Abstract
Leucyl-tRNA synthetase (LeuRS) has evolved an editing function to clear misactivated amino acids. An Escherichia coli-based assay was established to identify amino acids that compromise the fidelity of LeuRS and translation. Multiple nonstandard as well as standard amino acids were toxic to the cell when LeuRS editing was inactivated.
In the first step of protein synthesis, an aminoacyl-tRNA synthetase (aaRSs) is responsible for linking a single standard amino acid to its correct set of tRNA isoacceptors (11, 20). About half of the family of 20 aaRSs are challenged to distinguish among closely related amino acids and have evolved amino-acid-editing mechanisms to clear their mistakes (10). These editing mechanisms maintain translational fidelity by impeding the production of “statistical proteins,” which is hazardous to cell viability.
The fidelity of most aaRSs that edit is threatened by a limited number of noncognate standard amino acids. For example, isoleucyl-tRNA synthetase (IleRS) must distinguish between isoleucine and valine, which differ by a single missing methyl group (2, 5, 6, 9, 22, 23). Valyl-tRNA synthetase (ValRS) editing targets threonine, which has a hydroxyl group that is isosteric to the methyl moiety in valine (7, 8). Previously, we determined that leucyl-tRNA synthetase (LeuRS), which is homologous to IleRS and ValRS, misactivates a wide array of amino acids in vitro (19). Thus, we hypothesized that numerous structurally diverse amino acids might compete effectively for binding in the larger leucine-binding pocket of LeuRS (17) for aminoacylation to tRNALeu.
We sought to determine which of the amino acids that were misactivated by LeuRS might be detrimental to the cell if the editing activity of LeuRS was dysfunctional. This would also provide insight into which amino acids were actually targeted by the LeuRS editing activity in vivo to maintain the fidelity of translation. We employed the editing-defective TT/VV LeuRS mutant strain of Escherichia coli (26) to investigate the intracellular toxicity of LeuRS-misactivated amino acids. This mutated LeuRS has two conserved threonines that have been replaced by valines in the editing-active site (26). Aminoacylation is unaffected, but hydrolytic editing activity is abolished to stably produce mischarged Ile-tRNALeu in vitro.
We cotransformed the E. coli strain KL231, which has a temperature-sensitive LeuRS mutation (18) with plasmid pGP1-2, which carries the gene for T7 RNA polymerase (25), and plasmid pYZHAI3, which expresses the editing-defective TT/VV LeuRS mutant (26). Transformants were selected as described previously (12, 18). The editing-defective TT/VV LeuRS mutant enzyme as well as a wild-type LeuRS control (p15ec3-1 [19]) complemented the temperature-sensitive strain at 42°C (Fig. 1). In vitro aminoacylation assays indicated that mischarging by the TT/VV LeuRS mutant was slightly increased at 42°C compared to that at 30°C (data not shown), which could increase cell sensitivity to the editing defect at the higher temperature.
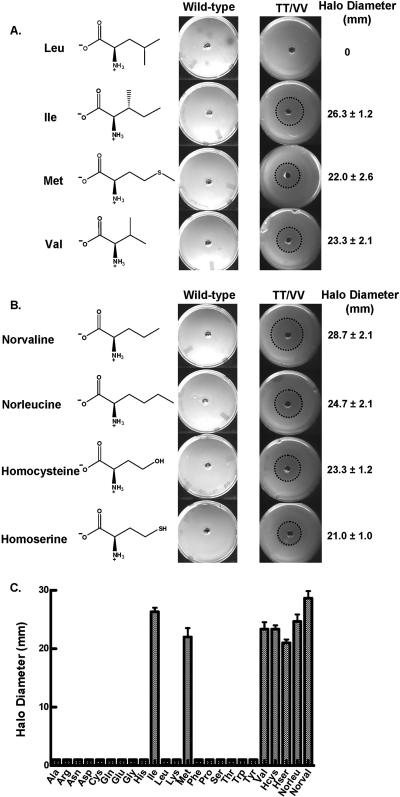
FIG. 1.
Amino acid toxicities to E. coli KL231 cells that were dependent on the editing-defective LeuRS TT/VV mutation. Halo assays were carried out as described previously (12). (A) Standard aliphatic amino acids. (B) Nonstandard aliphatic amino acids. (C) Histogram representing the halo diameters for all tested amino acids. Zones of inhibition or halos are marked by dashed circles. Standard deviations were based on the assay repeated at least in triplicate.
Previously, we showed that excess isoleucine was inhibitory to the growth of E. coli bacteria that were dependent on an editing-defective LeuRS (12). Herein, we expanded our toxicity studies to include all of the standard amino acids to identify those amino acids that threaten LeuRS fidelity in vivo. A zone of growth inhibition, or a halo, around a central well that contained concentrated nonleucine amino acids showed that among the standard amino acids, isoleucine, valine, and methionine were toxic to E. coli cells that were dependent on the editing-defective TT/VV LeuRS mutant for aminoacylation (Fig. 1A). We also tested a series of nonstandard amino acids that accumulate in metabolic pathways or act as signaling molecules within bacteria (3, 13-15, 24). Norvaline, norleucine, homoserine, and homocysteine yielded significant halos (Fig. 1B). In contrast, cells that were complemented by the wild-type LeuRS failed to produce a halo in the presence of high concentrations (near the limits of saturation) of any of these non-leucine amino acids. This demonstrates that the robust posttransfer editing function of LeuRS protects the cell from potential amino acid toxicities.
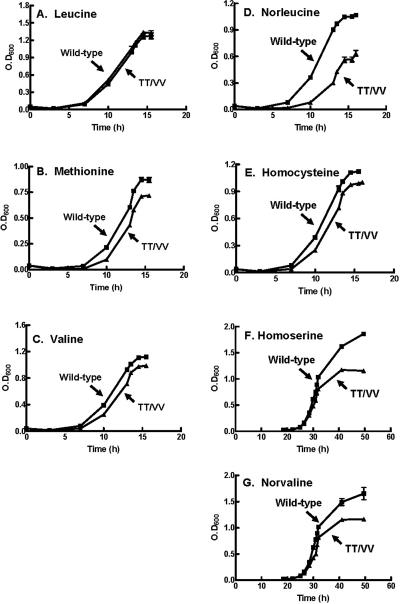
We also investigated the cell growth of E. coli KL231 cells harboring either the wild type or the editing-defective LeuRS TT/VV mutation in liquid minimal medium cultures. When the LeuRS TT/VV mutation was present, excess methionine or valine slowed growth rates in comparison to that of the cells complemented by the wild-type LeuRS (Fig. 2). In addition, these cells reached a lower plateau, indicating that growth was stunted, presumably as statistical protein mutations accumulated within the cell due to the LeuRS editing defect (Fig. 2). Likewise, liquid growth cultures showed that the nonstandard amino acids norleucine, norvaline, homocysteine, and homoserine reduced growth rates and lowered plateaus when E. coli was dependent on the editing-defective LeuRS (Fig. 2). These changes in growth patterns varied for different amino acids and between experiments due to the statistical nature of the accumulation of errors during protein synthesis.
FIG. 2.
Growth curves E. coli KL231 in the presence of toxic levels of nonleucine amino acids. Transformed E. coli KL231 was prepared, and its growth rates were measured as previously described (12). E. coli KL231 harboring wild-type (▪) or the TT/VV editing-defective mutant (▴) LeuRS was grown in liquid minimal medium that contained 16 mM of the following amino acids: (A) leucine, (B) methionine, (C) valine, (D) norleucine, (E) homocysteine, (F) homoserine, and (G) norvaline. Error bars are based on assays that were repeated in triplicate and are present for each point.
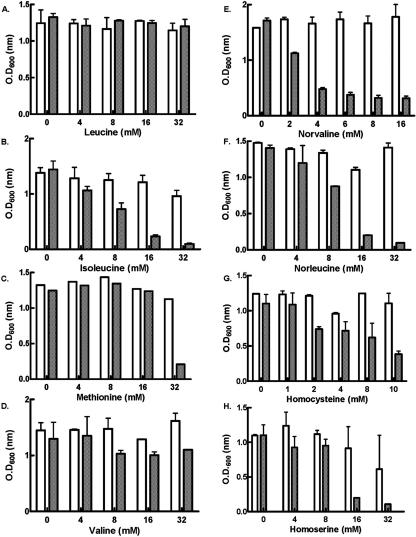
We carried out a dose-dependent analysis of the amino acids that were toxic to E. coli KL231 in the presence of an editing-defective LeuRS (Fig. 3). We also determined the 50% inhibitory concentration (IC50) values (± standard deviations) for the amino acids to quantitate their toxicities, as follows: 27.0 ± 4.7 mM for isoleucine, 36.6 ± 4.2 mM for methionine, 46.3 ± 25 mM for valine, and 14.1 ± 5.1 mM for homoserine. The unbranched, aliphatic norvaline and norleucine exhibited the most potent toxicities, with IC50 values of 4.2 ± 2.8 mM and 8.2 ± 0.5 mM, respectively. By comparison, the standard amino acid isoleucine had an IC50 value of 27.0 ± 4.7 mM. Despite repeated attempts, a reproducible IC50 value for homocysteine could not be measured because of heterogeneity in the sample and low solubility. Likewise, the IC50 value for valine varied significantly in repeated experiments because its IC50 value was high and approached the limits of solubility under the conditions used. In addition, potential IC50 values for effects on the wild-type enzyme could not be measured because of the relatively low solubility of each of these aliphatic amino acids.
FIG. 3.
Amino acid dose-dependent viability decrease of E. coli KL231 that is dependent on the wild type or the editing-defective TT/VV mutant LeuRS. E. coli KL231 harboring wild-type (open bars) or the TT/VV editing-defective mutant (solid bars) LeuRS was grown in liquid minimal medium containing increasing concentrations as indicated for the following amino acids: (A) leucine, (B) isoleucine, (C) methionine, (D) valine, (E) norvaline, (F) norleucine, (G) homocysteine, and (H) homoserine. Optical densities at 600 nm (OD600) were measured in triplicate after 24 h of growth to obtain standard deviations.
Overall, these combined results suggest that nonstandard amino acids might be a greater threat to the LeuRS-dependent fidelity of protein synthesis than standard amino acids. In the case of norvaline, it has been shown that intracellular levels of norvaline are low compared to those of leucine but can be significantly increased under conditions that induce high expression of recombinant proteins in E. coli (1). Interestingly, norvaline has been shown to at least partially bypass the LeuRS editing mechanism and to substitute for leucine under cell growth conditions that require high expression of recombinant proteins in E. coli (1). However, our results emphasize that norvaline is subject to LeuRS amino acid editing at levels that control its toxicity.
Our investigation supports the possibility that LeuRS has the complicated challenge of discriminating between multiple standard and nonstandard amino acids in vivo. As with other aaRSs (4, 21), LeuRS is required to block amino acid toxicity to the cell with high translational fidelity. While some aaRSs can achieve this level of discrimination within a single aminoacylation-active site to maintain accurate protein synthesis, LeuRS and other aaRSs have acquired a second active hydrolytic site to edit misactivated amino acids (10). Inactivation of the LeuRS editing activity clearly hinders cell growth. This is likely due to a steady accumulation of errors during translation that would yield misfolded and/or inactivated proteins (16). As these statistically generated protein mutations accumulate, intracellular processes would be compromised to lower cell viability and survival. These high fidelity requirements of LeuRS for protein synthesis are consistent with the early acquisition of its editing domain during evolution (27).
Acknowledgments
This work was supported by the National Institutes of Health (GM63789).
We thank A. M. Williams for technical assistance and advice.
Footnotes
Published ahead of print on 21 September 2007.
REFERENCES
- 1.Apostol, I., J. Levine, J. Lippincott, J. Leach, E. Hess, C. B. Glascock, M. J. Weickert, and R. Blackmore. 1997. Incorporation of norvaline at leucine positions in recombinant human hemoglobin expressed in Escherichia coli. J. Biol. Chem. 272:28980-28988. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A. N., and P. Berg. 1966. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem. 241:839-845. [PubMed] [Google Scholar]
- 3.Bogosian, G., B. N. Violand, E. J. Dorward-King, W. E. Workman, P. E. Jung, and J. F. Kane. 1989. Biosynthesis and incorporation into protein of norleucine by Escherichia coli. J. Biol. Chem. 264:531-539. [PubMed] [Google Scholar]
- 4.Döring, V., H. D. Mootz, L. A. Nangle, T. L. Hendrickson, V. de Crécy-Lagard, P. Schimmel, and P. Marliere. 2001. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science 292:501-504. [DOI] [PubMed] [Google Scholar]
- 5.Eldred, E. W., and P. R. Schimmel. 1972. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247:2961-2964. [PubMed] [Google Scholar]
- 6.Fersht, A. R. 1977. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16:1025-1030. [DOI] [PubMed] [Google Scholar]
- 7.Fersht, A. R., and C. Dingwall. 1979. Establishing the misacylation/deacylation of the tRNA pathway for the editing mechanism of prokaryotic and eukaryotic valyl-tRNA synthetases. Biochemistry 18:1238-1245. [DOI] [PubMed] [Google Scholar]
- 8.Fukai, S., O. Nureki, S. Sekine, A. Shimada, J. Tao, D. G. Vassylyev, and S. Yokoyama. 2000. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell 103:793-803. [DOI] [PubMed] [Google Scholar]
- 9.Fukunaga, R., and S. Yokoyama. 2006. Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J. Mol. Biol. 359:901-912. [DOI] [PubMed] [Google Scholar]
- 10.Hendrickson, T. L., V. de Crécy-Lagard, and P. Schimmel. 2004. Incorporation of nonnatural amino acids into proteins. Annu. Rev. Biochem. 73:147-176. [DOI] [PubMed] [Google Scholar]
- 11.Ibba, M., and D. Söll. 2000. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69:617-650. [DOI] [PubMed] [Google Scholar]
- 12.Karkhanis, V. A., M. T. Boniecki, K. Poruri, and S. A. Martinis. 2006. A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J. Biol. Chem. 281:33217-33225. [DOI] [PubMed] [Google Scholar]
- 13.Kisumi, M., M. Sugiura, and I. Chibata. 1976. Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J. Biochem. (Tokyo) 80:333-339. [DOI] [PubMed] [Google Scholar]
- 14.Kisumi, M., M. Sugiura, J. Kato, and I. Chibata. 1976. L-Norvaline and L-homoisoleucine formation by Serratia marcescens. J. Biochem. (Tokyo) 79:1021-1028. [DOI] [PubMed] [Google Scholar]
- 15.Lazzerini, P. E., P. L. Capecchi, E. Selvi, S. Lorenzini, S. Bisogno, M. Galeazzi, and F. Laghi Pasini. 2007. Hyperhomocysteinemia, inflammation and autoimmunity. Autoimmun. Rev. 6:503-509. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. W., K. Beebe, L. A. Nangle, J. Jang, C. M. Longo-Guess, S. A. Cook, M. T. Davisson, J. P. Sundberg, P. Schimmel, and S. L. Ackerman. 2006. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443:50-55. [DOI] [PubMed] [Google Scholar]
- 17.Lincecum, T. L., Jr., and S. A. Martinis. 2000. The tRNA synthetase proofreading and editing active sites: a novel antibiotic target. SAAS Bull. Biochem. Biotechnol. 13:25-33. [Google Scholar]
- 18.Low, B., F. Gates, T. Goldstein, and D. Söll. 1971. Isolation and partial characterization of temperature-sensitive Escherichia coli mutants with altered leucyl- and seryl-transfer ribonucleic acid synthetases. J. Bacteriol. 108:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinis, S. A., and G. E. Fox. 1997. Non-standard amino acid recognition by Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Symp. Ser. 36:125-128. [PubMed] [Google Scholar]
- 20.Martinis, S. A., and P. Schimmel. 1992. Aminoacyl-tRNA synthetases, p. 887-901. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), General features and relationships in Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 21.Nangle, L. A., V. De Crécy Lagard, V. Doring, and P. Schimmel. 2002. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 277:45729-45733. [DOI] [PubMed] [Google Scholar]
- 22.Nureki, O., D. G. Vassylyev, M. Tateno, A. Shimada, T. Nakama, S. Fukai, M. Konno, T. L. Hendrickson, P. Schimmel, and S. Yokoyama. 1998. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280:578-582. [DOI] [PubMed] [Google Scholar]
- 23.Schreier, A. A., and P. R. Schimmel. 1972. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry 11:1582-1589. [DOI] [PubMed] [Google Scholar]
- 24.Steindler, L., and V. Venturi. 2007. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266:1-9. [DOI] [PubMed] [Google Scholar]
- 25.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai, Y., and S. A. Martinis. 2005. Two conserved threonines collaborate in the Escherichia coli leucyl-tRNA synthetase amino acid editing mechanism. Biochemistry 44:15437-15443. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, M. W., R. Hao, J. F. Chen, F. Martin, G. Eriani, and E. D. Wang. 2003. Enzymes assembled from Aquifex aeolicus and Escherichia coli leucyl-tRNA synthetases. Biochemistry 42:7694-7700. [DOI] [PubMed] [Google Scholar]