Abstract
The Bacillus subtilis LiaRS two-component system (TCS) responds to perturbations of the cell envelope induced by lipid II-interacting antibiotics, such as vancomycin, ramoplanin, nisin, and bacitracin. Here, we characterize Tn7-generated mutations that induce the liaRS TCS. In addition to insertions in liaF, a known negative regulator of the LiaRS TCS, we identified two disruptions in the last two genes of the yydFGHIJ operon. This operon is predicted to encode a 49-amino-acid peptide (YydF), a modification enzyme (YydG), a membrane-embedded protease (YydH), and an ATP-binding cassette (ABC) transporter (YydIJ). Genome sequence comparisons suggest that the yydFGHIJ operon may have been acquired by horizontal transfer. Inactivation of the YydIJ transporter resulted in increased expression from the LiaR-dependent PliaI promoter only in the presence of the yydFGH genes. Cells harboring the complete yydFGHIJ operon induced LiaR activity in cocultured cells lacking either this transporter or the complete operon. These results suggest that this operon is involved in the synthesis and export of a modified peptide (YydF*) that elicits cell envelope stress sensed by the LiaRS TCS.
The Bacillus subtilis cell envelope consists of a thick peptidoglycan cell wall and the cell membrane. Conditions interfering with cell envelope function activate specific stress responses coordinated by extracytoplasmic function (ECF) σ factors and various two-component regulatory systems (TCS). The activity of ECF σ factors is often inhibited by a transmembrane anti-σ which is inactivated in response to extracytoplasmic stress (17). Similarly, TCS involve a membrane-located sensor kinase protein and a cytoplasmic response regulator (27). Thus, both systems couple changes in gene expression to conditions that affect the integrity or function of the cell envelope.
The soil is a competitive environment, and many soil microorganisms produce antibiotics to inhibit the growth of neighboring cells. Often, these antibiotics target the cell wall or cell membrane and, in response, bacteria have evolved mechanisms to monitor and counteract these attacks. Previously, we demonstrated that the σW regulon plays a major role in defense against antimicrobial peptides produced by B. subtilis and other Bacillus spp. (4). The LiaRS TCS is also strongly induced by cell-wall active antibiotics such as vancomycin, ramoplanin, bacitracin, and nisin (6, 28, 29) as well as by the human antimicrobial peptide LL37 (31). LiaRS is weakly induced by the antibiotics fosfomycin and tunicamycin (29), by detergents, ethanol, phenol and organic solvents (29), and under conditions of alkaline shock (37) or secretion stress (19). Weak induction is also observed during normal growth conditions in rich medium at the onset of stationary phase (21).
The LiaRS TCS activates expression of the liaIHGFSR operon and at least one other operon, yhcYZ-yhdA, which encodes a second TCS. Induction of the PliaI promoter results in a high level of expression of the LiaI and LiaH proteins. LiaI is a small (126-amino-acid [aa]) hydrophobic protein, predicted to be localized to the cell membrane. LiaH belongs to the PspA family of phage shock proteins and has been shown to act as a negative regulator of the yhcYZ-yhdA operon (20). While the expression of the liaIHGFSR operon in response to cell wall-active compounds is well documented, the role of LiaIH has remained elusive: deletion of this operon does not result in increased sensitivity to any of the compounds tested. In Escherichia coli and Yersinia entercolitica PspA binds to PspF, a transcriptional activator, under noninducing conditions and thereby prevents activation of the psp genes (8-10). Under inducing conditions (e.g., dissipation of the proton motive force), PspA interacts with the cytoplasmic membrane proteins PspB and PspC, freeing PspF to activate transcription of the psp genes. Under these conditions PspA is anchored to the inner surface of the cytoplasmic membrane, where it is proposed to contribute to maintenance of the proton motive force and membrane integrity (7). Homologs of the PspB, PspC, and PspF proteins have not been identified in B. subtilis. However, by analogy with the Psp system, Jordan and coworkers have proposed that LiaH may have a similar dual function and under stress conditions might act together with the membrane protein LiaI to maintain cell envelope integrity (20).
Homologs of the LiaRS proteins are found in many Bacillus species as well as in other firmicutes, such as Listeria monocytogenes (Lmo1021/Lmo1022) and various Staphylococcus species, including S. aureus (VraRS) (26). The VraRS system from S. aureus is also induced by cell wall-active antibiotics. This TCS is known to control genes involved in peptidoglycan biosynthesis, and deletion results in sensitivity toward β-lactam and glycopeptide antibiotics (25).
In this study, transposon mutagenesis was employed to search for genes which affect activity of the LiaRS TCS. In addition to liaF, a known negative regulator of the LiaRS TCS (20), we isolated several independent insertions in the yydIJ genes, encoding an ABC transporter. Further genetic analyses suggest that the YydIJ ABC transporter exports a modified YydF peptide (YydF*) and that defects in peptide export lead to cell envelope stress that is sensed by the LiaRS TCS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and primers used in this study are listed in Tables 1 and 2. Bacterial cultures were grown in liquid or solid Luria-Bertani (LB) (33), Difco sporulation medium (DSM) (15), or modified competence (MC) medium (24). Solid media contained 1.5% Bacto agar and, where indicated, 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Ampicillin (Amp) at a concentration of 100 μg/ml was used for selection of E. coli strains. Antibiotics for selection of various B. subtilis strains were used at the following concentrations: spectinomycin (Spec), 100 μg/ml; tetracycline (Tet), 20 μg/ml; chloramphenicol (Cm), 10 μg/ml; kanamycin (Kan), 15 μg/ml; neomycin (Neo), 8 μg/ml. For macrolide-lincomycin-streptogramin B (MLS) resistance, both lincomycin (25 μg/ml) and erythromycin (1 μg/ml) were added. Transformation of B. subtilis strains was carried out as previously described (15).
TABLE 1.
Strains used in this study
| B. subtilis strain | Genotype, description, and/or sequence | Reference or source |
|---|---|---|
| CU0165 | W168 trpC2 att SPβ | Laboratory strain |
| ZB703A | W168 SPβ 2Δ2::Tn917::pSK10Δ6 | 40 |
| HB6265 | CU1065 yydFGHIJ::spec | LFH-PCR → CU1065 |
| HB6266 | CU1065 yydHIJ::spec | LFH-PCR → CU1065 |
| HB6267 | CU1065 yydIJ::spec | LFH-PCR → CU1065 |
| HB0950 | CU1065 SPβ::PliaI-74-cat-lacZ | 29 |
| HB6296 | CU1065 yydIJ::spec SPβ::PliaI-74-cat-lacZ | Transduction, HB0950 phage → HB6267 |
| HB6297 | CU1065 yydHIJ::spec SPβ::PliaI-74-cat-lacZ | Transduction, HB0950 phage → HB6266 |
| HB6298 | CU1065 yydFGHIJ::spec SPβ::PliaI-74-cat-lacZ | Transduction, HB0950 phage → HB6265 |
| HB6301 | CU1065 SPβ | Transduction, ZB703A phage → CU1065 |
| HB6302 | CU1065 yydIJ::spec SPβ | Transduction, ZB703A phage → HB6267 |
| HB6303 | CU1065 yydHIJ::spec SPβ | Transduction, ZB703A phage → HB6266 |
| HB6304 | CU1065 yydFGHIJ::spec SPβ | Transduction, ZB703A phage → HB6265 |
| HB6274 | CU1065 ΔyydF | pBGB6 → CU1065 |
| HB6275 | CU1065 ΔyydG | pBGB7 → CU1065 |
| HB6305 | CU1065 ΔyydF SPβ | Transduction, ZB703A phage → HB6274 |
| HB6306 | CU1065 ΔyydG SPβ | Transduction, ZB703A phage → HB6275 |
| HB6307 | CU1065 PxylA-yydFGHIJ | pBGB8 → CU1065 |
| HB6192 | CU1065 amyE::PsigW-lacZ (Cm) | 4 |
| HB6299 | CU1065 amyE::PsigX-lacZ (Cm) | Lab collection |
| HB6300 | CU1065 amyE::PsigM-lacZ (Cm) | Lab collection |
| HB6314 | CU1065 amyE::PsigW-lacZ (Cm) yydIJ::spec | HB6192 Chr DNA → HB6267 |
| HB6315 | CU1065 amyE::PsigX-lacZ(Cm) yydIJ::spec | HB6299 Chr DNA → HB6267 |
| HB6316 | CU1065 amyE::PsigM-lacZ(Cm) yydIJ::spec | HB6300 Chr DNA → HB6267 |
| HB6317 | CU1065 amyE::PsigW-lacZ (Cm) yydFGHIJ::spec | HB6192 Chr DNA → HB6265 |
| HB6318 | CU1065 amyE::PsigX-lacZ(Cm) yydFGHIJ::spec | HB6299 Chr DNA → HB6265 |
| HB6319 | CU1065 amyE::PsigM-lacZ(Cm) yydFGHIJ::spec | HB6300 Chr DNA → HB6265 |
| HB6276 | CU1065 amyE::PyydF-lacZ (Cm) | pBGB9 (ScaI) → CU1065 |
| HB6308 | CU1065 amyE::PyydH-lacZ (Cm) | pBGB10 (ScaI) → CU1065 |
| HB6309 | CU1065 amyE::PyydI-lacZ (Cm) | pBGB11 (ScaI) → CU1065 |
| HB6278 | CU1065 amyE::PyydF-lacZ (Cm) rok::kan | LFH-PCR → HB6276 |
| HB6310 | CU1065 amyE::PyydH-lacZ (Cm) rok::kan | LFH-PCR → HB6308 |
| HB6311 | CU1065 amyE::PyydI-lacZ (Cm) rok::kan | LFH-PCR → HB6309 |
| HB6320 | CU1065 amyE::PyydF-lacZ (Cm) abrB::tet | SWV119 Chr DNA → HB6276 |
| HB6312 | CU1065 amyE::PyydH-lacZ (Cm) abrB::tet | SWV119 Chr DNA → HB6308 |
| HB6313 | CU1065 amyE::PyydI-lacZ (Cm) abrB::tet | SWV119 Chr DNA → HB6309 |
| HB6277 | CU1065 rok::kan | LFH-PCR → CU1065 |
| SWV119 | abrB::tet | Mark Strauch, Univ. Maryland Dental School, Baltimore |
TABLE 2.
Primers and probes used in this study
| Primer or probe | Description and sequencea |
|---|---|
| Primers for arbitrary PCR to map Tn7 insertions | |
| 2509 | BsHarb1, GGCCACGCGTCGACTAGTCA(N)(N)(N)(N)(N)(N)(N)(N)(N)(N)GATAT |
| 2501 | Tn7LHarb3, CCAGATAAGTGAAATCTAGTTCC |
| 2508 | BsHarb3, GGCCACGCGTCGACTAGTCA |
| 2502 | Tn7LHarb4, CGTATTAGCTTACGACGCTACACCC |
| Primers for LFH-PCR | |
| 2701 | yydF-up-fwd, CAACACGTGCTGGAATGCCT |
| 2702 | yydF-up-rev (spec), CGTTACGTTATTAGCGAGCCAGTCTTCATATTATCCCTCCTCC |
| 2703 | yydH-up-fwd, CACCAACTTCAACAACCAGG |
| 2704 | yydH-up-rev (spec), CGTTACGTTATTAGCGAGCCAGTCATAAATACGTTGTTTTGCAC |
| 2757 | yydI-up-fwd, AGTGCTTGTGCAAAACAACG |
| 2758 | yydI-up-rev (spec), CGTTACGTTATTAGCGAGCCAGTCCGCTATATTCATATACATACTCC |
| 2705 | yydJ-do-fwd (spec), CAATAAACCCTTGCCCTCGCTACGCTAGATGGATCAAAATGGG |
| 2706 | yydJ-do-rev, TCTTTCAGGTCAGAGGAAGC |
| 3125 | rok-up-for, GGACAGCTCCGTCACTTC |
| 3126 | rok-up-rev (kan), CCTATCACCTCAAATGGTTCGCTGCTAACCGCAAGCGCAAAGC |
| 3127 | rok-do-for (kan), CGAGCGCCTACGAGGAATTTGTATCGTCGAATCTGCAGAATCAGCAAACG |
| 3128 | rok-do-rev, CACTGCTTCAGGCAAAACAGC |
| Primers for the xylose-inducible yydFGHIJ operon | |
| 2872 | yydF-fwd-BamHI, CGCGGATCCGGTTTATATTAGAAAAGGAGG |
| 2873 | yydG-rev-EcoRI, AAAGAATTCATATCATGAAAGTACTCC |
| Primers for in-frame deletions with pMAD | |
| 2896 | yydF-up-for (BamHI), CGCGGATCCCAGATCACTGACAAAATGCTCG |
| 2895 | yydF-up-rev, CTCTCCTTTGTACCCCTCTAAATTATCCCTCCTCCTTTTCTAATATAAACC |
| 2894 | yydF-do-for, GGTTTATATTAGAAAAGGAGGAGGGATAATTTAGAGGGGTACAAAGGAGAG |
| 2897 | yydF-do-rev (NcoI), CATGCCATGGGCTTAATTCTAGTTTTAGCAGCGC |
| 2898 | yydG-up-for (BamHI), CGCGGATCCCAGCTTTTCAGTACAGGTTGG |
| 2900 | yydG-up-rev, GAAGTAGTTTATTTTTTCTGCAGAGTTGGTTGTTGAAGTTGGTGAACTAG |
| 2899 | yydG-do-for, CTAGTTCACCAACTTCAACAACCAACTCTGCAGAAAAAATAAACTACTTC |
| 2901 | yydG-do-rev (EcoRI), CCGGAATTCGACGAGAAAAGCATAAATGCC |
| Probes for Northern analysis | |
| 2906 | yydF-north-for, GGATAATATGAAAAAGGAAATCACTAAC |
| 2907 | yydF-north-rev, TTAATGACCACTTCCAAGAATCC |
| Primers for promoter-lacZ fusions | |
| 3066 | PyydF-for (EcoRI), CCGGAATTCGCTTAAAACAGCTTTTCAGTACAGG |
| 3067 | PyydF-rev (BamHI), CGCGGATCCCACAGTCTCATTGTTAGTGATTTCC |
| 3261 | PyydH-for (EcoRI), CCGGAATTCGCAGGAAATACCCTGATATCG |
| 3262 | PyydH-rev (BamHI), CGCGGATCCGCACAAGCACTTCATATTTTCC |
| 3263 | PyydI-for (EcoRI), GCAGTGAATTCAGTTGTACATTGG |
| 3264 | PyydI-rev (BamHI), CGCGGATCCCGGTGTCCTGAAGTAACG |
| Primers for 5′-RACE | |
| 3531 | yydF-GPS2, GGTCACAGGATGCATTACACC |
| 3526 | yydF-GPS1, GCAAATTCAGTTACTAACTCTC |
| 3530 | yydH-GPS2, GAACATGAGAGCCGTAAAGG |
| 3527 | yydH-GPS1, GTCCCAGTTCATGAAGCAC |
| 3529 | yydI-GPS2, CCTTAGAAACATCATTAGGTATATTCG |
| 3528 | yydI-GPS1, TTGATCAGAACTTTTCCATC |
Sequences complementary to the antibiotic resistance cassettes for LFH-PCR and sequences representing restriction enzyme sites are underlined.
Deletion of genes was performed by long flanking homology PCR (LFH-PCR) (28). Primers used to amplify the up and down fragments are listed in Table 2, and the primers and plasmids used for amplification of the antibiotic resistance cassettes can be found at http://www.micro.cornell.edu/cals/micro/research/labs/helmann-lab/supplements.cfm. Amplification of the fragments, as well as the joining reactions, was performed with the Expand Long Template PCR system (Roche). Five μl of the LFH-PCR product was introduced into the desired strain by transformation, and integration and deletion of the gene were confirmed by PCR.
In-frame deletions of yydF and yydG were created using the pMAD plasmid (3). Regions upstream and downstream of the gene to be deleted were amplified and joined by overlapping PCR using the Expand Long Template PCR system (Roche). The primers included restriction enzyme sites that allowed cloning of the joined fragment into pMAD, generating pBGB6 (pMAD-ΔyydF) and pBGB7 (pMAD-ΔyydG). Integration of this plasmid and generation of the clean deletion followed the published procedure (3).
Placement of the yydFGHIJ operon under PxylA control was performed using the pHTXyl plasmid (a pUC18-based vector containing the PxylA promoter and an erythromycin resistance gene [T. Msadek, unpublished]). A region including the predicted RBS site upstream of yydF, but lacking the promoter, and extending into the yydG was amplified by PCR using primers listed in Table 2 and the Expand Long Template PCR system (Roche). The resulting fragment was cloned into pHTXyl, placing the yydF gene downstream of the PxylA promoter (pBGB8). This plasmid was then transformed into CU1065, and a single integration event was selected for by MLSr and confirmed by PCR. A 2% xylose solution was used to induce expression of these genes.
The PliaI-74-cat-lacZ fusion (29) is harbored on a SPβ prophage, and introduction of this reporter into the desired strains was performed by transduction into the required CU1065-derived strains. CU1065 lacks the SPβ prophage but still contains the phage attachment site.
Quantification of β-galactosidase activity from cells grown on solid media was performed by spotting 5 μl of cells grown to mid-log phase on the desired solid medium poured into the wells of a Linbro 24-well tissue culture plate. After overnight incubation the cells were washed off the plate with Z-buffer, diluted to an optical density at 600 nm (OD600) of about 0.3, and lysed with 100 μg/ml of lysozyme at 37°C for 30 min, before determination of the β-galactosidase activity by standard methods (30).
PyydF-, PyydH-, and PyydI-lacZ fusions were created by amplification of the promoter regions (see Table 2 for primers) and cloning into the pDG1661 vector (13) containing a promoterless lacZ gene (creating plasmids pBGB9, pBGB10, and pBGB11, respectively). After sequencing, the resulting plasmids were digested with ScaI and introduced by transformation into CU1065. Integration into the amyE locus was selected by Cmr and confirmed by PCR. Expression from these promoter fusions was investigated on solid media containing X-Gal, or samples were removed throughout growth and the β-galactosidase activity was determined as previously described (30) with the following modifications: the cells were resuspended in Z-buffer and lysed with 100 μg/ml of lysozyme at 37°C for 30 min.
Tn7 mutagenesis.
Chromosomal DNA was isolated from B. subtilis and subjected to in vitro Tn7 mutagenesis using a modified Tn7 transposon carrying a Specr gene and an outward-facing xylose-inducible PxylA promoter (C. Bordi, A Hachmann, and J. D. Helmann, unpublished data). This DNA was transformed into B. subtilis, and the resulting transposants were grown in the presence of Spec (to select for transformation of the Tn7-mutagenized DNA) with and without added xylose (which results in expression of genes downstream of the transposon). Chromosomal DNA was prepared from these cultures.
This amplified library of chromosomal DNA was introduced by transformation into HB0950, a B. subtilis CU1065 strain carrying the SPβ prophage harboring a Plia74-cat-lacZ fusion known to have full bacitracin-inducible activity (29). We selected for Neor (confirming the presence of the PliaI-cat-lacZ fusion) and Specr (indicating the presence of Tn7) colonies based on increased Cmr and a blue color in the presence of X-Gal (indicating increased expression from the PliaI-cat-lacZ fusion). Colonies were selected on LB plus Spec plus Neo plates containing 1.5, 2, 2.5, or 3 μg/ml chloramphenicol and 40 μg/ml X-Gal. To confirm that the observed phenotypes were due to the presence of the transposon, chromosomal DNA was isolated from selected colonies using a GFX genomic blood DNA purification kit from Amersham (following the manufacturer's protocol for DNA purification from gram-positive bacteria) and retransformed into HB0950. Transformants were plated on LB plus Spec plus X-Gal and LB plus Cm plus X-Gal plates. If all colonies on the LB plus Spec plus X-Gal plates were blue and the numbers of transformants on the Cm and Spec plates were comparable, we considered the phenotype to be linked to the transposon disruption. From about 54,400 transformants, 26 blue Cmr colonies were isolated after 2 days of incubation at 37°C. Only 14 of the 26 upregulated phenotypes were found to be linked to the transposon disruption (see Table 3, below).
TABLE 3.
Tn7 disruptions resulting in increased expression from the PliaI-cat-lacZ promoter fusion
| Strain name(s) | Gene disrupted | Position in gene (size of gene) in bp |
|---|---|---|
| YPL3, YPL9, YPL14, YPL15 | yydI | 2 (267) |
| YPL6, YPL13 | yydJ | 257 (720) |
| YPL1, YPL25 | mutS | 398 (2,574) |
| BBL21 | mutS | 961 (2,574) |
| BBL5-3 | mutS | 1419 (2,574) |
| YPL16 | mutL | 467 (1,881) |
| YPL26, YPL18 | liaG | 226 (720) |
| BBL6 | liaF | NDa (723) |
ND, not determined.
Mapping transposon insertions.
The position of the Tn7 insertion in the chromosome was determined by arbitrary PCR. A first round of PCR was performed using the arbitrary primer (2509) and a specific primer (2501) complementary to Tn7 (Table 2). Primer 2509 consists of a 5-bp ATGCA sequence at the 3′ end followed by a random sequence of 10 nucleotides and a 5′ tail complementary to primer 2508. In order to amplify the PCR product containing the Tn7-chromosome junction, a second round of PCR using the 2508 (complementary to the 5′ tail of 2509) and 2502 (nested primer complementary to Tn7) primers with a 1:5 dilution of the product from the first-round PCR as the template was performed, and the product of this second PCR was sequenced using the 2502 primer by the Biotechnology Resource Center at Cornell University. All above PCRs were performed using 1.1× Thermo-start PCR Master Mix (ABgene) and purified with the QIAquick PCR purification kit (Qiagen).
Cocultivation experiments.
Strains to be mixed were grown to the same optical density (corresponding to mid-logarithmic growth) in liquid LB medium and mixed in equal volumes, and 5 μl of the mixture was spotted on solid LB, DSM, MC, or MM containing 80 μg/ml of X-Gal. These plates were incubated at 37°C overnight and photographed.
5′-RACE PCR and Northern analysis.
Total RNA was isolated from cells in mid-logarithmic- and stationary-phase growth using the RNeasy mini kit from Qiagen following the manufacturer's protocol for isolation of total RNA from bacteria with the following modification: the cells were lysed with 5 mg/ml of lysozyme for 10 min at 37°C. For 5′ rapid amplification of cDNA ends (RACE) PCR, the RNA was treated with DNase using Turbo DNA-free from Ambion followed by precipitation with 0.3 M sodium acetate and 70% ethanol. cDNA was synthesized using the Taqman reverse transcription reagents (Applied Biosystems) with the relevant GPS1 primers and either 2 μg of RNA isolated from log phase or 1.25 μg of RNA isolated from stationary phase. RNA was removed by treatment with 15 μl 1 M NaOH for 10 min at 70°C. This was neutralized by addition of 15 μl of 1 M HCl, and the cDNA was purified using QIAquick spin columns (Qiagen). This cDNA was dCTP tailed with terminal transferase (New England Biolabs) at 37°C for 30 min. PCR amplification of this tailed cDNA was performed with Thermo-start PCR Master Mix (ABgene) using the AAP primer complementary to the dCTP tail (3314) and the relevant GPS2 primer. A second PCR was performed to amplify the desired bands, which were then purified using the QIAquick spin columns (Qiagen) and sequenced by the Biotechnology Resource Center at Cornell University using the relevant GSP2 primer.
For Northern analysis, 8 μg and 10 μg of RNA isolated from log phase and stationary phase, respectively, were run on a 1% formaldehyde denaturing gel using NorthernMax denaturing gel buffer and running buffer from Ambion. The RNA was transferred to Zeta-Probe blotting membrane (Bio-Rad) using 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and prehybridized at 42°C for 1 h. The yydF, yydG, yydH, and yydI probes were amplified with the relevant primers (Table 2) using Thermo-start PCR Master Mix (ABgene), purified using the QIAquick spin columns (Qiagen), and labeled with the DECAprime II kit (Ambion) using [α-32P]dATP. Hybridization of the probe was performed with ULTRAhyb (Ambion) overnight at 42°C, and the membrane was washed twice with 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7; Ambion) low-stringency wash for 5 min at room temperature and twice with 0.1× SSPE high-stringency wash for 15 min at 42°C, before detection of the signal using a PhosphorImager (GE Healthcare).
RESULTS
Inactivation of yydIJ leads to increased Plia expression.
Expression from the LiaR-activated liaI promoter (PliaI) is known to be induced by the antibiotics ramoplanin, bacitracin, vancomycin, and nisin (29). However, neither the liaIHGFSR operon nor the LiaRS regulon appears to provide significant resistance to these antibiotics (29). Thus, the role of the LiaRS system has remained unclear. Here, we sought to identify transposon-induced mutations that result in upregulation of PliaI.
Mutagenesis was performed using a Tn7 transposon containing a Specr cassette and an outward-facing xylose-inducible PxylA promoter (to facilitate identification of genes whose induction leads to increased PliaI expression; see Materials and Methods). A library of Tn7-mutagenized chromosomal DNA was introduced into B. subtilis carrying the PliaI-74-cat-lacZ promoter fusion (HB0950) (29), thus allowing for both screening (β-galactosidase expression) and selection (Cmr). Fourteen transformants with elevated PliaI expression were characterized (Table 3). In all cases the increase in PliaI-74-cat-lacZ expression was not dependent on xylose, indicating that the effect was more likely due to gene disruption rather than increased expression of downstream genes.
Sequence analysis of these Tn7 insertions revealed three groups of transposition events (Table 3). The first group consists of insertions into the liaG and liaF genes. Previous experiments had shown that deletion of liaF results in constitutive expression from the PliaI promoter (20). These strains exhibit a dramatic, dark blue color after less than 1 day of incubation on X-Gal plates (data not shown). In contrast, the remaining isolates had to be incubated for at least 2 days in the presence of 80 μg/ml of X-Gal to see a faint blue color. The second group included four independent transposition events into the mutS and mutL genes (Table 3). The reason for the weak upregulation of PliaI under these conditions requires further investigation. The last group, and the subject of this study, consists of transposon insertions in the yydI and yydJ genes (Fig. 1A and Table 3). In both cases, the PxylA promoter carried on Tn7 is facing in the opposite direction to the gene, consistent with the hypothesis that PliaI upregulation is due to gene disruption. Analysis of the predicted protein products of these genes suggests that they form an ATP binding cassette (ABC) transporter (Table 4).
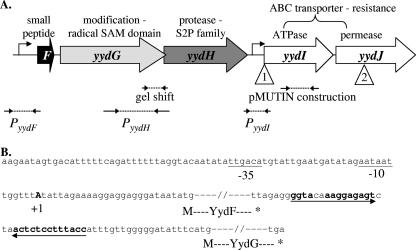
FIG. 1.
A. Arrangement of the yydFGHIJ operon. The schematic is drawn to scale, and predicted functions are shown above the genes. Large arrows represent the open reading frames, with the PyydF and PyydI promoters identified in this study marked. Open triangles show the positions of the Tn7 transposon insertions: 1, YPL3, YPL9, YPL14, and YPL15; 2, YPL6 and YPL13. Also indicated are the positions of the fragments used by Albano et al. for gel shift and pMUTIN construction (1) and the fragments used in this study to create lacZ fusions at the amyE locus. B. The yydF promoter region and yydF-yydG intergenic region. The −35 and −10 regions of a σA-type promoter are underlined, and the +1 start of transcription as determined by 5′-RACE PCR is shown in bold. The yydF and yydG open reading frames are indicated by dashes, and the inverted repeat is shown with arrows (with the complementary bases marked in bold).
TABLE 4.
Properties and functions of the proteins identified during Tn7 mutagenesis and selection for upregulation of PliaI
| Protein | Properties | Conserved domain(s)c | Closest homologd (%identity/%positive) | Function |
|---|---|---|---|---|
| YydF | 49 aa, +2.2 charge at pH 7.0, 34% hydrophobic, potential to form α-helix with 12 hydrophobic residues on the same surfacea | None | Streptococcus agalactiae strains (46-50/69) | Antimicrobial/signal/secreted peptidee |
| YydG | 319 aa, cytoplasmic protein | Predicted Fe-S oxidoreductase (COG0535), with 5′ region radical SAM superfamily (pfam 04055) | Streptococcus agalactiae COH1 (35/55) | Oxidoreductasee |
| YydH | 252 aa, 5 THMb | None | Streptococcus agalactiae COH1 (34/54) | Peptidasee (M50 peptidase familyf) |
| YydI | 209 aa, cytoplasmic protein | ABC_ATPase (cd00267) | Staphylococcus aureus subsp. aureus (61/77) | ABC transporter (ATPase subunit)e |
| YydJ | 240 aa, 6 THM2 | None | Staphylococcus aureus subsp. aureus (62/82) | ABC transporter (permease subunit)e |
| MutS | 858 aa | COG0249, MutS family | B. licheniformis (83/89) | DNA mismatch repair ATPase |
| MutL | 627 aa | COG0323, MutL | B. licheniformis (78/85) | DNA mismatch repair enzyme (predicted ATPase) |
Baesd on information in the Antimicrobial Peptide Database (http://aps.unmc.edu/AP/main.html).
THM, transmembrane helices, as determined using TMHMM (www.cbs.dtu.dk/services/TMHMM/) and TMPred (www.ch.embnet.org/software/TMPRED_form.html).
Based on the Conserved Domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Analysis with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Function predicted from sequence homology.
Based on information in MEROPS, the peptidase database (http://merops.sanger.ac.uk/).
The effect of the yydIJ deletion on PliaI expression is enhanced on solid competence medium (MC).
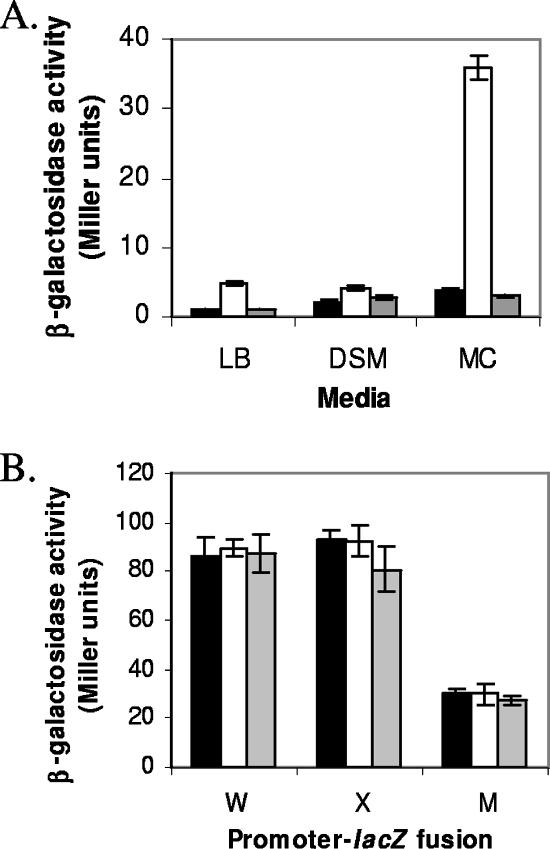
In order to confirm the data from the transposon mutagenesis, we constructed a yydIJ::spec deletion by LFH-PCR. As noticed with the transposon mutants, the induction of PliaI on LB medium in this strain was very weak. Since the yydHIJ genes were previously proposed to be controlled by Rok (1), and Rok is repressed by the regulator of competence ComK (18), we investigated whether the effects of the yydIJ mutation would be enhanced under different growth conditions, including growth on solid modified competence (MC) medium. Indeed, PliaI expression was significantly increased in the yydIJ::spec mutant on solid MC medium (close to 10-fold) as opposed to the more modest increase observed on solid LB and DSM (4.7- and 1.9-fold, respectively) (Fig. 2A). Three ECF σ factors, σW, σX, and σM, are also induced by stresses affecting the cell envelope (4-6). However, the activity of these three ECF σ factors was not detectably induced in the yydIJ mutant strain (Fig. 2B).
FIG. 2.
A. Induction of the PliaI-74-cat-lacZ fusion in WT (black bars), yydIJ::spec (white bars), and yydFGHIJ::spec (gray bars) strains on solid LB, DSM, and MC media. B. Activities of PsigW, PsigX, and PsigM-lacZ fusions in the same strains on MC medium only. After overnight incubation on solid medium, the cells were washed off and β-galactosidase activity was determined. Error bars represent the standard deviations between two independent experiments each assayed in duplicate.
Bioinformatic and genetic analyses suggest that the yydFGHIJ operon functions to synthesize and export a small peptide, YydF*.
Analysis of the yydIJ genes and their genomic context suggested that these genes might be part of a larger yydFGHIJ operon. Sequence analyses led us to hypothesize that the YydF peptide (49 aa) may be the precursor of an exported, biologically active peptide (designated YydF*) with antimicrobial or signaling properties (or both) (Table 4). YydG is a predicted radical SAM family oxidoreductase, and YydH is a predicted intramembrane protease (Table 4) that may modify and cleave the primary translation product of the yydF gene. The nature of these posttranslational modifications is not yet clear. The resulting active YydF* peptide is the presumed substrate for export by the YydIJ ABC transporter. In the absence of the YydIJ ABC transporter, we propose that YydF* elicits cell envelope stress that is sensed by the LiaRS TCS.
Genetic analyses suggest that yydF, yydG, and yydH are required for induction of PliaI in the absence of the YydIJ exporter. Induction of PliaI is lost if the entire yydFGHIJ operon is deleted (Fig. 2A) or if only yydH is additionally deleted (Table 5, column 1).
TABLE 5.
The product of the yydFGHIJ operon induces PliaI in a neighboring strain
| Reporter | β-Galactosidase activitya when cocultivated in equal amounts with:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| None | WT | yydIJ | yydHIJ | yydF-J | ΔyydF | ΔyydG |
Pxyl-yydF-J
|
||
| No xylose | 2% xylose | ||||||||
| WT | − | − | − | − | − | − | − | − | − |
| yydIJ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| yydHIJ | − | +++ | + | − | − | − | − | − | ++ |
| yydFGHIJ | − | +++ | + | − | − | − | − | − | ++ |
Reporter strains were plated on MC medium containing 80 μg/ml X-Gal. Reporter strains contained SPβ (PliaI-74-cat-lacZ). All strains mixed with reporter strains contained the SPβ prophage (with no reporter fusion) to ensure that effects observed were not due to sensitivity towards sublancin. −, no β-galactosidase activity observed (white color); +, β-galactosidase activity observed (blue color); the number of + signs indicates the intensity.
Wild-type cells induce Plia expression in neighboring strains lacking the yydFGHIJ operon.
Since the yydFGHIJ operon is postulated to synthesize and export a modified peptide (YydF*), we tested whether wild-type (WT) strains could induce PliaI in neighboring cells lacking yydFGHIJ. Strains containing the PliaI reporter were mixed with WT and various yyd operon mutants and spotted on the solid MC medium containing X-Gal (Table 5). As noted above, deletion of yydIJ (but not yydHIJ or yydFGHIJ) leads to activation of PliaI even in pure culture (Table 5, line 2). However, when mixed with wild-type strains, PliaI is induced in strains carrying yydHIJ and yydFGHIJ deletions (Table 5, column 2). The ability of the wild type to induce the reporter strain depends on yydF and yydG, as judged from the effects of in-frame deletions. Together, these genetic studies suggest that production of active YydF* requires yydF, yydG, and yydH.
Unexpectedly, weak induction was noted even when the producer strain was lacking the YydIJ ABC transporter (Table 5, column 3). We propose that under these conditions the active YydF* peptide is produced by the yydFGH genes but not efficiently exported from these cells, and the observed induction may result from small amounts of YydF* that are either exported from the cell via other methods or released by cell lysis. This weak induction is not observed when the reporter strain is mixed with a yydHIJ mutant, again suggesting that in the absence of the YydH protease, active YydF* peptide is not produced.
Induction of PliaI in response to exogenous YydF* peptide is only seen in cells lacking the YydIJ transporter. Induction is not observed in wild-type cells or in cells containing in-frame deletions of either yydF or yydG (data not shown). Thus, the YydIJ transporter prevents PliaI induction regardless of whether the peptide is produced by the same cell or a neighboring cell.
Induction during cocultivation is readily apparent on solid medium but not in liquid culture, suggesting that the concentrations of the active peptide in liquid culture may be too low for induction in trans. In order to test whether the YydF* peptide was released into the culture supernatant, we resuspended the reporter strains in cell-free medium collected from an overnight culture of wild-type or yydFGHIJ mutant cells grown in MC medium and tested for induction of PliaI by β-galactosidase assays (data not shown). However, no increase in Plia expression was apparent under the conditions tested. These results again suggest that the YydF* peptide does not accumulate to high enough levels in liquid culture to induce the LiaRS stress response.
The yydFGHIJ operon is expressed predominantly from a σA-type promoter upstream of yydF.
Previous Northern blot analysis had indicated that the yydFGHIJ genes were coexpressed at the onset of stationary phase in minimal media (as reported on the BSORF website [http://bacillus.genome.jp/bsorf.html]). However, a previous transcriptome study had found that expression of the yydHIJ genes was increased 1.7- to 3.1-fold in a rok mutant, and biochemical studies suggested that Rok bound to a region upstream of yydH (1). Inspection of the operon sequence reveals that the 3′ end of the yydG gene overlaps with the 5′ end of the yydH gene by 19 bp, making the region to which Rok binds within the yydG gene (Fig. 1A). On the other hand, there are 150 bp between the end of yydH and the start of yydI.
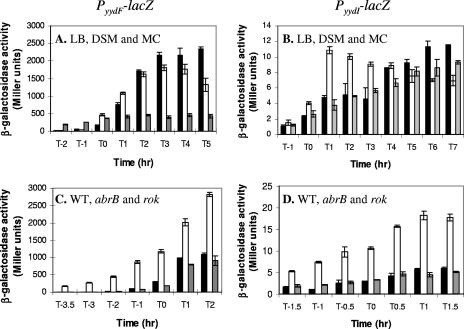
To further investigate the regulation and expression of this operon, and to test for the possible presence of promoters within the Rok-binding region and yydH-I intergenic region, we integrated lacZ reporter fusions (Fig. 1A) at the amyE locus and followed expression throughout growth (Fig. 3A and B). In LB and DSM, the PyydF promoter was strongly expressed after the transition to stationary phase (Fig. 3A). However, this postexponential induction was significantly lower in competence medium (MC). These data are somewhat surprising, since the effect of YydF* on PliaI expression was most apparent on MC medium (Fig. 2A). No activity was observed for the fusion with the fragment that included the Rok binding site upstream of yydH, suggesting that there is no promoter in this region (data not shown). Very low β-galactosidase activity (less than 12 Miller units) was seen with the PyydI-lacZ fusion in all media tested, but expression did appear to increase after transition to stationary phase (Fig. 3B). Thus, there may be a weak promoter within this intergenic region. These results suggest that the major promoter driving expression of the yydFGHIJ operon is upstream of yydF.
FIG. 3.
Expression from the PyydF and PyydI promoters. Expression levels of the PyydF-lacZ fusion (A and C) and PyydI-lacZ fusion (B and D) are shown. (A and B) Expression during growth in different media: rich medium (LB; black bars), sporulation medium (DSM; white bars), and MC medium (gray bars). (C and D) Expression from these promoters in the wild type (black bars) and abrB (white bars) and rok (gray bars) mutants grown in LB medium. Note that T0 corresponds to the transition from logarithmic growth to stationary phase. Each experiment was repeated at least twice, and a representative assay is shown here. Error bars represent standard deviations between triplicate β-galactosidase assays.
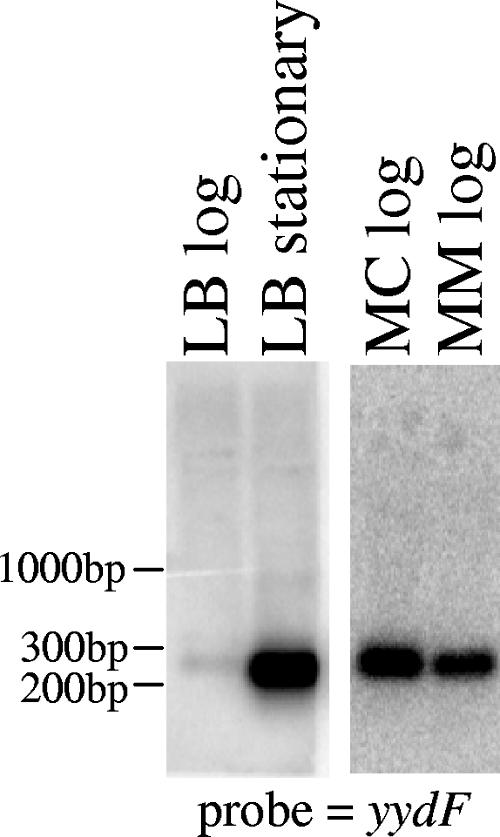
5′-RACE PCR was used to map potential transcriptional start sites upstream of the yydG, yydH, and yydI genes. The only observed product (with a yydG primer) mapped to an A residue 25 bp upstream of the yydF ATG (Fig. 1B). Northern analysis was performed with RNA isolated from cells growing in LB at log and stationary phase using probes to the yydF, yydG, yydH, and yydI genes. A strong signal was detected with the yydF probe corresponding to a small transcript of about 250 bp, and the amount of this transcript increased significantly when RNA was prepared from stationary-phase cells (Fig. 4). No full-length or other transcripts were detected with the other yyd probes (data not shown). This small transcript was also observed with RNA isolated from cells growing in MC and MM media and appeared to be increased compared to those in LB (Fig. 4). Analysis of the sequence downstream of yydF identified an imperfect inverted repeat between the yydF and yydG open reading frames (Fig. 1B). Since yydF has been found to be 1 of about 30 extremely stable B. subtilis mRNAs (14), we speculate that this repeat may form a secondary structure that stabilizes the yydF portion of the primary transcript.
FIG. 4.
Northern blot analysis showing that a small (approximately 250-bp) transcript corresponding to the yydF gene is transcribed in liquid LB, MC, and MM media and is increased in stationary phase. The blot was probed with a radiolabeled yydF fragment.
Expression of the yydFGHIJ operon does not appear to be under Rok control.
Since Albano et al. proposed that the yydHIJ genes were under the control of Rok (1), we tested expression from the PyydF and PyydI promoters in a rok mutant throughout growth in liquid medium. No increase in expression was observed in any of the media tested (LB [Fig. 3C and D] and MC and DSM [data not shown]). Contrary to the previously published results (1), deletion of rok did not increase expression from either of these promoters. It should be noted that we used ectopically located promoter fusions, while the previous study made use of an integrational plasmid (pMUTIN) inserted in the yydI gene. In our case it is possible that our promoter fusions are lacking cis elements required for Rok regulation, while in the latter case the pMUTIN integration will disrupt the YydIJ ABC transporter, which could also affect the results. Interestingly, when assayed using X-Gal on solid medium, expression from the PyydF promoter appeared to be slightly increased in a rok mutant (data not shown). However, an additional deletion of the sdpABC operon, also shown to be under Rok control (1), abolished this increase. We noticed that a rok mutant does not grow well on MC media (both liquid and solid); therefore, it is also possible that the increase in blue color observed on the solid medium could be due to increased lysis (presumably mediated by the toxic SdpC* peptide) and release of β-galactosidase from the cells.
Expression of the yydFGHIJ operon is negatively regulated by AbrB.
Previous transcriptional profiling studies compared expression of genes in WT and spo0A mutants (the master regulator of sporulation in B. subtilis) and revealed that expression of yydF is increased about 130-fold in a WT strain compared to a spo0A mutant at transition stage and about 30-fold 2 h later. However, expression of the downstream yydGHIJ genes was essentially unchanged under these conditions (11). Since Spo0A represses expression of the transition-state regulator AbrB, we tested whether this difference in expression was due to AbrB. Indeed, in an abrB mutant expression from the PyydF-lacZ fusion was increased throughout log growth and into stationary phase, confirming that AbrB normally represses this promoter during growth (Fig. 3C). AbrB repression of PyydI was also observed; however, the increase in expression in an abrB mutant was only about 4-fold, compared with the 20- to 90-fold derepression observed for PyydF (Fig. 3D). Thus, like many other operons encoding antimicrobial peptides, the yydFGHIJ operon is under negative control of the AbrB transition state regulator.
DISCUSSION
The LiaRS TCS is known to be strongly induced by various cell wall-active antibiotics and at least some antimicrobial peptides, but the physiological role of this stress response is not yet clear. Mutants lacking liaR, or the entire liaIHGFSR operon, do not display increased sensitivity to the known inducing compounds (29), suggesting that perhaps this system functions physiologically to protect cells against other, yet-unidentified stress conditions. Since the LiaRS-regulated LiaH protein is an ortholog of the phage shock protein, PspA, it is possible that this system functions to help maintain the integrity of the cell membrane under conditions that interfere with membrane function.
Here, we have used transposon mutagenesis to identify genes that affect the activity of the LiaRS TCS. Our results are consistent with a model in which the yydFGHIJ operon codes for the synthesis, modification, cleavage, and export of a small peptide, YydF*. Transposon insertions that disrupt either the yydI or yydJ genes, and therefore block peptide export, lead to elevated expression of the LiaR-activated PliaI promoter. This is dependent on presumed synthesis of the active YydF* peptide (product of the yydFGH genes), and production of this peptide can activate LiaR in neighboring cells lacking the yydFGHIJ operon.
The primary product of the yydF gene is a small (49-aa) peptide with a predicted positive charge at pH 7.0 and the ability to form an α-helix (Table 4), properties often associated with antimicrobial peptides. The YydG protein contains a radical SAM family domain (pfam 04055) (34). Members of this family include proteins involved in antibiotic biosynthesis, including AlbA, which is involved in biosynthesis of the bacteriocin subtilosin in B. subtilis (38). YydH is a member of the S2P protease family (M50) and is a weak paralog of another B. subtilis protein, SpoIVFB (39). This protein is a membrane-embedded metalloprotease involved in cleavage of pro-σK. Proteases of the S2P family are often involved in regulated intramembrane proteolysis (RIP), where a protease, embedded in the membrane, cleaves a membrane-spanning protein within or very close to the membrane to release a soluble truncated protein, often a signaling molecule or transcription regulator. YydH is predicted to have five transmembrane-spanning helices, with the conserved HExxH motif located in the second helix and DG residues within the fourth helix. In other S2P proteases, these two histidines and one aspartate residue are Zn ligands, while the glutamate is the catalytic residue (23). While many well-studied S2P family members are involved in the processing of regulators or their inhibitors, the metalloprotease Eep from Enterococcus faecalis is believed to play a role in the cleavage of a pheromone precursor (2). The final two genes of the operon, yydIJ, encode the ATPase and membrane subunits of an ABC transporter. As part of a previous inventory of the ABC transporters of B. subtilis, it was noted that the similarity of YydI with other ATP-binding proteins was too weak to allow for functional predictions (32). Based on our in silico analyses, we predict that the YydIJ ABC transporter functions in peptide export.
It is common for immunity towards peptide antibiotics to be provided by an ABC transporter that exports the toxic peptide from the cell. Therefore, inactivation of the yydIJ genes may result in sensitivity toward the active YydF peptide, which is sensed by the LiaRS system. However, in spot-on-lawn assays we noted that a yydFGHIJ mutant is not sensitive to a WT strain or to a strain overexpressing the yydFGHIJ operon under PxylA control, even when the liaIHGFSR operon is also deleted (data not shown). This suggests that YydF* is not toxic enough to cause visible lysis under these conditions, that there are other unidentified systems that provide resistance, or that YydF* is a signaling rather than an antimicrobial peptide. As yet we have been unsuccessful in isolating the active YydF from WT cell cultures or cell-free supernatants; therefore, we cannot confirm the structure and modifications of this peptide.
The yyd operon appears to be subject to complex regulation. Data presented here and in other studies (transcriptome data and BSORF) suggest that all five genes can be expressed as one transcript from PyydF. This strong promoter is activated during the transition phase, and this is likely due, in part, to relief of AbrB-mediated repression. In addition, we identified a weak promoter upstream of yydI. This may function to keep a small amount of the ABC transporter present in the cell for resistance to peptides produced by neighboring cells. Surprisingly, our Northern analyses detected a very strong signal corresponding to a small yydF transcript and no full-length transcript. The yydF mRNA was previously reported to be extremely stable, with a half-life of more than 15 min (14). Thus, this stable transcript may result from processing of the longer, primary transcript detected in previous analyses (BSORF).
Previous transcriptome analyses support the suggestion that some or all of the genes of the yydFGHIJ operon may be induced under stress conditions, including high salinity (35) and cold shock (22). In addition, the yydHIJ genes have been proposed to be under the control of Rok (repressor of ComK) (1). Rok was reported to bind immediately upstream of yydH, but we did not detect promoter activity from this region and we were unable to confirm previous suggestions that genes from within this operon are regulated by Rok. It is possible that binding of Rok to this region might affect transcription elongation, rather than initiation, and thereby induce a shift from production of full-length mRNA to shorter transcripts. It is interesting that Rok was also shown to repress several operons involved in the synthesis of bacteriocins: the sunAT-bdbA-yolJ-dbdB operon (which is involved in the production and secretion of the lantibiotic sublancin) and the sbo-alb operon (synthesis and export of subtilosin), as well as the sdpABC operon (encoding the antibiotic peptide SdpC). This suggests that competent B. subtilis cells may produce antimicrobial peptides to lyse neighboring cells (fratricide) to release chromosomal DNA for uptake, as has been shown in Streptococcus pneumonia (16). However, experimental support for this notion is not yet available.
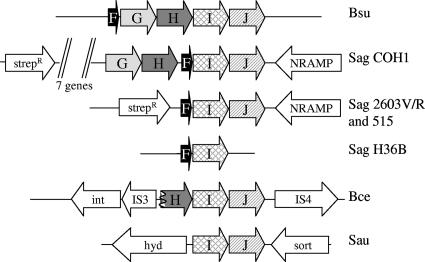
Phylogenomic comparisons suggest that the yydFGHIJ operon has likely been acquired by horizontal gene transfer. Homologous operons are present in B. subtilis 168 and its closest relatives (the recently sequenced wild-type isolates B. subtilis subsp. subtilis RO-NN-1 and B. subtilis subsp. spizizenii TU-B-10 [related to W23]), but not in other closely related bacilli, including Bacillus licheniformis and Bacillus amyloliquefaciens. Surprisingly, close homologs to all genes in the yydF operon are found in Streptococcus agalactiae. Many strains of S. agalactiae have been sequenced, and the “pan-genome” has been determined (36). The YydF homologs are only present in some of these strains and in all cases are found together with YydIJ homologs. Homologs of the YydGH proteins are only found in the S. agalactiae strain COH1 (Fig. 5). Homologs to the YydIJ ABC transporter are also found in Staphylococcus aureus and Bacillus cereus subsp. cytotoxis NVH 391-98. In the case of S. aureus there are no annotated small peptides or yydGH homologs in the region surrounding the yydIJ genes, while in B. cereus the homologous yydH gene is truncated and genes encoding a transposase and integrase are located on either side of the operon (Fig. 5). These observations support the previous prediction, based on the anomalous G+C content of these genes, that the yydFGHIJ region was horizontally transferred into the B. subtilis genome (Horizontal Gene Transfer Database [HGT-DB] at http://www.fut.es/∼debb/HGT/) (12).
FIG. 5.
Arrangement of yyd operon homologs in other bacteria. Open reading frames are not to scale. Homologs of the yydFGHIJ genes are shaded, while other surrounding genes are shown in white. Bsu, B. subtilis subsp. subtilis strains 168 and RO-NN-1 as well as B. subtilis subsp. spizizenii TU-B-10; Sag, Streptococcus agalactiae (strains indicated; genes encoding putative streptomycin resistance [Strepr] proteins and NRAMP family Mn2+/Fe2+ transporters [NRAMP] are also indicated); Bce, B. cereus subsp. cytotoxis NVH 391-98 (the yydH gene is truncated, and the upstream putative integrase [int] and flanking transposases of the IS3 and IS4 families are shown); Sau, Staphylococcus aureus subsp. aureus (predicted hydrolase [hyd] and sortase [sort] genes are also indicated). All sequences can be found at the National Center for Biotechnology website (http://www.ncbi.nlm.nih.gov), except for the unfinished genomes of B. subtilis sp. RO-NN-1 and TU-B-10.
Our results suggest that the yydFGHIJ operon encodes a modified and processed peptide (YydF*) that is exported from the cell via an ABC transporter. In the absence of the ABC transporter, YydF* activates the LiaRS TCS, perhaps by causing cell envelope stress. The human antimicrobial peptide LL-37 has been shown to activate several cell envelope stress responses in B. subtilis, including the LiaRS system (31). In this work we demonstrate that LiaRS additionally, and specifically, responds to at least one endogenous peptide produced by B. subtilis.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM-47446).
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Albano, M., W. K. Smits, L. T. Ho, B. Kraigher, I. Mandic-Mulec, O. P. Kuipers, and D. Dubnau. 2005. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 187:2010-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60:765-782. [DOI] [PubMed] [Google Scholar]
- 5.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 7.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 8.Darwin, A. J., and V. L. Miller. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429-444. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin, J., G. Jovanovic, and P. Model. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 11.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Vallve, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 14.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 15.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons, West Sussex, England.
- 16.Havarstein, L. S., B. Martin, O. Johnsborg, C. Granadel, and J. P. Claverys. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297-1307. [DOI] [PubMed] [Google Scholar]
- 17.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 18.Hoa, T. T., P. Tortosa, M. Albano, and D. Dubnau. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43:15-26. [DOI] [PubMed] [Google Scholar]
- 19.Hyyrylainen, H. L., M. Sarvas, and V. P. Kontinen. 2005. Transcriptome analysis of the secretion stress response of Bacillus subtilis. Appl. Microbiol. Biotechnol. 67:389-396. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, S., E. Rietkötter, M. A. Strauch, F. Kalamorz, B. G. Butcher, J. D. Helmann, and T. Mascher. 2007. LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology 153:2530-2540. [DOI] [PubMed] [Google Scholar]
- 22.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 23.Kinch, L. N., K. Ginalski, and N. V. Grishin. 2006. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 15:84-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunst, K., T. Msadek, and G. Rapoport. 1994. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis, p. 1-21. In P. J. Piggot, C. P. Moran, and P. Youngman (ed.), Regulation of bacterial differentiation. American Society for Microbiology, Washington, DC.
- 25.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 26.Mascher, T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133-144. [DOI] [PubMed] [Google Scholar]
- 27.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 29.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Pietiäinen, M., M. Gardemeister, M. Mecklin, S. Leskelä, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577-1592. [DOI] [PubMed] [Google Scholar]
- 32.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steil, L., T. Hoffmann, I. Budde, U. Volker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 38.Zheng, G., R. Hehn, and P. Zuber. 2000. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182:3266-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, R., and L. Kroos. 2005. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol. Microbiol. 58:835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]