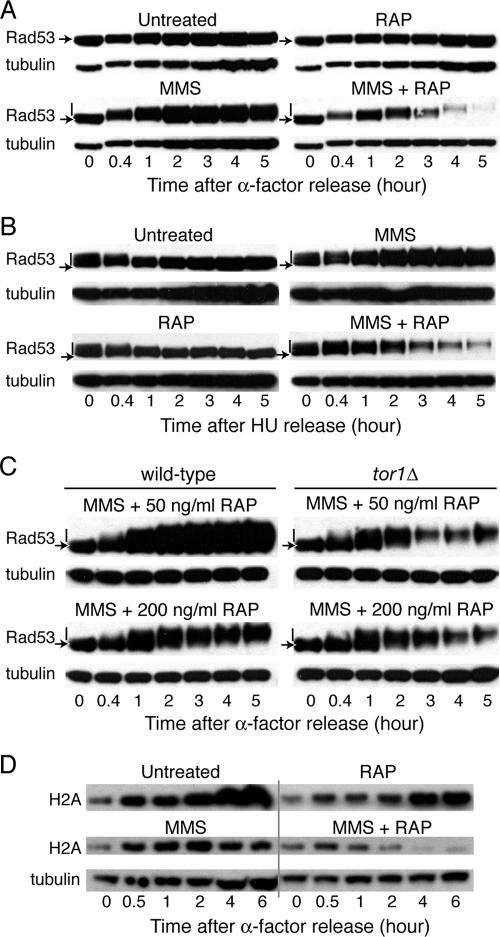
Inhibition of TORC1 increases MMS-induced Rad53 phosphorylation.
We then asked if TORC1 inhibition affected Rad53 signaling. Rad53 phosphorylation by Mec1 or Tel1 reflects the extent and duration of checkpoint activation (32). Following release of cells into S phase, RAP treatment alone failed to induce Rad53 phosphorylation or to affect Rad53 protein levels (Fig. 5A). In contrast, MMS treatment induced a shift in Rad53 mobility, which indicates Rad53 phosphorylation and checkpoint activation. Cotreatment with MMS+RAP produced an even more pronounced shift in Rad53 mobility and the downregulation of Rad53 protein levels. These data suggest that the inhibition of TORC1 signaling might increase the amount of DNA damage induced by MMS and, ergo, the hyperphosphorylated state of Rad53. However, the downregulation of Rad53 protein levels in the absence of TORC1 signaling might also impair cell viability in response to genotoxic stress.
FIG. 5.
RAP inhibition of TORC1 enhances MMS-induced Rad53 checkpoint activation. (A) Cells transformed with YCpT-RAD53-HA were released from α-factor into selective medium (control), RAP, MMS, or MMS+RAP. At the indicated times, TCA cell extracts were immunoblotted with HA and tubulin antibodies. Unphosphorylated (arrow) and phosphorylated (line) Rad53 proteins are indicated. (B) Cells transformed with YCpT-RAD53-HA were released from HU into selective medium (control), RAP, MMS, MMS+RAP, CHX, or MMS+CHX. At the times indicated, TCA cell extracts were immunoblotted with HA antibodies. (C) Isogenic wild-type and tor1Δ cells transformed with YCpT-RAD53-HA were released from α-factor into selective medium containing MMS+RAP (200 ng/ml) or MMS plus a low concentration of RAP (50 ng/ml). At the times indicated, TCA cell extracts were immunoblotted with HA and tubulin antibodies. The patterns of Rad53 mobility for the control, RAP (50 or 200 ng/ml), and MMS alone mirrored those shown in panel A. (D) As in panel A, TCA extracts of wild-type cells expressing histone H2A-HA (HTA2-HA) were immunoblotted with HA and tubulin antibodies.
To address these distinct possibilities, we considered the mechanism of Rad53 protein downregulation. First, cotreatment of cells with MMS+RAP and the proteasome inhibitor MG132 failed to stabilize Rad53 or any higher-molecular-weight forms of the protein (data not shown). Thus, increased protein turnover by ubiquitin-mediated proteolysis is unlikely. Indeed, MG132 cotreatment slightly increased the extent of Rad53 downregulation induced by MMS+RAP. However, since the cytotoxic activity of MMS+RAP was unaltered by MG132 (data not shown), these data suggested that Rad53 protein levels did not predict cell viability. Indeed, when cells were released into S phase in the presence of CHX (as shown in Fig. 3), Rad53 protein levels decreased, independent of phosphorylation status (see Fig. S3 in the supplemental material). While MMS+CHX induced a similar pattern of Rad53 downregulation, this drug combination failed to yield the increased phosphorylation of Rad53 observed with MMS+RAP (see Fig. S3 in the supplemental material). Moreover, CHX cotreatment also failed to enhance cell sensitivity to MMS (Fig. 3B). Taken together, these data suggest the downregulation of Rad53 protein at the level of translation in S phase. However, these data further indicate that it is the increased phosphorylation of Rad53, not relative Rad53 protein levels, which is relevant to the cytotoxic phenotype induced by MMS+RAP.
Several lines of evidence also suggest that the extent of Rad53 phosphorylation could not be attributed to RAP-induced alterations in the cell's ability to recover from checkpoint activation. First, cells treated with HU accumulate in early S phase due to Rad53 checkpoint activation. However, as shown in Fig. 5B, when cells were released from HU into medium containing RAP, the kinetics of cell recovery from checkpoint arrest, as assessed by increased Rad53 mobility relative to that at time zero, mirrored those of the untreated control. Yet the pattern of Rad53 phosphorylation in the presence of MMS or hyperphosphorylation induced by MMS+RAP resembled that observed following release from α-factor arrest. Second, cells deleted for ESC4 are more sensitive to MMS-induced DNA damage as a consequence of a failure to recover from checkpoint-induced S-phase arrest (38). However, these cells exhibited the same relative increase in cell death in the presence of MMS+RAP as did isogenic wild-type cells (data not shown).
The dependence of these events on RAP inhibition of TORC1 signaling is further supported by studies of wild-type and tor1Δ strains. The increased sensitivity of tor1Δ cells to MMS and lower concentrations of RAP, as shown in Fig. 4C, corresponds with an increase in Rad53 phosphorylation at lower drug concentrations in the tor1Δ, but not isogenic wild-type, strain (Fig. 5C). For both strains, the relative mobilities of Rad53 were similar in extracts of control and RAP- and MMS-treated cells (data not shown).
We then reasoned that any increase in checkpoint signaling induced by RAP treatment might be evident in downstream pathways. For instance, the Rad53 checkpoint coordinates histone biosynthesis with DNA synthesis, as excess free histones are toxic (19). When cells were released into S phase, histone H2A protein levels decreased in response to MMS-induced DNA damage; however, this effect was enhanced in the presence of MMS+RAP (Fig. 4C). RAP alone did not alter histone H2A protein levels. Thus, a direct effect of TOR signaling on histone H2A translation is unlikely. Rather, these data support a model of enhanced Rad53 checkpoint activation in response to the increase in MMS-induced DNA damage caused by TORC1 inhibition.
TOR signaling maintains Rad53 checkpoint-induced expression of RNR subunits Rnr1 and Rnr3, but not Rnr2 and Rnr4.
Perhaps the best-understood effector of the Rad53 checkpoint is RNR, which catalyzes the rate-limiting step in the production of deoxynucleoside triphosphates (dNTPs), the precursors of DNA synthesis (21, 28, 48). Eukaryotic RNR comprises an α2β2 tetramer. In yeast, the large subunit is typically a homodimer of Rnr1 (α1α1), while the small subunit is a heterodimer of Rnr2 and Rnr4 (ββ′) (28). A second large subunit, Rnr3, is highly induced by DNA damage and can complement an rnr1Δ mutation when overexpressed (16). However, in contrast to other RNR genes, RNR3 is nonessential, and no phenotype has been ascribed to rnr3Δ strains (12, 14).
In response to DNA damage, a six- to eightfold increase in intracellular dNTP pools is achieved by several mechanisms, including relaxed dATP feedback inhibition of RNR (12), Rad53-mediated transcriptional upregulation of RNR genes (21), degradation of the RNR inhibitor Sml1 (48), and subcellular relocalization of RNA small subunits (47). To ask if the phenotypes induced by TORC1 inhibition in MMS-treated cells resulted from alterations in Rad53 checkpoint regulation of RNR, we first determined whether deletion of the gene encoding the Sml1 inhibitor of RNR altered the cytotoxic activity of MMS+RAP in S phase. The viabilities of isogenic wild-type and sml1Δ cells exposed to MMS+RAP following α-factor release were identical (data not shown). Thus, the protective function of TORC1 signaling following MMS-induced checkpoint activation is not mediated by a direct effect on Sml1 regulation of RNR.
We next assessed the levels of individual RNR subunits in response to MMS+RAP. As shown in Fig. 6A, release from α-factor into RAP induced a progressive downregulation of Rnr1 protein levels relative to those in untreated controls, in part due to the accumulation of cells in the subsequent G1 phase (as shown in Fig. 1A). As previously reported (14), MMS-induced checkpoint activation produced a twofold increase in Rnr1 levels. However, this DNA damage-induced response was suppressed by RAP (MMS+RAP) (Fig. 6A), despite the fact that the majority of cells remained in S phase (Fig. 1A). This effect was even more pronounced with Rnr3. Although this RNR subunit is barely detectable in the absence of DNA damage, cotreatment with RAP dramatically suppressed the high levels of Rnr3 induced by exposure to MMS during S phase (Fig. 6A). Rnr2 and Rnr4 levels, on the other hand, were unaltered by RAP treatment (Fig. 6B; see Fig. S4 in the supplemental material).
FIG. 6.
TORC1 signaling maintains DNA damage-induced expression of Rnr1 and Rnr3. TCA extracts of wild-type or rfx1Δ cells expressing HA-tagged Rnr1 or Rnr3 (A) or of wild-type cells expressing HA-tagged Rnr4 (B), treated as described in the legend to Fig. 5A, were immunoblotted with HA or tubulin antibodies. (C) RNAs isolated from wild-type cells treated as described for panel A were subjected to Northern blot analysis with radiolabeled probes for RNR3, RNR1, and ACT1. (D) As described in the legend to Fig. 4B, rnr3Δ, rfx1Δ, and clb5Δ cells were released from α-factor into YPD alone (light gray bars), MMS (gray bars), or MMS+RAP (black bars), and the number of viable cells forming colonies at 220 min was plotted relative to that at time zero (n = 3). For comparison, wild-type cell viability data were reproduced from Fig. 4B (dashed lines). (E) RNR1-HA and GAL1 promoter-driven RNR1-HA strains were released from α-factor into galactose medium with no drug (control), RAP, MMS, or MMS+RAP. At the times indicated, cells were serially 10-fold diluted and spotted onto galactose plates. Colony formation was assessed at 30°C.
The initial increases of both Rnr1 and Rnr3 subunits in the presence of MMS+RAP (compare time zero with 0.5 and 1 h for Rnr1 and with 1 to 2 h for Rnr3) (Fig. 6A) coincide with checkpoint activation and Rad53 phosphorylation (as shown in Fig. 5A). At later times in MMS+RAP-treated cells, however, Rnr1 and Rnr3 decreases also parallel the temporal pattern of Rad53 downregulation (3 to 5 h in Fig. 6A and 5A, respectively). This transient accumulation of Rnr1 and Rnr3 suggests that the phenotypic consequences of RAP treatment are not restricted to translation inhibition. However, since DNA damage-induced RNR3 transcription is achieved by Rad53 inactivation of the Rfx1 (or Crt1) transcriptional repressor (21), we also considered that the decrease in Rad53 levels could alleviate Rfx1 inactivation to suppress RNR3 transcription. If this were the case, then an rfx1Δ mutation would abolish the RAP-induced downregulation of Rnr3 in MMS-treated cells. However, as shown in Fig. 6A, this was not the case. As reported (21), rfx1Δ cells exhibited little alteration in Rnr1 protein levels. In contrast, Rnr3 was overexpressed in rfx1Δ cells, independent of DNA damage. Although the absolute levels of Rnr3 differed substantially between wild-type and rfx1Δ strains, the kinetics of Rnr3 downregulation in the presence of MMS+RAP were comparable. Thus, the decreases in Rnr1 and Rnr3 induced by RAP inhibition of TORC1 were not due to alterations in Rad53 checkpoint regulation of Rfx1.
TORC1 signaling was also required to maintain the elevated RNR1 and RNR3 mRNA levels induced by Rad53 checkpoint activation. RNR1 expression is tightly cell cycle regulated (16). After release of cells from α-factor into medium (Fig. 6C, control panel), RNR1 mRNA levels increased by 15 min, when cells entered S phase; decreased at 30 min, when cells were in G2-M phase; and then increased again at 60 and 180 min as cells entered subsequent cell cycles. When cells were released into RAP, RNR1 transcript levels mirrored those of untreated controls as the cells progressed through S phase and then remained low as cells accumulated in the next G1 phase. In contrast, TOR signaling was required to sustain the increased expression of RNR1 and RNR3 in response to long-term MMS exposure. The increases in RNR1 and RNR3 transcripts induced by exposure to MMS were suppressed at 180 min in cells cotreated with RAP (Fig. 6C). This effect was more pronounced with RNR3 mRNA, consistent with the transient increase and then decrease in Rnr3 levels under the same conditions. These data indicate that TORC1 signaling is required to sustain checkpoint-induced expression of RNR1 and RNR3 transcripts and proteins.
Rnr3 protects cells from DNA damage in S phase.
We then considered the phenotypic consequences of RNR large-subunit downregulation in the face of persistent DNA damage. When TORC1 signaling is intact, RNR comprised of the Rnr1 large subunit (α1α1ββ′) apparently constitutes the major cellular complement of this essential enzyme, as deletion of RNR3 has no effect on cell viability in the presence or absence of MMS. For example, as shown in Fig. 6D, the viability of rnr3Δ cells exposed to MMS for 220 min following α-factor release is the same as that of wild-type cells. However, relative to wild-type RNR3 cells, rnr3Δ cells exhibited increased sensitivity to MMS when TORC1 signaling was inhibited. Thus, in RAP-treated cells, Rnr3 functions to increase the survival of cells treated with MMS. This surprising finding constitutes the first reported phenotype for rnr3Δ cells.
rfx1Δ cells exhibited a slight increase in MMS resistance (Fig. 6D) and higher basal levels of Rnr3 than that induced by MMS in wild-type cells (Fig. 6A). However, rfx1Δ cells were no more resistant to MMS+RAP than were isogenic wild-type strains. These data suggest that in the absence of a coordinate increase in Rnr1, increased levels of Rnr3 do not provide a survival advantage to cells exposed to MMS+RAP in S phase. To address the effect of Rnr1 overexpression, the galactose-inducible GAL1 promoter was inserted upstream of RNR1 by PCR-based homologous recombination. Since RNR1 is essential for cell viability, this approach necessitated culturing the cells in medium containing galactose, where the levels of Rnr1 in S phase exceed that of the wild-type control (see Fig. S5 in the supplemental material). This construct retained the 5′ untranslated region of the endogenous RNR1 gene, and thus a partial downregulation of Rnr1 by RAP was still evident. Nevertheless, as shown in Fig. 6E, the increased levels of Rnr1 partially suppressed the cytotoxic effects induced by RAP inhibition of TORC1 in MMS-treated cells.
Taken together, these findings suggest that the hyperphosphorylation of Rad53 in MMS+RAP-treated cells results from increased replication stress induced by decreased Rnr1 and Rnr3 levels. The analysis of replication intermediates in Fig. 2 indicates that early origins of replication fire efficiently in the presence of MMS+RAP but that the forks are unstable upon prolonged exposure to the drugs. A concomitant decrease in Rnr1/3 levels induced by MMS+RAP would also impact fork progression and stability. One prediction of these considerations is that strains exhibiting a prolonged S phase should be even more sensitive to MMS+RAP. In complex with Cdc28, Clb5-Cdk activates the firing of early and late origins in S phase, while Clb6-Cdk supports the firing of early origins only (15). As shown in Fig. 6D, deletion of the B-type cyclin Clb5 had no effect on cell sensitivity to MMS yet enhanced the cytotoxic activity of MMS in the presence of RAP. Restricting the initiation of DNA replication to early origins, and thereby decreasing the number of replication forks in the cell, increased the requirement for TORC1 signaling as a survival pathway following MMS activation of the S-phase checkpoint.
TORC1 signaling promotes MMS-induced mutagenesis.
Increased dNTP levels produced in response to DNA damage promote cell survival at the cost of increased mutation levels (12). Chabes et al. (12) posited that higher levels of dNTPs promote translesion DNA synthesis to bypass DNA lesions, as polymerases generally have a higher Km for binding nucleotides opposite a damaged base (36). We reasoned that the converse would also apply—that RAP- induced decreases in Rnr1/3 levels would fail to yield sufficient dNTP concentrations to promote translesion DNA synthesis, thereby decreasing cell viability and suppressing MMS-induced mutations. One measure of mutation frequency is the acquisition of canavanine resistance due to mutation of the arginine permease encoded by CAN1. Indeed, treatment with RAP completely suppressed the acquisition of canavanine resistance induced by MMS as well as reducing the frequency of spontaneous CAN1 mutants in the absence of MMS (Fig. 7). As shown in Fig. 3, this effect was not due to global effects on translation, as treating cells with CHX under the same conditions induced the downregulation of all RNR subunits yet failed to suppress the frequency of spontaneous or MMS-induced CAN1 mutants (Fig. 7 and data not shown). Thus, MMS-induced mutagenesis was selectively suppressed by RAP inhibition of TORC1.
FIG. 7.
TORC1 signaling promotes MMS-induced mutagenesis. Aliquots of wild-type cells released from α-factor into YPD medium (control), MMS, RAP, or MMS+RAP or released from HU into YPD medium (control), MMS, CHX, or MMS+CHX were plated onto YPD agar or onto synthetic medium without Arg and with canavanine at the indicated times. The number of canavanine-resistant colonies per total number of viable cells forming colonies on YPD was plotted relative to the spontaneous frequency of 2 × 10−6 canavanine-resistant mutants obtained at time zero.
DISCUSSION
TOR is a highly conserved serine/threonine kinase that functions as a central regulator of eukaryotic cell responses to environmental stresses, such as amino acid starvation, hypoxia, and growth factor deprivation (7, 8, 24, 33). Subsequent signaling through distinct downstream pathways regulates the cap-dependent translation of a subset of mRNAs as well as transcriptional stress responses, progression from G1 to S phase of the cell cycle, and cell survival. TOR is also a member of the family of phosphatidylinositol 3-kinase-related kinases, which include DNA-PK, ATM, ATR, and the yeast Mec1 and Tel1 kinases, all of which play important roles in DNA repair and/or checkpoint responses to DNA damage. The macrocyclic antibiotic RAP, in complex with an immunophilin (FKBP12), specifically targets TOR (8, 40), and several RAP analogs are currently in phase I to III oncology clinical trials (8).
Using yeast as a genetically tractable model, our studies reveal a novel role for TORC1 signaling as a determinant of cell survival in response to aberrations in DNA replication. RAP-sensitive TORC1 functions to promote S-phase transit and fork stability and to maintain cell viability following Rad53 checkpoint activation by DNA damage. Our studies further establish a critical function for Rnr3 in maintaining the viability of cells exposed to DNA damage in the absence of TORC1 signaling: while RAP-induced downregulation of Rnr1 and Rnr3 levels enhances MMS cytotoxicity, this effect is exacerbated in cells deleted for RNR3.
Taken together, our data support the model shown in Fig. 8. In S phase, RNR activity is regulated to a significant extent by cell cycle-dependent expression of RNR1, such that Rnr1 levels are maximal in S phase to produce sufficient dNTPs for DNA replication (16) (Fig. 8A). In the presence of DNA damage, Rad53 checkpoint activation enhances RNR activity, in part by increased RNR subunit gene transcription (21, 28, 48) (Fig. 6C and 8B). RNR3 is highly induced, yet rnr3Δ strains do not exhibit increased sensitivity to DNA-damaging agents. However, Rnr3 levels only reach 1/10 the Rnr1 levels, and the activity of α3α3ββ′ RNR is <1% that of α1α1ββ′ RNR (14). Thus, even though RNR comprised of Rnr1 and Rnr3 (α1α3ββ′) exhibits synergistic activity in vitro (14), the contribution of this complex to the net increase in dNTPs would be negligible. Instead, RNR composed of α1α1ββ′ would predominate, with relaxed feedback inhibition generating sufficient dNTPs for cell survival (12).
FIG. 8.
Model for RAP-induced alterations in RNR, cell survival, and mutation rates in response to Rad53 checkpoint activation (see the text for details).
This situation contrasts with the downregulation of both Rnr1 and Rnr3 when TORC1 signaling is inhibited in the presence of MMS-induced DNA damage (Fig. 8C). In this case, diminished RNR (α1α1ββ′) levels would adversely impact dNTP production, such that the synergistic activity of even limiting amounts of RNR comprised of α1α3ββ′ might promote cell survival. This model predicts that limiting dNTPs in the presence of MMS+RAP would preclude efficient translesion DNA synthesis, thereby suppressing MMS-induced mutations. Indeed, RAP inhibition of TORC1 suppressed the frequencies of spontaneous and MMS-induced CAN1 mutations, as shown in Fig. 7.
Another prediction of this model is that deletion of RNR3 would selectively enhance MMS+RAP cytotoxicity. However, alterations in Rnr3 levels should not affect cell survival when TORC1 signaling is intact, since the normal complement of Rnr1 would suffice to maintain dNTP levels. Indeed, while rnr3Δ cells were no more sensitive to MMS than wild-type cells in S phase, cells lacking Rnr3 were hypersensitive to MMS+RAP (Fig. 6D). This constitutes a novel phenotype for rnr3Δ cells and demonstrates that in the absence of TORC1 signaling, Rnr3 function is necessary to maintain the viability of MMS-treated cells.
This model also explains the phenotypes of rfx1Δ cells. Since the basal Rnr3 level in rfx1Δ cells exceeded that induced by MMS in wild-type cells (Fig. 6A), the slight increase in MMS resistance of rfx1Δ cells (Fig. 6D) may result from increased levels of RNR composed of α1α3ββ′. However, the model holds that unless Rnr1 levels are also sustained, Rnr3 overexpression simply increases α3α3ββ′ RNR levels, which would fail to suppress the increased cytotoxicity induced by MMS+RAP. Indeed, Rnr1 levels were similar in wild-type and rfx1Δ cells (Fig. 6A), as were wild-type and rfx1Δ cell sensitivities to MMS+RAP (Fig. 6D). Thus, in the absence of TORC1 signaling, Rnr1 and Rnr3 both function to promote cell viability in the presence of DNA damage.
In summary, we have shown that TOR functions in S phase to regulate cell survival in response to a variety of genotoxic stresses. Our data support a model whereby the TOR pathway is required to sustain the DNA damage-mediated induction of RNR1 and RNR3, thereby ensuring sufficient RNR activity to generate the high levels of dNTPs necessary for translesion DNA synthesis to bypass MMS-induced DNA lesions. The regulation of translesion synthesis of DNA adducts by this mechanism could also explain the increased sensitivity of mammalian cells to cisplatin induced by RAP (5). One implication of our work is that clinically acquired drug resistance may be a consequence of TOR-dependent mutations resulting from S-phase checkpoint activation. We consider this unique finding pertinent at a time when molecularly targeted agents are being combined with classical cytotoxic drugs. The potential to block drug-induced mutations that confer resistance represents a unique application of RAP analogs with potential clinical importance for the treatment of adult and pediatric malignancies.
Supplementary Material
Acknowledgments
We thank Peter Houghton and members of the Bjornsti lab for helpful discussions and Carol Newlon and Jim Theis for help with 2-D gel methodology.
This work was supported by Public Health Service grant CA23099 (to M.-A.B) from the National Cancer Institute, by NCI Cancer Center Core grant CA21765, and by the American Lebanese Syrian Associated Charities.
Footnotes
Published ahead of print on 13 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abraham, R. T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amsterdam) 3:883-887. [DOI] [PubMed] [Google Scholar]
- 2.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 3.Aronova, S., K. Wedaman, S. Anderson, J. Yates III, and T. Powers. 2007. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2779-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuvink, I., A. Boulay, S. Fumagalli, F. Zilbermann, S. Ruetz, T. O'Reilly, F. Natt, J. Hall, H. A. Lane, and G. Thomas. 2005. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell 120:747-759. [DOI] [PubMed] [Google Scholar]
- 6.Bjornsti, M. A. 2002. Cancer therapeutics in yeast. Cancer Cell 2:267-273. [DOI] [PubMed] [Google Scholar]
- 7.Bjornsti, M. A., and P. J. Houghton. 2004. Lost in translation: dysregulation of cap-dependent translation and cancer. Cancer Cell 5:519-523. [DOI] [PubMed] [Google Scholar]
- 8.Bjornsti, M. A., and P. J. Houghton. 2004. The TOR pathway: a target for cancer therapy. Nat. Rev. Cancer 4:335-348. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D. J., and D. Church. 1991. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3691-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cafferkey, R., M. M. McLaughlin, P. R. Young, R. K. Johnson, and G. P. Livi. 1994. Yeast TOR (DRR) proteins: amino-acid sequence alignment and identification of structural motifs. Gene 141:133-136. [DOI] [PubMed] [Google Scholar]
- 11.Calzada, A., B. Hodgson, M. Kanemaki, A. Bueno, and K. Labib. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein, and L. Thelander. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112:391-401. [DOI] [PubMed] [Google Scholar]
- 13.Cobb, J. A., T. Schleker, V. Rojas, L. Bjergbaek, J. A. Tercero, and S. M. Gasser. 2005. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19:3055-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domkin, V., L. Thelander, and A. Chabes. 2002. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 277:18574-18578. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 16.Elledge, S. J., and R. W. Davis. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4:740-751. [DOI] [PubMed] [Google Scholar]
- 17.Fiorani, P., R. J. Reid, A. Schepis, H. R. Jacquiau, H. Guo, P. Thimmaiah, P. Benedetti, and M. A. Bjornsti. 2004. The deubiquitinating enzyme Doa4p protects cells from DNA topoisomerase I poisons. J. Biol. Chem. 279:21271-21281. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 19.Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537-549. [DOI] [PubMed] [Google Scholar]
- 20.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926-1945. [DOI] [PubMed] [Google Scholar]
- 21.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 22.Huang, S., L. N. Liu, H. Hosoi, M. B. Dilling, T. Shikata, and P. J. Houghton. 2001. p53/p21(CIP1) cooperate in enforcing rapamycin-induced G(1) arrest and determine the cellular response to rapamycin. Cancer Res. 61:3373-3381. [PubMed] [Google Scholar]
- 23.Huang, S., L. Shu, M. B. Dilling, J. Easton, F. C. Harwood, H. Ichijo, and P. J. Houghton. 2003. Sustained activation of the JNK cascade and rapamycin-induced apoptosis are suppressed by p53/p21(Cip1). Mol. Cell 11:1491-1501. [DOI] [PubMed] [Google Scholar]
- 24.Inoki, K., H. Ouyang, Y. Li, and K. L. Guan. 2005. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 69:79-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquiau, H. R., R. C. van Waardenburg, R. J. Reid, M. H. Woo, H. Guo, E. S. Johnson, and M. A. Bjornsti. 2005. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J. Biol. Chem. 280:23566-23575. [DOI] [PubMed] [Google Scholar]
- 26.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078-1083. [DOI] [PubMed] [Google Scholar]
- 28.Kolberg, M., K. R. Strand, P. Graff, and K. K. Andersson. 2004. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta 1699:1-34. [DOI] [PubMed] [Google Scholar]
- 29.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 30.Longhese, M. P., M. Clerici, and G. Lucchini. 2003. The S-phase checkpoint and its regulation in Saccharomyces cerevisiae. Mutat. Res. 532:41-58. [DOI] [PubMed] [Google Scholar]
- 31.Lopes, M., C. Cotta-Ramusino, G. Liberi, and M. Foiani. 2003. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell 12:1499-1510. [DOI] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 33.Martin, D. E., and M. N. Hall. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17:158-166. [DOI] [PubMed] [Google Scholar]
- 34.Mulet, J. M., D. E. Martin, R. Loewith, and M. N. Hall. 2006. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281:33000-33007. [DOI] [PubMed] [Google Scholar]
- 35.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 36.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 37.Reid, R. J., P. Fiorani, M. Sugawara, and M. A. Bjornsti. 1999. CDC45 and DPB11 are required for processive DNA replication and resistance to DNA topoisomerase I-mediated DNA damage. Proc. Natl. Acad. Sci. USA 96:11440-11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouse, J. 2004. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 23:1188-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarbassov, D. D., S. M. Ali, and D. M. Sabatini. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596-603. [DOI] [PubMed] [Google Scholar]
- 40.Sawyers, C. L. 2003. Will mTOR inhibitors make it as cancer drugs? Cancer Cell 4:343-348. [DOI] [PubMed] [Google Scholar]
- 41.Shi, Y., A. Frankel, L. G. Radvanyi, L. Z. Penn, R. G. Miller, and G. B. Mills. 1995. Rapamycin enhances apoptosis and increases sensitivity to cisplatin in vitro. Cancer Res. 55:1982-1988. [PubMed] [Google Scholar]
- 42.Tabuchi, M., A. Audhya, A. B. Parsons, C. Boone, and S. D. Emr. 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26:5861-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 44.Tourriere, H., G. Versini, V. Cordon-Preciado, C. Alabert, and P. Pasero. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol. Cell 19:699-706. [DOI] [PubMed] [Google Scholar]
- 45.Wullschleger, S., R. Loewith, and M. N. Hall. 2006. TOR signaling in growth and metabolism. Cell 124:471-484. [DOI] [PubMed] [Google Scholar]
- 46.Wullschleger, S., R. Loewith, W. Oppliger, and M. N. Hall. 2005. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280:30697-30704. [DOI] [PubMed] [Google Scholar]
- 47.Yao, R., Z. Zhang, X. An, B. Bucci, D. L. Perlstein, J. Stubbe, and M. Huang. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. USA 100:6628-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, X., and R. Rothstein. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99:3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.