Abstract
Two-component oxygenases catalyze a wide variety of important oxidation reactions. Recently we characterized a novel arylamine N-oxygenase (PrnD), a new member of the two-component oxygenase family (J. Lee et al., J. Biol. Chem. 280:36719-36728, 2005). Although arylamine N-oxygenases are widespread in nature, aminopyrrolnitrin N-oxygenase (PrnD) represents the only biochemically and mechanistically characterized arylamine N-oxygenase to date. Here we report the use of bioinformatic and biochemical tools to identify and characterize the reductase component (PrnF) involved in the PrnD-catalyzed unusual arylamine oxidation. The prnF gene was identified via sequence analysis of the whole genome of Pseudomonas fluorescens Pf-5 and subsequently cloned and overexpressed in Escherichia coli. The purified PrnF protein catalyzes reduction of flavin adenine dinucleotide (FAD) by NADH with a kcat of 65 s−1 (Km = 3.2 μM for FAD and 43.1 μM for NADH) and supplies reduced FAD to the PrnD oxygenase component. Unlike other known reductases in two-component oxygenase systems, PrnF strictly requires NADH as an electron donor to reduce FAD and requires unusual protein-protein interaction with the PrnD component for the efficient transfer of reduced FAD. This PrnF enzyme represents the first cloned and characterized flavin reductase component in a novel two-component arylamine oxygenase system.
For oxygenases that require NAD(P)H, the catalyzed reaction can be separated into two steps, i.e., the oxidation of NAD(P)H to generate two reducing equivalents and the oxygenation of substrates. Most of the monooxygenases catalyzing the oxygenation are flavoprotein enzymes that carry out the two reactions on a single polypeptide chain (31). However, two-component monooxygenases where NAD(P)H oxidation and the oxygenation reaction are catalyzed by separate polypeptides linked by an electron transport chain also have been described (12, 17, 29). Bacterial two-component monooxygenase systems that utilize reduced flavin as a substrate are also continually being identified (3, 9, 13, 39, 47). Although these enzyme systems catalyze distinct reactions, a central theme in the two-component oxygenase family is the presence of a flavin-dependent reductase involved in flavin reduction followed by the transfer of reduced flavin to the oxygenase component.
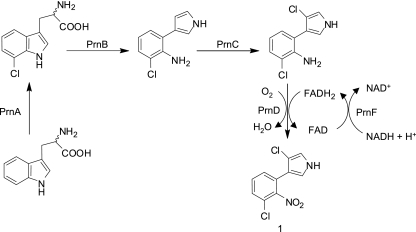
Recently we reported the characterization of a Rieske N-oxygenase, aminopyrrolnitrin oxygenase (PrnD), that catalyzes unusual arylamine oxidation (23). Although arylamine oxygenases seem to be widely distributed and used in a variety of metabolic reactions, PrnD represents the only characterized example of arylamine N-oxygenases involved in arylnitro-group formation. PrnD is involved in the biosynthesis of the antibiotic pyrrolnitrin (compound 1 in Fig. 1), which is produced by many pseudomonads and has broad-spectrum antifungal activity (5, 22, 36). In the proposed biosynthetic pathway of pyrrolnitrin (Fig. 1), PrnD catalyzes the oxidation of the amino group of aminopyrrolnitrin to a nitro group, forming pyrrolnitrin (41). In addition, we have obtained direct evidence for the involvement of hydroxylamine and nitroso intermediates in the PrnD-catalyzed arylamine oxygenation reaction, substantiating a two-component monooxygenase-type catalytic mechanism for the conversion of arylamine to arylnitro compounds (24). This may be the primary mechanism by which arylamines are oxygenated to give arylnitro compounds in biological processes. The other arylamine-oxygenating enzyme, AurF, was recently identified as a monooxygenase based on whole-cell experiments, not on in vitro experiments with the purified enzyme (37, 42).
FIG. 1.
Proposed biosynthetic pathway for pyrrolnitrin.
Although PrnD, a Rieske N-oxygenase, has been characterized and the chemical mechanism regarding arylamine oxygenation has been suggested, little information is available about the coupled flavin reductase, the enzyme responsible for the supply of the reduced flavin. Flavin reductase catalyzes the reduction of flavins, such as flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), and riboflavin by NAD(P)H to form reduced flavins that are required to activate oxygen by the terminal oxygenases. It has been reported that flavin reductases from Vibrio harveyi and Photobacterium fischeri stimulated the enzyme activities of various monooxygenases from different microorganisms (43, 44). Luciferase (40), styrene monooxygenase (30), 4-hydroxyphenylacetate 3-monooxygenase (12), pyrrole-2-carboxylate monooxygenase (2), nitrilotriacetate monooxygenase (21, 45), and EDTA monooxygenase (34) have been characterized as two-component monooxygenases. Other members of the two-component monooxygenase class have also been described, including the monooxygenases involved in biosynthesis of antibiotics such as pristinamycin IIA and valanimycin (32, 39).
The elucidation of the Pseudomonas fluorescens Pf-5 genome sequence in 2005 opened the door for genome-scale research with this important microbial strain (33). Therefore, we decided to identify the flavin reductase to complete the two-component arylamine oxygenase system for P. fluorescens Pf-5. Because the genes involved in the same antibiotic biosynthetic pathway are usually clustered on the chromosome (15, 28), we started our study by carrying out sequence analysis around a previously identified prnD gene and chose a gene (prnF) as a candidate encoding the flavin reductase based on the bioinformatics studies. Here we describe the cloning of prnF from P. fluorescens Pf-5 and the characterization of the recombinant protein. We also provide experimental evidence that PrnF is the flavin:NADH reductase component of the two-component arylamine oxygenase system in P. fluorescens Pf-5. The characterization of the PrnD/PrnF arylamine oxygenase system adds a new and interesting member to the family of two-component monooxygenases and gives a better understanding of the formation of arylnitro compounds in nature.
MATERIALS AND METHODS
Materials.
The pMal-c2x expression vector, malE primer, factor Xa, amylose resin, Taq DNA polymerase, T4 DNA ligase, DNase I, and restriction endonucleases were purchased from New England Biolabs (Beverly, MA). Plasmid pQE30Xa was obtained from Qiagen (Valencia, CA). The bicinchoninic acid protein assay kit, AMP, NADH, NADPH, NAD+, FAD, FMN, riboflavin, lumiflavin, 4-aminobenzyl amine (pABA), and 4-nitrobenzyl amine (pNBA) were from Sigma-Aldrich (St. Louis, MO). Materials for PCR product purification, gel extraction, and plasmid preparation were obtained from Qiagen. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA).
Bacterial strains and growth conditions.
Escherichia coli BL21(DE3) and DH5α were obtained from Novagen (Madison, WI) and the University of Illinois Biochemistry Department's Cell and Media Facility (Urbana, IL), respectively. Pseudomonas fluorescens Pf-5 (ATCC no. BAA-447) was purchased from the American Type Culture Collection (Manassas, VA). E. coli strains DH5α and BL21(DE3) were grown aerobically at 37°C or 30°C in Luria-Bertani (LB) medium with constant shaking (220 rpm). When necessary, kanamycin was added at 50 μg/ml, ampicillin at 100 μg/ml, and chloramphenicol at 25 μg/ml. Solid media were prepared by addition of 1.5% (wt/vol) agar.
Construction of pQE30Xa-prnF expression plasmid.
The prnF gene was amplified by PCR from P. fluorescens Pf-5 genomic DNA using two oligonucleotide primers, 5′-GGGGATCCATGAATGCTGCCACCGAAAC-3′ (BamHI restriction site is underlined) and 5′-GGAAGCTTCTATTCGTGCTTGAGGACGC-3′ (HindIII restriction site is underlined). The PCR amplification was conducted using PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) to minimize potential point mutations introduced by PCRs under standard conditions. The PCR program was as follows: 2 min at 96°C, followed by 30 cycles of 1 min at 94°C, 1 min at 52°C, 2 min at 72°C, and a final elongation of 10 min at 72°C. The PCR products were cleaved by BamHI and HindIII and purified using the Qiaex II gel purification kit (Qiagen). The purified product was cloned into the BamHI- and HindIII-digested expression vector pQE30Xa. The result, pQE30Xa-prnF, is under the control of the T5 promoter and expresses PrnF as a protein fused to the N terminus of a His6 tag. The cloned prnF gene was confirmed to be free of point mutations by DNA sequencing at the Biotechnology Center of the University of Illinois using the BigDye Terminator sequencing method and an ABI PRISM 3700 sequencer (Applied Biosystems, Foster City, CA).
Expression and purification of PrnF and PrnD proteins.
E. coli BL21(DE3) cells (Invitrogen) overexpressing PrnF were grown in LB medium supplemented with ampicillin (100 μg/ml). Cells were grown at 30°C until the absorbance at 600 nm reached ∼0.6. Then, protein expression was induced with 100 μM isopropyl β-d-thiogalactopyranoside (IPTG) and grown for an additional 6 h at 20°C. Cells were harvested by centrifugation at 4°C for 20 min at 6,000 × g, rinsed with phosphate-buffered saline, and frozen and stored at −20°C. The yield was approximately 3 g bacterial wet weight/liter of culture. For the purification of the His6-tagged PrnF protein, 3 g of cells were resuspended in 10 ml of binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.5) supplemented with 25 μg/ml DNase I. A bacterial lysate was prepared by thawing and resuspending cells; this suspension was treated with 10 mg of lysozyme for 30 min. After clarification by centrifugation for 30 min at 20,000 × g and 4°C, the volume of the crude extract was adjusted to 10 ml with binding buffer. The crude extract was then loaded at a flow rate of 25 ml/h onto a 2.5-ml HisBind resin column (Novagen), and the PrnF protein was purified according to the manufacturer's protocol. An additional wash step with a buffer containing 100 mM imidazole was performed for 6 column volumes prior to PrnF elution with 250 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, and 20% glycerol, pH 7.5. For cleavage with factor Xa, the purified His6-PrnF fusion protein was incubated with factor Xa (1 μg/200 μg of fusion protein) for 16 h at 4°C. The His6 tag was removed after cleavage by a second adsorption to the nickel-nitrilotriacetic acid (Ni-NTA) column. Fractions from the Ni-NTA column were then concentrated with Centricon-10 ultrafiltration units (Amicon) and adjusted to 100 mM NaCl, 20 mM Tris-HCl, and l mM dithiothreitol, pH 7.5. The PrnD enzyme was expressed, purified, and reconstituted as described elsewhere (23).
Construction of a recombinant E. coli strain coexpressing prnD and prnF genes.
The PCR product obtained by using the same oligonucleotide primers with BamHI and HindIII restriction sites was cleaved by BamHI and HindIII and purified using the Qiaex II gel purification kit (Qiagen). The purified product was ligated into the plasmid pACYC-Duet (Novagen), which had been treated with the same restriction endonucleases. The resulting plasmid, pACYC-prnF, was transformed into E. coli strain XL1-Blue (Stratagene, La Jolla, CA). The plasmid was reisolated and transformed into E. coli strain BL21(DE3) harboring the plasmid pTKXb-prnD (23). The strain was cultured in LB medium containing chloramphenicol (25 μg/ml) and kanamycin (50 μg/ml). Recombinant E. coli cells carrying pTKXb-prnD and pACYC-prnF or pTKXb-prnD only were cultivated at 30°C until the absorbance at 600 nm reached ∼0.6 and then induced with 100 μM IPTG and grown for an additional 12 h at 20°C. The cells were harvested and washed as described above and suspended in 50 mM Tris-HCl (pH 7.0). The optical density at 600 nm of the cell suspension was adjusted to 20. One hundred microliters of a 10 mM pABA stock solution was added to 15-ml screw-cap test tubes containing 0.9 ml of the cell suspension to a final concentration of 1 mM pABA. Resting cell reactions were carried out with shaking at 100 rpm for 2 h. pABA and its N-oxygenated product, pNBA, were determined using high-performance liquid chromatography (HPLC) as described previously (24).
Enzyme assay.
Flavin reductase activity was determined by measuring the decrease of the absorbance at 340 nm (ɛ340 = 6.22 mM−1 cm−1) due to the oxidation of NADH, using a Varian Cary 100 Bio UV-Vis spectrophotometer (Varian, Palo Alto, CA) (10, 26). Appropriate amounts of PrnF were incubated at 30°C in a reaction mixture (final volume, 1 ml) containing 200 μM NADH and 30 μM FAD in 50 mM NaCl, 20 mM Tris-HCl, pH 7.0. Reactions were started by addition of enzyme to the reaction mixture. An assay mixture without FAD was used as a blank. One unit was defined as the amount of enzyme catalyzing the oxidation of 1 μmol of NADH/min at 30°C under aerobic conditions. PrnD oxygenase activity was assayed as described previously (23).
Analytical methods.
Optical spectra were recorded on a Varian Cary 100 Bio UV-Vis spectrophotometer. Enzyme reaction products were analyzed by using an Agilent 1100 Series HPLC system. The sample was eluted on a Zorbax SB-C8 column (4.6 by 150 mm; Agilent). HPLC parameters were as follows: 25°C; solvent A, 1% acetic acid in water; solvent B, methanol; gradient, 5% B for 2 min, followed by 100% B for 18 min and finally maintenance at 100% B for 2 min; flow rate, 1.0 ml/min; detection by UV spectroscopy at 254 nm. The reaction was also analyzed by HPLC coupled to an electrospray ionization mass spectrometer (TSQ Quantum; Thermo-Finnigan, San Jose, CA) in positive-ion mode. Promoter scan analysis was done using the PROSCAN Version 1.7 suite of programs developed by Dan Prestridge at the Advanced Biosciences Computing Center, University of Minnesota. Gel filtration chromatography was performed using a Superdex 200 gel filtration column (Amersham Bioscience, Inc., Uppsala, Sweden) mounted on an AKTA fast-performance liquid chromatography system (Amersham Bioscience, Inc.). The column was preequilibrated with 20 mM Tris-HCl buffer (pH 7.0)-50 mM NaCl. Gel filtration standards were purchased from Bio-Rad (Hercules, CA).
Fluorescence measurements.
Registration of changes in fluorescence was carried out on a Perkin Elmer MPF-44 fluorescence spectrophotometer (Perkin Elmer, Wellesley, MA). To follow the change in fluorescence, emission intensity measurements were made at 530 nm with excitation at 450 nm; excitation and emission slit widths were 15 and 20 nm, respectively (1).
Fluorometric titration of FAD with PrnF.
Binding of FAD to PrnF was determined by spectrofluorometric titrations, monitoring the decrease in the relative intensity of the FAD emission due to fluorescence quenching of the flavin upon binding to PrnF. A 0.04 μM FAD solution in Tris-HCl buffer, pH 7.0, at 25°C was titrated with increasing concentrations of PrnF (0.01 to 0.4 μM) (1, 17).
Fluorometric titration of FAD-bound PrnF with PrnD.
To investigate direct binding of PrnD to PrnF, the effect of PrnD binding on FAD-bound PrnF was examined. Briefly, 0.4 μM PrnF was titrated with increasing PrnD concentrations (0.004 to 0.5 μM). In PrnD-induced PrnF fluorescence-enhancing experiments, the number of binding sites (n) was estimated by fitting the binding curve to a Hill plot according to the equation y = axb/(cb + xb), where a is Fmax, b is the number of binding sites (n), and c is the dissociation constant (Kd) (17, 35).
RESULTS
Identification of genes involved in pyrrolnitrin biosynthesis.
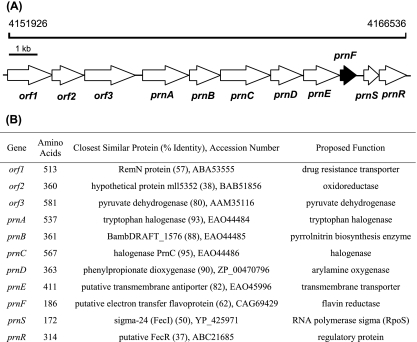
The sequence analysis of the whole genome of P. fluorescens Pf-5 suggested the presence of a pyrrolnitrin biosynthetic gene cluster (Fig. 2), including four open reading frames (ORFs) (prnA to -D), which have been previously described as a complete operon for pyrrolnitrin biosynthesis (14, 41). All ORFs were in the same orientation from prnA (the first ORF) to the eighth ORF. The ORFs from the fifth to the eighth were annotated as a Na+/H+ antiporter transmembrane transporter, a putative flavin:NAD(P)H reductase, an RNA polymerase sigma factor, and a sigma factor regulatory protein, respectively, suggesting that the sixth ORF might encode a flavin reductase to supply the reduced flavin for the PrnD component. Accordingly, four ORFs were named prnE, prnF, prnS, and prnR, respectively. As to the three ORFs in front of prnA in the pyrrolnitrin biosynthetic gene cluster, their deduced products also showed similarities to proteins of known functions (Fig. 2B). Although there is a possibility that orf1 and orf2 are involved in the pyrrolnitrin biosynthetic gene cluster as a resistance gene and an oxidoreductase gene, the deduced product of orf3, pyruvate dehydrogenase, is apparently unrelated to pyrrolnitrin biosynthesis. In addition, prnF is much closer to prnD than orf2 in the genome. Further promoter scan analysis strongly suggested the presence of a promoter in front of prnA, indicating that prnD and prnF are very likely to be in the same operon while prnD and orf2 are not. Thus, based on these bioinformatics studies, we considered PrnF a better candidate reductase for PrnD than ORF2.
FIG. 2.
Proposed pyrrolnitrin biosynthetic gene cluster. (A) Genetic organization of the proposed pyrrolnitrin biosynthetic gene cluster including prnF, encoding flavin reductase. (B) Proposed functions for individual ORFs.
Characterization of the prnF gene encoding a flavin reductase.
The prnF gene encodes a polypeptide of 160 amino acids, with a calculated molecular mass of 17,101 Da and an overall GC content of 62%, which is similar to that of the chromosomes of Pseudomonas species (60 to 66%) (27). The deduced prnF gene product contains the pfam01613 flavin reductase-like domain present in flavin reductases and various monooxygenase components. In the C-terminal region of PrnF, a highly conserved GDH motif, which plays a role in NAD(P)H binding in Fre (16), was found.
A homology search revealed that the deduced prnF gene product showed 31.5, 28.6, 26.4, 25.6, and 25.5% amino acid identity with PheA2 (8, 20), SnaC (39), VlmR (32), ActVB (18), and HpaC (12, 46), respectively. The deduced product of prnF belongs to a family of flavin:NAD(P)H reductases, the majority of which are part of two-component monooxygenase systems. VlmR is a flavin reductase that functions in a two-component enzyme system to provide isobutylamine N-hydroxylase with reduced flavin and is involved in the synthesis of valanimycin. ActVB is also a flavin:NADH reductase that participates in the last step of actinorhodin biosynthesis. In analogy to these two-component flavin monooxygenases, PrnF and PrnD are proposed to form a two-component oxygenase system in which PrnF supplies the reduced flavin that PrnD needs to function (Fig. 1).
Heterologous expression of prnF gene and identification of flavin:NADH reductase.
In order to check its proposed function, prnF was cloned in the T5 RNA polymerase-based plasmid pQE30Xa to give pQE30Xa-prnF and heterologously expressed in E. coli BL21(DE3). The pQE30Xa vector encodes an N-terminal His tag and a 16-amino-acid spacer that contains a factor Xa cleavage site (GSGSGSGIERGPYNGT). Analyses carried out with the extracts of E. coli BL21(DE3) harboring pQE30Xa-prnF revealed the presence of a high level of flavin:NADH oxidoreductase activity compared with the control extracts of E. coli BL21(DE3) cells harboring the plasmid pQE30Xa. To ascertain that FAD reduction in the presence of NADH was carried out specifically by the PrnF protein and not by another enzyme induced in the host cell as a consequence of the overexpression of the prnF gene, the PrnF enzyme was purified. Total protein extracts from E. coli BL21(DE3) transformed with pQE30Xa-prnF or with pQE30Xa as a control were analyzed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A 19-kDa protein, which was in agreement with the predicted molecular mass for the PrnF protein, could be identified in total and soluble extracts only from cells harboring pQE30Xa-prnF and induced by IPTG (see Fig. S1 in the supplemental material). This protein was assigned as PrnF, and it accounted for ∼5% of total protein based on the specific activity in the cell extracts. The enzyme was purified by Ni-NTA affinity chromatography about 25-fold to near-homogeneity. The N-terminal His6 tag was then removed by factor Xa. The factor-Xa-treated PrnF protein was separated from the fusion protein and His6 tag peptide fragment using an Ni-NTA column. The purified PrnF reductase was colorless, and the UV-visible spectrum showed no evidence of any chromogenic cofactor. Oxidation of NAD(P)H to NAD(P) was monitored by the decrease in absorbance at 340 nm. In the presence of FAD, PrnF oxidized NADH with a kcat value of 65 s−1 (Km = 3.2 μM for FAD and 43.1 μM for NADH). NADPH was not accepted for oxidation by PrnF. These findings strongly supported the assumption that the reductase activity observed in crude extracts of E. coli BL21(DE3) harboring pQE30Xa-prnF corresponded to that of the PrnF protein. By using gel filtration chromatography, the molecular mass of native PrnF was determined to be ∼39 kDa, showing that the protein is homodimeric in solution. The characterization of PrnF as a NADH-dependent flavin reductase then allowed for the investigation of its participation in the two-component oxygenase reaction.
Formation of arylnitro compound by PrnD in the presence of PrnF.
Although we have observed that PrnF was able to produce reduced FAD in vitro in the absence of PrnD, it was necessary to investigate whether such activity could participate in the oxygenation of arylamines. An assay was performed with FAD, NADH, and purified PrnD and PrnF. pABA was used as the substrate to detect the appearance of new products derived from that arylamine compound (23, 24). Analysis by HPLC revealed a new product peak that was formed when PrnF-PrnD was incubated with the substrate pABA, FAD, and NADH. The product coeluted with an authentic standard of pNBA with the same retention time (3.86 min). This compound was further identified as pNBA by high-resolution (HR) electron ionization mass spectrometry (M+ calculated, 152.0586; found, 152.0587). This product was absent if heat-denaturated PrnD was used or if pABA was omitted from the enzyme assay. This indicated that the product was derived enzymatically from pABA. In the absence of PrnF, PrnD showed almost no oxygenating activity. However, rates of product formation by PrnD (kcat = 11.3 min−1; Km pABA = 398 μM) were dramatically enhanced with the addition of PrnF. The observation that PrnD activity was absolutely dependent on the presence of PrnF supports the hypothesis that both components are required for arylamine oxygenation. When the ratio of PrnF to PrnD was varied, PrnD activity exhibited saturation above a 0.1:1 ratio. This might be ascribed to the fact that PrnF (kcat = 65 s−1) has much higher activity than PrnD (kcat = 11.3 min−1). Subsequent kinetic characterization of PrnF in the presence of PrnD was conducted at this ratio of protein components.
Substrate specificity.
PrnF enzyme activity was measured using various electron acceptors in place of FAD at fixed concentrations of the second substrates. When flavin compounds were added as electron acceptors, FMN, riboflavin, and lumiflavin were 12, 9, and 6% as effective as FAD, respectively (Table 1). In addition to flavin compounds, the enzyme acted on artificial electron acceptors, such as methylene blue and ferricyanide, but these compounds were less than 5% as effective as FAD. This behavior appears to be uncommon for flavin reductases that do not contain a flavin as a prosthetic group, since they reduce FMN, FAD, and riboflavin with similar efficiencies (10, 12, 20, 30). As an electron donor or the second substrate, PrnF preferred NADH over NADPH. The fact that FAD is a true substrate rather than a tightly bound cofactor for the enzyme was demonstrated by the complete reduction of an excess of FAD by the enzyme in the presence of excess NADH under anaerobic conditions (18). When oxygen was readmitted to the incubation mixture, the absorption at 450 nm due to FAD reappeared.
TABLE 1.
Substrate specificities of the flavin:NADH reductase (PrnF)a
| Substrate | Second substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|---|
| FAD | NADH | 65.0 ± 8.2 | 3.2 ± 0.6 | 20.3 ± 3.45 |
| FMN | NADH | 72.4 ± 9.0 | 28.6 ± 3.6 | 2.53 ± 0.27 |
| Riboflavin | NADH | 68.2 ± 7.7 | 38.3 ± 5.4 | 1.78 ± 0.25 |
| Lumiflavin | NADH | 66.5 ± 9.4 | 58.4 ± 7.8 | 1.14 ± 0.16 |
| NADH | FAD | 62.6 ± 7.6 | 43.1 ± 5.7 | 1.45 ± 0.19 |
| NADPH | FAD |
Enzyme activity was measured as described in Materials and Methods at various concentrations of substrates in 50 mM Tris-HCl (pH 7.0) at 30°C. Km is the Michaelis constant for the organic substrate.
Optimum pH and thermal stability.
The optimum pH for reduction of FAD by purified PrnF was 7.0, with 73, 90, 56, and 81% of maximum activity at pH 6.0, 6.5, 7.5, and 8.0, respectively. The isoelectric point of PrnF was pH 6.5 as determined by isoelectric focusing. This value agrees with the theoretical value (6.3) estimated from the amino acid sequence. The stability of PrnF was tested at pH 7.0 in standard 50 mM Tris-HCl buffer containing 1 mM dithiothreitol. Preparations were incubated at 4, 16, 30, 42, and 50°C and retained 50% of their initial activities after 12 day, 5 days, 1 day, 30 min, and 1 min, respectively.
Flavin binding stoichiometry and dissociation constant.
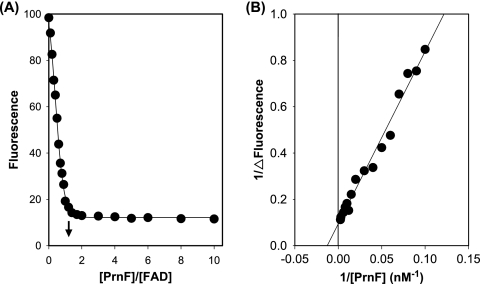
Upon binding of FAD to PrnF, the intensity of the fluorescence emission of FAD was significantly decreased. This quenching of FAD fluorescence by PrnF was used to determine the stoichiometry and the dissociation constant of FAD binding by PrnF. FAD (40 nM) was titrated with various amounts of PrnF (10 to 400 nM), and the quenching of the FAD fluorescence emission at 530 nm was monitored (Fig. 3A) (1). Higher degrees of quenching were observed at higher concentrations of PrnF. The extrapolated breakpoint of such a fluorescence titration curve corresponded to a binding of 1.02 PrnF per FAD (Fig. 3A). The reciprocal of ΔFluorescence (ΔFluorescence is defined as the fluorescence intensity of the FAD in the absence of PrnF minus that in the presence of a designated amount of PrnF) was plotted against the reciprocal of the PrnF concentration (Fig. 3B), and the intercept at the abscissa allows the calculation of a Kd of 78 nM for PrnF binding to FAD (38).
FIG. 3.
Fluorometric titrations of FAD with PrnF enzyme. (A) FAD (40 nM) was titrated with various concentrations of PrnF (10 to 400 nM). Flavin fluorescence emissions at 530 nm were measured using 450-nm excitation. The arrow points to the extrapolated breakpoint of maximal fluorescence decrease. (B) Reciprocals of ΔFluorescence (defined as the fluorescence in the absence of PrnF minus that in the presence of PrnF) at various concentrations of PrnF were plotted against the reciprocals of PrnF concentrations. All measurements were carried out in 50 mM Tris-HCl buffer (pH 7.0) at 25°C.
Kinetic parameters of PrnF in the presence of the PrnD component.
The mechanism of flavin transfer could occur either through a diffusion mechanism or by direct flavin transfer involving protein interactions between the reductase and oxygenase components. Steady-state kinetic analysis was performed to determine if the kinetic parameters of PrnF were altered in the presence of PrnD (10, 26). For these experiments, either NADH or FAD was held constant at saturating levels while varying the alternate PrnF substrate in the presence of PrnF (0.01 mM), PrnD (0.01 mM), and saturating pABA substrate. Specific-activity data with various concentrations of FAD at the fixed concentration of NADH (10-fold greater than the corresponding Km) were fitted with the Michaelis-Menten equation to give a Km value for FAD of 39.2 μM and a kcat value of 70.6 s−1 (23). A 12-fold increase in the Km value for FAD in the presence of PrnD and the pABA substrate suggests that the reaction mechanism of PrnF is modified.
Protein-protein interaction between PrnF and PrnD.
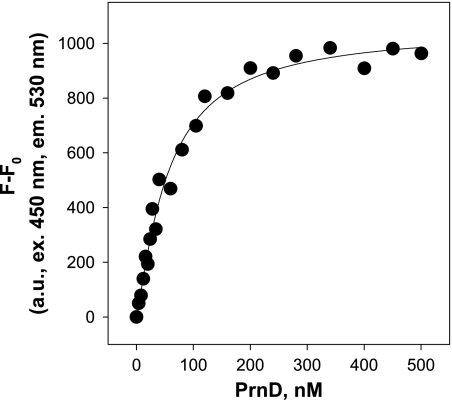
To verify whether direct contact between the flavin reductase PrnF and the oxygenase PrnD is necessary, the two protein components were separated by a dialysis membrane permeable to compounds smaller than 12 kDa (12). Under these conditions, oxygenation of pABA occurred only with about 5% (4.9 U/mg protein compared to 95 U/mg protein) of the efficiency seen under normal conditions. In addition, the binding parameters of FAD-bound PrnF for PrnD were determined. The increase of FAD-bound PrnF emission at 530 nm was examined upon titration with increasing concentrations of PrnD. The fluorescence emission of FAD-bound PrnF was increased by increasing concentrations of PrnD. Upon excitation of FAD-bound PrnF at 450 nm, the binding curve of PrnD to PrnF demonstrated saturation binding (Fig. 4). The number of binding sites was resolved by fitting the binding curve to a Hill plot, as described in Materials and Methods. The Hill plot yielded an n value of 1.20, i.e., one binding site with a Kd value of 56 nM. Taken together, measurement of the increase of FAD-bound PrnF fluorescence by PrnD binding indicated that PrnF bound PrnD with nanomolar affinity at one binding site. The complex formation of PrnD and PrnF was further demonstrated by gel filtration studies. PrnD and PrnF were mixed together in Tris-HCl buffer (pH 7.0) and subjected to gel filtration chromatography. Gel filtration studies showed that PrnD comigrated with PrnF as a complex in solution. The apparent molecular mass of the PrnD-PrnF complex appeared to be approximately 120 kDa, as assessed from the elution time of the protein complex peak compared with that of the protein standards, confirming a 1:1 stoichiometry for binding of the two proteins, PrnD (86 kDa) and PrnF (39 kDa), to form a catalytic complex.
FIG. 4.
Fluorometric titrations of FAD-bound PrnF with PrnD. The binding curve was plotted as the PrnD concentration versus the increase in flavin fluorescence of FAD-bound PrnF. FAD-bound PrnF (400 nM) was titrated with various concentrations of PrnD (4 to 500 nM). Flavin fluorescence emissions at 530 nm were measured using 450-nm excitation. All measurements were carried out in 50 mM Tris-HCl buffer (pH 7.0) at 25°C.
In combination with the change in the kinetic parameter in the presence of PrnD, it can be concluded that direct protein-protein interaction between the reductase PrnF and the oxygenase PrnD is necessary for electron transfer or oxygenation of arylamine. All these data suggest that PrnF reduces FAD to FADH2, which is then directly transferred to PrnD, where it is used by PrnD to catalyze the oxygenation of arylamine. Since the PrnD oxygenase component does require a direct interaction with the PrnF reductase component to oxygenate arylamine, other flavin reductases present in the host cell would not supplant the role of PrnF. Indeed, the turnover rate of PrnD in the presence of PrnF (kcat = 11.3 min−1) was almost two times higher than that in the presence of the E. coli flavin SsuE reductase (kcat = 6.8 min−1). And even after compartmentation, PrnD showed ∼70% oxygenation of pABA in the presence of SsuE, in contrast to that in the presence of PrnF, indicating that the interaction between PrnD and SsuE is not necessary.
Coexpression of prnF with prnD in E. coli.
The pACYC-prnF plasmid was constructed as described in Materials and Methods. The low-copy-number plasmid pACYC is compatible with plasmids of the Col-E1 group due to its P15A replicon (6). This pACYC-prnF plasmid construct was transformed into the recombinant strain harboring pTKXb-prnD engineered for expression of a recombinant E. coli PrnD protein (23). The gene prnF was coexpressed with prnD in E. coli. Resting cells of E. coli BL21(DE3) carrying both prnF and prnD exhibited about 15-fold-higher N oxygenation activity than that of E. coli BL21(DE3) cells carrying only prnD. This result indicates that coexpression of prnF with prnD is critical for high N oxygenation ability and the background reductases are not enough for the optimum PrnD activity.
Characterization of ORF2.
The deduced product of orf2 showed similarities to many FAD-dependent oxidoreductases, such as ABB08681 from Burkholderia sp. strain 383, EAN18270 from Frankia sp. strain EAN1pec, and EAM75868 from Kineococcus radiotolerans SRS30216, which suggested the possibility of the deduced product of orf2 as the second candidate reductase for the two-component arylamine oxygenase system. In order to check this possibility, orf2 was cloned and heterologously expressed in E. coli. The purified oxidoreductase enzyme in combination with PrnD N-oxygenated pABA with a less than 10% conversion rate, supporting the hypotheses that the PrnF flavin reductase is the best candidate for coupling with the PrnD oxygenase component and that the protein-protein interaction between the PrnD and PrnF components is unusual. Indeed, the N-oxygenating activity of PrnD was only slightly affected even after it was separated from the ORF2 oxidoreductase by a membrane, indicating the reduced flavin can be released from the ORF2 oxidoreductase and then diffused into the active site of PrnD.
DISCUSSION
Given the prominence of arylamine oxygenation among natural aromatic products, there has been intense interest in understanding the mechanisms by which an arylnitro functional group is incorporated during natural product biosynthesis (4, 19, 24, 42). The first flavin-dependent arylamine oxygenase was recently characterized from a strain producing pyrrolnitrin, P. fluorescens Pf-5 (23). Despite the many representatives of this N-oxygenase class and its role in the biosynthesis of important N-oxygenated aromatic natural products, only PrnD has been characterized (23, 24). Our previous work on PrnD has shown several basic features of this oxygenase (23, 24). First, a separate flavin reductase is required to catalyze the initial reduction of flavin by NAD(P)H. Second, it depends on flavin cofactors in the reduced state. A third feature is the requirement for molecular oxygen. In the current work, we established the requirement for a separate flavin reductase to provide reduced FADH2 for the PrnD oxygenase and identified the PrnF flavin reductase, in analogy with two-component flavin monooxygenase systems (18, 20, 32).
Based on the analysis of the genome sequence of P. fluorescens Pf-5, a flavin reductase-encoding gene (prnF) was proposed as encoding a flavin reductase coupling with the arylamine N-oxygenase (PrnD) involved in pyrrolnitrin biosynthesis. The identified prnF gene was cloned from P. fluorescens Pf-5 and overexpressed in E. coli, and it was confirmed that this gene product, PrnF, exhibited flavin reductase activity. Since the prnF gene is located in close proximity to the gene encoding PrnD on the P. fluorescens Pf-5 chromosome (Fig. 2), it appears likely that the function of the PrnF protein is to provide reduced FAD to PrnD. This hypothesis is supported by the fact that a mixture of PrnF and PrnD efficiently catalyzes the oxygenation of arylamine to the arylnitro compound. Since O18 incorporation experiments strongly support that hydroxylamine and nitroso compounds are the intermediates in pyrrolnitrin biosynthesis (24), it is reasonable to assume that both the prnF and prnD gene products are involved in the consecutive monooxygenation for pyrrolnitrin biosynthesis as a novel two-component monooxygenase system. In addition, coexpression of prnF with the N-oxygenation gene prnD from P. fluorescens Pf-5 was critical for high N-oxygenating activity, resulting in about 15-fold-higher N-oxygenating activity than that of E. coli BL21 carrying only prnD. The low N-oxygenation activity of E. coli BL21 carrying only prnD was attributed to the low level and inefficiency of the native flavin reductases existing in E. coli, such as Fre (11) and NADPH-sulfite oxidoreductase (7), two reported housekeeping flavin reductases in E. coli.
Flavin frequently acts as an electron carrier between NADH and O2. In two-component flavin oxygenases, FADH2 shuttles electrons from a reductase that oxidizes NAD(P)H to a second protein. However, the use of two proteins, one to generate FADH2 and the other to catalyze oxidation of the substrate, creates the liability that diffusing FADH2 will be intercepted by O2 adventitiously. Thus, reduced flavin transfer between the flavin reductase and the oxygenase component should be tightly controlled. Direct flavin transfer from the flavin reductase to the monooxygenase enzyme through protein-protein interactions would protect the flavin from oxidation. Only two examples of protein-protein interactions in the two-component oxygenase family have been identified, between Vibrio harveyi luciferase and NADPH-preferring flavin reductase (Frp) (26) and between E. coli SsuD and SsuE (1). Thus, the PrnF/PrnD two-component system is the third example exhibiting protein-protein interactions for the transfer of reduced flavins.
The flavin:NAD(P)H reductases can be classified into several families according to their sequence similarities and biochemical properties. One of these families (class I) is comprised of the flavoprotein reductases that contain a tightly bound flavin as a prosthetic group, e.g., the sulfite reductase from E. coli (7) and the Frp reductase from Vibrio harveyi (25). Another family (class II) is represented by those enzymes that do not contain a flavin as a prosthetic group and thus cannot be considered flavoproteins. Instead, they use flavins as substrates, with rather broad substrate specificity. The PrnF flavin reductase from P. fluorescens Pf-5 does not show the yellow color and fluorescence typical of flavin-containing enzymes, suggesting that this enzyme belongs to class II. A homology search shows that PrnF has significant sequence identity (up to 31.5%) with the class II flavin reductase components of the two-component oxygenases PheA2, SnaC, VlmR, ActVB, and HpaC. These proteins and PrnF all exist as homodimers and utilize flavins as substrates rather than as cofactors. However, PrnF is unique in that it prefers FAD and is specific for NADH. In addition, PrnF is believed to transfer electrons to the oxygenase component via protein-protein interactions. No sequence similarities were found between any other known flavoproteins and PrnF.
In summary, the prnF gene encoding a flavin:NADH reductase was cloned from the pyrrolnitrin producer P. fluorescens Pf-5 and overexpressed in soluble form. The purified PrnF protein exhibited the expected flavin reductase enzymatic activity, thereby confirming the identity of the protein. The proximity of the prnF gene to the prnD gene, coding for aminopyrrolnitrin oxygenase, on the P. fluorescens Pf-5 chromosome and evidence from enzymology and bioinformatics experiments strongly suggest that these two genes are components of a two-component arylamine oxygenase system. Compared to other known two-component oxygenases, this PrnD/PrnF system has several unique features. (i) This system catalyzes the unusual arylamine oxidation. With the completion of this work, this system now represents the only characterized example of oxidoreductases involved in the conversion of arylamines to arylnitro compounds. (ii) Compared to other reductases involved in two-component oxygenases, PrnF is unique in that it prefers FAD and is specific for NADH. (iii) PrnF transfers electrons to PrnD through protein-protein interactions via reduced flavins, representing the third example involved in such an electron transfer mechanism. The successful overexpression and characterization of the PrnD/PrnF two-component oxygenase allows us to complete the functional characterization of all the genes for arylamine oxygenation in P. fluorescens Pf-5 and now sets the stage for more-detailed investigations of this novel two-component arylamine oxygenase system, such as X-ray crystallography and protein-protein interaction studies.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Office of Naval Research (N00014-02-1-0725) and partially supported by the Institute of Biomedical Science and Technology, Konkuk University.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdurachim, K., and H. R. Ellis. 2006. Detection of protein-protein interactions in the alkanesulfonate monooxygenase system from Escherichia coli. J. Bacteriol. 188:8153-8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, D., T. Schrader, and J. R. Andreesen. 1997. Two-component flavin-dependent pyrrole-2-carboxylate monooxygenase from Rhodococcus sp. Eur. J. Biochem. 249:739-747. [DOI] [PubMed] [Google Scholar]
- 3.Beltrametti, F., A. M. Marconi, G. Bestetti, C. Colombo, E. Galli, M. Ruzzi, and E. Zennaro. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl. Environ. Microbiol. 63:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buddha, M. R., T. Tao, R. J. Parry, and B. R. Crane. 2004. Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase. J. Biol. Chem. 279:49567-49570. [DOI] [PubMed] [Google Scholar]
- 5.Burkhead, K. D., D. A. Schisler, and P. J. Slininger. 1994. Pyrrolnitrin production by biological-control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 60:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from P15a cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coves, J., V. Niviere, M. Eschenbrenner, and M. Fontecave. 1993. NADPH-sulfite reductase from Escherichia coli—a flavin reductase participating in the generation of the free-radical of ribonucleotide reductase. J. Biol. Chem. 268:18604-18609. [PubMed] [Google Scholar]
- 8.Duffner, F. M., and R. Muller. 1998. A novel phenol hydroxylase and catechol 2,3-dioxygenase from the thermophilic Bacillus thermoleovorans strain A2: nucleotide sequence and analysis of the genes. FEMS Microbiol. Lett. 161:37-45. [DOI] [PubMed] [Google Scholar]
- 9.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 10.Filisetti, L., M. Fontecave, and V. Niviere. 2003. Mechanism and substrate specificity of the flavin reductase ActVB from Streptomyces coelicolor. J. Biol. Chem. 278:296-303. [DOI] [PubMed] [Google Scholar]
- 11.Fontecave, M., R. Eliasson, and P. Reichard. 1987. NAD(P)H-flavin oxidoreductase of Escherichia-coli—a ferric iron reductase participating in the generation of the free-radical of ribonucleotide reductase. J. Biol. Chem. 262:12325-12331. [PubMed] [Google Scholar]
- 12.Galan, B., E. Diaz, M. A. Prieto, and J. L. Garcia. 2000. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182:627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gisi, M. R., and L. Y. Xun. 2003. Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:flavin adenine dinucleotide oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Bacteriol. 185:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer, P. E., D. S. Hill, S. T. Lam, K. H. VanPee, and J. M. Ligon. 1997. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 63:2147-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopwood, D. A., M. J. Bibb, K. F. Chater, G. R. Janssen, F. Malpartida, and C. Smith. 1986. Regulation of gene expression in antibiotic-producing Streptomyces. Cambridge University Press, Cambridge, United Kingdom.
- 16.Ingelman, M., S. Ramaswamy, V. Niviere, M. Fontecave, and H. Eklund. 1999. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry 38:7040-7049. [DOI] [PubMed] [Google Scholar]
- 17.Jeffers, C. E., J. C. Nichols, and S. C. Tu. 2003. Complex formation between Vibrio harveyi luciferase and monomeric NADPH:FMN oxidoreductase. Biochemistry 42:529-534. [DOI] [PubMed] [Google Scholar]
- 18.Kendrew, S. G., S. E. Harding, D. A. Hopwood, and E. N. G. Marsh. 1995. Identification of a flavin-NADH oxidoreductase involved in the biosynthesis of actinorhodin—purification and characterization of the recombinant enzyme. J. Biol. Chem. 270:17339-17343. [DOI] [PubMed] [Google Scholar]
- 19.Kers, J. A., M. J. Wach, S. B. Krasnoff, J. Widom, K. D. Cameron, R. A. Bukhalid, D. M. Gibson, B. R. Crane, and R. Loria. 2004. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429:79-82. [DOI] [PubMed] [Google Scholar]
- 20.Kirchner, U., A. H. Westphal, R. Muller, and W. J. H. van Berkel. 2003. Phenol hydroxylase from Bacillus thermoglucosidasius A7, a two-protein component monooxygenase with a dual role for FAD. J. Biol. Chem. 278:47545-47553. [DOI] [PubMed] [Google Scholar]
- 21.Knobel, H. R., T. Egli, and J. R. vanderMeer. 1996. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J. Bacteriol. 178:6123-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert, B., F. Leyns, L. Vanrooyen, F. Gossele, Y. Papon, and J. Swings. 1987. Rhizobacteria of maize and their antifungal activities. Appl. Environ. Microbiol. 53:1866-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. K., M. Simurdiak, and H. M. Zhao. 2005. Reconstitution and characterization of aminopyrrolnitrin oxygenase, a Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 280:36719-36728. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. K., and H. M. Zhao. 2006. Mechanistic studies on the conversion of arylamine to arylnitro compounds by aminopyrrolnitrin oxygenase: identification of intermediates and kinetic studies. Angew. Chem. Int. Ed. 45:622-625. [DOI] [PubMed] [Google Scholar]
- 25.Lei, B. F., M. Y. Liu, S. Q. Huang, and S. C. Tu. 1994. Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing, and overexpression of the gene and purification and characterization of the cloned enzyme. J. Bacteriol. 176:3552-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei, B. F., and S. C. Tu. 1998. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry 37:14623-14629. [DOI] [PubMed] [Google Scholar]
- 27.Mandel, M. 1966. Deoxyribonucleic acid base composition in the genus Pseudomonas. J. Gen. Microbiol. 43:273-292. [DOI] [PubMed] [Google Scholar]
- 28.Martin, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 29.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto, K., K. Hofstetter, M. Rothlisberger, B. Witholt, and A. Schmid. 2004. Biochemical characterization of StyAB from Pseudomonas sp. strain VLB120 as a two-component flavin-diffusible monooxygenase. J. Bacteriol. 186:5292-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palfey, B. A., D. P. Ballou, and V. Massey. 1995. Oxygen activation by flavins and pterins, p. 37-83. In J. S. Valentine, C. S. Foote, A. Greenburg, and J. F. Lieberman (ed.), Active oxygen: reactive oxygen species in biochemistry. Blackie Academic & Professional, London, United Kingdom.
- 32.Parry, R. J., and W. Y. Li. 1997. An NADPH:FAD oxidoreductase from the valanimycin producer, Streptomyces viridifaciens—cloning, analysis, and overexpression. J. Biol. Chem. 272:23303-23311. [DOI] [PubMed] [Google Scholar]
- 33.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. A. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. H. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. W. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson, L. S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne, J. W., H. Bolton, J. A. Campbell, and L. Y. Xun. 1998. Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1. J. Bacteriol. 180:3823-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrescu, A. D., R. Hertz, J. Bar-Tana, F. Schroeder, and A. B. Kier. 2002. Ligand specificity and conformational dependence of the hepatic nuclear factor-4 alpha (HNF-4 alpha). J. Biol. Chem. 277:23988-23999. [DOI] [PubMed] [Google Scholar]
- 36.Pfender, W. F., J. Kraus, and J. E. Loper. 1993. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici repentis in wheat-straw. Phytopathology 83:1223-1228. [Google Scholar]
- 37.Simurdiak, M., J. K. Lee, and H. M. Zhao. 2006. A new class of arylamine oxygenases: evidence that p-aminobenzoate N-oxygenase (AurF) is a di-iron enzyme and further mechanistic studies. ChemBioChem 7:1169-1172. [DOI] [PubMed] [Google Scholar]
- 38.Tang, C. K., C. E. Jeffers, J. C. Nichols, and S. C. Tu. 2001. Flavin specificity and subunit interaction of Vibrio fischeri general NAD(P)H-flavin oxidoreductase FRG/FRase I. Arch. Biochem. Biophys. 392:110-116. [DOI] [PubMed] [Google Scholar]
- 39.Thibaut, D., N. Ratet, D. Bisch, D. Faucher, L. Debussche, and F. Blanche. 1995. Purification of the 2-enzyme system catalyzing the oxidation of the d-proline residue of pristinamycin-Iib during the last step of pristinamycin-Iia biosynthesis. J. Bacteriol. 177:5199-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu, S. C. 1982. Isolation and properties of bacterial luciferase intermediates containing different oxygenated flavins. J. Biol. Chem. 257:3719-3725. [PubMed] [Google Scholar]
- 41.Vanpee, K. H., O. Salcher, and F. Lingens. 1980. Formation of pyrrolnitrin and 3-(2-amino-3-chlorophenyl)pyrrole from 7-chlorotryptophan. Angew. Chem. Int. Ed. 19:828-829. [Google Scholar]
- 42.Winkler, R., and C. Hertweck. 2005. Sequential enzymatic oxidation of aminoarenes to nitroarenes via hydroxylamines. Angew. Chem. Int. Ed. 44:4083-4087. [DOI] [PubMed] [Google Scholar]
- 43.Witschel, M., S. Nagel, and T. Egli. 1997. Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103. J. Bacteriol. 179:6937-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi, L., C. H. Squires, D. J. Monticello, and J. D. Childs. 1997. A flavin reductase stimulates DszA and DszC proteins of Rhodococcus erythropolis IGTS8 in vitro. Biochem. Biophys. Res. Commun. 230:73-75. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Y. R., M. W. Mortimer, T. S. Fisher, M. L. Kahn, F. J. Brockman, and L. Y. Xun. 1997. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH:flavin mononucleotide oxidoreductase. J. Bacteriol. 179:1112-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xun, L. Y., and E. R. Sandvik. 2000. Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xun, L. Y., and C. M. Webster. 2004. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Bacteriol. 279:6696-6700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.