Abstract
Chronic granulomatous disease (CGD) is an inherited immune deficiency characterized by increased susceptibility to infection with Staphylococcus, certain gram-negative bacteria, and fungi. Granulibacter bethesdensis, a newly described genus and species within the family Acetobacteraceae, was recently isolated from four CGD patients residing in geographically distinct locales who presented with fever and lymphadenitis. We sequenced the genome of the reference strain of Granulibacter bethesdensis, which was isolated from lymph nodes of the original patient. The genome contains 2,708,355 base pairs in a single circular chromosome, in which 2,437 putative open reading frames (ORFs) were identified, 1,470 of which share sequence similarity with ORFs in the nonpathogenic but related Gluconobacter oxydans genome. Included in the 967 ORFs that are unique to G. bethesdensis are ORFs potentially important for virulence, adherence, DNA uptake, and methanol utilization. GC% values and best BLAST analysis suggested that some of these unique ORFs were recently acquired. Comparison of G. bethesdensis to other known CGD pathogens demonstrated conservation of some putative virulence factors, suggesting possible common mechanisms involved in pathogenesis in CGD. Genotyping of the four patient isolates by use of a custom microarray demonstrated genome-wide variations in regions encoding DNA uptake systems and transcriptional regulators and in hypothetical ORFs. G. bethesdensis is a genetically diverse emerging human pathogen that may have recently acquired virulence factors new to this family of organisms.
Chronic granulomatous disease (CGD) is due to a genetic defect in phagocyte superoxide formation. Patients with CGD develop recurrent life-threatening infections with Staphylococcus aureus, Serratia marcescens, Burkholderia cepacia complex, Nocardia species, and Aspergillus species (33). We recently described a novel member of the Acetobacteraceae, Granulibacter bethesdensis, infecting three CGD patients from geographically distinct locations who presented with fever and lymphadenitis (11, 12).
The Acetobacteraceae are a family of alphaproteobacteria that include the acetic acid bacteria, which are organisms that oxidize their carbon sources incompletely, such as in the conversion of alcohol to acetic acid. Acetobacteraceae members currently include the genera Acetobacter, Gluconobacter, Acidomonas, Gluconacetobacter, Swaminathania, Neoasaia, Saccharibacter, Asaia, and Kozakia (42). They are found throughout the world and have been isolated from diverse vegetables, fruits, and flowers (32, 36). The acetic acid bacteria have many industrial applications, including the production of vinegar and various drugs. The genome of the industrially important acetic acid bacterium Gluconobacter oxydans was recently published (26). Despite the presence of acetic acid bacteria in human industry and food production, Granulibacter bethesdensis is the first to be identified as an agent of invasive human disease. To identify factors that have allowed this first member of the family to invade humans and to identify virulence factors potentially specific to CGD, we sequenced the entire genome to completion. We also developed a custom, full-genome, high-density microarray to look at whether the “emergence” of this pathogen in these four patients was due to the same strain of G. bethesdensis.
The pathology caused by G. bethesdensis in a CGD mouse model is similar to that observed in humans (11). In contrast, G. oxydans caused little pathology in the CGD mouse model, indicating that the virulence factors present in G. bethesdensis are not likely to be present in the Acetobacteraceae family as a whole (11). Since the factors that unite CGD pathogens are unknown, we compared G. bethesdensis to other sequenced CGD bacterial pathogens, with a focus on putative virulence.
MATERIALS AND METHODS
G. bethesdensis isolates and in vitro cultivation.
Four CGD patients from geographically distinct locations, presenting with fever and lymphadenitis, were identified. The patients were referred to as NIH1, NIH2, NIH3, and NIH4. Both the patient number and the number of times G. bethesdensis was retrieved in a clinical specimen designate the isolates. Hence, for the four separate isolates grown from lymph node biopsies from patient NIH1 collected on separate occasions before and during treatment, the organisms are designated NIH1.1, NIH1.2, NIH1.3, and NIH1.4. The single isolates from the other patients are designated NIH2.1, NIH3.1, and NIH4.1. Isolated bacteria were cultured to either the log phase or stationary phase of growth in 50 ml SOC (tryptone, 2% [wt/vol]; yeast extract, 0.5% [wt/vol]; NaCl, 8.6 mM; KCl, 2.5 mM; MgSO4, 20 mM; glucose, 20 mM) at 37°C with shaking, as indicated.
Genome sequencing and analysis.
Isolate NIH1.1 was used for genomic DNA isolation and whole-genome sequencing. Genomic DNA was isolated by standard protocols (Gentra Systems, Minneapolis, MN), sheared, size fractionated, and used to construct libraries in both plasmids (2- to 3-kb inserts in pGEM3) and cosmids (30- to 35-kb inserts using Lorist6) (9). Whole-genome shotgun sequencing was performed to 7× coverage, and the sequence was assembled using established PHRED/PHRAP/CONSED and proprietary algorithms as previously described (6, 14, 17, 25). Manual analysis, primer walking, and PCR fragment generation were used to close gaps as previously described (6, 14, 17). Sequencing was carried out to average Q40 (1/10,000 error-to-base-quality ratio) throughout.
Open reading frame (ORF) calling was performed using public and proprietary algorithms, with a minimum length cutoff of 40 amino acids, as previously described (6, 14, 17). Truncated or frameshifted ORFs were analyzed empirically at the raw chromatogram and assembly stage. The ORFs were entered into the ERGO bioinformatics suite (Integrated Genomics, Chicago, IL) for genome and functional metabolic reconstruction. Comparative genome analysis was performed using frequently updated sequences in ERGO (Integrated Genomics, Chicago, IL) as previously described (25), using BLAST algorithms. Based on similarity and other criteria, ORFs were assigned to preexisting orthologous clusters, whereupon functions were ascribed. Manual analysis of every ORF was performed using sequence similarity tools, such as the motif/pattern databases Pfam, Prosite, Prodom, and COGs, and a number of proprietary tools (21), including but not limited to algorithms for the identification of fusion clusters, chromosomal clusters, and regulatory ORF clusters. GC skew was calculated as (C − G)/(G + C), with a 20-kb sliding window moving in 500-bp incremental steps. GC% was calculated using 2-kb and 20-kb sliding windows and compared to total GC% for the entire chromosome. Once functions were assigned to the ORFs, they were connected to the corresponding pathway(s). Finally, metabolic reconstructions were derived by extrapolating interconnections for the entire set of pathways. The origin of replication was calculated by Oriloc (8) and denoted position 0. Identification of best BLAST hits for G. bethesdensis protein sequences was performed based upon the P scores of the BLAST hits.
Electron microscopy and microbiology studies.
Freshly grown subcultures of the reference strain CGDNIH1.1 were inoculated onto tryptic soy agar with 5% sheep blood agar (Remel, Lenexa, KS) and tryptic soy agar with 5% horse blood agar (BBL, Becton Dickinson Microbiology Systems, Cockeysville, MD). Plates were incubated at 35°C in 5% CO2 and air incubators. Thermanox coverslips (Nalgene Nunc International, Rochester, NY) were coated with 0.01% poly-l-lysine in water, air dried, and placed onto colonies of G. bethesdensis. After 5 seconds, the coverslips were lifted off the agar, immersed into Karnovsky's fixative (Electron Microscopy Sciences, Hatfield, PA) containing 0.01% ruthenium red and 0.1 M sodium cacodylate, pH 7.2, and maintained overnight at 4°C. The samples were washed in cacodylate buffer and postfixed for 1 h with a mixture of 1% osmium tetroxide and 0.8% potassium ferrocyanide in cacodylate buffer. Samples were washed and then stained for 1 h with 1% aqueous uranyl acetate. After further washes, the samples were dehydrated in ethanol and embedded in ultra-low-viscosity resin (Polysciences, Inc., Warrington, PA). Thin sections were examined at 80 kV. For scanning electron microscopy, 30 μl of bacterial suspension was allowed to settle for 10 min onto polylysine-coated silicon chips. Adsorbed bacteria were fixed for 30 min in Karnovsky's fixative in 0.1 M sodium phosphate, pH 7.2. Following two washes with phosphate buffer, the samples were postfixed for 30 min with 1% osmium tetroxide in phosphate buffer. The samples were washed once with phosphate buffer and twice with water, dehydrated in ethanol, and critical point dried through carbon dioxide. The chips were lightly sputtered with chromium before examination.
Custom high-density microarray.
A Custom GeneChip high-density oligo-DNA array was manufactured at Affymetrix (Santa Clara, CA) as described previously (10) and denoted RMLchip3a520351F. This array consists of 23,015 probe sets, which represent 18 bacterial genomes, and 28,880 putative ORFs. The Granulibacter bethesdensis portion of the multigenome array consists of 2,468 probe sets (37 are redundant), each of which represents a single putative ORF.
DNA-DNA hybridization to high-density custom array.
Bacterial genomic DNAs were extracted from stationary-phase cultures by using a QIAGEN DNeasy kit (Valencia, CA) following the manufacturer's recommendations. A total of 10 mg of genomic DNA was digested with 0.005 units of DNase I (Roche Diagnostics) in 1× DNase I buffer (10 mM Tris, pH 7.5, 2.5 mM MgCl2, 0.1 mM CaCl2) at 37°C for 10 min and then denatured at 98°C for 10 min. The fragmented genomic DNA, ranging from 50 bp to 200 bp, was 3′ end labeled with biotin-ddUTP, using a Bioarray terminal labeling kit (Enzo Diagnostics, Farmingdale, NY), for 1 h at 37°C according to the manufacturer's instructions. Prior to hybridization to the RMLchip3a array, the genomic DNA samples were placed in hybridization buffer (100 mM morpholineethanesulfonic acid [MES], 1 M [Na+], 20 mM EDTA, 0.01% Tween 20), heated to 95°C for 5 min, and then immediately placed on ice for 5 min. Hybridization was carried out for 16 h at 60°C. Chips were then washed and scanned following standard Affymetrix chip protocols.
DNA array hybridization analysis.
Affymetrix GeneChip operating software (GCOS v1.4) was used to perform the preliminary analysis of the custom chips at the probe set and probe level. For the probe set analysis, all *.cel files were scaled to a trimmed mean of 500 by using a scale mask consisting of only the Granulibacter bethesdensis probe sets to produce the *.chp files. A pivot table for all samples, including calls, P values, and signal intensities, was created. The pivot table was then imported into GeneSpring GX 7.3, where hierarchical clustering (condition tree) using a Pearson correlation similarity measure with average linkages was used to produce the dendrogram.
Targeted PCR analysis.
Forty ORF sequences were selected among the total number of ORFs or probe sets demonstrating no signal on the custom chips. These ORFs were analyzed by targeted PCR and DNA sequence analysis. ORF sequences along with 200 bases 5′ and 3′ of the coding regions were collected, and PCR oligonucleotides were designed within 150 to 200 bases upstream and downstream of the coding regions. PCR was performed under the following amplification conditions: 25 cycles of 95°C for 1 min, 58°C for annealing, and 72°C for 2 min for extension. PCR fragments were visualized on 8% agarose gels. Positive PCR fragments, defined as single, dominant bands at or within 1 kb of the estimated size range, were sequenced with the same amplification primers. Sequence data were searched by BLAST analysis against the whole genome sequence, and results were analyzed to confirm that amplicons were from the targeted regions.
RNA extraction and quantitative RT-PCR analysis.
Granulibacter bethesdensis strain NIH1.1 was cultured at both 26°C and 35°C in yeast extract-peptone-glucose medium as described before (12), and bacteria were harvested at the mid-exponential, late-exponential, and stationary phases of growth. RNAs were extracted using the standard RNeasy protocol (QIAGEN, Valencia, CA) and treated with DNase I as described previously (39) to remove contaminating genomic DNA. Reverse transcription-PCR (RT-PCR) was performed as previously described (39). Briefly, Platinum Q-PCR SuperMix-UDG RT-PCRs (Invitrogen Corp.) were carried out in 20-μl reaction mixtures containing 1× Platinum Q-PCR SuperMix-UDG mix, 6 mM MgCl2, 1× ROX reference dye (1.25 μM 5-carboxy-X-rhodamine, succinimidyl ester), 300 nM VIC dye- and minor groove binding quencher (MGB; Applied Biosystems, Foster City, CA)-labeled tufA probe, a 200 nM concentration (each) of target forward and reverse primers, and 300 nM of the target TaqMan oligonucleotide under the following conditions: 50°C for 2 min, 95°C for 2 min, and 55 cycles of 95°C for 15 s and 60°C for 1 min. All TaqMan oligonucleotide probes were labeled with 6-carboxyfluorescein at the 5′ end and with the MGB nonfluorescent quencher at the 3′ end. TaqMan primer and probe sequences used in this effort are provided in Table S7 in the supplemental material. The comparative cycle threshold method was used to determine the ratio of target and endogenous control (Applied Biosystems) values as previously described (39).
Signal sequence analysis of G. bethesdensis ORFs.
All ORFs of the genome were extracted from ERGO (Integrated Genomics, Chicago, IL) and analyzed using the public web servers SignalP 3.0 and LipoP 1.0 (http://www.cbs.dtu.dk/services/) (2, 16). The two sets of ORFs were inspected manually to evaluate functional assignments. In nine instances, ORFs were removed from the original SignalP set because although a related motif was found, the location of the motif and annotation for the ORF suggested a more likely cytoplasmic location or function.
Nucleotide sequence accession number.
The complete genome sequence of G. bethesdensis has been deposited in GenBank under accession number CP000394.
Microarray data accession number.
Microarray data were deposited at http://www.ebi.ac.uk under accession number E-TABM-148.
RESULTS
G. bethesdensis genome has unique genes and potential virulence-encoding capabilities.
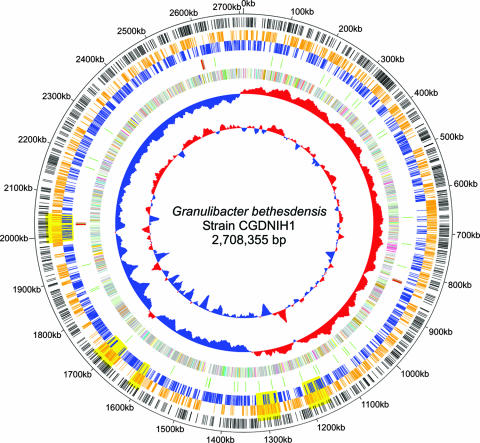
The genome of the G. bethesdensis isolate NIH1.1 (strain CGDNIH1T; ATCC BAA-1260T) was sequenced to entirety. It is composed of a single circular chromosome of 2,708,355 bp with a calculated total G+C content of 59.07% (Table 1; Fig. 1). While G. oxydans contains five plasmids (26), none were found in G. bethesdensis during genome assembly, plasmid DNA isolation analysis, or plasmid replication gene searches. A total of 2,437 protein-encoding ORFs were predicted, and 1,832 (75.17%) ORFs were found to have an assigned function (Table 1). Of the 605 ORFs without an assigned function, 453 had homology to genes in other organisms, while 152 ORFs were considered unique. The annotated functional categories of all ORFs in G. bethesdensis are provided in Table S2 in the supplemental material.
TABLE 1.
General features of the Granulibacter bethesdensis genome
| Parameter | Value or description |
|---|---|
| G. bethesdensis parameters | |
| Size of chromosome (bp) | 2,708,355 |
| Coding region (%) | 92.91 |
| G+C content (%) | 59.07 |
| rRNAs | 5S, 16S, 23S (3×) |
| No. of tRNAs | 52 |
| No. of ORFs | |
| Total | 2,437 |
| With assigned function | 1,832 |
| With no assigned function | 605 |
| With no function, but with similarity | 453 |
| With no function or similarity | 152 |
| Unique to G. bethesdensis | 152 |
| With signal peptides (SIGNALP) | 353 |
| With signal peptides (LIPOP) | 486 |
| With lipoprotein signal peptides | 34 |
| With peptidase domains | 16 |
| Potentially frameshifted | 123 |
| G. oxydans parameters | |
| Size of chromosome (bp) | 2,702,173 |
| Size (sum) of five plasmids (bp) | 220,211 |
| Total no. of ORFs (with plasmids) | 2,664 |
| Comparative parameters | |
| No. of G. bethesdensis ORFs shared with G. oxydans | 1,470 |
| No. of G. oxydans ORFs shared with G. bethesdensis | 1,573 |
| No. of unique G. bethesdensis ORFs compared to G. oxydans | 967 |
| No. of unique G.oxydans ORFs compared to G. bethesdensis | 1,091 |
FIG. 1.
Genome circle of Granulibacter bethesdensis. The genome is 2,708,355 base pairs long, and the rings, starting inside and moving out, show the following: GC content with sliding 20-kb window; GC skew; ORFs colored by metabolic category (virulence = turquoise, cell wall metabolism = light yellow, stress = light orange, phage = orange, signal transduction = purple, cellular processing = yellow, secondary metabolism = blue, membrane transport = cyan, secretion = teal, motility and chemotaxis = pink, information processing = sage, phosphorus metabolism = olive, nitrogen metabolism = tangerine, coenzyme and cofactor metabolism = saffron, sulfur metabolism = light green, bioenergetics = lime, one-carbon metabolism = beige, aromatic compound degradation = light purple, nucleotide metabolism = light blue, lipid metabolism = lemon, carbohydrate metabolism = rust, amino acid metabolism = peach); RNAs (tRNAs are shown in green, and rRNAs are shown in red); ORFs colored and positioned by orientation (orange, positive strand; and blue, negative strand); yellow shaded areas representing regions undergoing DNA changes across the four isolates, based on chip hybridization data; ORFs present in G. bethesdensis that are unique compared to G. oxydans (black bars); and numeric markers.
A BLAST hit analysis of all the G. bethesdensis ORFs found that the G. oxydans genome (26) had the most ORFs with best-hit similarities (Table 1). Of a total of 2,286 G. oxydans ORFs, 737 (32.2%) showed best-hit homologues in G. bethesdensis (Table 1; see Table S3 in the supplemental material). One thousand ten protein families (1,470 ORFs) are shared between G. oxydans and G. bethesdensis, with 901 protein families (967 ORFs) unique to G. bethesdensis and 964 protein families (1,091 ORFs) unique to G. oxydans (Table 1). A graphical example of unique ORFs for G. bethesdensis compared to G. oxydans is shown in the outer ring of Fig. 1. Whole-genome comparisons of G. bethesdensis to the published genomes of the CGD pathogens Burkholderia cenocepacia, Chromobacterium violaceum, Nocardia farcinica, and methicillin-susceptible Staphylococcus aureus were done to determine genetic factors shared among the bacteria that commonly infect CGD patients (see Table S4 in the supplemental material).
G. bethesdensis contains unique dehydrogenases and a respiratory chain not present in G. oxydans.
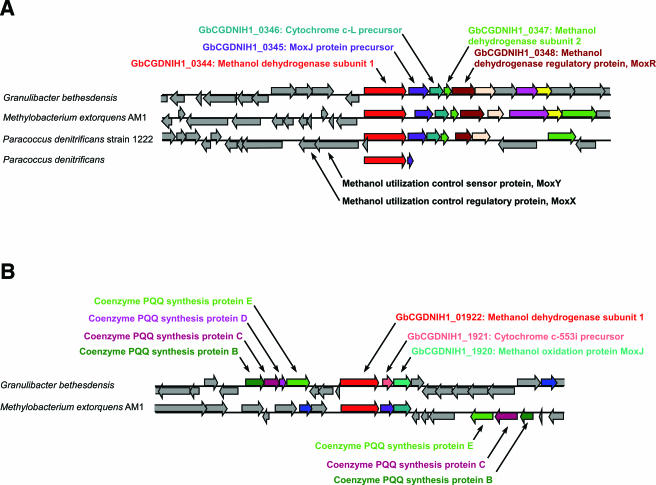
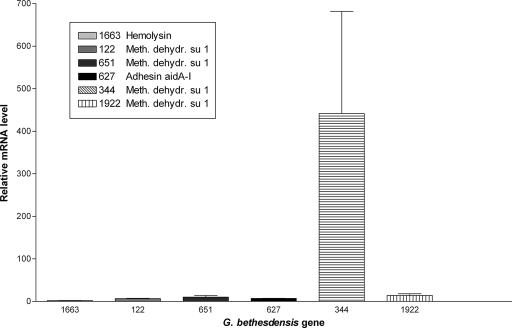
Both G. bethesdensis and G. oxydans are capable of converting alcohols to acetic acid (12). They both contain genes that encode the enzymes necessary to carry out these reactions, including membrane-bound acetaldehyde dehydrogenase (GbCGDNIH1_0793-0795), which converts acetaldehyde to acetate (26). However, G. bethesdensis contains unique dehydrogenase genes not found in G. oxydans, such as GbCGDNIH1_0122, GbCGDNIH1_0651, GbCGDNIH1_0344, and GbCGDNIH1_1922, each of which encodes a methanol dehydrogenase subunit 1. Signal sequence analysis indicated that these four ORFs contain secretion signals, suggesting a periplasmic location. GbCGDNIH1_0344 seems to be part of an operon with downstream ORFs showing homology to the genes for a MoxJ precursor (GbCGDNIH1_0345; GC%, 52.57%), a cytochrome c-L precursor (GbCGDNIH1_0346), a methanol dehydrogenase subunit 2 (GbCGDNIH1_0347), and the methanol dehydrogenase regulatory protein MoxR (GbCGDNIH1_0348), all of which are critical to a functional methanol dehydrogenase complex (38). GbCGDNIH1_0344 shows a best BLAST hit to the methanol dehydrogenase gene of another methylotrophic bacterium, Methylobacterium extorquens AM1, and shares a locational context identical to that of orthologous clusters in both M. extorquens AM1 and Paracoccus denitrificans strain 1222 (Fig. 2A). GbCGDNIH1_1922 is adjacent to ORFs encoding methanol utilization functions, including the coenzyme PQQ synthesis proteins B (GbCGDNIH1_1929), C (GbCGDNIH1_1928), D (GbCGDNIH1_1927), and E (GbCGDNIH1_1926) and a MoxJ-like methanol oxidation protein (GbCGDNIH1_1920). The GbCGDNIH1_1922 gene cluster also demonstrates locational conservation with a homologous locus in M. extorquens (Fig. 2B). G. bethesdensis is capable of growth on methanol as a sole carbon source (12). It is not known at this time whether methanol dehydrogenase serves the function of converting both ethanol and methanol to acetaldehyde. In addition, there appeared to be an immune response to the GbCGDNIH1_0344 protein in at least one of the patients (11). Multiplex quantitative PCR was used to measure the transcripts of the four methanol dehydrogenase subunit 1 ORFs (GbCGDNIH1_0122, GbCGDNIH1_0651, GbCGDNIH1_0344, and GbCGDNIH1_1922) under six different growth conditions, namely, 26°C and 35°C during mid-exponential, late-exponential, and stationary phases of growth in YPG medium (12), and their signals were normalized to the mRNA signal from the transcript for the protein translation elongation factor TU (tufA) gene. In all cases, the GbCGDNIH1_0344 gene showed the highest expression relative to tufA and the other three genes tested (data not shown). Figure 3 shows the expression data for the four methanol dehydrogenase subunit 1 ORFs in addition to two other ORFs tested (GbCGDNIH1_1663 and GbCGDNIH1_0627) during the stationary phase of growth at 35°C, which is representative of the other conditions. Therefore, G. bethesdensis is both an acetic acid bacterium and a methylotroph; this bifunctional phenotype has previously been described for the genus Acidomonas. Given the possible immune recognition of this protein in at least one patient, GbCGDNIH1_0344 must be expressed under in vivo and in vitro growth conditions.
FIG. 2.
Comparative genomic context of Granulibacter bethesdensis methanol dehydrogenase genes. A comparative region analysis of two methanol dehydrogenase subunit 1 ORFs in G. bethesdensis compared to other best-hit genes and genomes is shown. (A) GbCGDNIH1_0344 and surrounding genes and their relationships to those in other methylotrophic organisms. (B) GbCGDNIH1_1922 and surrounding genes needed for methanol utilization and their relationship to the methylotrophic bacterium Methylobacterium extorquens AM1.
FIG. 3.
Quantitative PCR analysis of six different Granulibacter bethesdensis gene mRNAs at 35°C in the stationary phase of growth. Bacteria were cultured in YPG broth at 35°C to stationary phase. Multiplex quantitative PCR was performed to measure the transcripts from six different G. bethesdensis genes, and their signals were normalized to the mRNA signal from the transcript for the protein translation elongation factor TU (tufA) gene. The y axis shows relative mRNA levels for each of the six genes compared to tufA, and the x axis shows the six individual genes tested.
Respiratory systems generate proton motive force, regenerate NAD+, thereby maintaining intracellular redox balance, and scavenge oxygen. G. oxydans lacks the genes encoding NADH complex 1. Prust et al. suggested that this may be related to its incomplete oxidative lifestyle, resulting in a lower energy transducing efficiency, which may in part be responsible for G. oxydans lower growth yields (26). In contrast, G. bethesdensis contains a full complement of NADH-quinone oxidoreductase chain genes (GbCGDNIH1_1289 to -1302) but has slower growth and lower yields than G. oxydans on solid media. Additional studies and analysis will be needed to elucidate why and how the presence or absence of this system in these two related organisms contributes to growth and survival.
LPS and capsule biosynthesis genes.
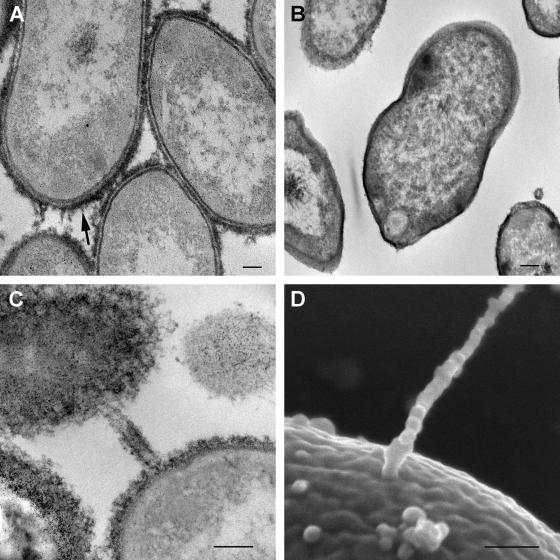
Lipopolysaccharide (LPS) and cell surface capsular polysaccharides are known to be important factors in the pathogenesis of gram-negative bacterial infections (24). G. bethesdensis and G. oxydans appear to contain similar complements of genes for the biosynthesis of LPS as well as cell surface capsular polysaccharides (see Table S4 in the supplemental material). However, G. bethesdensis has a unique glucosyltransferase (GbCGDNIH1_0741) (see Table S4 in the supplemental material) relative to G. oxydans. Light microscopy with India ink (data not shown) in conjunction with transmission electron microscopy (ruthenium red and negative staining) demonstrated capsule-like material surrounding G. bethesdensis (Fig. 4A and B).
FIG. 4.
Ultrastructural evidence of extracellular polysaccharide capsule and conjugative pilus-like structures. (A and B) Transmission electron micrographs of G. bethesdensis NIH1.1 prepared with (A) and without (B) ruthenium red for preserving and contrasting extracellular polysaccharides. The arrow in panel A points out the capsular material on the surface. (C and D) Transmission (C) and scanning (D) electron micrographs of structures resembling conjugative pili. Bars, 100 nm.
Iron utilization genes.
Iron is important in many enzymatic reactions, and scavenging it from the host is a widely adopted strategy for pathogen survival. A variety of genes involved in iron utilization were discovered in G. bethesdensis but not in G. oxydans, including a heme oxygenase gene (GbCGDNIH1_0284; GC%, 52.64%) and an iucABCD (iron uptake chelate) gene cluster for aerobactin synthesis (GbCGDNIH1_2378 to -75). Heme oxygenase degrades heme into iron and biliverdin, and aerobactin is a hydroxamate siderophore prevalent in many species of Shigella (27). Interestingly, GbCGDNIH1_2376 demonstrated a best BLAST hit to a Shigella flexneri homolog (54.6% GC) (see Table S3 in the supplemental material). An ORF encoding a transporter protein (GbCGDNIH1_2379; GC%, 60.97%) followed by a ferric hydroxymate reductase ORF (GbCGDNIH1_2380; GC%, 62.57%) was found nearby. The transporter protein may facilitate transfer of the iron-siderophore complex back into the cell. The presence of the heme oxygenase and the iucABCD aerobactin gene cluster in G. bethesdensis, but not G. oxydans, suggests that G. bethesdensis is equipped for iron uptake within mammalian systems. Development of a genetic system and knockout mutants of these loci will help to determine the roles of these pathways in G. bethesdensis pathogenesis.
Adhesin-, hemagglutinin-, and hemolysin-encoding genes.
AIDA (adhesin involved in diffuse adherence) genes were originally described for diffusely adherent Escherichia coli and appear to be integral to bacterial binding to human and nonhuman cells (35). There are many unique AIDA adhesins in G. bethesdensis which are not present in G. oxydans, notably, GbCGDNIH1_1601, GbCGDNIH1_0627, GbCGDNIH1_0615, GbCGDNIH1_1413, and GbCGDNIH1_1465 (see Table S4 in the supplemental material). Comparison to other CGD pathogens shows that homologs of GbCGDNIH1_0627 and GbCGDNIH1_0615 exist in the Burkholderia cepacia complex, with the former also found in the virtually pathognomonic CGD pathogen Chromobacterium violaceum (see Table S4 in the supplemental material). A G. bethesdensis locus of AIDA adhesins (GbCGDNIH1_1601 to -1605) has five ORFs showing GC% values ranging from −5.8% to +0.8% lower or higher than the genome average. Three of these adhesins (GbCGDNIH1_1603 to -1605) have best BLAST hits to AIDA homologs in diarrheagenic E. coli (3) (see Table S3 in the supplemental material). Several large hemagglutinin-related protein genes not present in G. oxydans and without full-length homologues in the database were also found in G. bethesdensis (GbCGDNIH1_1408, GbCGDNIH1_1474, and GbCGDNIH1_1135). These genes potentially encode proteins of 283 to 379 kDa and demonstrate similarity with AIDA adhesins and the filamentous hemagglutinin of Bordetella pertussis (29).
G. bethesdensis encodes several hemolysins (GbCGDNIH_1663, GbCGDNIH1_1661, and GbCGDNIH1_1660) not found in G. oxydans (see Table S4 in the supplemental material) which have been identified previously or implicated in the lysis of host cells or red blood cells. The two adjacent ORFs, GbCGDNIH_1664 (GC%, 48.32%) and GbCGDNIH_1663 (GC%, 50.56%), are of particular interest, in that they encode a hemolysin activator protein similar to ShlB in the CGD pathogen Serratia marcescens (40) and a hemolysin similar to the filamentous hemagglutinin FhaB protein in Bordetella pertussis (29), respectively. Both genes encode proteins involved in the two-partner secretion pathway (15), a process for the extracellular transport of large virulence proteins. Homologous genes also exist in the CGD pathogen Chromobacterium violaceum (see Table S4 in the supplemental material). G. bethesdensis also contains a putative operon absent from G. oxydans encoding a type 1 protein secretion system that includes hemolysins as potential secretion substrates (see Table S4 in the supplemental material). This system consists of an ATP-binding protein/transmembrane protein-encoding subunit (GbCGDNIH1_1527), an ATPase-encoding component (GbCGDNIH1_1528), and an adaptor-encoding protein (GbCGDNIH1_1526). GbCGDNIH1_1527 homologs exist in Burkholderia cepacia, Chromobacterium violaceum, Nocardia farcinica, and Staphylococcus aureus, while a GbCGDNIH1_1526 homolog is present in C. violaceum and N. farcinica only (see Table S4 in the supplemental material).
The hemolysin gene data suggest several components that may be responsible for the α-hemolysis seen with G. bethesdensis, but not with G. oxydans (data not shown), on sheep blood agar (12). Hemolysins and their secretion may be critical factors in bacterial pathogenesis in CGD.
Oxidative and acid stress response-related genes.
The G. bethesdensis and G. oxydans genomes contain several genes important for tolerating oxidative stress, such as the superoxide dismutase gene (sod) (GbCGDNIH_0954) and several catalase-encoding genes (GbCGDNIH1_1969, GbCGDNIH1_1598, and GbCGDNIH1_1677). A peroxidase/catalase-encoding ORF (GbCGDNIH_1677) was unique to G. bethesdensis (see Table S4 in the supplemental material) and demonstrated a best BLAST hit to a homolog in Burkholderia vietnamiensis (see Table S3 in the supplemental material). The presence of these genes may help to protect G. bethesdensis from environments containing reactive oxygen species.
G. bethesdensis converted urea to ammonia and carbon dioxide in three of four patient isolates (12). Chromosomal loci in G. bethesdensis that can contribute to these functions are the ureDABCEFG locus (GbCGDNIH_2161 to -67), an amide-urea transport locus (GbCGDNIH_1831 to -36), and an allophanate hydrolase and urea carboxylase locus (GbCGDNIH_1744 and -45). The last locus is present in G. oxydans, but the ureDABCEFG and amide-urea transport loci are missing (see Table S4 in the supplemental material). Therefore, G. bethesdensis, in contrast to G. oxydans, may be able to alter and survive in low-pH environments by conversion of urea to ammonia. This process of environmental pH neutralization using these two clusters of genes has been described previously for other organisms, notably Helicobacter pylori, in which the conversion of urea to ammonia and carbon dioxide facilitates H. pylori survival in acidic environments, including the human stomach (28). B. cepacia, N. farcinica, and S. aureus also contain all three loci (see Table S4 in the supplemental material). These data suggest that G. bethesdensis may be capable of tolerating acidic vacuoles, such as those found in macrophages and neutrophils.
Antibiotic resistance genes.
G. bethesdensis is resistant to multiple antibiotics (11). Many antimicrobial-related resistance genes in G. bethesdensis have homologs in G. oxydans (see Table S4 in the supplemental material). However, two beta-lactamases (GbCGDNIH_0107 and GbCGDNIH_1173) were unique to G. bethesdensis (see Table S4 in the supplemental material). More work remains to be performed with regards to determining exactly which genes confer the multiple-antibiotic resistance phenotype seen for G. bethesdensis. Clinical isolates of Serratia marcescens and Burkholderia cepacia often have some degree of drug resistance (13).
DNA uptake and horizontal transfer-related genes.
The trb and tra operons encode type IV DNA conjugation systems that deliver DNA or protein into bacterial or eukaryotic target cells and are therefore considered virulence factors (4). Unlike G. oxydans, G. bethesdensis contains multiple ORFs encoding Trb and Tra components (see Table S4 in the supplemental material). G. bethesdensis has only four of the genes described for the Trb system (20), i.e., trbF (GbCGDNIH_1062), trbG (GbCGDNIH_1063), trbI (GbCGDNIH_1064), and trbL (GbCGDNIH_1061), for which GC% ranges are 69 to 72%. GbCGDNIH_1064 and GbCGDNIH_1061 encode inner membrane channel-forming proteins and GbCGDNIH_1063 encodes the outer membrane channel-forming protein, all of which potentially facilitate the transport of macromolecules. TrbF is thought to be a minor pilus component (31). Pilus-like structures were visualized on G. bethesdensis by both transmission electron microscopy and scanning electron microscopy (Fig. 4C and D). G. oxydans contains a full complement of flagellum-encoding genes, but G. bethesdensis does not. Efforts to determine if these are true pilus-like structures on the surface of G. bethesdensis and to identify the gene(s) that encodes them are under way.
The Tra-DNA conjugation system is essential for DNA translocation (20). Of the genes described for the Tra-DNA conjugation system, G. bethesdensis contains traD, encoding the inner membrane type IV DNA complex coupler (GbCGDNIH1_1512), and traI (GbCGDNIH1_1514), encoding the helicase. While a type IV ATPase gene was not found in the G. bethesdensis genome, a type II secretion ATPase (GbCGDNIH_1756) with high homology to a type IV system ATPase in Rhizobium leguminosarum was identified (data not shown).
G. oxydans has two regions with ORFs showing similarity to phage ORFs (26). G. bethesdensis contains one region likely representing a phage integration event, spanning 39 ORFs. Nine of these ORFs have significant similarities to annotated phage proteins, while 12 of the 39 ORFs have best BLAST hits to Haemophilus influenzae phage ORFs (see Table S3 in the supplemental material). However, we have not determined whether this phage element is functional.
In addition, G. bethesdensis harbors a 25-kb region that contains an unusually large number of ORFs encoding transposases (11 of the total of 16 for the entire genome). The GC content for this region is well above the genome average, by 5 to 10%, and it therefore potentially represents a locus of newly transferred or acquired DNA. Given that the ORFs are primarily transposon elements, this could be a hot spot for recombination events (21).
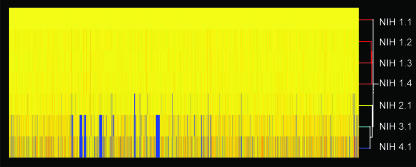
Whole-genome hybridization/genotyping of G. bethesdensis isolates.
Comparative genome hybridization (CGH) facilitates rapid genotyping of bacterial isolates (1). Using a custom high-density expression array containing nearly all ORFs for the CGDNIH1.1 genome sequence, we performed CGH with genomic DNAs from all of the G. bethesdensis isolates. The four isolates from patient NIH1 (NIH1.1, NIH1.2, NIH1.3, and NIH1.4) showed identical, complete hybridization to all array ORFs (Fig. 5) by both probe set and probe-level analyses, suggesting no variation within the patient by this method of detection. In contrast, each of the other three patient isolates (NIH2.1, NIH3.1, and NIH4.1) had unique hybridization patterns relative to that for patient NIH1, with NIH4.1 and NIH3.1 diverging most from NIH1.1 (Fig. 5). A maximum of 174 ORFs in at least one of the three isolates had neither probe set signals nor individual probe signals and were therefore designated as absent or altered in sequence significantly enough to fail to give a hybridization signal. Interestingly, the loss of signal appeared to be fairly conserved with regards to regions of difference (RODs) across isolates NIH4.1 and NIH3.1. The genes in these RODs include the trb locus, traD-traI loci, transcriptional regulator genes, LPS synthesis genes, protein secretion genes, transposase genes, and hypothetical or hypothetically secreted protein-encoding genes.
FIG. 5.
Dendrogram of DNA hybridization of seven isolates with NIH1.1. A DNA-DNA hybridization microarray-based investigation of gene distributions among G. bethesdensis isolates was performed. The absence of genes is shown in blue, and the presence of genes is shown in yellow and red. A dendrogram is shown with the strain names listed. The analysis was performed with GeneSpring software.
To confirm the CGH absence calls and to determine whether DNA had been acquired or lost in the RODs, primers were designed and PCR performed against flanking sequences for 40 selected ORFs called present in NIH2.1 but absent in either NIH3.1 or NIH4.1 The microarray data (see Table S5 in the supplemental material) were confirmed in that genes called present by the microarray provided amplified fragments, while those called absent did not. Failure to be amplified by PCR could have been due to divergent DNA sequences relative to NIH1.1-based primers (see Table S6 in the supplemental material). Sequence data from the PCR-amplified fragments were BLAST searched against the G. bethesdensis genome to confirm the targeted loci (data not shown). Large RODs representing multiple successive ORFs that show no hybridization are shown in yellow in the genome circle in Fig. 1.
DISCUSSION
Granulibacter bethesdensis is a newly discovered genus and species of the acetic acid bacteria in the family Acetobacteraceae and is the first member of the Acetobacteraceae reported to cause invasive human disease (11, 12). Members of the acetic acid bacteria are characterized by the ability to incompletely oxidize a wide range of carbohydrates and alcohols, and they can grow in highly concentrated sugar or alcohol solutions. Thus, their natural habitats are often associated with the flowers and fruits of plants. The process of periplasm-associated incomplete oxidation coupled with rapid product excretion into the medium and low biomass production makes these organisms valuable in industrial applications. The process of incomplete oxidation can result in heat production (7), suggesting that the acetic acid bacteria may create warmer conditions than their ambient environs. Both G. oxydans and G. bethesdensis contain the ORFs necessary for capsule synthesis, and electron microscopy shows an apparent capsule surrounding G. bethesdensis. For Zymomonas mobilis, a capsule may facilitate its resistance to osmotic pressure at high concentrations of sugar and alcohol (34).
CGD is a rare genetic disease of the phagocyte NADPH oxidase system characterized by defective production of toxic oxygen metabolites (33). Patients develop recurrent life-threatening infections by catalase-producing organisms, ranging from the common Staphylococcus aureus to unusual and virtually pathognomonic invasive infections with Chromobacterium violaceum (22), Burkholderia gladioli (30), and Francisella philomiragia (41). Because these organisms are environmental but cause infection almost exclusively in CGD, it seems likely that some common pathological features must connect these otherwise disparate organisms. It was appreciated early on that the bacteria and fungi that infect CGD patients are almost invariably catalase producing. It was supposed that microbial catalase scavenged hydrogen peroxide produced by the ingested organism, thus depriving the CGD neutrophil of the hydrogen peroxide that could be used to create bleach (18). However attractive this hypothesis was, mutants of Staphylococcus aureus and Aspergillus nidulans deleted for catalase had undiminished virulence in CGD mice (5, 23). Furthermore, numerous catalase-producing organisms do not cause clinical infection in CGD, indicating that catalase is neither necessary nor sufficient for virulence in CGD (37). Therefore, the bacterial properties that make for virulence in CGD are still undetermined. Since infection due to G. bethesdensis had never been described and since it belonged to a family of organisms that had never been recognized as mammalian pathogens, we undertook whole-genome sequencing to identify potential bacterial genetic virulence factors in CGD.
G. bethesdensis has several methanol dehydrogenase genes that appear to have been acquired recently from Methylobacterium extorquens, an organism widespread in terrestrial and aquatic habitats and in the phyllosphere (19). DNA uptake systems in G. bethesdensis, such as the trb and tra systems, which are lacking in G. oxydans, may have facilitated the horizontal transfer of genes from Methylobacterium or other species. Virulence-related ORFs from other bacteria may also have been acquired by G. bethesdensis through a similar process.
Heme oxygenase is important in heme, hemoglobin, and haptoglobin utilization. The accretion of genes coding for transporter function (GbCGDNIH1_2379; GC%, 60.97%), ferric hydroxymate reductase, and aerobactin synthesis suggests that G. bethesdensis may survive in iron-poor environments. The GC percentages for these genes differ from the G. bethesdensis genome average, and best BLAST hits identified homologs in Shigella. As with methanol dehydrogenase, iron acquisition genes may have been obtained by horizontal transfer, potentially facilitating adaptation to a pathogenic lifestyle.
AIDA genes encode adhesins, which promote bacterial binding to mammalian cells, autoaggregation via intercellular recognition, and highly efficient biofilm formation (35). G. bethesdensis displayed autoaggregation during cultivation (data not shown). For one locus of five AIDA genes, GC percentages were up to ∼6% lower than the G. bethesdensis genome average. Best BLAST hits for three of the five genes were for AIDA genes from diarrheagenic E. coli. Three unique, large, hemagglutinin-like genes with homology to AIDA and Bordetella filamentous hemagglutinin were discovered in G. bethesdensis, as well as shlB and fhaB hemolysin gene homologs. Many of these ORFs have homology to ORFs identified in previously known CGD pathogens, such as C. violaceum, S. marcescens, and the B. cepacia complex. Therefore, G. bethesdensis contains a unique repertoire of adhesins and hemolysins, which are not present in G. oxydans, that are also found in known CGD pathogens. The presence of a capsule, the ability to tolerate low pH and heat, and the acquisition of virulence-related ORFs, some of which are found in other CGD pathogens, may account for the movement of this organism from an exclusively environmental niche into susceptible human hosts.
Whole-genome sequencing frequently identifies genes of unknown function as well as many genes unique to the specific organism. In G. bethesdensis, of the 605 ORFs without assigned functions, 152 were unique to G. bethesdensis and may represent signature sequences. These G. bethesdensis-specific sequences will facilitate broadcast PCR studies in the environment to identify the reservoir of G. bethesdensis as well as the discrete sources of infection for our patients.
Genomic hybridization of the four G. bethesdensis isolates showed regions of variance or plasticity between the isolates, suggesting microheterogeneity in G. bethesdensis. The reasons for the sudden emergence of these strains in four CGD patients remain unclear. The other members of the Acetobacteraceae are found in numerous tropical environments, including fruits and flowers. It is reasonable to hypothesize that there is an environmental nonhuman niche for G. bethesdensis as well. What might allow an organism to transition from a nonhuman environment to a human one must require an important set of genetic traits, both evolved endogenously and acquired from other organisms, encoding pathogenicity and virulence-related functions. We have identified numerous genetic factors that may be associated with this ability in G. bethesdensis.
This is the first published genome sequence for an organism isolated directly from a patient with CGD. Combined with the fact that this is the first member of the Acetobacteraceae to be an invasive human pathogen, we hope that further study of Granulibacter genomics will shed light not only on what makes an acetic acid bacterium pathogenic but on which characteristics are important for pathogenicity in CGD. Understanding CGD-specific virulence in this novel member of the Acetobacteraceae family may lead us to the critical minimum genetic requirements for bacterial pathogenesis in CGD.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIAID.
We thank David Welch for providing us with the fourth patient isolate, NIH4.1. We also thank Kol Zarember for critically reviewing the manuscript.
Footnotes
Published ahead of print on 7 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Beare, P. A., J. E. Samuel, D. Howe, K. Virtaneva, S. F. Porcella, and R. A. Heinzen. 2006. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J. Bacteriol. 188:2309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. C., B. H. Segal, S. M. Holland, G. F. Miller, and K. J. Kwon-Chung. 1998. Virulence of catalase-deficient Aspergillus nidulans in p47(phox)−/− mice. Implications for fungal pathogenicity and host defense in chronic granulomatous disease. J. Clin. Investig. 101:1843-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deppenmeier, U., M. Hoffmeister, and C. Prust. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60:233-242. [DOI] [PubMed] [Google Scholar]
- 8.Frank, A. C., and J. R. Lobry. 2000. Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics (Oxford, England) 16:560-561. [DOI] [PubMed] [Google Scholar]
- 9.Gibson, T. J., A. Rosenthal, and R. H. Waterston. 1987. Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene 53:283-286. [DOI] [PubMed] [Google Scholar]
- 10.Graham, M. R., K. Virtaneva, S. F. Porcella, W. T. Barry, B. B. Gowen, C. R. Johnson, F. A. Wright, and J. M. Musser. 2005. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 166:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg, D. E., L. Ding, A. M. Zelazny, F. Stock, A. Wong, V. L. Anderson, G. Miller, D. E. Kleiner, A. R. Tenorio, L. Brinster, D. W. Dorward, P. R. Murray, and S. M. Holland. 2006. A novel bacterium associated with lymphadenitis in a patient with chronic granulomatous disease. PLoS Pathog. 2:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, D. E., S. F. Porcella, F. Stock, A. Wong, P. S. Conville, P. R. Murray, S. M. Holland, and A. M. Zelazny. 2006. Granulibacter bethesdensis gen. nov., sp. nov., a distinctive pathogenic acetic acid bacterium in the family Acetobacteraceae. Int. J. Syst. Evol. Microbiol. 56:2609-2616. [DOI] [PubMed] [Google Scholar]
- 13.Guide, S. V., F. Stock, V. J. Gill, V. L. Anderson, H. L. Malech, J. I. Gallin, and S. M. Holland. 2003. Reinfection, rather than persistent infection, in patients with chronic granulomatous disease. J. Infect. Dis. 187:845-853. [DOI] [PubMed] [Google Scholar]
- 14.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 15.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 16.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klebanoff, S. J., and L. R. White. 1969. Iodination defect in the leukocytes of a patient with chronic granulomatous disease of childhood. N. Engl. J. Med. 280:460-466. [DOI] [PubMed] [Google Scholar]
- 19.Laukel, M., M. Rossignol, G. Borderies, U. Volker, and J. A. Vorholt. 2004. Comparison of the proteome of Methylobacterium extorquens AM1 grown under methylotrophic and nonmethylotrophic conditions. Proteomics 4:1247-1264. [DOI] [PubMed] [Google Scholar]
- 20.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 22.Macher, A. M., T. B. Casale, J. I. Gallin, H. Boltansky, and A. S. Fauci. 1983. Chromobacterium violaceum infectious and chronic granulomatous disease. Ann. Intern. Med. 98:259. [DOI] [PubMed] [Google Scholar]
- 23.Messina, C. G., E. P. Reeves, J. Roes, and A. W. Segal. 2002. Catalase negative Staphylococcus aureus retain virulence in mouse model of chronic granulomatous disease. FEBS Lett. 518:107-110. [DOI] [PubMed] [Google Scholar]
- 24.Miller, S. I., R. K. Ernst, and M. W. Bader. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3:36-46. [DOI] [PubMed] [Google Scholar]
- 25.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prust, C., M. Hoffmeister, H. Liesegang, A. Wiezer, W. F. Fricke, A. Ehrenreich, G. Gottschalk, and U. Deppenmeier. 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat. Biotechnol. 23:195-200. [DOI] [PubMed] [Google Scholar]
- 27.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183:4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radosz-Komoniewska, H., T. Bek, J. Jozwiak, and G. Martirosian. 2005. Pathogenicity of Helicobacter pylori infection. Clin. Microbiol. Infect. 11:602-610. [DOI] [PubMed] [Google Scholar]
- 29.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross, J. P., S. M. Holland, V. J. Gill, E. S. DeCarlo, and J. I. Gallin. 1995. Severe Burkholderia (Pseudomonas) gladioli infection in chronic granulomatous disease: report of two successfully treated cases. Clin. Infect. Dis. 21:1291-1293. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt-Eisenlohr, H., M. Rittig, S. Preithner, and C. Baron. 2001. Biomonitoring of pJP4-carrying Pseudomonas chlororaphis with Trb protein-specific antisera. Environ. Microbiol. 3:720-730. [DOI] [PubMed] [Google Scholar]
- 32.Seearunruangchai, A., S. Tanasupawat, S. Keeratipibul, C. Thawai, T. Itoh, and Y. Yamada. 2004. Identification of acetic acid bacteria isolated from fruits collected in Thailand. J. Gen. Appl. Microbiol. 50:47-53. [DOI] [PubMed] [Google Scholar]
- 33.Segal, B. H., T. L. Leto, J. I. Gallin, H. L. Malech, and S. M. Holland. 2000. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 79:170-200. [DOI] [PubMed] [Google Scholar]
- 34.Seo, J. S., H. Chong, H. S. Park, K. O. Yoon, C. Jung, J. J. Kim, J. H. Hong, H. Kim, J. H. Kim, J. I. Kil, C. J. Park, H. M. Oh, J. S. Lee, S. J. Jin, H. W. Um, H. J. Lee, S. J. Oh, J. Y. Kim, H. L. Kang, S. Y. Lee, K. J. Lee, and H. S. Kang. 2005. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 23:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokollek, S. J., C. Hertel, and W. P. Hammes. 1998. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int. J. Syst. Bacteriol. 48:935-940. [DOI] [PubMed] [Google Scholar]
- 37.Speert, D. P., M. Bond, R. C. Woodman, and J. T. Curnutte. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524-1531. [DOI] [PubMed] [Google Scholar]
- 38.Van Spanning, R. J., C. W. Wansell, T. De Boer, M. J. Hazelaar, H. Anazawa, N. Harms, L. F. Oltmann, and A. H. Stouthamer. 1991. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J. Bacteriol. 173:6948-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker, G., R. Hertle, and V. Braun. 2004. Activation of Serratia marcescens hemolysin through a conformational change. Infect. Immun. 72:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wenger, J. D., D. G. Hollis, R. E. Weaver, C. N. Baker, G. R. Brown, D. J. Brenner, and C. V. Broome. 1989. Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognized human pathogen. Ann. Intern. Med. 110:888-892. [DOI] [PubMed] [Google Scholar]
- 42.Yukphan, P., T. Malimas, W. Potacharoen, S. Tanasupawat, M. Tanticharoen, and Y. Yamada. 2005. Neoasaia chiangmaiensis gen. nov., sp. nov., a novel osmotolerant acetic acid bacterium in the alpha-Proteobacteria. J. Gen. Appl. Microbiol. 51:301-311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.