Abstract
Although flavin-dependent ThyX proteins show thymidylate synthase activity in vitro and functionally complement thyA defects in heterologous systems, direct proof of their cellular functions is missing. Using insertional mutagenesis of Rhodobacter capsulatus thyX, we constructed the first defined thyX inactivation mutant. Phenotypic analyses of the obtained mutant strain confirmed that R. capsulatus ThyX is required for de novo thymidylate synthesis. Full complementation of the R. capsulatus thyX::spec strain to thymidine prototrophy required not only the canonical thymidylate synthase ThyA but also the dihydrofolate reductase FolA. Strikingly, we also found that addition of exogenous methylenetetrahydrofolate transiently inhibited the growth of the different Rhodobacter strains used in this work. To rationalize these experimental results, we used a mathematical model of bacterial folate metabolism. This model suggests that a very low dihydrofolate reductase activity is enough to rescue significant thymidylate synthesis in the presence of ThyX proteins and is in agreement with the notion that intracellular accumulation of folates results in growth inhibition. In addition, our observations suggest that the presence of flavin-dependent thymidylate synthase X provides growth benefits under conditions in which the level of reduced folate derivatives is compromised.
The folate cycle plays a central role in cell metabolism. Folate-dependent enzymes are required for methionine synthesis, numerous methylation reactions, and synthesis of purine and pyrimidine nucleotides. As the different loops of the folate cycle are interconnected, a mathematical model of this cycle has been described recently for eukaryotic cells (21). This basic model has the qualitative behavior seen in a variety of experimental studies on folate homeostasis in the cytosol of human cells. Moreover, it predicts that the activities of folate-dependent enzymes depend on the size of the total folate pool in a nonlinear fashion (23). For instance, actively dividing cells require large quantities of the DNA precursor thymidylate (dTMP). In human cells, the thymidylate synthase ThyA (EC 2.1.1.45) catalyzes the reductive methylation of dUMP to dTMP, using 5,10-methylenetetrahydrofolate (CH2THF) as a donor of one-carbon units and as a reductant (2). The resulting 7,8-dihydrofolate (DHF) is reduced to tetrahydrofolate (THF) by the dihydrofolate reductase (DHFR) FolA (Fig. 1). As formation of dTMP is rate limiting for DNA replication, in human cells the thyA gene is up-regulated by the transcription factor E2-F (5). This greatly enhances dTMP synthesis, whereas other branches of folate metabolism are scarcely affected (21).
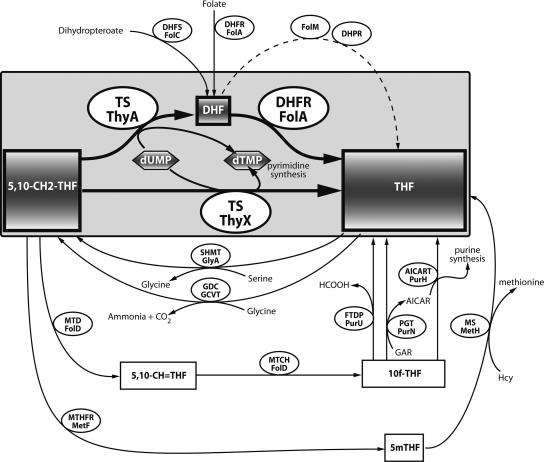
FIG. 1.
Schematic representation of different folate metabolic pathways identified in Bacteria. The ThyX and ThyA pathways for de novo thymidylate synthesis are highlighted by the large gray box. Although both thymidylate synthases produce dTMP, they use different reductive mechanisms. Genetic inactivation of FolA or inhibition of its activity by trimethoprim leads to the accumulation of DHF in ThyA-containing organisms, thus resulting in growth inhibition (see Fig. 5). In contrast, ThyX proteins produce THF directly as a reaction product. GlyA (serine hydroxymethyltransferase) can be found in both ThyX- and ThyA-containing organisms. All the reactions were considered for simulations. Substrates are enclosed in rectangles, and enzymes are enclosed in ellipses. In several cases both the eukaryotic and bacterial abbreviations for the enzymes are shown (redox active compounds participating in many of the reactions are not shown). Abbreviations: 5mTHF, 5-methyltetrahydrofolate; 5,10-CH2-THF, 5,10-methylenetetrahydrofolate; 5,10-CH=THF, 5,10-methenyltetrahydrofolate; 10f-THF, 10-formyltetrahydrofolate; AICART, aminoimidazolecarboxamide ribotide transformylase; TS, thymidylate synthase; MTD, 5,10-methylenetetrahydrofolate dehydrogenase; MTCH, 5,10-methylenetetrahydrofolate cyclohydrolase; PGT, phosphoribosyl glycinamidetransformylase; SHMT, serine hydroxymethyltransferase; MTHFR, 5,10-methylenetetrahydrofolate reductase; MS, methionine synthase; GAR, glycinamide ribotide; AICAR, aminoamidazolecarboxamide ribotide; GDC, glycine decarboxylase; DHPR, dihydropteridine reductase; DHFS, dihydrofolate synthase; GCVT, glycine cleavage system aminomethyltransferase; FTDP, formyltetrahydrofolate deformylase; Hcy, homocysteine.
In contrast to the human thymidylate synthase ThyA, the members of the novel ThyX family of thymidylate synthases (EC 2.1.1.148) are NAD(P)H oxidases that use flavin adenine nucleotide to mediate hydride transfer (1, 10, 11, 20). Therefore, although both ThyA and ThyX catalyze the formation of thymidylate in vitro, their reductive mechanisms are dramatically different (Fig. 1). ThyX catalysis results in the formation of THF, not DHF, as the product of the methylation reaction (12, 19), but virtually nothing is known to date about how the activity of the flavin-dependent thymidylate synthase ThyX influences the different folate-dependent branches of bacterial metabolism. This is of particular interest as, e.g., Mycobacterium and Corynebacterium species contain thyA and thyX genes, but to date why both genes are maintained in these organisms is poorly understood.
Previous genetic studies on thyX either were performed using poorly defined genetic backgrounds (7) or were based upon multicopy heterologous complementation systems using either bacterial (9, 12, 20, 29) or viral (10, 11) thyX genes.
In order to obtain direct evidence for the in vivo role of ThyX enzymes, we inactivated the thyX gene from the purple bacterium Rhodobacter capsulatus by insertion of an antibiotic cartridge. The thyX::Specr mutant obtained was viable only when thymidine was provided in the growth medium, proving that ThyX proteins indeed function in de novo thymidylate synthesis. We also showed that functional complementation of the thyX::Specr mutant requires not only thyA but also folA, whereas earlier studies showed that thyX alone complements a thyA deletion strain. This growth defect in the absence of FolA cannot be rescued by addition of exogenous folates, which unexpectedly results in growth inhibition. Mathematical modeling of folate metabolism for ThyX-containing bacteria in the presence and absence of ThyA indicated that ThyA is a critical determinant of reduced folates in actively dividing bacterial cells. In particular, in organisms with only the thymidylate synthase ThyX, a very low level of DHFR activity is sufficient to rescue significant thymidylate synthesis. These findings suggest that the presence of ThyX proteins is beneficial when the steady-state level of reduced folate molecules is compromised (for instance, upon exposure to antifolates).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains and plasmids used in this study and their relevant characteristics are listed in Table 1. R. capsulatus strains were grown at 35°C under chemoheterotrophic or photoheterotrophic conditions using enriched MPYE medium (3 g peptone per liter, 3 g yeast extract per liter, 1.6 mM MgCl2, 1.0 mM CaCl2) or on Sistrom's minimal medium (27a). For photoheterotrophic growth on solid medium, anaerobic jars and H2/CO2 generator envelopes from BBL GasPak were used. Respiratory doubling times under aerobic growth conditions in liquid medium were calculated from semilogarithmic growth curves obtained at 630 nm. CH2THF and THF were kindly provided by R. Moser (Merck Eprova AG). Where necessary, antibiotics, nucleotides, and folate derivatives were added at the following final concentrations: tetracycline, 2 μg/ml; spectinomycin, 10 μg/ml; rifampin, 75 μg/ml; thymidine, 1 or 50 μg/ml; and folates, 50 or 100 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this work
| Strain, plasmid, or oligonucleotide | Relevant characteristics | Source or reference |
|---|---|---|
| R. capsulatus strains | ||
| MT1131 | Parental strain | F. Daldal |
| DL1 | thyX::Specr | This study |
| R. sphaeroides Ga | crt | F. Daldal |
| E. coli strains | ||
| SURE | e14− (McrA−) Δ(mcrCB-hsdSMR-mrr)171 endA1 supE44 thi-1 gyrA96 relA1 lac recB recJ sbcC umuC::Tn5 (Kanr) uvrC [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| HB101 | F−proA2 hsdS20 recA13 ara-14 lacY xyl-5 galK2 rpsL20 supE44 rplsL20 supE44 proA2 mtl-1 | ATCC |
| S17-1 | thi pro ara recA StrepRP4 2-Tet::Mu-Kan::Tn7 | 27 |
| Plasmids | ||
| pGEM-T Easy | Ampr | Promega |
| pBluescript II KS+ | Ampr | Stratagene |
| pRK415 | Tetr broad-host-range plasmid | 13 |
| pRK2013 | Kanr helper | 6 |
| pHP45Ω | Specr Ampr | 22 |
| pSUP202 | Ampr Cmr Tetr, suicide plasmid | 27 |
| pDL5 | R. capsulatus thyX in pGEM-T Easy | This study |
| pDL7 | 3.5-kb PstI fragment with thyX::Specr allele in pSUP202 | This study |
| pDL9 | 1.6-kb NotI-BglII fragment with thyX gene in pBluescript II KS+ | This study |
| pDL11 | 1.4-kb PstI fragment with R. capsulatus thyX gene in pRK415 | This study |
| pDL12 | 1.7-kb SpeI-EcoRI fragment with R. sphaeroides thyA and folA in pRK415 | This study |
| pDL14 | 1.4-kb XbaI-EcoRI fragment with R. sphaeroides thyA in pRK415 | This study |
| Oligonucleotides | ||
| ORF311-A | 5′-ATGTCGCTTACCCCAGACC | This study |
| ORF311-B | 5′-ACCCCACTCCTCCACAAACT | This study |
| ORF311-C | 5′-CATGCGGTAGGCGGTATAG | This study |
| ORF311-E | 5′-AATCTCCAAGCGTTTGCTTC | This study |
| OLI-SPEC | 5′-CTCGGTTTTCTGGAAGGCGA | This study |
| THYA-DR-A | 5′-CACAACTCCTGGGGATCAAG | This study |
| THYA-DR-B | 5′-GACCTGCATGTTGTCCCTTC | This study |
Chromosomal inactivation of the R. capsulatus thyX gene by insertion mutagenesis.
All oligonucleotides used in this work were obtained from Sigma-Genosys. The plasmid constructs obtained were confirmed by restriction analyses and DNA sequencing. To construct pDL5 (Table 1), the chromosomal region carrying the R. capsulatus strain MT1131 thyX gene was amplified by PCR using primers ORF311-C and ORF311-E (Table 1). The amplified 1.7-kb DNA fragment was inserted into the pGEM-T Easy vector (Promega), and its correct orientation was verified by restriction analyses. The construct was digested with NotI and BglII, and a 1.6-kb fragment containing the R. capsulatus thyX gene was ligated into the NotI and BamHI sites of pBluescript II KS+ (Stratagene) to generate pDL9 (Table 1). A 2-kb BamHI fragment containing an Smr-Specr resistance cartridge from pHP45Ω (22) was inserted into the unique BamHI site of pDL9 in order to interrupt the R. capsulatus thyX gene. A 3.5-kb PstI fragment of the plasmid obtained was subsequently ligated into the suicide vector pSUP202, yielding pDL7.
pDL7, carrying the R. capsulatus thyX::Specr allele, is not capable of replication in Rhodobacter strains. This plasmid was transferred into R. capsulatus strain MT1131 (Rifr Tets Specs) from Escherichia coli S17-1 (Rifs) by diparental conjugation as described previously (4). As inactivation of thyX was expected to result in thymidine auxotrophy, transconjugants were selected for spectinomycin resistance on enriched MPYE solid medium containing thymidine and rifampin added as described above. A total of 235 Specr colonies were screened for tetracycline resistance (single recombination) or sensitivity (double recombination). Fourteen tetracycline-sensitive clones, corresponding to ∼6% of the total number of transconjugants, were maintained. The expected chromosomal location of the spectinomycin resistance cartridge and the absence of the thyX wild-type gene in strain DL1 were confirmed by PCR using primers ORF311-A, ORF311-B, and OLI-SPEC (Fig. 2).
FIG. 2.
Physical maps of the chromosomal regions carrying the thyX gene of R. capsulatus, as well as of the different plasmids used in this work. (A) ORF310, ORF312, and ORF313 are putative genes around R. capsulatus thyX (ORF311). (B) Disruption of R. capsulatus thyX by insertion of an Smr-Specr cartridge. The resistance cartridge inserted at the unique BamHI site is transcribed in the direction opposite that of thyX. (C to E) Schematic representation of plasmids used in the complementation studies, including the approximate locations of the primers used in cloning (see Table 1). Note that R. sphaeroides thyA and folA form a gene cluster and are likely translationally coupled (not shown). R. sph, R. sphaeroides; R. caps, R. capsulatus; S/X, SpeI/XbaI hybrid site.
The pDL11 vector carrying the wild-type allele of R. capsulatus thyX (ORF311) (Fig. 2) was obtained by insertion of a 1.4-kb PstI fragment from pDL9 into the low-copy-number vector pRK415 at the unique PstI site (Table 1). In Rhodobacter sphaeroides, the thyA and folA genes form a gene cluster on the chromosome, which allows simultaneous cloning of them. The R. sphaeroides thyA and folA genes were amplified using primers THYA-DR-A and THYA-DR-B. The 1.7-kb DNA fragment obtained was inserted into pGEM-T Easy (Promega), and its orientation was confirmed by restriction analyses. The construct was digested with SpeI and EcoRI, and a 1.7-kb fragment containing both the thyA and folA genes was ligated into the XbaI and EcoRI sites of pRK415, yielding pDL12. The pDL14 vector carrying only R. sphaeroides thyA was obtained using primers THYA-DR-A and THYA-DR-B. The pRK415 derivatives pDL11, pDL12, and pDL14 were transferred by triparental conjugation from E. coli HB101 (Rifs) into R. capsulatus DL1 (Rifr) with the helper plasmid pRK2013 carrying the RK2 transfer genes (6). Tetr transconjugants were selected on enriched MPYE medium plates in the presence of added thymidine and rifampin.
In vivo labeling of chromosomal DNA with [5,6-3H]uracil.
Isotope labeling experiments with R. capsulatus cells were performed using MPYE liquid medium containing [5,6-3H]uracil (49.0 Ci/mmol; Amersham Biosciences). Radioactively labeled DNA was isolated from early-stationary-phase cultures using cesium chloride gradients. Prior to centrifugation, the refraction indices of the cesium chloride gradients were adjusted to 1.3980, corresponding to a density of 1.6850 g/ml. Uracil incorporated into RNA was not detected, as the gradients under the conditions described separate RNA and DNA.
Dialyzed DNA was hydrolyzed to free nucleobases by boiling it in 50 mM perchloric acid for 1 h. Samples were neutralized with potassium hydroxide, and the free nucleobases were separated by high-performance liquid chromatography (Beckman Gold system) using reverse-phase chromatography (Ultrasphere octyldecyl silane; column dimensions, 4.6 mm by 25 cm). Isocratic elution was performed using 10 mM potassium phosphate buffer (pH 4) at a flow rate of 1 ml/min. Isotope incorporation into deoxycytidine (dC) and deoxythymine (dT) was monitored by using an online radioactive flow detector (LabLogic β-RAM) with FlowLogic SafeScint scintillation liquid. The identities of the radioactive peaks were confirmed by simultaneous monitoring of known nucleobase standards. Data analyses were performed using Laura Light 3 software from LabLogic.
Mathematical model of bacterial folate metabolism.
We have estimated that the intracellular concentration of the total folate pool in Bacteria is approximately 50 μM (17). The general model of Nijhout et al. (21) for mammalian hepatic folate metabolism was modified to make it more appropriate for bacteria. This involved adding several reactions and modifying the kinetic parameters of many of the enzymes involved. The values used for simulations are included in the supplemental material. The reaction scheme that we modeled is shown in Fig. 1 (which shows the features relevant for thymidylate metabolism). We have added a pathway, catalyzed by ThyX, that converts CH2THF to THF. We have also added an additional pathway from DHF to THF catalyzed by dihydropteridine reductase and/or FolM. It was demonstrated that these enzymes have been shown to possess DHFR activity in Thermus thermophilus (28) and E. coli (8), respectively, and are believed to be an alternative pathway from DHF to THF in other bacteria as well. The available data indicate that the catalytic efficiencies of these alternative enzymes with DHFR activity are relatively low (8, 28), as their reported Vmax values appear to correspond to approximately to 10 to 25% of the Vmax values measured for a number of bacterial FolA proteins (see the supplemental material). Our earlier in vitro measurements suggested that the dTMP-forming activity of ThyX proteins is significantly less than that of ThyA proteins. Also shown in Fig. 1 is the glycine cleavage system (modeled as the glycine decarboxylation reaction), since it exists in some bacteria but not in the cytosol of eukaryotic cells. We removed the nonenzymatic conversion of THF to CH2THF from the bacterial model of folate metabolism. Formylmethionyl tRNAfMet is essential for initiation of protein synthesis in bacterial systems but not in eukaryotic systems and cannot be provided exogenously. Consequently, we added a new substrate, 5-formyltetrahydrofolate, and the reactions that interconvert it with 10-formyltetrahydrofolate and methylenetetrahydrofolate. These reactions are catalyzed by the multifunctional FolD protein in bacteria (3). All of these enzymes are assumed to have Michaelis-Menten kinetics. Kinetic constants used in simulations were collected from the BRENDA database (http://www.brenda.uni-koeln.de/).
RESULTS
In silico analysis of the genome sequence of R. capsulatus SB1003.
Strains with thyA or thyX deleted are expected to be either nonviable or thymidine auxotrophic, depending on whether they also carry the tdk gene coding for thymidine kinase, which is required for salvage of extracellular thymidine. Consequently, studies on the in vivo functions of flavin-dependent thymidylate synthase X are facilitated by the use of strains that contain tdk in addition to thyX. Using manual similarity searches and the pedant database (http://pedant.gsf.de/) to access the genome sequence of R. capsulatus SB1003, we identified several bacterial and archaeal species that apparently contain thyX and tdk genes, and R. capsulatus is one of these species. Importantly, thyA and folA were not found in the R. capsulatus genome sequence. Genetic tools allowing efficient manipulation of Rhodobacter species have been developed (15, 18). Interestingly, in the closely related species R. sphaeroides thyA and folA are present, whereas thyX is absent. Therefore, R. capsulatus is an excellent organism for heterologous complementation studies designed to understand the in vivo role(s) of ThyX proteins.
Inactivation of R. capsulatus thyX results in thymidine auxotrophy.
To inactivate R. capsulatus thyX, we inserted an Smr-Specr antibiotic resistance cartridge into the unique BamHI site of this gene as described in Materials and Methods (Fig. 2). The R. capsulatus thyX::Specr allele obtained was transferred by diparental conjugation into R. capsulatus MT1131. Spectinomycin-resistant transconjugants were selected on enriched MPYE medium containing thymidine in the presence of rifampin to eliminate the E. coli donor strain. The absence of a wild-type thyX copy in the retained tetracycline-sensitive strain was confirmed by PCR. Phenotypic testing of the thyX::Specr strain obtained, designated DL1, indicated that it grew on enriched MPYE or minimal medium plates supplemented with 1 μg/ml thymidine (Table 2), whereas in the absence of thymidine, growth was not detected. The observed thymidine auxotrophy did not result from polar effects due to insertion of the Smr-Specr cartridge into the chromosomal thyX gene, as the thyX::Specr strain after complementation in trans by R. capsulatus thyX had a doubling time similar to that of the parental strain MT1131 (Table 2). These genetic data provide the first direct indication of the in vivo function of ThyX proteins in de novo synthesis of dTMP.
TABLE 2.
Growth phenotypes of R. capsulatus strains used in this worka
| Strain | Genotype | Growth in:
|
Doubling time (min) withb:
|
||||
|---|---|---|---|---|---|---|---|
| MPYE medium + thymidine | Minimal medium MA | Minimal medium MA + thymidine | No CH2THF | 50 μg/ml CH2THF | 100 μg/ml CH2THF | ||
| MT1131 | thyX+ (wild type) | +++ | +++ | +++ | 122 | 180 | 225 |
| DL1 | thyX::Specr | +++ | − | +++ | NMc | NM | NM |
| DL1/pDL7 | thyX +/thyX::Specr | +++ | +++ | +++ | NM | NM | NM |
| DL1/pDL12 | thyA+ folA+/thyX::Specr | +++ | +++ | +++ | 133 | 198 | 371 |
| DL1/pDL14 | thyA+/thyX::Specr | +++ | ±d | +++ | 287 | 325 | 346 |
Growth phenotypes of the strains were scored after 2 days (MPYE medium plates) or 3 days (minimal medium MA plates) of incubation at 35°C. Similar results were obtained under respiratory and photosynthetic growth conditions.
Doubling times were determined using MPYE medium.
NM, not measured.
Some growth without formation of individual colonies was visible after extended incubation.
Nonorthologous functional replacement of thyX by R. sphaeroides thyA or thyA and folA genes.
Heterologous complementation tests have shown that E. coli thyA can be functionally replaced by thyX genes from a variety of sources (9, 11, 20, 29). To investigate whether the reciprocal nonorthologous replacement is possible, we constructed transferable plasmids carrying either the R. sphaeroides thyA gene alone (pDL14) or the R. sphaeroides thyA and folA genes (pDL12) (Fig. 2). pDL12 and pDL14 were transferred independently into R. capsulatus thyX::Specr strain DL1 by triparental conjugation. On solid medium, only the strain complemented by plasmid pDL12 (carrying R. sphaeroides thyA and folA) formed colonies in a thymidine-independent manner (Table 2), whereas no growth was observed in the presence of pDL14 carrying only the thyA gene. This observation indicates that thyA alone cannot efficiently replace R. capsulatus thyX and that both thyA and folA are necessary for full functional complementation of a thyX defect under these conditions.
To quantitatively address the level of functional complementation, the doubling times for the strains described above were measured using liquid cultures in minimal medium MA and MA containing thymidine (50 μg/ml). In the presence of thymidine, the doubling times of all strains were essentially identical to that of the wild-type strain. In agreement with phenotypic observations on solid minimal medium, MT1131 and DL1/pDL12 had similar doubling times (103 and 108 min, respectively), whereas the negative control strain DL1/pRK415 was unable to grow under these conditions (Table 2). Strain DL1/pDL14, which grew only very poorly on solid minimal medium (Table 2), had a doubling time of 310 min in liquid minimal medium, a value approximately three times higher than the value measured for the wild-type strain. The slow growth in the presence of ThyA but in the absence of FolA presumably shows that reduced folate derivatives are exhausted during the exponential growth and that the oxidized forms produced, like DHF, are not efficiently regenerated. This results in a substantial decrease in the growth rate and cell yield (Fig. 3). During the later stages of growth, another activity could replenish the folate pool at a low rate in the latter strain. Altogether, these observations indicate that the absence of FolA limits cellular growth when ThyA is used for thymidylate synthesis.
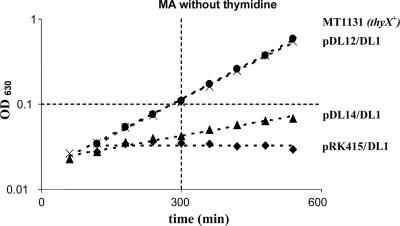
FIG. 3.
Semilogarithmic growth curves for strains MT1131 (thyX+), DL1/pDL12 (thyX/thyA+ folA+), DL1/pDL14 (thyX/thyA+), and DL1/pRK415 (thyX) on minimal medium MA without thymidine. OD630, optical density at 630 nm.
FolA and ThyA are functionally coupled during thymidylate synthesis.
To extend the results of the genetic complementation studies described above, we investigated whether R. capsulatus strain DL1 is indeed unable to synthesize thymidylate de novo. To this end, R. capsulatus chromosomal DNA was radioactively labeled with [5,6-3H]uracil and subsequently hydrolyzed by boiling in perchloric acid. The use of [5,6-3H]uracil results in labeling of dC at positions 5 and 6 of the pyrimidine ring and in labeling of dT at position 6 (the label from position 5 is lost during ThyX/ThyA catalysis). Free nucleobases were separated by high-pressure liquid chromatography using isocratic elution.
The negative control strain DL1/pRK415 produced only one peak corresponding to dC, whereas the same strain carrying pDL12 was able to synthesize de novo both dC (peak at 4 min) and dT (peak at 11 min) (Fig. 4). An intermediate result was obtained for the DL1/pDL14 strain carrying only R. sphaeroides thyA on the plasmid. Although the latter strain was able to synthesize dT, its thymidylate synthesis capability was reduced by 70% compared to that of DL1/pDL12 (Fig. 4), as indicated by quantitative analyses that were normalized using the total DNA quantity.
FIG. 4.
Incorporation of tritium from [5,6-3H]uracil into dC and dT of DL1/pDL12 (thyX::Specr/thyA+ folA+), DL1/pDL14 (thyX::Specr/thyA+), and DL1/pRK415/DL1 (thyX). Elution positions of dC and dT separated by isocratic high-performance liquid chromatography are indicated by arrows. Peak areas were quantified using Laura Light software and were normalized using the total DNA quantity.
Extracellular THF and CH2THF inhibit the growth of R. capsulatus.
Some bacteria, like Lactobacillus casei (26) and cyanobacteria (14), actively transport folates, raising the possibility that the growth defect that we observed for R. capsulatus DL1/pDL14 (thyA::Specr/thyA+) could be rescued by adding various folates to the growth medium. To test this hypothesis, we cultivated this strain in the presence of THF. We observed that, instead of restoring growth as we expected, addition of THF resulted in marked growth inhibition of this strain (Fig. 5). This phenotype could indicate either that strain DL1/pDL14 (thyA::Specr/thyA+) is specifically inhibited by THF or, alternatively, that the observed phenotype is a characteristic feature of all R. capsulatus strains, including the wild type. Consequently, we cultivated the various strains used in this work in the presence and absence of various folates (Fig. 5). Strikingly, we found that addition of THF and CH2THF markedly inhibited, in a dose-dependent manner, the growth of all strains used. In this respect it noteworthy that in Eukarya extracellular folate compounds inhibit the folate-dependent enzymes through product inhibition (23).
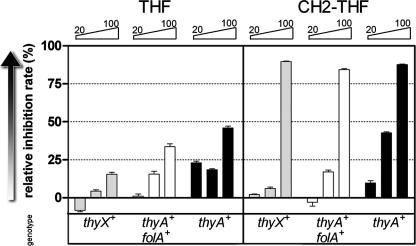
FIG. 5.
Growth inhibition of R. capsulatus by folate derivatives. Growth was scored after 12 h of incubation in MPYE media to which various folate derivatives were added at concentrations of 20, 50, and 100 μg/ml. Gray bars, strain MT1131 (thyX+); open bars, strain DL1/pDL12 (thyX::Specr/thyA+ folA+); black bars, strain DL1/pDL14 (thyX::Specr/thyA+).
Figure 5 also shows that the inhibitory effect of CH2THF on the growth of all R. capsulatus strains (including the wild type) was particularly pronounced, prompting us to measure the doubling times of these strains in the presence of 50 and 100 μg/ml of CH2THF (Table 2). The results obtained indicate that addition of CH2THF significantly increases the doubling times of the strains used. In addition to the product inhibition, increased CH2THF levels could specifically reverse the reaction catalyzed by serine hydroxymethyltransferase shown in Fig. 1 (25). The observed growth defect was transient, as prolonged cultivation even in the presence of 100 μg/ml of CH2THF eventually resulted in growth similar to that observed in the absence of this compound (data not shown).
Mathematical modeling of thymidylate metabolism in Bacteria.
The experimental results indicated that in thymidylate synthase ThyA-containing bacteria, which account for approximately two-thirds of the microbial species whose complete genome sequences are known, DHFR FolA is a key determinant of folate-dependent metabolic pathways (Fig. 1). Without efficient DHFR activity, ThyA catalysis would result in depletion of the THF pool in rapidly growing bacteria. Evidence for this can be seen in the simulation results shown in Fig. 6. For bacteria containing ThyA and FolA, the simulation shows that a decrease in the Vmax of FolA leads to a situation where the steady-state concentration of THF decreases progressively (Fig. 6A). In agreement with this notion, recent isotope ratio-based profiling studies of E. coli folate pools in trimethoprim-treated cells have revealed decreased THF levels and increased DHF (or more oxidized compound) levels in comparison to the levels in nontreated cultures (17). Consequently, when the level of FolA activity is low, the intracellular concentration of the ThyA substrate CH2THF declines rapidly to a very low level, resulting in a decreased rate of the ThyA reaction. This predicts that when FolA activity is inhibited, alternative ways to reduce DHF (8, 28) are not efficient enough to take on the DHF production in fast-growing ThyA-containing bacteria, explaining why these bacteria are highly sensitive to trimethoprim, a specific inhibitor of bacterial FolA. The mathematical simulation model also predicts that a Vmax of FolA of at least 120 μM/h is necessary to support maximal thymidylate synthesis activity in bacteria containing ThyA and FolA (simulation not shown). The presence of ThyX and the absence of ThyA render the “DHF trap” nonfunctional, as ThyX proteins do not catalyze the net oxidation of folate molecules (Fig. 1). Low levels of DHFR activity could be enough to maintain the total folate pool sufficiently reduced to ensure correct ThyX-dependent DNA replication. Genome analyses also show that approximately 10% of bacterial species contain flavin-dependent thymidylate synthase ThyX and FolA. As our results indicate that thymidylate synthesis in ThyX-containing organisms is essentially FolA independent and cells are resistant to trimethoprim (data not shown), the predicted function of FolA in the latter subset of organisms is to increase the intracellular concentration of reduced folate derivatives for other folate-dependent pathways in RNA and protein metabolism.
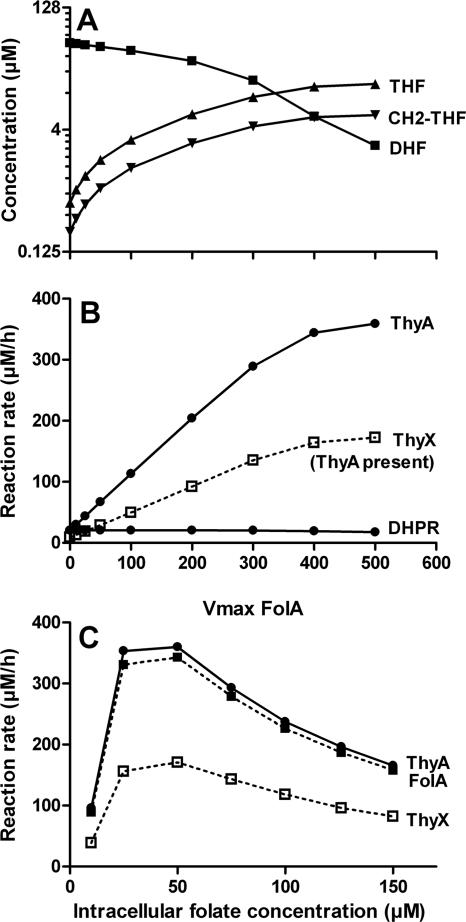
FIG. 6.
(A) Effects of variation in the activity of FolA (DHFR) on selected steady-state concentrations of folate compounds in bacteria containing only ThyA. The model indicates that the decrease in the Vmax of FolA results in depletion of the THF pool in the cell. CH2-THF, methylenetetrahydrofolate. (B) Rate of thymidylate synthesis (for ThyA and ThyX) as a function of the FolA reaction rate. The graph also indicates that the dihydropteridine reductase (DHPR) rate is expected to be independent of the FolA rate. The simulations predicted that the relatively high ThyA activity renders the thymidylate synthesis activity dependent on FolA, even in the presence of ThyX. For these simulation results the model assumed that FolA and ThyA are upregulated, thus representing a rapidly dividing cell. (C) Effect of variation in the intracellular folate concentration on the activity of enzymes involved in folate metabolism. The model indicates that an increase in the folate pool results in inhibition of several enzymes, such as ThyA, FolA, and ThyX.
Interestingly, the model also shows that in bacterial systems with ThyX, ThyA, and FolA (approximately 5% of bacterial systems, including those of Mycobacterium species), DNA synthesis is still dependent on FolA, and consequently, the cells are sensitive to FolA inhibitors (Fig. 6B). In this case, the “DHF trap” would reduce the intracellular concentration of the ThyX substrate, CH2THF, to a very low level (Fig. 6B) that would be insufficient to sustain a high rate of ThyX-driven dTMP synthesis. In support of this prediction, we have observed that E. coli wild-type cells overexpressing ThyX proteins in trans are still sensitive to trimethoprim (data not shown). It is noteworthy that according to this model, folate concentrations cannot be maintained at a high level as this would result in growth inhibition (Table 2 and Fig. 6C).
In slowly growing bacteria, if FolA is absent (but ThyA and ThyX are both present), the “DHF trap” does not necessarily exist; in this case, alternative enzymes catalyzing THF formation, like E. coli FolM (8) or T. thermophilus dihydropteridine reductase (DHTt) (28), could convert enough DHF to THF to keep the remainder of the folate cycle operating at near-normal rates (simulations not shown). This situation could naturally occur in some slowly growing bacteria, for instance, Crocosphaera watsonii (doubling time, 35 h) and Dehalococcoides species (doubling time, 19 h), which contain ThyA and ThyA/ThyX, respectively, but appear to lack FolA and Tdk.
DISCUSSION
In agreement with earlier biochemical and heterologous complementation studies, our studies have proven that flavin-dependent ThyX proteins play an important cellular role in de novo thymidylate synthesis. We demonstrate here that the genetic complementation of an R. capsulatus thyX::Specr strain requires not only thyA but also folA. We also show that the absence of folA in a thyA+ strain results in a decrease in dTMP production, resulting in a DNA replication defect. This is presumably due to the fact that the DHF resulting from ThyA catalysis is not reduced at a sufficiently high rate by alternative folate reductases that must be present in ThyX-containing species, including R. capsulatus (19). We also show that the addition of THF and particularly CH2THF can markedly inhibit bacterial growth. To our knowledge, this growth inhibition, presumably resulting from product inhibition of the folate-dependent enzymes (Fig. 5 and 6C), has not been previously described.
Our mathematical model predicts that in thyX+ strains even a low dihydrofolate-reducing activity is sufficient for thymidylate synthesis. This suggests that different oxidoreductases belonging to a large family of short-chain alcohol dehydrogenases that have been described as suppressors of folA, like FolM (8) and DHTt (28), could act as promiscuous DHFRs. These enzymes possess trimethoprim-insensitive DHFR activity in vitro and can complement folA defects in E. coli when they are overexpressed from high-copy-number plasmids. We detected at least 19 sequences showing sequence similarity to genes encoding FolM and/or DHTt in the R. capsulatus and R. sphaeroides genome sequences. Although we have not investigated the possible physiological roles of any of these dehydrogenases in bacterial folate metabolism, our experiments indicate that the alternative pathways for formation of reduced folates are not sufficient for the recycling of oxidized folates formed by R. sphaeroides ThyA in the absence of FolA activity during DNA replication. Note also that we cannot exclude the possibility that additional novel enzymes with THF-forming activities have not been discovered yet.
As ThyA proteins perform the only currently known cellular reaction catalyzing the net oxidation of THF, these enzymes function as a critical determinant of reduced folate levels. In this respect, mycobacteria that contain both thyA and thyX genes provide a particularly interesting case. Systematic transposon mutagenesis of Mycobacterium bovis BCG has indicated that thyX encodes essential functions even in the presence of thyA, whereas the accumulation of mutations in the thyA gene represents a mechanism of developing resistance to drugs targeting folate metabolism (24). Our simulations also predict that the presence of thyX in mycobacteria provides a molecular basis for resistance for antifolates targeting FolA. It is also noteworthy that the availability of distinct redox cofactors required by ThyA and ThyX proteins might differ during the different stages of DNA replication, suggesting additional differences in the coordination of cellular metabolism in thyA- and thyX-containing species.
In conclusion, our results demonstrated that ThyX proteins are required for de novo thymidylate synthesis. The thymidylate synthase ThyA and some enzymes involved in folate metabolism, like DHFRs and dihydropteroate synthases, are well-characterized therapeutic targets in anticancer and antimicrobial treatments (16). The essential role of the thymidylate synthase ThyX in bacterial survival and the presence of this protein in several pathogens therefore offer an attractive opportunity to design novel antibacterial drugs.
Supplementary Material
Acknowledgments
We thank F. Daldal for necessary Rhodobacter strains and plasmids that were required for this research.
This work was supported by a grant from the CNRS Programme Microbiologie Fondamentale (to U.L. and H.M.). H.M also acknowledges financial support from the INSERM Bioavenir program and the Fondation Bettencourt Schuller. F.E. and D.L. received a fellowship from the French Ministry of Research. F.N. and M.R. acknowledge support from NIH grant 5RO1-CA105437 and NSF grants DMS-0109872 and DMS-061670.
Footnotes
Published ahead of print on 21 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agrawal, N., S. A. Lesley, P. Kuhn, and A. Kohen. 2004. Mechanistic studies of a flavin-dependent thymidylate synthase. Biochemistry 43:10295-10301. [DOI] [PubMed] [Google Scholar]
- 2.Carreras, C. W., and D. V. Santi. 1995. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 64:721-762. [DOI] [PubMed] [Google Scholar]
- 3.D'Ari, L., and J. C. Rabinowitz. 1991. Purification, characterization, cloning, and amino acid sequence of the bifunctional enzyme 5,10-methylenetetrahydrofolate dehydrogenase/5,10-methenyltetrahydrofolate cyclohydrolase from Escherichia coli. J. Biol. Chem. 266:23953-23958. [PubMed] [Google Scholar]
- 4.Davis, J., T. J. Donohue, and S. Kaplan. 1988. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J. Bacteriol. 170:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGregori, J., T. Kowalik, and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dynes, J. L., and R. A. Firtel. 1989. Molecular complementation of a genetic marker in Dictyostelium using a genomic DNA library. Proc. Natl. Acad. Sci. USA 86:7966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giladi, M., N. Altman-Price, I. Levin, L. Levy, and M. Mevarech. 2003. FolM, a new chromosomally encoded dihydrofolate reductase in Escherichia coli. J. Bacteriol. 185:7015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giladi, M., G. Bitan-Banin, M. Mevarech, and R. Ortenberg. 2002. Genetic evidence for a novel thymidylate synthase in the halophilic archaeon Halobacterium salinarum and in Campylobacter jejuni. FEMS Microbiol. Lett. 216:105-109. [DOI] [PubMed] [Google Scholar]
- 10.Graziani, S., J. Bernauer, S. Skouloubris, M. Graille, C. Z. Zhou, C. Marchand, P. Decottignies, H. van Tilbeurgh, H. Myllykallio, and U. Liebl. 2006. Catalytic mechanism and structure of viral flavin-dependent thymidylate synthase ThyX. J. Biol. Chem. 281:24048-24057. [DOI] [PubMed] [Google Scholar]
- 11.Graziani, S., Y. Xia, J. R. Gurnon, J. L. Van Etten, D. Leduc, S. Skouloubris, H. Myllykallio, and U. Liebl. 2004. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J. Biol. Chem. 279:54340-54347. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, J., C. Roshick, E. Iliffe-Lee, and G. McClarty. 2005. Catalytic mechanism of Chlamydia trachomatis flavin-dependent thymidylate synthase. J. Biol. Chem. 280:5456-5467. [DOI] [PubMed] [Google Scholar]
- 13.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 14.Klaus, S. M., E. R. Kunji, G. G. Bozzo, A. Noiriel, R. D. de la Garza, G. J. Basset, S. Ravanel, F. Rebeille, J. F. Gregor III, and A. D. Hanson. 2005. Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 280:38457-38463. [DOI] [PubMed] [Google Scholar]
- 15.Koch, H.-G., H. Myllykallio, F. Daldal, and M. Lee. 1998. Using genetics to explore cytochrome function and structure in Rhodobacter. Methods Enzymol. 297:81-94. [Google Scholar]
- 16.Kompis, I. M., K. Islam, and R. L. Then. 2005. DNA and RNA synthesis: antifolates. Chem. Rev. 105:593-620. [DOI] [PubMed] [Google Scholar]
- 17.Lu, W., Y. K. Kwon, and J. D. Rabinowitz. 2007. Isotope ratio-based profiling of microbial folates. J. Am. Soc. Mass Spectrom. 18:898-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrs, B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 71:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myllykallio, H., D. Leduc, J. Filee, and U. Liebl. 2003. Life without dihydrofolate reductase FolA. Trends Microbiol. 11:220-223. [DOI] [PubMed] [Google Scholar]
- 20.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105-107. [DOI] [PubMed] [Google Scholar]
- 21.Nijhout, H. F., M. C. Reed, P. Budu, and C. M. Ulrich. 2004. A mathematical model of the folate cycle: new insights into folate homeostasis. J. Biol. Chem. 279:55008-55016. [DOI] [PubMed] [Google Scholar]
- 22.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Reed, M. C., H. F. Nijhout, M. L. Neuhouser, J. F. Gregory III, B. Shane, S. J. James, A. Boynton, and C. M. Ulrich. 2006. A mathematical model gives insights into nutritional and genetic aspects of folate-mediated one-carbon metabolism. J. Nutr. 136:2653-2661. [DOI] [PubMed] [Google Scholar]
- 24.Rengarajan, J., C. M. Sassetti, V. Naroditskaya, A. Sloutsky, B. R. Bloom, and E. J. Rubin. 2004. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol. Microbiol. 53:275-282. [DOI] [PubMed] [Google Scholar]
- 25.Schirch, V., S. Hopkins, E. Villar, and S. Angelaccio. 1985. Serine hydroxymethyltransferase from Escherichia coli: purification and properties. J. Bacteriol. 163:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shane, B., and E. L. Stokstad. 1975. Transport and metabolism of folates by bacteria. J. Biol. Chem. 250:2243-2253. [PubMed] [Google Scholar]
- 27.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 27a.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 22:778-785. [DOI] [PubMed] [Google Scholar]
- 28.Wilquet, V., M. Van de Casteele, D. Gigot, C. Legrain, and N. Glansdorff. 2004. Dihydropteridine reductase as an alternative to dihydrofolate reductase for synthesis of tetrahydrofolate in Thermus thermophilus. J. Bacteriol. 186:351-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong, J., S. Skouloubris, Q. Dai, H. Myllykallio, and A. G. Barbour. 2006. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J. Bacteriol. 188:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.