Abstract
Randomly moving but self-propelled agents, such as Escherichia coli bacteria, are expected to fill a volume homogeneously. However, we show that when a population of bacteria is exposed to a microfabricated wall of funnel-shaped openings, the random motion of bacteria through the openings is rectified by tracking (trapping) of the swimming bacteria along the funnel wall. This leads to a buildup of the concentration of swimming cells on the narrow opening side of the funnel wall but no concentration of nonswimming cells. Similarly, we show that a series of such funnel walls functions as a multistage pump and can increase the concentration of motile bacteria exponentially with the number of walls. The funnel wall can be arranged along arbitrary shapes and cause the bacteria to form well-defined patterns. The funnel effect may also have implications on the transport and distribution of motile microorganisms in irregular confined environments, such as porous media, wet soil, or biological tissue, or act as a selection pressure in evolution experiments.
Motility enhances the chances of survival in a changing environment, and thus it is an important part of the competition strategies of many different organisms. For this reason, research on bacterial motility has been very active over the past 3 decades (see reference 3 for a review). Both the basic physical aspects (stochasticity and hydrodynamics) and the coupling to biological and biochemical processes (e.g., chemotaxis) have been studied extensively. On the other hand, very little has been done to put the gathered knowledge to use and to gain control over the motion of microorganisms. In this paper, we demonstrate that properly shaped microstructures can interfere with swimming bacteria and guide, concentrate, and arrange populations into arbitrary patterns.
Escherichia coli bacteria swim by means of rotating helical filaments, i.e., the flagella. When rotating counterclockwise, the flagella assemble into a bundle and the cell runs straight forward. Periodically, the flagella switch to a clockwise rotation, which induces the disassembly of the bundle and reorients the cell. The alternation of runs and tumbles results in a random walk, with a given step size being equal to the run length. As a consequence, in the absence of chemical gradients, E. coli cells are expected to distribute homogeneously in a volume. However, when they are geometrically constrained, the interaction of bacteria with boundaries has some interesting properties. Near flat surfaces, bacteria become hydrodynamically trapped and keep swimming along the surface, although (as a consequence of the rotation of the cell body) they do so in circles instead of straight runs (4, 6, 11, 15). This preferential turning offers an interesting way to manipulate the bacteria (5), but we believe that it does not play an important role in our experiments. Instead, we simply rely on the behavior of bacteria swimming along walls. The mechanism of cell concentration we demonstrate in this paper is shown schematically in Fig. 1A. Consider a wall and a swimmer running into the wall. When a swimmer driven by a propulsive force hits the wall, it will be trapped to follow the wall by the combination of the transverse force, which forces it against the wall, and the parallel component, which slides it along the wall. Traces 1 and 2 show that the odds are roughly 1:1 that the bacteria will be led through the funnel hole or away from it. On the narrow opening side, however, the wall diverts the swimmers away from the opening in either case (traces 3 and 4). Thus, the chance of getting through the funnel wall depends on the side of the wall approached. This is expected to lead to a density difference between the two sides with time. While patterned surfaces of molecular motors can rectify the motion of microtubules (7, 14) and geometry-induced asymmetric diffusion has been observed in a mixture of suitably sized particles (12), that technique requires a specialized active surface (13). The funnel mechanism depicted here is due only to mechanical and hydrodynamic interactions of actively moving (gliding or swimming) objects with passive (geometrical) restrictions. Thus, we expect it to have more general implications.
FIG. 1.
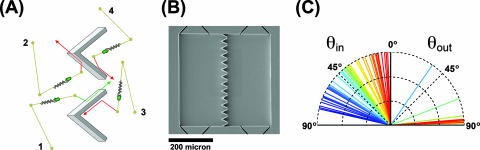
Microstructures with funnel walls. (A) Schematic drawing of the interaction of bacteria with the funnel opening. Bacteria on the left side may (trace 1) or may not (trace 2) get through the gap, depending on the angle of attack. On the right, all bacteria colliding with the wall are diverted away from the gap (traces 3 and 4). (B) Scanning electron micrograph of the device. (C) Distribution of incoming and outgoing angles for bacteria colliding with a wall. Data were taken for 70 events.
MATERIALS AND METHODS
We used microlithography and reactive ion etching to create microfluidic enclosures of 400 μm by 400 μm on a silicon wafer, which were divided into two 200-μm by 200-μm sections by an array of 13 funnel-like structures. The sides of the funnels were 27 μm long and formed a 60° angle. Gaps (3.8 μm wide) were situated at the apexes of the funnels. The enclosures were etched to a depth of 20 μm, creating 200-μm by 200-μm by 20-μm-deep volumes in which the bacteria could swim. The chips were sealed (by oxygen plasma treatment) to microscope slides with a thin (20 μm) film of cured polydimethylsiloxane (GE Silicones) on them. The gas-permeable polydimethylsiloxane allowed enough oxygen supply to enter the volume such that high densities of bacteria could be maintained (8).
We used E. coli strains RP 437/pGFPμ2, RP 437 cheAW/pGFPμ2, and DH5α/pRFP in the experiments {F− thr-1(Am) leuB6 his-4 metF159(Am) eda-50 rpsL136 [thi-1 ara-14 lacYI mtl-1 xyl-5 tonA31 tsx-78]}. Bacteria were grown in LB broth (with 50 μg/ml ampicillin) at 24°C to an optical density at 600 nm of 0.6. We then filled the chips with the culture. We used a Nikon Eclipse 90i microscope equipped with a mercury lamp (Nikon Inc.) and a Retiga 1300 charge-coupled device camera (QImaging Corp.) to observe the fluorescing bacteria in the chip. The images were captured at a low magnification (×4) and a 50-μm depth of field so that all bacteria in the 20-μm-deep device were in focus. Analysis of the bacterial populations was done using Matlab (Mathworks Inc.).
RESULTS AND DISCUSSION
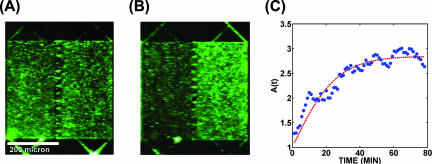
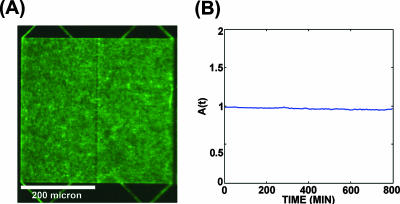
Our swimmers were green fluorescent protein (GFP)-expressing motile (E. coli) bacteria (strain RP 437/pGFPμ2). They were initially uniformly spread in both compartments filled with LB medium. Individual bacteria were tracked as they approached and left the internal walls of the chamber, far removed from the funnels. Figure 1C shows that for the 70 tracks examined, the impinging distribution (angles θout) was dramatically different from the distribution of the angles of incidence (θin). The latter was effectively a uniform random distribution over the 0°-to-80° range (measured with respect to the surface normal; we discarded all tracks with θin values of >80° for reasons of ambiguity). The outgoing angles were strongly confined to θout values of >80°. This indicates that the bacteria practically follow walls and lose information about their initial angle of attack. They keep this direction during an entire straight run, even if the wall ends. Thus, near the walls, the motion of bacteria is not a random walk but instead correlates with the constraining geometry. We indeed observed a concentration of swimming bacteria, as shown in Fig. 2, supporting the mechanism depicted in Fig. 1A. After a uniform initial distribution (Fig. 2A), the E. coli cells became increasingly concentrated with time on the restricted exit side of the funnel array (Fig. 2B). In about an hour, there were three times more cells on the right side than on the left. As a control, we filled the chip with an aqueous solution of 100-nm-diameter fluorescent polystyrene beads, which remained uniformly distributed during a 24-h period, and thus this population imbalance occurs only if the objects actively swim, as opposed to spreading due to diffusion (data not shown). Since bacteria communicate with each other (1) and (moreover) move towards one another (9, 10), it is possible that such quorum-chemotaxis processes could strongly influence the results shown in Fig. 2. We did control experiments to show that in this case the concentration was due to swimming motility and was not a result of bacterial chemotaxis (data not shown). A motile strain with the chemosensing network knocked out (RP 437 cheAW/pGFPu2) showed the same concentration increase with time, thus showing that the process is not due to chemotaxis. A flat wall with evenly spaced openings but no funnels showed no development of asymmetry in cell density, demonstrating the necessity for broken symmetry of the funnel wall (Fig. 3).
FIG. 2.
Distribution of bacteria in a structure with a funnel wall. (A) Uniform distribution after injection. (B) Steady-state distribution after 80 min. (C) Ratios of densities in the left and right compartments versus time. The blue circles are experimental data, and the dashed red line is a fit of equation A2 from the Appendix.
FIG. 3.
Distribution of bacteria in a structure with a flat wall. (A) Steady-state distribution after 80 min. (B) A(t) versus time.
We used the average fluorescence intensity in the two compartments as a measure of the cell density. Figure 2C shows how the density ratio [A(t) = ρR/ρL] changes with time (with ρR and ρL being the densities on the right and left, respectively).
A simple model (see Appendix) with two differential equations (equations A1) describing the changes in the density of cells due to growth and transfer between the compartments can be used to characterize the kinetics of the system. The two parameters are the fractions of the populations on the two sides that cross the funnel wall in unit time (cLR for crossing left to right and cRL for crossing right to left). The solution of the differential equations (equation A2) fits well with the experimental data, as shown in Fig. 2C. In equilibrium, equal numbers of bacteria cross in either direction, and thus we get the equation  = A(t = ∞) = ρR/ρL = cLR/cRL. From the fit in Fig. 2C, we get an  value of 3 for our experiment. Since the compartments in our chip are identical (same geometry and dimensions), the same fraction of the bacteria is exposed to the wall in a given time. Thus, the equilibrium density ratio is determined by the probabilities of these bacteria getting through the opening in either direction. The probabilities are determined by the geometry of the funnel.
As a first approximation, the only way to cross from right to left is to aim right for the opening from any direction. This gives a chance of 13 × 3.8 μm/400 μm = 0.13, which is the total width of gaps divided by the length of the wall. To get from left to right, a swimmer might just move straight through or hit the wall under a suitable angle so that it is diverted towards the opening. Considering the 60° angle between the wall segments, it turns out that only bacteria approaching in a 120° range get reflected towards the gap, so the probability on this side is 180°/120° = 0.66. As a result, we get an  value of 0.66/0.13 = 5, which is higher than what we observed. One reason for the lower measured ratio might be that the fabrication process makes the edges of the obstacles rounded. This can give rise to a funnel effect on the restricted side, increasing the effective gap size to a value 50% greater than the original size.
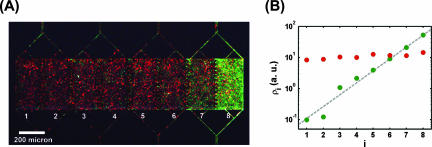
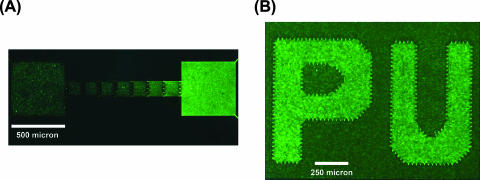
There are several interesting applications of this idea of swimmer self-concentration. For example, one can stack the funnel walls serially. Each funnel wall gives rise to a fractional increase in concentration (Â), and n funnel walls will yield a total concentration increase of Ân. The series thus acts to exponentially funnel motile agents to one side, and this staged funneling can be used to evacuate with a high yield virtually all motile bacteria in a volume if Ân is large enough. To show this multistage pumping effect, we used a chip with eight 400-μm-high, 200-μm-wide, and 20-μm-deep compartments and seven funnel walls in between. We introduced motile GFP-expressing bacteria (green) and nonmotile red fluorescent protein (RFP)-expressing bacteria (DH5α/pRFP) evenly into all compartments and monitored the cell density versus time with two color channels. After about 80 min, the motile bacteria showed an exponential increase in density throughout the chip from left to right, while the nonmotile bacteria showed no change in differential density (Fig. 4). During a time of 80 min, we would expect a nonmotile bacterium to diffuse a mean distance, <x2>1/2, of ∼2Dt1/2 (16), where D is the diffusion coefficient of an E. coli cell, i.e., kBT/6πηR, where kBT is the thermal energy, η is the viscosity of water, and R is the average value of the radius of E. coli, roughly 0.5 μm. We found <x2>1/2 to be ∼200 μm in 80 min, a distance similar to the combined 400-μm length of both chambers, so the nonmotile bacteria should sample all of the volume and would concentrate if there were an effect over that we discussed here. Thus, the effect is probably due to motility, and the device can be used to greatly concentrate and separate motile bacteria from an initially uniform mixture of motile and nonmotile cells.
FIG. 4.
Serial wall chip with eight compartments inoculated with motile (green) bacteria and nonmotile (red) bacteria. (A) After 80 min, motile bacteria were concentrated on the far right and nonmotile ones remained homogeneously dispersed. (B) Densities of motile (green circles) and nonmotile (red circles) bacteria in the compartments (i). The dashed gray line represents an exponential fit to the densities of the motile cells.
When such a structure with serial arrays is placed in a channel connecting two reservoirs, the wall series acts as an effective pump that empties out the bacteria from one tank and concentrates them into the other, relying purely on cell motility (Fig. 5A). In another application, we created closed corrals formed of funnel walls. As bacteria congregated on one side of the funnels, they assembled into patterns defined by the corrals. An example is shown in Fig. 5B, where cells are forming letters. One could change the geometry of the funnels to tune the densities of bacteria in the patterns, and the use of multiple walls for the same purpose is also possible.
FIG. 5.
Practical applications of structures with a funnel wall. (A) A series of funnel arrays can function as an effective pump to remove motile organisms from one reservoir and concentrate them in another one. (B) Spontaneous aggregation of motile bacteria inside a corral formed in the shape of letters.
Our experimental results show that manipulation of populations of motile microorganisms is possible by using asymmetric obstacles, in our case funnels. It is known that motility has a great impact on the transport of bacteria through wet soil or fractured bedrock (2). In such irregular porous media, asymmetric boundaries can exist and can cause the organisms to concentrate in or escape from certain cavities due to the funnel effect. This can have important implications for the microecology of such environments.
The funnel mechanism of concentration relies solely on motility and the interaction with boundaries. This suggests a wide variety of possible uses of similar designs. By rectifying their motion in certain directions, microorganisms can be separated, concentrated from suspensions, and guided into specific chambers. Microstructures can be engineered so that bacteria are arranged into well-defined patterns in them. No flow or moving parts are necessary in any of these devices, since any change in cell distribution is spontaneous.
Once the patterns are formed, one can study, for example, how they change in response to various external effects (e.g., changes in nutrient distribution, local delivery of antibiotics, temperature changes, etc.). Such engineered patterns can be useful for studying different aspects of cell-to-cell communication. Perhaps the most intriguing idea is to use such structures in evolution experiments (8) to imply a selective pressure on the organisms and test if they can develop a strategy to counteract the funnel walls.
Acknowledgments
We thank Peter Wolanin for supplying us with the E. coli strain RP 437 cheAW/pGFPu2 and Cees Dekker for helpful comments.
This work was supported by DARPA (W911NF-05-1-0392), the AFOSR, NIH (HG01506), NSF Nanobiology Technology Center (BSCECS9876771). It was also performed in part at the Cornell Nano-Scale Science and Technology Facility (CNF), which is supported by the National Science Foundation under grant ECS-9731293, by its users, by Cornell University, and by Industrial Affiliates.
APPENDIX
The following equations describe the changes in the density of bacteria in the two compartments:
 |
(A1) |
ρL and ρR are the densities of bacteria on the left and right sides, respectively. cLR is the fraction of the population on the left going to the right in unit time, and cRL gives the fraction that crosses in the opposite direction. The growth rate is denoted by r. After solving these equations with uniform distribution as the initial condition, we can write the following formula for the ratio A(t) = ρR/ρL:
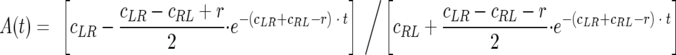 |
(A2) |
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 2.Becker, M. W., D. W. Metge, S. A. Collins, A. M. Shapiro, and R. W. Harvey. 2003. Bacterial transport experiments in fractured crystalline bedrock. Ground Water 41:682-689. [DOI] [PubMed] [Google Scholar]
- 3.Berg, H. C. 1993. Random walks in biology. Princeton University Press, Princeton, NJ.
- 4.Berg, H. C., and L. Turner. 1990. Chemotaxis of bacteria in glass-capillary arrays. Escherichia coli, motility, microchannel plate and light scattering. Biophys. J. 58:919-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiLuzio, W. R., L. Turner, M. Mayer, P. Garstecki, D. B. Weibel, H. C. Berg, and G. M. Whitesides. 2005. Escherichia coli swim on the right-hand side. Nature 435:1271-1274. [DOI] [PubMed] [Google Scholar]
- 6.Frymier, P. D., R. M. Ford, H. C. Berg, and P. T. Cummings. 1995. 3-Dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA 92:6195-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiratsuka, Y., T. Tada, K. Oiwa, T. Kanayama, and T. Q. P. Uyeda. 2001. Controlling the direction of kinesin-driven microtubule movements along microlithographic tracks. Biophys. J. 81:1555-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keymer, J. E., P. Galajda, C. Muldoon, S. Park, and R. H. Austin. 2006. Bacterial metapopulations in nanofabricated landscapes. Proc. Natl. Acad. Sci. USA 103:17290-17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park, S., P. M. Wolanin, E. A. Yuzbashyan, H. Lin, N. C. Darnton, J. B. Stock, P. Silberzan, and R. H. Austin. 2003. Influence of topology on bacterial social interaction. Proc. Natl. Acad. Sci. USA 100:13910-13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park, S., P. M. Wolanin, E. A. Yuzbashyan, P. Silberzan, J. B. Stock, and R. H. Austin. 2003. Motion to form a quorum. Science 301:188. [DOI] [PubMed] [Google Scholar]
- 11.Ramia, M., D. L. Tullock, and N. Phan-Thien. 1993. The role of hydrodynamic interaction in the locomotion of microorganisms. Biophys. J. 65:755-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw, R. S., N. Packard, M. Schröther, and H. L. Swinney. 2007. Geometry-induced asymmetric diffusion. Proc. Natl. Acad. Sci. USA 104:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabó, B., J. Szöllösi, B. Gönci, Z. Jurányi, D. Selmeczi, and T. Vicsek. 2006. Phase transition in the collective migration of tissue cells: experiment and model. Phys. Rev. E 74:061908. [DOI] [PubMed] [Google Scholar]
- 14.van den Heuvel, M. G. L., C. T. Butcher, R. M. M. Smeets, S. Diez, and C. Dekker. 2005. High rectifying efficiencies of microtubule motility on kinesin-coated gold nanostructures. Nano Lett. 5:1117-1122. [DOI] [PubMed] [Google Scholar]
- 15.Vigeant, M. A. S., and R. M. Ford. 1997. Interactions between motile Escherichia coli and glass in media with various ionic strengths, as observed with a three-dimensional-tracking microscope. Appl. Environ. Microbiol. 63:3474-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Y. M., R. H. Austin, and E. C. Cox. 2006. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 97:048302. [DOI] [PubMed] [Google Scholar]