Abstract
Many gram-negative bacterial pathogenicity factors that function beyond the outer membrane are secreted via a contact-dependent type III secretion system. Two types of substrates are predestined for this mode of secretion, namely, antihost effectors that are translocated directly into target cells and the translocators required for targeting of the effectors across the host cell membrane. N-terminal secretion signals are important for recognition of the protein cargo by the type III secretion machinery. Even though such signals are known for several effectors, a consensus signal sequence is not obvious. One of the translocators, LcrV, has been attributed other functions in addition to its role in translocation. These functions include regulation, presumably via interaction with LcrG inside bacteria, and immunomodulation via interaction with Toll-like receptor 2. Here we wanted to address the significance of the specific targeting of LcrV to the exterior for its function in regulation, effector targeting, and virulence. The results, highlighting key N-terminal amino acids important for LcrV secretion, allowed us to dissect the role of LcrV in regulation from that in effector targeting/virulence. While only low levels of exported LcrV were required for in vitro effector translocation, as deduced by a cell infection assay, fully functional export of LcrV was found to be a prerequisite for its role in virulence in the systemic murine infection model.
Type III secretion (T3S) is employed by diverse gram-negative bacteria to translocate proteins into eukaryotic cells, resulting in commensalistic, mutualistic, or parasitic outcomes (14, 38). Pathogenic Yersinia spp. all utilize a T3S system (T3SS) to parasitize host cells (7). Translocating an armory of toxins, termed Yop (Yersinia outer protein) effectors (60), into immune cells allows Yersinia cells to avoid phagocytosis and to replicate extracellularly (17, 24, 49). Yop effector translocation requires the secretion of a second set of type III substrates, the translocators, which include LcrV, YopB, and YopD (11, 13, 16, 39, 47, 55). YopB and YopD are believed to form a translocon pore in the target eukaryotic cell plasma membrane (6, 15, 16, 27, 35, 58), through which effectors may gain access to the eukaryotic cell interior.
LcrV resides at the T3S needle tip emanating from the bacterial envelope (33). From here, LcrV promotes assembly of the translocon pore in the eukaryotic cell plasma membrane (6, 15, 27). Since antibodies specific for LcrV also prevent effector translocation and protect animals from challenge with virulent yersiniae (8, 18, 32, 62), LcrV is being pursued as a vaccine candidate against plague and other Yersinia infections (59). LcrV also impacts the positive regulatory loop needed for Yop synthesis. This is evident inside bacteria, where LcrV can remove the LcrG gating mechanism (36), or it might transduce an activating signal through the needle after it interacts with the surfaces of host cells, perhaps via Toll-like receptor 2 (TLR2) (52). Interaction with TLR2 also permits LcrV to modulate tumor necrosis factor alpha, gamma interferon, and interleukin-10 (IL-10) cytokine production (32, 34, 50-52). Hence, LcrV is a multifunctional protein that exerts its biological effects at three distinct locations, i.e., inside the bacterium, at the tip of the T3S needle complex, and as an exported protein. It is difficult to envisage how this multitasking by LcrV is coordinated, but controlled secretion could be one possibility.
The T3S effector substrate N terminus is an important secretion signal (24, 25, 40, 43, 51, 54). Reporter fusion studies with Yop substrates indicated that fewer than the first 10 residues are sufficient for T3S. However, no consensus sequence is evident, even though reciprocal substrate secretion and translocation between functionally distinct systems can occur. The genetic makeup of the signal may actually constitute both mRNA (for example, see references 1, 2, and 44) and amino acids (for example, see references 21, 26, and 61). However, the molecular makeup of translocator substrate secretion signals is essentially unknown.
With the idea to scrutinize the significance of secretion for the different proposed biological functions of LcrV, we set out to determine if the LcrV N terminus, similar to that of Yop effectors, contains the secretion motif recognized by the T3SS of Yersinia pseudotuberculosis. Secretion was impaired for several LcrV variants with mutations within the first 15 amino acids of the N terminus: amino acids 2 to 4 and 11 to 13 were essential for LcrV secretion, while amino acids 5 to 10 were important but not absolutely required. Some LcrV variants were also affected in general type III substrate secretion, yop regulation, and Yop effector translocation. However, the role of LcrV in regulation could clearly be separated from its ability to be secreted. In addition, we investigated the impact of altered LcrV secretion on the pathogenicity of Yersinia. While only low-level LcrV secretion was needed for functional T3S, as measured by an in vitro cell infection assay, wild-type levels of LcrV secretion were required for full virulence in the mouse infection model. This highlights the important role of export for the full biological function of LcrV in pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise indicated, bacteria were routinely cultivated in Luria-Bertani (LB) agar or broth at either 26°C (Y. pseudotuberculosis) or 37°C (Escherichia coli), with aeration. Where appropriate, the following antibiotics were added at the indicated final concentrations: ampicillin (Amp; 100 μg per ml), chloramphenicol (Cm; 20 μg per ml), spectinomycin (Sp; 20 μg per ml), and kanamycin (Km; 50 μg per ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7679 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S17-1 λpir | recA thi pro hsdRM+ Smr <RP4:2-Tc:Mu:Ku:Tn7> Tpr | 48 |
| Y. pseudotuberculosis strains | ||
| YPIII/pIB102 | yadA::Tn5 (wild type) Kmr | 5 |
| YPIII/pIB10201 | pIB102; lcrV with a +1 frameshift mutation incorporating codons 4 to 13; Kmr | This study |
| YPIII/pIB10202 | pIB102; lcrV with a −1 frameshift mutation incorporating codons 2 to 15; Kmr | This study |
| YPIII/pIB10203 | pIB102; lcrV with wobble base mutations between codons 1 and 15, altering only the mRNA sequence (Scramble); Kmr | This study |
| YPIII/pIB10204 | pIB102; lcrV in-frame deletion of codons 2 to 20; Kmr | This study |
| YPIII/pIB10205 | pIB102; lcrV in-frame deletion of codons 3 to 20; Kmr | This study |
| YPIII/pIB10206 | pIB102; lcrV in-frame deletion of codons 5 to 20; Kmr | This study |
| YPIII/pIB10207 | pIB102; lcrV in-frame deletion of codons 7 to 20; Kmr | This study |
| YPIII/pIB10208 | pIB102; lcrV in-frame deletion of codons 9 to 20; Kmr | This study |
| YPIII/pIB10209 | pIB102; lcrV in-frame deletion of codons 11 to 20; Kmr | This study |
| YPIII/pIB10210 | pIB102; lcrV in-frame deletion of codons 13 to 20; Kmr | This study |
| YPIII/pIB10211 | pIB102; lcrV in-frame deletion of codons 15 to 20; Kmr | This study |
| YPIII/pIB10212 | pIB102; lcrV in-frame deletion of codons 17 to 20; Kmr | This study |
| YPIII/pIB10213 | pIB102; lcrV in-frame deletion of codons 19 and 20; Kmr | This study |
| YPIII/pIB10214 | pIB102; lcrV in-frame deletion of codons 25 to 40; Kmr | This study |
| YPIII/pIB10215 | pIB102; lcrV in-frame deletion of codons 2 to 4; Kmr | This study |
| YPIII/pIB10216 | pIB102; lcrV in-frame deletion of codons 5 to 7; Kmr | This study |
| YPIII/pIB10217 | pIB102; lcrV in-frame deletion of codons 8 to 10; Kmr | This study |
| YPIII/pIB10218 | pIB102; lcrV in-frame deletion of codons 11 to 13; Kmr | This study |
| YPIII/pIB10219 | pIB102; lcrV in-frame deletion of codons 14 to 16; Kmr | This study |
| YPIII/pIB10220 | pIB102; lcrV in-frame deletion of codons 17 to 18; Kmr | This study |
| YPIII/pIB19 | pIB102; lcrV in-frame deletion of codons 10 to 313; Kmr | 39 |
| YPIII | Virulence plasmid-cured strain | 4 |
| YPIII/pIB26 | pIB102; lcrQ in-frame deletion; Kmr Spr/Smr | 40 |
| YPIII/pIB1926 | pIB19; lcrQ in-frame deletion; Kmr Spr/Smr | 19 |
| YPIII/pIB1020126 | pIB10201; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020226 | pIB10202; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020326 | pIB10203; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020426 | pIB10204; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020526 | pIB10205; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020626 | pIB10206; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020726 | pIB10207; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020826 | pIB10208; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1020926 | pIB10209; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021026 | pIB10210; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021126 | pIB10211; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021226 | pIB10212; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021326 | pIB10213; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021426 | pIB10214; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021526 | pIB10215; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021626 | pIB10216; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021726 | pIB10217; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021826 | pIB10218; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1021926 | pIB10219; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| YPIII/pIB1022026 | pIB10220; in-frame deletion of lcrQ; Kmr Spr/Smr | This study |
| Plasmids | ||
| pCR4-TOPO | TA cloning vector; Kmr Ampr | Invitrogen |
| pDM4 | Suicide plasmid carrying sacBR; Cmr | 31 |
| pRN53 | pDM4 in which the entire lcrQ allele is replaced by an Sp/Sm resistance gene; Cmr Spr/Smr | 40 |
| pJEB357 | pDM4 encoding LcrVΔ2-20; Cmr | This study |
| pJEB358 | pDM4 encoding LcrVΔ3-20; Cmr | This study |
| pJEB359 | pDM4 encoding LcrVΔ5-20; Cmr | This study |
| pJEB360 | pDM4 encoding LcrVΔ7-20; Cmr | This study |
| pJEB361 | pDM4 encoding LcrVΔ9-20; Cmr | This study |
| pJEB362 | pDM4 encoding LcrVΔ11-20; Cmr | This study |
| pJEB363 | pDM4 encoding LcrVΔ13-20; Cmr | This study |
| pJEB364 | pDM4 encoding LcrVΔ15-20; Cmr | This study |
| pJEB365 | pDM4 encoding LcrVΔ17-20; Cmr | This study |
| pJEB366 | pDM4 encoding LcrVΔ19-20; Cmr | This study |
| pJEB367 | pDM4 encoding LcrVΔ25-40; Cmr | This study |
| pJEB368 | pDM4 encoding LcrV with a +1 frameshift mutation incorporating codons 4 to 13; Cmr | This study |
| pJEB369 | pDM4 encoding LcrV with a −1 frameshift mutation incorporating codons 2 to 15; Cmr | This study |
| pJEB370 | pDM4 encoding LcrV with wobble base mutations of codons 2 to 15; Cmr | This study |
| pJEB378 | pDM4 encoding LcrVΔ2-4; Cmr | This study |
| pJEB379 | pDM4 encoding LcrVΔ5-7; Cmr | This study |
| pJEB380 | pDM4 encoding LcrVΔ8-10; Cmr | This study |
| pJEB381 | pDM4 encoding LcrVΔ11-13; Cmr | This study |
| pJEB382 | pDM4 encoding LcrVΔ14-16; Cmr | This study |
| pJEB383 | pDM4 encoding LcrVΔ17-18; Cmr | This study |
Construction of lcrV mutations by allelic exchange.
Amplified DNA fragments used for constructing in-frame deletion mutants, frameshift mutants, and the wobble base mutant were generated by overlap PCR (20), using Y. pseudotuberculosis YPIII/pIB102 as a template. The primer combinations used to create the mutants are listed in Table 2. Each fragment containing a sequence flanking the specific lcrV deletion was confirmed by sequence analysis (MWG Biotech AG, Martinsried, Germany), which was facilitated by initial cloning into the pCR4-TOPO TA cloning vector (Invitrogen AB, Stockholm, Sweden). The fragments were then cloned into XhoI-SacI-digested suicide mutagenesis vector pDM4 (31). E. coli S17-1 λpir was used as the donor strain in conjugal mating experiments with parental Y. pseudotuberculosis YPIII/pIB102. To construct double lcrV lcrQ mutants, the mutagenesis plasmid pRN53, in which the entire lcrQ allele is replaced by an Sp/streptomycin (Sm) resistance gene, was used (40). For selection of the appropriate allelic exchange events, we used established methods (31). All of the resulting mutants listed in Table 1 were verified by PCR and DNA sequencing.
TABLE 2.
Oligonucleotides used for construction of lcrV mutants in cis
| LcrV mutanta | Oligonucleotide pair (sequence [5′-3′)b |
|---|---|
| Deletion mutants | |
| V1 (2-20) | pLcrVdelA (GCA TGT CTC GAG CAG AAC TGG CAA TAG CCG AC) and pLcrVdel2-20b (CAT ATT AAA TAA TTT GCC CTC GCA TC) |
| pLcrVdel2-20C (GCA AAT TAT TTA ATA TGG TGG AAC AAC TTA CTG GTC ATG G) and pLcrVdelD1 (GCA TGT GAG CTC ACT CGC TTG ATG CCA TTT TGC AG) | |
| V2 (3-20) | pLcrVdelA and pLcrVdel3-20b (AAT CAT ATT AAA TAA TTT GCC CTC GC) |
| pLcrVdel3-20c (ATT ATT TAA TAT GAT TGT GGA ACA ACT TAC TGG TCA TGG) and pLcrVdelD1 | |
| V3 (5-20) | pLcrVdelA and pLcrVdel5-20b (GGC TCT AAT CAT ATT AAA TAA T) |
| pLcrVdel5-20c (TAA TAT GAT TAG AGC CGT GGA ACA ACT TAC TGG TCA TGG) and pLcrVdelD1 | |
| V4 (7-20) | pLcrVdelA and pLcrVdel7-20b (TTC GTA GGC TCT AAT CAT ATT) |
| pLcrVdel7-20c (ATT AGA GCC TAC GAA GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V5 (9-20) | pLcrVdelA and pLcrVdel9-20b (GTT TTG TTC GTA GGC TCT AAT) |
| pLcrVdel9-20c (GCC TAC GAA CAA AAC GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V6 (11-20) | pLcrVdelA and pLcrVdel11-20b (TTG TGG GTT TTG TTC GTA GGC T) |
| pLcrVdel11-20c (CGA ACA AAA CCC ACA AGT GGA ACA ACT TAC TGG TCA TGG) and pLcrVdelD1 | |
| V7 (13-20) | pLcrVdelA and pLcrVdel13-20b (AAA TGT TGT GGG TTT TGT TCG) |
| pLcrVdel13-20c (AAC CCA CAA CAT TTT GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V8 (15-20) | pLcrVdelA and pLcrVdel15-20b (CTC AAT AAA ATG TTG TGG GTT) |
| pLcrVdel15-20c (CAA CAT TTT ATT GAG GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V9 (17-20) | pLcrVdelA and pLcrVdel17-20b (TAG ATC CTC AAT AAA ATG TTG TG) |
| pLcrVdel17-20c (TTT ATT GAG GAT CTA GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V10 (19-20) | pLcrVdelA and pLcrVdel19-20b (TTT TTC TAG ATC CTC AAT AAA ATG) |
| pLcrVdel19-20c (GAG GAT CTA GAA AAA GTG GAA CAA CTT ACT GGT CAT GG) and pLcrVdelD1 | |
| V11 (25-40) | pLcrVdelA and pLcrVdel25-40b (AAG TTG TTC CAC CCT AAC TT) |
| pLcrVdel25-40c (AGG GTG GAA CAA CTT GAT AAA AAT ATA GAT ATT TCC ATT A) and pLcrVdelD2 (GCA TGT GAG CTC CTA CGG GCA TCA CCA TGA TGA T) | |
| V12 (2-4) | pLcrVdelA and pLcrVdel2-20b |
| pLcrVdel2-4c (GCA AAT TAT TTA ATA TGT ACG AAC AAA ACC CAC AAC ATT) and pLcrVdelD1 | |
| V13 (5-7) | pLcrVdelA and pLcrVdel5-20b |
| pLcrVdel5-7c (TAA TAT GAT TAG AGC CAA CCC ACA ACA TTT TAT TGA GG) and pLcrVdelD1 | |
| V14 (8-10) | pLcrVdelA and pLcrVdel8-10b (TTG TTC GTA GGC TCT AAT CAT) |
| pLcrVdel8-10C (TTA GAG CCT ACG AAC AAC ATT TTA TTG AGG ATC TAG AA) and pLcrVdelD1 | |
| V15 (11-13) | pLcrVdelA and pLcrVdel11-20b |
| pLcrVdel11-13c (CGA ACA AAA CCC ACA AGA GGA TCT AGA AAA AGT TAG G) and pLcrVdelD1 | |
| V16 (14-16) | pLcrVdelA and pLcrVdel14-16b (AAT AAA ATG TTG TGG GTT TTG TTC G) |
| pLcrVdel14-16c (CCC ACA ACA TTT TAT TGA AAA AGT TAG GGT GGA ACA ACT T) and pLcrVdelD1 | |
| V17 (17-18) | pLcrVdelA and pLcrVdel17-20b |
| pLcrVdel17-18c (TTT ATT GAG GAT CTA GTT AGG GTG GAA CAA CTT ACT GGT) and pLcrVdelD1 | |
| Frameshift and wobble base mutants | |
| FS +1 (4-13) | pLcrVdelA and pLcrV + 1(4-13)b (TTG TGG GTT TTG TTC GTA GGC TTC TAA TCA TAT TAA ATA A) |
| pLcrV + 1(4-13)c (CTA CGA ACA AAA CCC ACA ACA TTT TAT GAG GAT CTA GAA AAA GTT AGG GT) and pLcrVdelD1 | |
| FS −1 (2-15) | pLcrVdelA and pLcrV-1(2-15)b (TTG TGG GTT TTG TTC GTA GGC TCT AAC ATA TTA AAT AAT TTG CCC TCG CA) |
| pLcrV-1(2-15)c (CGA ACA AAA CCC ACA ACA TTT TAT TGA GGA TTC TAG AAA AAG TTA GGG TG) and pLcrVdelD1 | |
| Scramble (2-15) | pLcrVdelA and pLcrVmRNAmutb (CTG CGG ATT CTG CTC ATACGC CCT GAT CAT ATT AAA TAA TTT GCC CTC GCA T) |
| pLcrVmRNAmutc (TAT GAG CAG AAT CCG CAG CAC TTC ATC GAA GAC CTA GAA AAA GTT AGG GTG GAA CA) and pLcrVdelD1 |
Numbers in parentheses indicate the internal amino acids deleted or altered in LcrV (316 amino acids).
Italic type indicates the incorporated XhoI and SacI restriction sites used for cloning of the PCR-amplified DNA fragments. Underlining indicates the complementary overlap between the corresponding B and C primers. Bold type indicates altered or inserted oligonucleotides.
Analysis of mRNA secondary structure.
The secondary structures of lcrV mRNA sequences were predicted with the program Mfold, version 3.2 (29, 64), using the online Mfold server at http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi. The default parameters (37°C, 1 M NaCl, no divalent ions, and 5% suboptimality) were used for the predictions. The sequences tested incorporated the 42-bp region encoding residues 2 to 15, which were systematically mutated in the scramble mutant YPIII/pIB10203. Upstream (U) and downstream (D) flanking regions of a total of 100 bp were included in the analysis as follows: 100 U (1), 80 U-20 D (2), 60 U-40 D (3), 50 U-50 D (6), 40 U-60 D (4), 20 U-80 D (5), and 100 D (7). This generated a total of seven sequences each for wild-type LcrV and the Scramble mutant, which were used in the structure prediction analysis.
Growth phenotype assessment.
The growth phenotype was assessed after culturing cells in liquid TMH medium under high- and low-Ca2+ conditions at 37°C (12, 57). In short, overnight cultures grown in TMH at 26°C were diluted in fresh TMH or TMH supplemented with 2.5 mM CaCl2 to an optical density at 600 nm (OD600) of 0.1. Duplicates of each sample were made; one was kept at 26°C, while the other was incubated at 37°C. During a 10-h period, samples were taken every 2 hours for OD measurements. Parental Yersinia strains displayed calcium-dependent (CD) growth, while the lcrV full-length deletion mutant grew equally well regardless of the Ca2+ concentration, a phenotype referred to as calcium-independent (CI) growth. The growth phenotypes of individual lcrV mutants are summarized in Table 3.
TABLE 3.
lcrV mutants in cis and their phenotypesa
| LcrV variant | LcrV phenotype
|
Yop phenotype
|
Growth phenotyped |
lcrQ::sp-sm effect
|
Translocatione | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stabilityb | Synthesisc | Secretionc | Synthesisc | Secretionc | LcrV secretionc | Yop secretionc | |||
| Parent | ↓ | ↓ | ↓ | ↓ | ↓ | CD | ↑ (plus Ca2+) | ↓ | ↓ |
| LcrVnull | NA | NA | NA | ↓↓ | ↓↓ | CI | NA | ↓ | ↓↓ |
| Frame +1 | ↔ | ↔ | ↔ | ↔ | ↔ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| Frame −1 | ↔ | ↔ | ↔ | ↔ | ↔ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| Scramble | ↔ | ↔ | ↔ | ↔ | ↔ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| V1 (Δ2-20) | ↓ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V2 (Δ3-20) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V3 (Δ5-20) | ↔ | ↑ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↑ (plus Ca2+) | ↓↓ |
| V4 (Δ7-20) | ↔ | ↑ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V5 (Δ9-20) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V6 (Δ11-20) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V7 (Δ13-20) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↑ (plus Ca2+) | ↓↓ |
| V8 (Δ15-20) | ↔ | ↔ | ↓ | ↓ | ↓ | CD-like | ↑ (plus Ca2+) | ↔ | ↔ |
| V9 (Δ17-20) | ↔ | ↔ | ↓ | ↓ | ↓ | CD | ↑ (plus Ca2+) | ↑ (translocators)f | ↔ |
| V10 (Δ19-20) | ↔ | ↔ | ↓ | ↔ | ↔ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| V11 (Δ25-40) | ↔ | ↔ | ↓ | ↓ | ↓ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| V12 (Δ2-4) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD-like | ↓↓g | ↔ | ↓↓ |
| V13 (Δ5-7) | ↔ | ↔ | ↓ | ↔ | ↔ | CD | ↑ (plus Ca2+) | ↑ (translocators)f | ↔ |
| V14 (Δ8-10) | ↔ | ↔ | ↓ | ↓ | ↓ | CD | ↑ (plus Ca2+) | ↔ | ↔ |
| V15 (Δ11-13) | ↔ | ↔ | ↓↓g | ↓ | ↓ | CD | ↓↓g | ↔ | ↓↓ |
| V16 (Δ14-16) | ↔ | ↔ | ↔ | ↔ | ↔ | CD | ↔ | ↔ | ↔ |
| V17 (Δ17-18) | ↔ | ↔ | ↔ | ↔ | ↔ | CD | ↔ | ↔ | ↔ |
↑, the mutant achieved a better functional level than did the parent; ↔, the mutant is functionally equivalent to the parent; ↓, the mutant partially lost function compared to the parent; ↓↓, the mutant has a severe phenotypic defect; NA, not available.
Protein stability was measured by sensitivity to endogenous proteases (37).
Analysis of Yop production and secretion was assessed following growth in BHI medium without Ca2+ (T3S inducing) or with Ca2+ (T3S repressing).
The Yersinia growth phenotype was calculated by monitoring growth at 37°C in TMH medium alone (minus Ca2+) or supplemented with 2.5 mM CaCl2 (plus Ca2+) (57).
The YopE-dependent HeLa cell cytotoxicity assay measured the extent of altered cell monolayer morphology (46).
In an lcrQ background, these lcrV mutants selectively secreted translocator proteins constitutively.
Loss of LcrV secretion.
Analysis of Yop synthesis and secretion.
Yop synthesis and secretion were induced as previously described (12, 13). In short, overnight cultures of Yersinia strains grown at 26°C were diluted (1:13) in fresh medium (brain heart infusion [BHI] plus Ca2+ or BHI without Ca2+) and grown for 1 h at 26°C, after which they were shifted to 37°C for a further 3 h. Total protein levels were assessed by sampling directly from the bacterial culture suspension, containing a mix of proteins secreted into the culture medium and those associated with intact bacteria. Protein levels associated only with intact bacteria were assessed by analysis of pelleted bacteria concentrated 5 times by centrifugation. Finally, sampling of cleared culture supernatants permitted scrutiny of secreted protein levels. All protein fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then subjected to immunoblotting. Sample volumes were adjusted according to the OD600 of the culture prior to loading. Detection of specific proteins on a membrane support was achieved by the use of antisera specifically recognizing YopH, YopB, LcrV, YopD, and YopE or an antiserum raised against all secreted Yops. The Yop expression/secretion patterns of individual lcrV mutants are represented in Table 3.
Cultivation and infection of HeLa cells.
The human epithelial cell line HeLa was used in all in vitro infection experiments. Culture maintenance and infections with Yersinia followed our standard methods (13, 46). In short, bacteria grown overnight in LB broth at 26°C were diluted in RPMI 1640 with Glutamax I (Gibco BRL, Life Technologies) to an OD600 of 0.2 and incubated for 30 min at 26°C and then for 1 h at 37°C. Infections were initiated by adding the bacteria to the wells to an OD600 of 0.02. The cytotoxicity of infected HeLa cells was monitored by light microscopy, and images were collected at successive time points. Parental Y. pseudotuberculosis (YPIII/pIB102) was included as the positive control, whereas the lcrV full-length null mutant (YPIII/pIB19), which is unable to translocate YopE, was included as the negative control. See Table 3 for a summary of the translocation capacities of individual lcrV mutants.
Intraperitoneal infection of mice.
YPIII/pIB102 (parental) and the Ysc/Yop T3SS-negative YPIII (virulence plasmid-cured) strains were used as controls for comparison to infections with the newly generated mutants YPIII/pIB10215 (LcrVΔ2-4) and YPIII/pIB10217 (LcrVΔ8-10). Bacteria grown overnight in LB broth at 26°C were harvested and resuspended in phosphate-buffered saline. Dilutions were made to correspond to ∼107 to 104 bacteria/ml. For each concentration used, 0.1 ml was injected intraperitoneally into five C57BL/6 (Scanbur BK) female mice. Aliquots of the diluted cultures were also plated on Km-containing plates to determine the number of CFU injected. The mice were monitored at least two times daily for 13 days. Mice that showed severe symptoms, such as hunched carriage, tousled fur, listlessness, and loss of appetite, were sacrificed according to the instructions of the Local Ethical Committee on Laboratory Animals, Umeå, Sweden, which was also responsible for approving this study.
RESULTS
Targeted mutagenesis of the LcrV N terminus to generate nonsecreted variants.
In order to dissect the significance of extracellular localization for the multiple biological functions assigned to LcrV, we targeted the N-terminal region of LcrV for mutagenesis with the assumption that it would contain a putative secretion signal. This strategy was chosen since the signal necessary for Yop effector secretion has been localized to the N termini of various effector substrates (25, 43, 54). Based on previous work, two different theories regarding the nature of the signal have emerged; one proposes that the signal is located within the mRNA, while the other suggests it to be of protein origin (25, 43, 54).
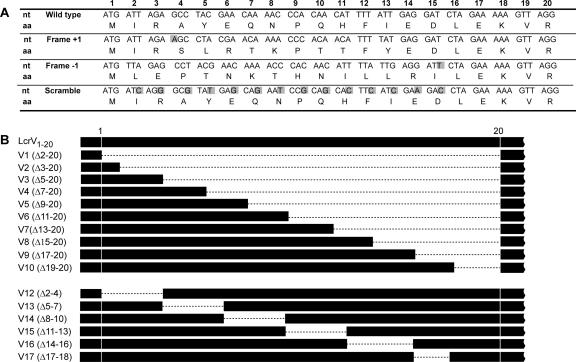
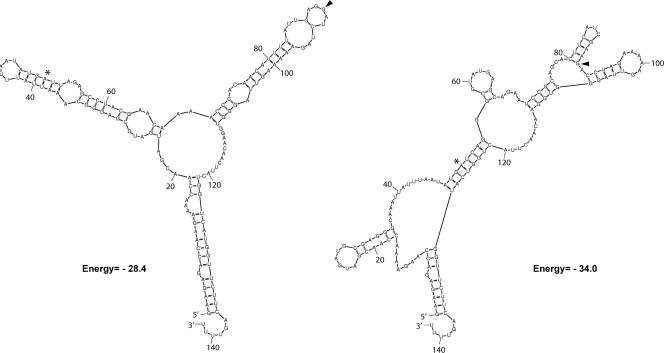
To analyze the contributions of mRNA and amino acid signals to LcrV secretion, two frameshift mutations affecting the protein sequence and one wobble mutation affecting the mRNA sequence were made in cis on the virulence plasmid (Fig. 1A). Since insertion of a nucleotide directly after the translational start would result in a premature stop codon, the nucleotide A was inserted immediately after the ninth nucleotide of LcrV. This insertion was compensated by the removal of a T at position 39 to restore the reading frame after codon 13 (mutant Frame +1). A second frameshift mutation was generated by omission of the fourth nucleotide (an A) and insertion of T after nucleotide 45 of codon 15 (mutant Frame −1). Because both constructs carry significantly altered amino acid sequences with only subtle changes to the mRNA sequence, these were designed to test if the LcrV secretion signal is protein based. Another mutant (designated the Scramble mutant) incorporated mutations within the wobble nucleotides of the coding region of the 5′ end of lcrV. This resulted in 14 substitutions of a possible 42 in the mRNA encoding residues 2 to 15, without alterations to the amino sequence (Fig. 1A). We used the web-based program Mfold to analyze the effects that these mutations may have on the secondary structures of the mRNA (for details, see Materials and Methods). The results suggest that the 5′ part of the mRNA encoding wild-type LcrV could potentially form two stem-loop structures that might play a role in substrate recognition and secretion by the Yersinia T3SS. These predicted structures were disrupted or changed in the Scramble mutant, in which 14 substitutions were introduced (Fig. 2).
FIG. 1.
Schematic representation of N-terminal mutagenesis of LcrV. The Frame +1, Frame −1, and Scramble mutants are shown in panel A, while the in-frame deletion mutants V1 to V17 are displayed in panel B. All mutants were introduced in cis on the Ysc-Yop virulence plasmid (pIB102) to avoid any copy number effects. (A) Numbers 1 to 20 indicate the nucleotide triplets positioned with respect to the start codon of LcrV. Altered or inserted residues are shaded in gray. (B) Residues missing in the deletion mutants are indicated within parentheses and are illustrated by broken lines. Mutant V11 (Δ25-40) was not included.
FIG. 2.
RNA structures predicted for the 5′ ends of wild-type and mutated lcrV mRNAs. RNA structures were predicted using the program Mfold at http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi. The first and the last codons of the 42-bp region mutated in the Scramble mutant (Fig. 1A) are indicated with an asterisk and an arrowhead, respectively. Shown here are upstream and downstream flanking regions of 50 bp, but regions of 20, 40, 60, 80, and 100 bp were also used in the analysis (for details, see Materials and Methods). The two stem-loops predicted to span this region in wild-type LcrV (left) were conserved in all but one of the potential structures obtained (data not shown) but were clearly altered in the Scramble mutant (right).
We utilized our frameshift and scramble mutations to determine if LcrV, similar to the effector class of substrates, contains an N-terminal secretion signal. Parental and mutant Yersinia cells were grown in BHI under T3S-inducing (no Ca2+) or noninducing (plus Ca2+) conditions at 37°C. Fractions containing secreted proteins, those associated with bacteria, or a mix of the two were isolated and analyzed by SDS-PAGE and Western blotting. All mutants efficiently synthesized and secreted Yop substrates, such as YopH, when grown under T3S-inducing conditions (Fig. 3A, compare lanes a and b with lanes c to h). Moreover, despite Frame +1 and Frame −1 harboring 8/13 and 14/15 different amino acids in their N termini, respectively, and Scramble containing multiple alterations in the mRNA sequence and potentially the mRNA structure, these mutated variants were produced and secreted at levels similar to those observed for the parental strain (Fig. 3B, compare lanes a and b with lanes c to h). Hence, if the LcrV N terminus does function as a secretion signal, our results suggest its molecular makeup to be different from that of previously characterized Yersinia T3S substrates.
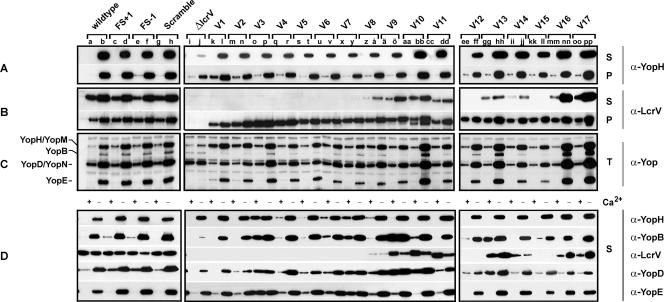
FIG. 3.
Analysis of Yop synthesis and secretion by lcrV mutants of Y. pseudotuberculosis. Bacteria were grown in BHI medium, with (+) or without (−) Ca2+. Proteins were separated by SDS-PAGE and identified by immunoblot analysis, using polyclonal rabbit anti-YopH, anti-YopB, anti-LcrV, anti-YopD, and anti-YopE antisera or an antiserum recognizing all secreted Yops (α-Yop). Bacterium-associated protein levels were determined using pelleted bacteria (P). Total sample (T) refers to a mixture of proteins secreted into the culture medium and contained within intact bacteria, while supernatant samples (S) contained only secreted proteins. The experiment was repeated at least two times, with reproducible results. (A to C) Yop synthesis and secretion by lcrV mutants still containing the negative regulatory element LcrQ. Lanes: a and b, YPIII/pIB102 (parent); c and d, YPIII/pIB10201 (FS+1); e and f, YPIII/pIB10202 (FS−1); g and h, YPIII/pIB10203 (Scramble); i and j, YPIII/pIB19 (ΔlcrV null mutant lacking codons 10 to 313); k and l, YPIII/pIB10204 (V1 [Δ2-20]); m and n, YPIII/pIB10205 (V2 [Δ3-20]); o and p, YPIII/pIB10206 (V3 [Δ5-20]); q and r, YPIII/pIB10207 (V4 [Δ7-20]); s and t, YPIII/pIB10208 (V5 [Δ9-20]); u and v, YPIII/pIB10209 (V6 [Δ11-20]); x and y, YPIII/pIB10210 (V7 [Δ13-20]); z and å, YPIII/pIB10211 (V8 [Δ15-20]); ä and ö, YPIII/pIB10212 (V9 [Δ17-20]); aa and bb, YPIII/pIB10213 (V10 [Δ19-20]); cc and dd, YPIII/pIB10214 (V11 [Δ25-40]); ee and ff, YPIII/pIB10215 (V12 [Δ2-4]); gg and hh, YPIII/pIB10216 (V13 [Δ5-7]); ii and jj, YPIII/pIB10217 (V14 [Δ8-10]); kk and ll, YPIII/pIB10218 (V15 [Δ11-13]); mm and nn, YPIII/pIB10219 (V16 [Δ14-16]); oo and pp, YPIII/pIB10220 (V17 [Δ17-18]). (D) Proteins secreted from the equivalent strains differing only by the disruption of lcrQ via allelic exchange with an sp-sm resistance cartridge.
Contributions of N-terminal residues to the secretion of LcrV.
Failure to detect defects in LcrV secretion by generating frameshift or scramble mutations led us to analyze the LcrV N terminus by using a different approach. Several independent in-frame deletions within the first 20 residues of LcrV were generated in cis on the virulence plasmid. For simplicity, these mutants were designated V1 to V10 (Fig. 1B, upper panel). Protein synthesis and secretion from these new mutants were determined by Western blot analysis of bacterially associated protein (bacterial pellets) and secreted protein (cleared supernatant fractions), first using an antiserum specifically recognizing LcrV. Significantly, the amount of each LcrV deletion variant associated with pelleted bacteria was similar to or somewhat higher than that of wild-type LcrV (Fig. 3B, compare lanes a and b with lanes k to dd). In parallel, using suspended bacterial cultures (a mixture of LcrV associated with bacteria and that secreted into the medium) to assess the relative amounts of the individual LcrV deletion mutants, all were found to be produced at levels similar to that of the wild type, with the exception of V1, which was noticeably less abundant (data not shown). Moreover, each variant migrated according to its expected size, although for V10 (Δ19-20) a lower-molecular-weight band of unknown origin was also observed. Interestingly, however, the stable variants V1 (Δ2-20), V2 (Δ3-20), V3 (Δ5-20), V4 (Δ7-20), V5 (Δ9-20), V6 (Δ11-20), and V7 (Δ13-20) were barely, if at all, detected in the bacterium-free culture medium (Fig. 3B, lanes k to y). Furthermore, the variants V8 (Δ15-20), V9 (Δ17-20), and V10 (Δ19-20) were secreted, but at somewhat lower levels than that of wild-type LcrV (Fig. 3B, compare lanes a and b with lanes z to bb). As a secretion control, mutant V11 was constructed to contain a deletion of codons 25 through 40 of LcrV. This region has previously been earmarked as a minor secretion signal of LcrV from Yersinia pestis (53). Indeed, a small secretion defect was observed for this Y. pseudotuberculosis mutant (Fig. 3B, compare lanes a and b with lanes cc and dd). Defects in secretion of V1 to V7 were not due to a general defect in T3S, because the YopH (Fig. 3A, compare lanes a and b with lanes i to y) and YopE (data not shown) effectors were secreted by all mutants during bacterial growth under no-Ca2+ conditions, albeit at slightly lower levels than those for parental Yersinia. Notably, however, a moderate reduction in Yop effector secretion was also observed for mutants able to efficiently secrete LcrV, such as V9 (Fig. 3A, lanes ä to ö). In all cases, the small defect in effector secretion coincided with slightly smaller amounts of Yops still associated with pelleted bacteria (Fig. 3A, compare lanes a and b with lanes z to ö and lanes cc and dd).
This mutational analysis suggested that a region incorporating one or more of residues 2 to 13 is essential for LcrV secretion. In an effort to further pinpoint the critical residues, we generated another set of deletion mutants, removing two or three amino acids at a time. This resulted in LcrV mutants V12 (Δ2-4), V13 (Δ5-7), V14 (Δ8-10), V15 (Δ11-13), V16 (Δ14-16), and V17 (Δ17-18) (Fig. 1B, lower panel). These additional mutants were analyzed for LcrV production and secretion. All variants were found to be produced at levels generally equivalent to that of wild-type LcrV (Fig. 3B, compare lanes a and b with lanes ee to pp). Interestingly, no V12 or V15 variant was visible in the cleared Yersinia culture supernatant (Fig. 3B, lanes ee, ff, kk, and ll), and only low levels of V13 and V14 variants were detected (Fig. 3B, lanes gg to jj). In contrast, the levels of mutants V16 (Δ14-16) and V17 (Δ17-18) were similar to that of wild-type LcrV. The absence of certain LcrV variants in the culture medium could result from them being more prone to aggregation and then sticking to the bacterial surface. However, a more likely interpretation is that within the first 18 residues of LcrV, amino acids 2 to 4 and 11 to 13 are absolutely critical for the actual secretion of LcrV, while residues 5 to 10 are involved but play a lesser role. Thus, by using a deletion mutagenesis approach, a secretion signal within the N terminus of LcrV that is absolutely essential for LcrV export could be identified.
LcrV secretion is not required for control of Yop synthesis.
LcrV plays an important role in the regulation of yop expression. Increased intracellular levels of LcrV are proposed to bind and titrate away LcrG, the cytoplasmic gating mechanism of the Yersinia T3SS (9, 30, 36). As such, Yersinia cells devoid of functional LcrV are impaired in Yop synthesis, particularly during growth in T3S-permissive media (39, 42, 53). Thus, we wanted to assess our lcrV mutants for the impact that LcrV secretion has on yop regulatory control. Total Yop levels derived from suspended bacterial cultures (a total mixture of Yops associated with bacteria and those secreted into the medium) were examined by immunoblotting. In parallel, the regulatory status of each mutant was interpreted from its growth behavior at 37°C in the presence or absence of Ca2+.
Parental Yersinia and the mutants Frame +1, Frame −1, Scramble, V10, V13, V16, and V17 all showed similar patterns and levels of Yop synthesis, with distinct induction occurring in medium devoid of Ca2+ (Fig. 3C). In addition, they displayed CD growth identical to that of the parental strain YPIII/pIB102 when grown in defined TMH medium, with or without calcium (Table 3; data not shown). Hence, this mutant group was unaffected in yop regulatory control. Another mutant class, consisting of V9, V11, V14, and V15, also behaved identically to wild-type Yersinia with respect to growth (Table 3; data not shown), although these mutants produced slightly lower Yop levels under T3S-permissive conditions (Fig. 3C). We believe that these mutants are also regulation competent. A similar reduction in Yop synthesis was seen for the remaining mutants, V1 to V8 and V12, under T3S-permissive conditions (Fig. 3C). However, these mutants also displayed a more intermediate growth phenotype in that more growth occurred under no-Ca2+ conditions than was evident for parental bacteria. Thus, while parental Yersinia cells did not exceed an OD600 of ∼0.4 at 10 h post-temperature upshift, mutants V1 to V8 and V12 reached OD600 values of between 0.6 and 1.0 (data not shown). Moreover, for unknown reasons, V3 to V8 and V12 never reached densities as high as those of parental Yersinia after 10 h at 37°C under plus-Ca2+ conditions (OD600 values of 1.8 to 2.5 versus 2.5 to 3.0) (data not shown), although no growth differences were distinguishable at 26°C (data not shown). Significantly, no mutants displayed the same CI growth as the full-length lcrV deletion mutant YPIII/pIB19 (OD600 of between 2.6 and 2.8, regardless of the Ca2+ concentration) (data not shown), nor were any as defective in Yop synthesis induction caused by Ca2+ depletion (Fig. 3C). Thus, mutants V1 to V8 and V12 apparently exhibit only a minor defect in yop regulatory control. Taken together, the data show that reduced LcrV secretion in mutants V10 (Δ19-20) and V13 (Δ5-7) or its complete absence in mutants V12 and V15 occurred without a significant loss of yop regulatory control. Thus, unadulterated LcrV secretion is not a prerequisite for yop regulatory control, consistent with an earlier observation (53).
The absence of LcrQ does not influence the fate of nonsecreted LcrV but can exhibit dramatic effects on Yop secretion.
LcrQ is a negative regulatory element of Yop synthesis. Its presence in the bacterial cytoplasm somehow prevents Yop induction, while LcrQ depletion derepresses Yop synthesis (40, 45, 56). Moreover, indications are that LcrQ can also promote ordered substrate secretion through the T3SS (63). An lcrQ mutant is also known to specifically secrete LcrV under nonpermissive conditions, while secretion of other substrates occurs only under permissive conditions (45, 56). We therefore wondered whether our N-terminal mutants could be recognized and secreted more efficiently in the absence of LcrQ. As expected, introduction of an lcrQ::sp-sm mutation into our mutant backgrounds resulted in derepression of Yop synthesis (data not shown). Despite constitutive Yop synthesis regardless of the Ca2+ concentration, the loss of LcrQ did not allow V1 to V7, V12, and V15 mutant bacteria to secrete LcrV (Fig. 3D). Moreover, the limited LcrV secretion by V8 and V14 bacteria was not enhanced by removal of LcrQ (Fig. 3D). In fact, the only notable change occurred for V9 to V11 mutants devoid of LcrQ—these preferentially secreted LcrV in T3S-restrictive medium (plus Ca2+) (Fig. 3D). Hence, the fate of nonsecreted LcrV is not significantly affected by the absence of LcrQ.
Next, we utilized our lcrQ lcrV double mutants to investigate the pattern of Yop secretion by Western blotting with rabbit polyclonal antisera specifically recognizing YopH, YopB, YopD, and YopE. Curiously, the double mutants involving V3 (Δ5-20) and V7 (Δ13-20) displayed constitutive secretion of Yops regardless of the calcium concentration (Fig. 3D, lanes o, p, x, and y). This might also be true of the double mutants involving V4 to V6 and V8, although lower substrate levels were secreted during bacterial growth under plus-Ca2+ conditions (Fig. 3D, lanes q, r, u, v, z, and å). Interestingly, another mutant set, primarily incorporating V9 (Δ17-20) and V13 (Δ5-7) but also including, to a lesser extent, V10, Frame +1, and Frame −1, reproducibly and selectively secreted the pore-forming translocon components YopB and YopD, also under noninducing conditions (plus Ca2+), while maintaining CD control of Yop effector secretion (Fig. 3D, lanes c to f, ä to bb, gg, and hh). To the best of our knowledge, such an LcrV-dependent phenotype has never been described before. Furthermore, it is distinct from our lcrV lcrQ::sp-sm double mutant, which actually secretes less YopB and YopD. This suggests a cross talk between LcrQ and the N terminus of LcrV that can preferentially affect YopB and YopD translocator secretion.
Functional Yop effector translocation requires only low levels of LcrV secretion.
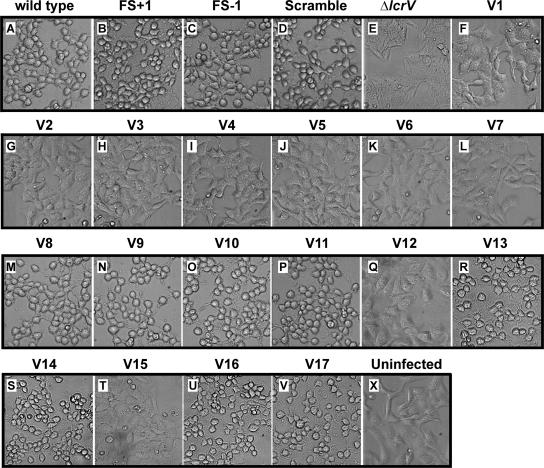
To determine whether our mutants were affected in the ability to translocate Yop proteins, we used the YopE cytotoxicity assay. When translocated, the intracellular YopE GTPase-activating protein will depolymerize actin, causing a general rounding up of infected cells (46). However, a full-length lcrV mutant was unable to translocate YopE and induce a cytotoxic response (Fig. 4, compare panels E and X) (39). In contrast, the mutants Frame +1, Frame −1, Scramble, V8 to V11, V13, V14, V16, and V17 efficiently caused a cytotoxic effect on infected cells, at rates comparable to that of parental bacteria (Fig. 4), suggesting that they are clearly competent for Yop translocation. As expected, the V1 to V7, V12, and V15 mutants, which were unable to secrete detectable levels of LcrV, were also incapable of translocating the YopE cytotoxin (Fig. 4). This scenario was not altered by prolonged infection times or by the introduction of an lcrQ::sp-sm mutation that raised Yop levels (data not shown). Of special interest is the observation that poorly secreted variants also induced cytotoxicity. For instance, V14 was secreted at very low levels in vitro but still induced a YopE-mediated cytotoxic response on infected HeLa cells. From these observations, it is clear that only meager amounts of secreted LcrV are needed to establish a functional translocon.
FIG. 4.
Infection of HeLa cells by different strains of Y. pseudotuberculosis. At 2 h of infection, the effect of the bacteria on HeLa cells was recorded by phase-contrast microscopy. The experiment was repeated at least three times. Note the extensive rounding up of the YopE-dependent, cytotoxically affected HeLa cells (A to D, M to P, R, S, U, and V). Shown are phase-contrast images. The strain designations are identical to those used in Fig. 1. Panel X is an uninfected HeLa cell monolayer control.
N-terminal lcrV mutants are attenuated for virulence in mice.
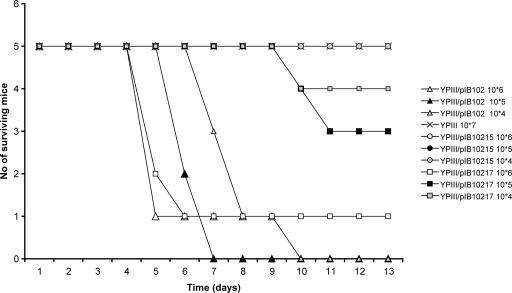
Finally, we wanted to address the importance of LcrV secretion levels for virulence in the mouse infection model. We therefore included the two LcrV variants V12, for which neither detectable LcrV secretion nor functional translocation was seen, and V14, which secreted low levels of LcrV with retained functional translocation, in these studies. As controls, the parental wild-type strain YPIII/pIB102 and the virulence plasmid-cured strain YPIII were used. The outcome and kinetics of infection for the different strains after intraperitoneal infection of C57BL/6 mice are shown in Fig. 5. Like the plasmid-cured strain, the V12 mutant was essentially avirulent, unable to cause a lethal infection even at the highest dose used (Fig. 5). The calculated infection doses (i.e., dose estimated to give lethal infection in 50% of the mice) were >2.1 × 107 (no mice infected with highest dose died) for the plasmid-cured strain and 7.0 × 106 for the V12 mutant strain. In contrast, the parental strain caused a lethal infection at the lowest dose used (Fig. 5), and in this case, the calculated infection dose was 1.4 × 103. Mutant V14, however, displayed an intermediate phenotype. At the lowest infection dose tested, only one of five mice died, and this did not occur until day 10. Also, for the two higher doses, the infection kinetics was slower for this strain, and one mouse also survived the highest dose. The calculated infection dose for the V14 mutant was 3.3 × 105, which is >200-fold higher than that for the parental strain. This 200-fold virulence attenuation can clearly be linked to the reduced secretion of LcrV, as this strain phenotypically resembled the wild type in all other aspects investigated (growth and Yop expression and secretion). Mutant V12 was almost as attenuated as a virulence plasmid-cured strain. The in vitro growth phenotype of this strain deviated somewhat from that of the wild-type strain and mutant V14, displaying slightly enhanced growth in the absence of calcium and slightly suppressed growth in medium with calcium (Table 3; data not shown). Yet attenuation in this case also most likely reflects a lack of LcrV secretion: while the growth phenotype was similar to that of the wild-type strain, importantly, the pattern of Yop expression and secretion also resembled that of the wild-type strain with retained Ca2+ regulation. Thus, while low levels of LcrV were sufficient for functional translocation in vitro, it is clear that for in vivo infection, LcrV secretion levels need to be significantly higher.
FIG. 5.
Intraperitoneal infections of mice. Survival curves were determined for mice infected with parental YPIII/pIB102, mutants YPIII/pIB10215 (V12 [Δ2-4]) and YPIII/pIB10217 (V14 [Δ8-10]), or the virulence plasmid-cured strain YPIII. Mice were infected intraperitoneally with successive 10-fold dilutions (104 to 107 bacteria/ml) of a bacterial suspension and monitored at least twice daily for 13 days. The exact doses for each strain were determined by viable counts, and the dose for YPIII was 2.1 × 107, that for YPIII/pIB102 was 3.1 × 107 to 3.1 × 104, that for YPIII/pIB10215 was 3.25 × 107 to 3.25 × 105, and that for YPIII/pIB10217 was 2.4 × 106 to 2.4 × 104.
DISCUSSION
LcrV is a multifunctional protein exerting its different functions at three discrete locations, namely, inside the bacterium, as a secreted protein, and at the tip of the T3S needle complex. With the aim of dissecting its different biological functions, we devised a strategy involving the generation of specific secretion-incompatible variants of LcrV. Using this strategy, we verified that secretion is dispensable for LcrV's role in regulation, while high levels of secretion are required for full virulence in the systemic mouse infection model. Interestingly, only low levels of LcrV secretion were needed for functional translocation of YopE in the in vitro cell infection model. This highlights the need to include in vivo infection models to validate the function of an important virulence mechanism such as T3S as well as for determining the direct significance of LcrV secretion for its function in virulence.
Our targeted mutagenesis approach verified that LcrV secretion is dependent on N-terminal domains. Removal of codons encompassing positions 2 to 4 and 11 to 13 ablated secretion, while taking away the codons positioned at residues 5 to 10 also affected secretion levels. In contrast, +1 and −1 frameshift mutations within the first 4 to 13 and 2 to 15 amino acids, respectively, or mutation of the wobble nucleotides of codons 2 to 15 had no effect on LcrV secretion. In the latter case, mutation of the wobble nucleotides was also predicted to result in significant changes in the secondary structure of the mRNA. Therefore, it is still unclear whether LcrV targeting is mediated via an N-terminal amino acid signal or an mRNA signal.
Prior to this work, a signal supporting LcrV secretion had been mapped to a region encompassing residues 108 to 125 and, to a lesser extent, residues 25 to 40 (53). These deletion variants accumulated inside bacteria, suggesting that protein stability was sound. However, analysis of the LcrV crystal structure indicated that such deletions would likely disturb the structural integrity of the protein (10). Nevertheless, we have corroborated this finding in part by proving that at least the N terminus is a secretion signal. N-terminal signals are believed to be inherently unstructured and perhaps amphipathic in nature, which are general features necessary for promoting recognition of a diverse array of substrates by the T3SSs (25). It is assumed that the N terminus of LcrV functions similarly. Conversely, exactly how residues 108 to 125 contribute to LcrV secretion is not understood. LcrG, the intracellular gating protein of the T3SS, interacts with LcrV. As well as performing important regulatory functions inside the cell (36), this interaction improves the efficiency of LcrV secretion (9). As a result, LcrG could be considered a potential chaperone for LcrV (23). Although the region of amino acids 108 to 125 is not known to bind LcrG (22), it might still aid in generating a secondary LcrG chaperone-dependent secretion signal for LcrV secretion.
At this stage, we are not able to definitively conclude that the N-terminal secretion signal of LcrV is significantly different from that of the Yop effector proteins of Yersinia. Nevertheless, unlike the case for the Yop effector substrates (1, 2, 44), neither scramble nor frameshift mutations affected the secretion efficiency of full-length LcrV, which might indicate a different secretion signal. Another feature of this study lends credence to the notion of different secretion signals between effector proteins and translocators. In the absence of LcrQ, some lcrV mutations caused distinct secretion phenotypes; one collection secreted all substrates constitutively, regardless of whether the T3SS was induced, while a second group freely permitted secretion of only the translocator substrates. The very fact that a subset of mutants somehow caused the T3SS to discriminate between the two substrate classes is an intriguing observation. This is not without support; for example, an lcrQ mutant preferentially secreted both LcrV and YopD under T3S-restrictive conditions (45, 56), while an lcrH mutant constitutively secreted LcrV (12, 53). Our interpretation of all these data is that the translocators LcrV, YopB, and YopD are recognized by the Ysc T3SS differently from secreted Yop effectors. We assume that the basis of this discrimination is imposed by a unique signal housed within the translocators (or their specific chaperones) that is lacking in the effectors (or their chaperones). Unfortunately, the molecular cause of this phenomenon is unclear, as is its relevance to Yersinia infections.
LcrV knockout mutants are impaired for induction of yop expression in response to depletion of calcium in vitro (36) and, presumably, also in response to target cell contact under more in vivo-like conditions. This regulatory phenotype is a likely consequence of the C terminus of LcrV binding to and displacing the negative regulator LcrG, which gates the secretion machinery from inside the bacterium. It also bears some resemblance to the phenotype of secretion knockout mutants, where the downregulation is believed to be a consequence of a negative feedback control mechanism. However, LcrV knockout mutants are still secretion competent, although they are defective for translocation. Moreover, the localization of LcrV at the tip of the needle complex suggests that it could also be involved in sensing cell contact; by relaying this signal into the bacterial interior, T3S could be induced. In this work, we engineered various LcrV mutants and identified variants with only minor changes in the N-terminal part that were not secreted. Since these variants would be expected to interact with LcrG, this could explain why some of the secretion-negative variants, such as V12 (Δ2-4), are very similar to the wild type with respect to induction of Yop production—an LcrG interaction would relieve the secretion block—a notion compatible with the proposed LcrG gating mechanism. In contrast, however, it would argue against a role for LcrV in sensing cell contact to induce expression, since secretion is a presumed prerequisite for localization at the tip of the needle complex. However, the molecular mechanisms and protein-protein interactions that determine the localization of LcrV at the needle complex are still not completely understood.
LcrV is multifunctional and very central to the virulence of pathogenic Yersinia species. The contributions of these different functions to virulence are further complicated by the fact that they are exerted at three discrete locations. Here we have used a strategy of engineering LcrV variants that are secretion defective to address the significance of secretion for LcrV's function. We have shown that LcrV secretion, as such, does not have a major impact on regulation, while it is a requirement for localization of LcrV at the tip of the needle complex to exert its function in Yop effector targeting. However, secretion is also a likely requirement for the reported role of LcrV in immunosuppression. While LcrV-TLR2 interactions are clearly linked to virulence in Yersinia enterocolitica, for which it has also been verified that TLR2 knockout mice are less susceptible to infection than wild-type mice (50), the situation is less clear for Y. pestis and Y. pseudotuberculosis. Two recent studies failed to verify any difference in susceptibility to Y. pestis and Y. pseudotuberculosis infection of TLR2 knockout mice compared to isogenic wild-type mice (3, 41). There are, however, some conflicting results, as one of these studies also failed to verify any decreased susceptibility of TLR2 knockout mice to Y. enterocolitica infections (3). With our LcrV variants, in combination with robust in vitro assays and good mouse models, we are now in a position to prize apart the direct contribution of secreted LcrV to Yop targeting to immune cells (28) and the potential consequences of interactions with TLR2 (50-52) during active Yersinia infections. Determination of the relative contributions of effector targeting and immunosuppression remains an important area in future studies of how this multifunctional protective antigen promotes infection.
Acknowledgments
This work was supported by grants from the Carl Tryggers Foundation for Scientific Research (M.S.F.), the Swedish Research Council (M.S.F. and Å.F.), the Foundation for Medical Research at Umeå University (M.S.F.), and the Swedish Medical Association (M.S.F.).
We thank Solveig Ericsson for excellent assistance with the animal infection studies and Yngve Östberg, together with Jörgen Johansson, for valuable help with the mRNA structure prediction analysis.
Footnotes
Published ahead of print on 14 September 2007.
REFERENCES
- 1.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 3.Auerbuch, V., and R. R. Isberg. 2007. Growth of Yersinia pseudotuberculosis in mice occurs independently of Toll-like receptor 2 expression and induction of interleukin-10. Infect. Immun. 75:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bölin, I., L. Norlander, and H. Wolf-Watz. 1982. Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect. Immun. 37:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bölin, I., and H. Wolf-Watz. 1984. Molecular cloning of the temperature-inducible outer membrane protein 1 of Yersinia pseudotuberculosis. Infect. Immun. 43:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bröms, J. E., C. Sundin, M. S. Francis, and Å. Forsberg. 2003. Comparative analysis of type III effector translocation by Yersinia pseudotuberculosis expressing native LcrV or PcrV from Pseudomonas aeruginosa. J. Infect. Dis. 188:239-249. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan, C., A. V. Philipovskiy, C. R. Wulff-Strobel, Z. Ye, and S. C. Straley. 2005. Anti-LcrV antibody inhibits delivery of Yops by Yersinia pestis KIM5 by directly promoting phagocytosis. Infect. Immun. 73:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBord, K. L., V. T. Lee, and O. Schneewind. 2001. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica. J. Bacteriol. 183:4588-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derewenda, U., A. Mateja, Y. Devedjiev, K. M. Routzahn, A. G. Evdokimov, Z. S. Derewenda, and D. S. Waugh. 2004. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure (Cambridge) 12:301-306. [DOI] [PubMed] [Google Scholar]
- 11.Fields, K. A., M. L. Nilles, C. Cowan, and S. C. Straley. 1999. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 67:5395-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 13.Francis, M. S., and H. Wolf-Watz. 1998. YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol. Microbiol. 29:799-813. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goure, J., P. Broz, O. Attree, G. R. Cornelis, and I. Attree. 2005. Protective anti-V antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J. Infect. Dis. 192:218-225. [DOI] [PubMed] [Google Scholar]
- 16.Håkansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 17.Hanski, C., U. Kutschka, H. P. Schmoranzer, M. Naumann, A. Stallmach, H. Hahn, H. Menge, and E. O. Riecken. 1989. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect. Immun. 57:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, J., S. E. Leary, K. F. Griffin, E. D. Williamson, and R. W. Titball. 1997. Regions of Yersinia pestis V antigen that contribute to protection against plague identified by passive and active immunization. Infect. Immun. 65:4476-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmström, A., J. Olsson, P. Cherepanov, E. Maier, R. Nordfelth, J. Pettersson, R. Benz, H. Wolf-Watz, and Å. Forsberg. 2001. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol. Microbiol. 39:620-632. [DOI] [PubMed] [Google Scholar]
- 20.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson (ed.), Directed mutagenesis: a practical approach. Oxford University Press, New York, NY.
- 21.Karavolos, M. H., A. J. Roe, M. Wilson, J. Henderson, J. J. Lee, D. L. Gally, and C. M. Khan. 2005. Type III secretion of the Salmonella effector protein SopE is mediated via an N-terminal amino acid signal and not an mRNA sequence. J. Bacteriol. 187:1559-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawton, D. G., C. Longstaff, B. A. Wallace, J. Hill, S. E. Leary, R. W. Titball, and K. A. Brown. 2002. Interactions of the type III secretion pathway proteins LcrV and LcrG from Yersinia pestis are mediated by coiled-coil domains. J. Biol. Chem. 277:38714-38722. [DOI] [PubMed] [Google Scholar]
- 23.Lee, V. T., C. Tam, and O. Schneewind. 2000. LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275:36869-36875. [DOI] [PubMed] [Google Scholar]
- 24.Lian, C. J., W. S. Hwang, and C. H. Pai. 1987. Plasmid-mediated resistance to phagocytosis in Yersinia enterocolitica. Infect. Immun. 55:1176-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd, S. A., Å. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 27.Marenne, M. N., L. Journet, L. J. Mota, and G. R. Cornelis. 2003. Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb. Pathog. 35:243-258. [DOI] [PubMed] [Google Scholar]
- 28.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 309:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 30.Matson, J. S., and M. L. Nilles. 2001. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J. Bacteriol. 183:5082-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motin, V. L., R. Nakajima, G. B. Smirnov, and R. R. Brubaker. 1994. Passive immunity to yersiniae mediated by anti-recombinant V antigen and protein A-V antigen fusion peptide. Infect. Immun. 62:4192-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller, C. A., P. Broz, S. A. Muller, P. Ringler, F. Erne-Brand, I. Sorg, M. Kuhn, A. Engel, and G. R. Cornelis. 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310:674-676. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima, R., V. L. Motin, and R. R. Brubaker. 1995. Suppression of cytokines in mice by protein A-V antigen fusion peptide and restoration of synthesis by active immunization. Infect. Immun. 63:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neyt, C., and G. R. Cornelis. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971-981. [DOI] [PubMed] [Google Scholar]
- 36.Nilles, M. L., A. W. Williams, E. Skrzypek, and S. C. Straley. 1997. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J. Bacteriol. 179:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsson, J., P. J. Edqvist, J. E. Bröms, Å. Forsberg, H. Wolf-Watz, and M. S. Francis. 2004. The YopD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J. Bacteriol. 186:4110-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallen, M. J., S. A. Beatson, and C. M. Bailey. 2005. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol. Rev. 29:201-229. [DOI] [PubMed] [Google Scholar]
- 39.Pettersson, J., A. Holmström, J. Hill, S. Leary, E. Frithz-Lindsten, A. von Euler-Matell, E. Carlsson, R. Titball, Å. Forsberg, and H. Wolf-Watz. 1999. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol. Microbiol. 32:961-976. [DOI] [PubMed] [Google Scholar]
- 40.Pettersson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf-Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231-1233. [DOI] [PubMed] [Google Scholar]
- 41.Pouliot, K., N. Pan, S. Wang, S. Lu, E. Lien, and J. D. Goguen. 2007. Evaluation of the role of LcrV-Toll-like receptor 2-mediated immunomodulation in the virulence of Yersinia pestis. Infect. Immun. 75:3571-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, S. B., C. Cowan, R. D. Perry, and S. C. Straley. 1991. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J. Bacteriol. 173:2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramamurthi, K. S., and O. Schneewind. 2003. Substrate recognition by the Yersinia type III protein secretion machinery. Mol. Microbiol. 50:1095-1102. [DOI] [PubMed] [Google Scholar]
- 44.Ramamurthi, K. S., and O. Schneewind. 2003. Yersinia yopQ mRNA encodes a bipartite type III secretion signal in the first 15 codons. Mol. Microbiol. 50:1189-1198. [DOI] [PubMed] [Google Scholar]
- 45.Rimpiläinen, M., Å. Forsberg, and H. Wolf-Watz. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J. Bacteriol. 174:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosqvist, R., Å. Forsberg, and H. Wolf-Watz. 1991. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect. Immun. 59:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosqvist, R., C. Persson, S. Håkansson, R. Nordfeldt, and H. Wolf-Watz. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib. Microbiol. Immunol. 13:230-234. [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:787-796. [Google Scholar]
- 49.Simonet, M., S. Richard, and P. Berche. 1990. Electron microscopic evidence for in vivo extracellular localization of Yersinia pseudotuberculosis harboring the pYV plasmid. Infect. Immun. 58:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sing, A., D. Reithmeier-Rost, K. Granfors, J. Hill, A. Roggenkamp, and J. Heesemann. 2005. A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence. Proc. Natl. Acad. Sci. USA 102:16049-16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sing, A., A. Roggenkamp, A. M. Geiger, and J. Heesemann. 2002. Yersinia enterocolitica evasion of the host innate immune response by V antigen-induced IL-10 production of macrophages is abrogated in IL-10-deficient mice. J. Immunol. 168:1315-1321. [DOI] [PubMed] [Google Scholar]
- 52.Sing, A., D. Rost, N. Tvardovskaia, A. Roggenkamp, A. Wiedemann, C. J. Kirschning, M. Aepfelbacher, and J. Heesemann. 2002. Yersinia V-antigen exploits Toll-like receptor 2 and CD14 for interleukin 10-mediated immunosuppression. J. Exp. Med. 196:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skrzypek, E., and S. C. Straley. 1995. Differential effects of deletions in lcrV on secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J. Bacteriol. 177:2530-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorg, J. A., N. C. Miller, and O. Schneewind. 2005. Substrate recognition of type III secretion machines—testing the RNA signal hypothesis. Cell. Microbiol. 7:1217-1225. [DOI] [PubMed] [Google Scholar]
- 55.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 56.Stainier, I., M. Iriarte, and G. R. Cornelis. 1997. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol. Microbiol. 26:833-843. [DOI] [PubMed] [Google Scholar]
- 57.Straley, S. C., and W. S. Bowmer. 1986. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tardy, F., F. Homble, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Titball, R. W., and E. D. Williamson. 2001. Vaccination against bubonic and pneumonic plague. Vaccine 19:4175-4184. [DOI] [PubMed] [Google Scholar]
- 60.Viboud, G. I., and J. B. Bliska. 2005. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59:69-89. [DOI] [PubMed] [Google Scholar]
- 61.Warren, S. M., and G. M. Young. 2005. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J. Bacteriol. 187:6075-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weeks, S., J. Hill, A. Friedlander, and S. Welkos. 2002. Anti-V antigen antibody protects macrophages from Yersinia pestis-induced cell death and promotes phagocytosis. Microb. Pathog. 32:227-237. [DOI] [PubMed] [Google Scholar]
- 63.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43:411-423. [DOI] [PubMed] [Google Scholar]
- 64.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]