Abstract
The Rhodobacter capsulatus genome contains three genes (olsA [plsC138], plsC316, and plsC3498) that are annotated as lysophosphatidic acid (1-acyl-sn-glycerol-3-phosphate) acyltransferase (AGPAT). Of these genes, olsA was previously shown to be an O-acyltransferase in the second step of ornithine lipid biosynthesis, which is important for optimal steady-state levels of c-type cytochromes (S. Aygun-Sunar, S. Mandaci, H.-G. Koch, I. V. J. Murray, H. Goldfine, and F. Daldal. Mol. Microbiol. 61:418-435, 2006). The roles of the remaining plsC316 and plsC3498 genes remained unknown. In this work, these genes were cloned, and chromosomal insertion-deletion mutations inactivating them were obtained to define their function. Characterization of these mutants indicated that, unlike the Escherichia coli plsC, neither plsC316 nor plsC3498 was essential in R. capsulatus. In contrast, no plsC316 olsA double mutant could be isolated, indicating that an intact copy of either olsA or plsC316 was required for R. capsulatus growth under the conditions tested. Compared to OlsA null mutants, PlsC316 null mutants contained ornithine lipid and had no c-type cytochrome-related phenotype. However, they exhibited slight growth impairment and highly altered total fatty acid and phospholipid profiles. Heterologous expression in an E. coli plsC(Ts) mutant of either R. capsulatus plsC316 or olsA gene products supported growth at a nonpermissive temperature, exhibited AGPAT activity in vitro, and restored phosphatidic acid biosynthesis. The more vigorous AGPAT activity displayed by PlsC316 suggested that plsC316 encodes the main AGPAT required for glycerophospholipid synthesis in R. capsulatus, while olsA acts as an alternative AGPAT that is specific for ornithine lipid synthesis. This study therefore revealed for the first time that some OlsA enzymes, like the enzyme of R. capsulatus, are bifunctional and involved in both membrane ornithine lipid and glycerophospholipid biosynthesis.
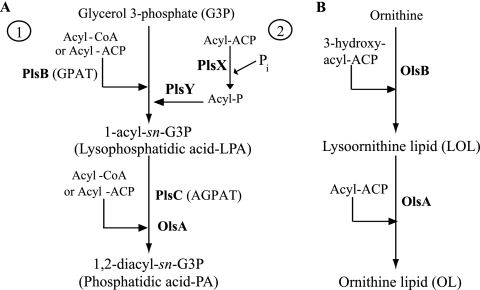
In many organisms, phosphatidic acid (PA) is a key intermediate in de novo synthesis of glycerophospholipids and in signal transduction (9). Two different pathways are known for the formation of PA: the glycerol-3-phosphate (G3P) pathway and the dihydroxyacetone phosphate pathway. Whereas the G3P pathway of PA synthesis is present in prokaryotes, plants, Saccharomyces cerevisiae, and mammalian cells, the dihydroxyacetone phosphate pathway seems to be restricted to yeast and mammalian cells (2). In the G3P pathway, PA is synthesized by two sequential acylation reactions of G3P. In some bacteria like Escherichia coli, the first step is catalyzed by the membrane-bound G3P acyltransferase (sn-G3P acyltransferase [GPAT]) encoded by plsB. GPAT transfers a fatty acyl chain from either acyl-coenzyme A (acyl-CoA) or acyl-acyl carrier protein (acyl-ACP) to the sn-1 position of G3P to produce lysophosphatidic acid (LPA) (13, 38) (Fig. 1A, step 1). GPAT is not a widespread enzyme as many bacteria lack a plsB homologue and, instead, use the recently identified two-step (PlsX/PlsY) pathway to form LPA (33). In this route, the acyl-phosphate intermediate derived from acyl-ACP by PlsX is transferred to G3P by PlsY to produce LPA (Fig. 1A, step 2). The second step of the G3P pathway is well conserved among bacteria and is catalyzed by an LPA acyltransferase (1-acyl-sn-G3P acyltransferase [AGPAT]) enzyme, encoded by plsC. In this step, AGPAT acylates the sn-2 hydroxyl group of LPA to generate PA (Fig. 1A) (2, 11, 12, 17, 38), which is subsequently converted via the central intermediate CDP-diacylglycerol to membrane glycerophospholipids such as phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (2). In addition to playing a vital role in phospholipid synthesis, AGPATs are also involved in cell signaling pathways and apoptosis in certain eukaryotic tumor cells (5).
FIG. 1.
PA and OL biosynthesis pathway in bacteria. (A) The first step for PA biosynthesis from G3P can be carried out by two different routes. In some bacteria, like E. coli, GPAT (PlsB) acylates the sn-1 position of G3P using either acyl-ACP or acyl-CoA to form LPA (step 1). In other bacteria, a recently identified route uses the soluble PlsX to convert acyl-ACP to acyl-phosphate (acyl-P), followed by the membrane-associated PlsY transferring the acyl chain to G3P (step 2). In all bacteria, the second step for PA biosynthesis is catalyzed by the membrane-associated AGPAT (PlsC) enzyme, which transfers an acyl chain from either acyl-ACP or acyl-CoA to LPA to yield PA. In R. capsulatus OlsA is alternative AGPAT enzyme for production of PA. (B) During OL biosynthesis, the first enzyme OlsB catalyzes the formation of an amide linkage (N-acyltransferase) between the α-amino group of ornithine and the carboxyl group of a 3-hydoxy fatty acid, forming LOL. The second enzyme, OlsA, catalyzes the formation of an ester linkage (O-acyltransferase) between the 3-hydroxy group of the fatty acyl group and the carboxyl of a second fatty acid, converting LOL to OL.
Several membrane-associated AGPATs have been cloned and expressed from many bacteria, yeast, various plant species, and several mammals (6, 7, 8, 12, 14, 22, 27, 29, 37, 42, 44, 45, 49). The well-studied bacterium E. coli possesses only one AGPAT, and a deficiency in this enzyme is lethal, resulting in the accumulation of the LPA intermediate (11, 12). Thus, E. coli plsC mutants are temperature-sensitive and can be complemented for growth by heterologous expression of plant and mammalian AGPAT homologues (7, 22, 49). Unlike E. coli, certain bacteria such as Neisseria meningitidis, Neisseria gonorrhoeae, Pseudomonas fluorescens, and Pseudomonas aeruginosa have multiple functional plsC homologues that function in diverse environments (4, 14, 42, 45). Multiple AGPAT isozymes expressed in different tissues have been identified in eukaryotes, such as Limnanthes douglasii, Arabidopsis thaliana, human, and mouse as well (1, 8, 20, 26, 31, 32, 44, 49, 50). In bacteria, the AGPATs also play a role in regulating lipid acyl composition through their substrate specificities (14, 42). Inactivation of one of the multiple plsC genes often alters fatty acid profiles of phospholipids and their membrane properties (14, 42).
Prior to this study, despite the broad importance of AGPATs, only limited knowledge was available on these enzymes, especially those from photosynthetic purple bacteria, including Rhodobacter species (C. Benning, personal communication). Earlier, we had isolated Rhodobacter capsulatus mutants that are defective in maintaining optimal steady-state levels of c-type cytochromes (cyt) (28). Studies of these mutants led us to the identification of olsA and olsB genes responsible for the biosynthesis of membrane ornithine lipid (OL) in R. capsulatus (3) (Fig. 1B). Initially, olsA was misannotated as plsC138 encoding an AGPAT homologue based on its high degree of similarity to acyl-acyltransferases (http://www.ergo-light.com). Mutants lacking an active olsA (or olsB) were unable to produce OL, but they contained a full complement of membrane glycerophospholipids, including PE, PG, and phosphatidylcholine (PC) (3). Thus, PA production must be carried out by an unknown enzyme distinct from OlsA. A whole-genome survey revealed that the R. capsulatus chromosome contained two additional open reading frames (ORFs), annotated plsC316 and plsC3498, as candidates for an AGPAT enzyme involved in PA biosynthesis. In this work, we demonstrate that the plsC316 product is specific for only PA and not OL biosynthesis and that plsC3498 is involved in neither of these two pathways. We also show that the R. capsulatus olsA product is a bifunctional O-acyltransferase involved in both OL and PA biosynthesis. Furthermore, our findings indicate that while R. capsulatus plsC316 is likely to encode the primary AGPAT enzyme involved in PA biosynthesis, OL synthesis-specific olsA can also act as an alternate AGPAT to ensure glycerophospholipid production.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. E. coli strains were grown aerobically in LB medium (35), and R. capsulatus strains were grown at 35°C in either minimal medium A (MedA) (43) or enriched (MPYE) medium supplemented with appropriate antibiotics, as described previously (36). The ability of various R. capsulatus genes to complement the growth defect of a temperature-sensitive E. coli PlsC(Ts) mutant (11) was tested by monitoring growth at 42°C on LB plates supplemented with ampicillin (100 mg/ml) and 0 to 2% l-arabinose, as appropriate. The ability of R. capsulatus genes to complement an E. coli PlsB− mutant was tested by monitoring at 37°C the G3P auxotrophy of appropriate derivatives of strain SJ22 (plsB26 plsX50) (39) on minimal medium E (35) plates supplemented with l-arabinose (2%), ampicillin (100 mg/ml), and 0.04% G3P, as needed.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Relevant characteristic(s) | Reference or source |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr)rpsL20 xyl-5 mtl-1 recA13 | Strr; rB− mB− | 40 |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Strr; cloning host | Invitrogen |
| XL-1 Blue | recA1endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′proAB lacIqΔM15 Tn10] | Tetr; cloning host | Stratagene |
| SM2-1 | plsC1metC162::Tn10 thr-1 ara-14 Δ(gal-attλ)hisG4 rpsL136 xyl-5 mtl-1 lacY1 tsx-78 eda-50 rfbD1 thi-1 | plsC(Ts) mutant | 11 |
| SJ22 | plsB26plsX50 panD2 zac-220::Tn10 glpD3 glpR glpKi relA1 spoT1 pit-10 phoA8 ompF627 fluA22 fadL701 | Auxotrophic for G3P on medium E | 39 |
| R. capsulatus | |||
| MT1131 | crtD121 Rifr | Wild type | 28 |
| Y262 | GTA overproducer | 51 | |
| SA4 | Δ(olsA::spe) | Sper | 3 |
| SA11 | plsC3498::gm | Gmr | This study |
| SA12 | plsC3498::gm Δ(olsA::spe) | Gmr Sper | This study |
| SA13 | Δ(plsC316::kan) | Kanr | This study |
| SA14 | plsC3498::gm Δ(plsC316::kan) | Gmr Kanr | This study |
| SA15 | Δ(olsA::spe) Δ(plsC316::kan) harboring intact olsA on the plasmid | Sper Kanr Tetr | This study |
| SA16 | Δ(olsA::spe) Δ(plsC316::kan) harboring intact plsC316 on the plasmid | Sper Kanr Tetr | This study |
| Plasmids | |||
| pRK2013 | tra+ (RK2) | Kanr; conjugative helper | 18 |
| pRK415 | Tetr; broad-host-range vector | 19 | |
| pBSII | pBluescript II (KS+) | Ampr | Stratagene |
| pMA117 | ΩKan | Kanr | 15 |
| pCHB::Gmr | Gmr Tetr | K. Zhang and F. Daldal | |
| pBAD/Myc-His A | Ampr; arabinose-inducible vector | Invitrogen | |
| pMRC | 6-kb chromosomal EcoRI fragment containing olsA in pLAFR1 | Tetr | 28 |
| pSEM11 | Δ(olsA::spe) | Tetr Sper | 3 |
| pDML1 | 657-bp PCR product containing plsC3498 cloned into NcoI-EcoRI sites of pBAD/Myc-HisA | Ampr | This study |
| pDML3 | 768-bp PCR product containing plsC3498 cloned into XbaI-KpnI sites of pBSII | Ampr | This study |
| pDML4 | XbaI-HindIII-ΩGm of pCHB::Gm inserted into unique SmaI site of plsC3498 on pDML3 | Ampr Gmr | This study |
| pSEM17 | 828-bp PCR product containing olsA cloned into NcoI-EcoRI sites of pBAD/Myc-HisA | Ampr | This study |
| pSEM18 | NsiI-cut pSEM17 ligated into PstI-cut pRK415 | Tetr Ampr | This study |
| pSEM21 | 1.9-kb PCR product containing plsC316 cloned into XbaI-KpnI sites of pBSII | Ampr | This study |
| pSEM24 | 1.9-kb XbaI-KpnI fragment of pSEM21 cloned into XbaI-KpnI sites of pRK415 | Tetr | This study |
| pSEM25 | 819-bp PCR product containing plsC316 cloned into NcoI-SfuI sites of pBAD/Myc-HisA | Ampr | This study |
| pSEM26 | NsiI-cut pSEM25 ligated into PstI-cut pRK415 | Tetr Ampr | This study |
| pSEM27 | NsiI-cut pDML1 ligated into PstI-cut pRK415 | Tetr Ampr | This study |
| pSEM31 | 578-bp Tth111I-RrsII fragment of plsC316 on pSEM21 replaced with SalI-Ωkan of pMA117 | Ampr Kanr | This study |
| pSEM32 | 2.2-kb BglI fragment of pSEM31 ligated to HindIII-KpnI sites of pRK415 | Tetr Kanr | This study |
| pSEM35 | 1,870-bp PshAI-KpnI fragment of pDML4 cloned into KpnI sites of pRK415 | Tetr Gmr | This study |
Abbreviations of antibiotic resistances are as follows: Amp, ampicillin; Gm, gentamicin; Kan, kanamycin; Rif, rifampin; Spe, spectinomycin; Str, streptomycin; Tet, tetracycline.
Molecular genetic techniques.
Standard molecular biological techniques were performed according to Sambrook et al. (40) and Daldal et al. (15). Homology searches and amino acid sequence alignments were done using MacVector (Accelerys, San Diego, CA) and appropriate software programs as described earlier (36).
The plsC316 gene (annotated RRC00316 on the R. capsulatus genome) was cloned by PCR amplification using chromosomal DNA and the primers 5′-AAGTCTAGATTCGGCGCCGCCCGATCAGATGGAAA-3′ and 5′-CACCGGTACCCGCGTTCGACCGAAAAATGCCT-3′ containing the XbaI and KpnI sites (in boldface) at positions 689 bp 5′ upstream and 497 bp 3′ downstream of the start and stop sites of plsC316, respectively. The 1.9-kb PCR product thus generated was digested with XbaI and KpnI and cloned into the identical restriction sites of the pBluescript II KS (Stratagene Inc., La Jolla, CA) and to pRK415 (19) (Table 1) to yield pSEM21 and pSEM24, respectively (Table 1). Similarly, the plsC3498 gene (annotated RRC03498 on the R. capsulatus genome) was cloned using genomic DNA as a template and the primers 5′-GGTCAATCTAGATCAGCAGTTGCGCG-3′ and 5′-AAGATCGGTACCAAAGCAGAATCC-3′ containing the XbaI and KpnI sites (in boldface) at positions 54 bp 5′ upstream and 58 bp 3′ downstream of the start and stop sites of plsC3498, respectively. The 768-bp PCR product thus obtained was digested with XbaI and KpnI and ligated into the corresponding sites of the pBluescript II KS to generate pDML3 (Table 1).
Construction of mutant alleles of plsC316 and plsC3498.
Interposon mutagenesis, using either the Kanr gene of pMA117 (15) or Gmr gene of pCHB::Gmr (K. Zhang and F. Daldal, unpublished data), was performed using the gene transfer agent (GTA) (51), as described earlier (15). First, an insertion-deletion allele of plsC316 was obtained by replacing the 578-bp Tth111I-RsrII blunt-ended fragment of pSEM21 with a 1.6-kb blunt-ended SalI fragment of pMA117 carrying the Kanr cartridge to yield pSEM31 (Table 1). Similarly, an insertion allele of plsC3498 was obtained by ligating the 1.16-kb HindIII and XbaI blunt-ended Gmr cartridge from pCHB::Gmr into the unique SmaI site of plsC3498 carried by pDML3 to yield pDML4 (Table 1). Derivatives of the transferable plasmid pRK415 carrying Δ(plsC316::kan) and plsC3498::gm deletion-insertion and insertion alleles of plsC316 and plsC3498, respectively, were constructed by cloning the 2.2-kb blunt-ended BglI fragment of pSEM31 between the HindIII and KpnI sites of pRK415 and the 1.87-kb blunt-ended PshAI and KpnI fragment of pDML4 into the KpnI site of pRK415 to generate pSEM32 and pSEM35, respectively (Table 1). The latter plasmids were conjugated into the GTA overproducer strain Y262, and following appropriate GTA crosses into the wild-type strain MT1131, the single mutants SA11 (plsC3498::gm) and SA13 [Δ(plsC316::kan)] (Table 1) were obtained. Similarly, the double mutants SA12 [Δ(olsA::spe) plsC3498::gm] and SA14 [Δ(plsC316::kan) plsC3498::gm] were obtained by using either SA4 [Δ(olsA::spe)] (3) or SA13 [Δ(plsC316::kan)] single mutants instead of the wild-type strain MT1131 (Table 1).
Expression of olsA, plsC316, and plsC3498 in E. coli.
The olsA gene was PCR amplified using the plasmid pMRC (28) (Table 1) as a template and the primer pairs olsA-NcoI (5′-GGACGCCCATGGCACGACCGATCTGG-3′) and olsA-EcoRI (5′-CTGCGCGAATTCCGCGACCGCTGACC-3′) containing the NcoI and EcoRI restriction enzymes sites (in boldface), respectively. The plsC316 gene was amplified using the plasmid pSEM21 (Table 1) as a template and the primers plsC316-NcoI (5′-ATTCGCCCATGGTCGTTTGGCAATAC-3′) and plsC316-SfuI (5′-GGCTGACCTTCGAACCGATCTTCATCAGC-3′) containing the NcoI and SfuI (isochizomer BstBI) restriction enzyme sites (in boldface), respectively. The plsC3498 gene was amplified using genomic DNA as a template and the primers plsC3498-NcoI (5′-CCGGCGCCATGGCGGGGCTGACGCGG-3′) and plsC3498-EcoRI (5′-AGGCCGAATTCCGCGCCGCCCCCAGC-3′) containing the NcoI and EcoRI restriction enzymes sites (in boldface), respectively. The PCR products obtained were digested with appropriate restriction enzymes, cloned into the corresponding sites of the expression vector pBAD/Myc-His A (Invitrogen Inc., Carlsbad, CA), yielding pSEM17 (olsA), pSEM25 (plsC316), and pDML1 (plsC3498), respectively (Table 1). The resulting plasmids were sequenced to confirm that olsA, plsC316, and plsC3498 were in frame with the vector's translation start site and were epitope tagged at their carboxyl termini. Automated DNA sequencing with a BigDye terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, CA) was used with the primers pBAD-Seq-F (5′-ATGCCATAGCATTTTTATCC-3′) and pBAD-Seq-R (5′-GATTTAATCTGTATCAGG-3′). Derivatives of the transferable plasmid pRK415 carrying olsA, plsC316, or plsC3498 were constructed by cloning the 4.9-kb NsiI fragment of pSEM17, the 4.9-kb NsiI fragment of pSEM25, and 4.8-kb NsiI fragment of pDML1 into the PstI site of pRK415 to generate pSEM18, pSEM26, and pSEM27, respectively. Conjugal transfer of all plasmids from E. coli to R. capsulatus was carried out as described earlier (18).
Expression of R. capsulatus plsC homologues in either E. coli or R. capsulatus.
To monitor the expression of olsA, plsC316, or plsC3498, the plasmids pSEM17 (olsA), pSEM25 (plsC316), or pDML1 (plsC3498), respectively, were transformed into the E. coli strain SM2-1 [plsC(Ts)] (11). Appropriate derivatives of SM2-1 were grown to an optical density at 600 nm (OD600) of ∼0.5, and cultures were induced for 4 h with increasing amounts (0 to 2%) of l-arabinose. In each case 1 ml of cell culture was collected by centrifugation, and the whole-cell pellets were resuspended in 100 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer and boiled for 4 min. To monitor the expression of olsA, plsC316, or plsC3498 in R. capsulatus, the plasmids pSEM18 (olsA), pSEM26 (plsC316), and pSEM27 (plsC3498) were conjugated into the mutants SA4 (ΔolsA::spe), SA13 (ΔplsC316::kan), and SA11 (plsC3498::gm), respectively. The resulting transconjugants were grown on MPYE plates with or without 1% l-arabinose for 2 days under aerobic conditions. In each case, two isolated colonies were dispersed in 10 μl of distilled H2O, centrifuged, and resuspended in 10 μl of sample loading buffer. Cells were lysed by incubation at 35°C for 7 min. E. coli and R. capsulatus cell extracts were separated using 15% SDS-PAGE (30) and transferred to polyvinylidene difluoride membranes. OlsA, PlsC316, or PlsC3498 produced in E. coli was detected using monoclonal anti-Myc1-9E10 antibody (at a dilution of 1:1,000) (Cell Center, University of Pennsylvania) as a primary antibody, and the antigen-antibody complexes were detected with horseradish peroxidase-conjugated sheep anti-mouse antibody (at a 1:2,000 dilution) (GE Healthcare Bio-Sciences, Buckinghamshire, United Kingdom) as a secondary antibody, with diaminobenzidine staining enhanced with NiCl2 (25). Similarly, OlsA, PlsC316, or PlsC3498 produced in R. capsulatus was detected using the same anti-Myc1-9E10 primary antibody, except that alkaline phosphatase-conjugated goat anti-mouse antibodies (at a 1:2,000 dilution) (Bio-Rad, Hercules, CA) were used as secondary antibodies with the chromogenic substrate 4-nitroblue tetrazolium-5-bromo-4-cloro-3-indolyl phosphate (Sigma Inc., St. Louis, MO).
Analysis of c-type cyt.
R. capsulatus intracytoplasmic membrane vesicles (chromatophores) were prepared using a French pressure cell as described earlier (3). Membrane proteins (100 μg per lane) were incubated at 37°C for 10 min in the sample loading buffer prior to loading, and after separation by 16.5% (wt/vol) tricine-SDS-PAGE (41), the c-type cyt were visualized via their peroxidase activities using tetramethylbenzidine and H2O2 (46). cbb3-Cox (cyt c oxidase) activity of R. capsulatus mutants was determined by using Nadi staining as previously described (28).
Determination of GPAT and AGPAT activities using purified E. coli or R. capsulatus membranes.
Combined GPAT and AGPAT activities of E. coli or R. capsulatus strains were measured by using a filter paper disk assay (21). The assay mixture contained 0.1 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol, 0.5 mM G3P, 0.005 μCi of [U-14C]G3P, and 7 μM cis-vaccenyl-ACP (see the supplemental material for a description of cis-vaccenyl-ACP synthesis) in a reaction volume of 20 μl. This mixture was further supplemented with 5 mM Na3VO4 as a phosphatase inhibitor when R. capsulatus membranes were used. The enzymatic assay, initiated by the addition of membrane particles (see the supplemental material for a description of membrane particle preparation), was carried out for incubations of 0, 1, 2, 5, 10, 15, and 20 min at 35°C. At each time point, 18.5 μl of the reaction mixture was removed and deposited onto Whatmann 3 MM filter paper. Filter papers were washed for 20 min in 10%, 5%, and 1% ice-cold trichloroacetic acid and then dried; the radioactivity retained was determined using a scintillation counter (Beckman LS-9000; Fullerton, CA). A scaled-up version of the same assay (60-μl reaction mixture with a 5-min incubation at 35°C) was also run to monitor LPA and PA production using thin layer chromatography (TLC). At the end of the incubation period, 2 ml of chloroform:methanol (1:1, vol/vol), 0.19 ml of distilled H2O, and 1 ml of 0.1N KCl were added to the assay mixture. After vigorous vortexing, samples were centrifuged at 8,000 × g for 15 min, the lower chloroform phase containing the lipids was evaporated under a stream of argon, and extracted lipids were dissolved in 100 μl of chloroform. Extracts (7,000 and 2,000 total cpm for E. coli and R. capsulatus extracts, respectively) were applied to a preheated silica gel G60 TLC plate (EMD Chemicals Inc., Gibbstown, PA) and developed with chloroform:methanol:glacial acetic acid (39:9:3, vol/vol/vol) in one dimension. Radiolabeled lipids were visualized using a phosphorimager (Typhoon 9410; Amersham Biosciences, Arlington Heights, IL) and quantitated with ImageQuant software (Amersham Biosciences). Spots corresponding to LPA and PA were identified based on their comigration with unlabeled LPA and PA standards (Avanti Polar Lipids, Alabaster, AL).
Total lipid and fatty acid analyses.
For total lipid analyses, R. capsulatus cells were labeled for 24 h in 1 ml of MedA or MPYE medium supplemented with 2 μCi of [1-14C]acetate (60 mCi mmol−1 specific activity). Labeled cells were analyzed as described previously (3) by using two-dimensional (2D)-TLC, and radiolabeled (60,000 total cpm) lipids were deposited on heat-activated silica gel G60 plates. Plates were developed with chloroform:methanol:water (14:6:1, vol/vol/vol) and chloroform-methanol-glacial acetic acid (13:5:2, vol/vol/vol) for the first and second dimensions, respectively (16). Radiolabeled lipids were visualized, identified, and quantified as described above. Fatty acid compositions of appropriate R. capsulatus strains were determined using approximately 30 mg of wet cell pellets grown in MPYE medium, and fatty acid methyl ester analysis was carried out by MIDI Inc. (Newark, DE).
Chemicals, reagents, and enzymes.
Restriction enzymes, oligonucleotide primers, [U-14C]G3P (150 mCi mmol−1 specific activity) and [1-14C]acetate (60 mCi mmol−1 specific activity) were purchased from New England Biolabs, the Cell Center facility of the University of Pennsylvania, American Radiolabeled Chemicals Inc., and NEN Life Science Products, respectively. ACP was obtained from either Sigma Chemical Co. or Invitrogen Inc. cis-Vaccenic acid, G3P, and LPA were from Sigma Chemical Co.; PA was from Avanti Polar Lipids; and DEAE cellulose (DE52) was from Whatmann. All other chemicals were from commercial sources and of highest available purity.
RESULTS
Identification of two additional plsC homologues in R. capsulatus genome.
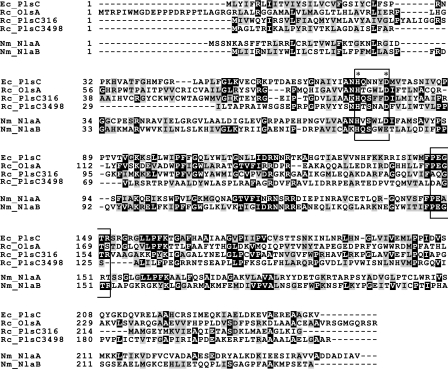
Our previous work established that OlsA null mutants lack only OL; are not lethal, unlike an E. coli PlsC mutant; and still produce adequate levels of PE, PC, and PG (3). These findings indicated that in the absence of olsA, R. capsulatus must have other means of producing PA, which is an essential intermediate for membrane glycerophospholipid biosynthesis. A survey of the R. capsulatus genome (http://www.ergo-light.com) revealed two additional AGPAT candidates in addition to RRC00138, which was initially annotated as plsC138 but subsequently renamed olsA, acting as O-acyltransferase engaged in OL synthesis (3). The ORFs RRC00316 (plsC316) and RRC03498 (plsC3498) were annotated as AGPAT homologues, and, like OlsA, they exhibited high degrees of similarities to the E. coli PlsC. They contained a conserved acyltransferase (pfam01553/COG0204) motif and a highly conserved (HX4D) sequence thought to be common to GPAT and AGPAT enzymes (24) (Fig. 2).
FIG. 2.
Comparison of various AGPAT homologues of R. capsulatus. The R. capsulatus (Rc) AGPAT homologues were aligned with the E. coli (Ec) and N. meningitidis (Nm) AGPAT sequences using the program ClustalW and presented using the BOXSHADE, version 3.21, software. Identical residues are shaded in black, and similar residues are shaded in gray. The catalytic (HX4D) motif (24) and the substrate-binding (PEGTR) motif of GPATs and AGPATs are boxed and indicated by asterisks.
On the R. capsulatus chromosome, the two plsC homologues are located at different regions distant from each other and from olsA. plsC316 is 819 bp in length, encodes 273 amino acids, and is surrounded by the ORFs RRC00314, RRC00315, RRC00701, and RRC00317, corresponding to mlcA, accA, ftsX, and ftsE, respectively; plsC3498 is 657 bp in length, encodes 219 amino acids, and is surrounded by the ORFs RRC03495, RRC03496, RRC03497, and RRC03498 corresponding to acoB, acoC, cdsA, and cysE, respectively (see Fig. S1 in the supplemental material and the legend for a functional description of these genes). Multiple alignments of these ORFs illustrated their similarities to the E. coli PlsC and to each other (Fig. 2). For example, R. capsulatus OlsA, PlsC316, and PlsC3498 show 18%, 19%, and 19% identity and 24%, 32%, and 26% similarity to the E. coli PlsC, respectively. Note that the highest degree of similarity is seen between the R. capsulatus PlsC316 and E. coli PlsC. Moreover, plsC316 is also flanked by cell division-related genes ftsX and ftsE (http://www.ergo-light.com), like the E. coli plsC located between sufI involved in cell division and parC encoding a topoisomerase involved in chromosome partitioning (12). No similar synteny between E. coli and R. capsulatus was observed for olsA or plsC3498, which are located immediately downstream of olsB, encoding an N-acyltransferase involved in OL biosynthesis (3), or cdsA, encoding phosphatidate cytidylytransferase (RRC03497) converting PA to CDP-diacylglycerol, respectively (see Fig. S1 in the supplemental material).
Insertional inactivation of R. capsulatus plsC homologues and characterization of ensuing mutants.
The R. capsulatus AGPAT homologues plsC316 and plsC3498 were cloned, and their mutant alleles were constructed using interposon mutagenesis, as described in Materials and Methods, in order to define which one of them is responsible for PA biosynthesis in R. capsulatus. The single mutants lacking an active PlsC316 (SA13 [Δ(plsC316::kan)]) or PlsC3498 (SA11 [plsC3498::gm]) were obtained readily and compared with a mutant lacking an active OlsA (SA4 [Δ(olsA::spe)]). Unlike the E. coli PlsC− mutants that are lethal, neither plsC316 nor plsC3498 was essential for growth of R. capsulatus under the photosynthetic or respiratory conditions on MPYE or MedA growth medium. However, it was noted that the PlsC316− mutant formed slightly smaller colonies than the OlsA− or the PlsC3498− mutants under all growth conditions, indicating a slight growth defect (Fig. 3A). The doubling time of wild-type, OlsA−, and PlsC316− strains that were grown in liquid MPYE medium were 100, 122, and 131 min, respectively.
FIG. 3.
Characterization of plsC mutants. (A) Growth of wild-type (wt), olsA (SA4), plsC316 (SA13), and plsC3498 (SA11) null mutants on MPYE medium at 35°C under aerobic conditions after 2 days of incubation. (B) Growth of plsC316 mutant harboring a plasmid with (SA13/pMRC) or without (SA13/pRK415) olsA under the same conditions as described for panel A. (C) Comparison of the c-type cyt profiles of R. capsulatus plsC316 and olsA mutants. Membrane fractions were isolated from cells grown at 35°C in MPYE medium, proteins were separated by using 16.5% tricine-SDS-PAGE, and the c-type cyt were revealed using tetramethylbenzidine, as described in Materials and Methods. The c-type cyt subunits of the cbb3-Cox (cp and co), the cyt bc1 complex (c1), and the electron carrier cyt cy (cy) are indicated on the left together with the 32.5- and 25-kDa molecular size markers.
Double mutants with all possible combinations of olsA, plsC316, and plsC3498 were then sought to probe any possible functional redundancy between these genes. Like the single mutants, the PlsC316− PlsC3498− (SA14) and the OlsA− PlsC3498− (SA12) double mutants were readily obtained. These mutants were able to grow on all media tested and exhibited the corresponding single-mutant phenotypes (slow growth and OL deficiency, respectively). Thus, combined inactivation of plsC316 with plsC3498 or of olsA with plsC3498 had no deleterious growth effect, indicating that the function of plsC3498 was not redundant with either of the two other genes. In contrast, despite many attempts under various conditions, inactivation of both olsA and plsC316 was impossible. The inability to obtain an OlsA− PlsC316− double mutant strongly suggested that an intact copy of either olsA or plsC316 was required to support growth of R. capsulatus under the conditions tested. This observation was further confirmed by using olsA or plsC316 diploid strains (SA15 and SA16) as recipients for interposon mutagenesis (Table 1). These diploid strains carried a copy of a given gene on the chromosome and another copy of the same gene on an autonomously replicating plasmid. Using these strains, mutants carrying inactive chromosomal copies of both olsA and plsC316 but complemented by plasmid-borne copies of either of these genes were readily obtained. The genetic data therefore indicated that an intact copy of either plsC316 or olsA was required for growth of R. capsulatus. That OlsA and PlsC316 had overlapping functions was further suggested by the fact that a PlsC316− mutant regained wild-type-like growth properties when it harbored a plasmid-borne copy of olsA (Fig. 3B). However, an OlsA− mutant carrying an intact copy of plsC316 was still devoid of OL.
The cyt c profiles and membrane polar lipid and fatty acid compositions of R. capsulatus mutants lacking various plsC homologues.
Considering that OL and, hence, its biosynthetic genes olsA and olsB are required for the presence of normal steady-state amounts of several c-type cyt and cbb3-Cox activity in R. capsulatus (3), we examined the effect of plsC316 inactivation on the c-type cyt content of R. capsulatus. Analyses of various plsC316 (Fig. 3C, lane 3) and also plsC3498 (data not shown) single or double mutants indicated that, unlike OlsA− mutants, these mutants produced wild-type levels of membrane-bound (Fig. 3C, lane 1) and soluble c-type cyt and had cbb3-Cox activities (data not shown).
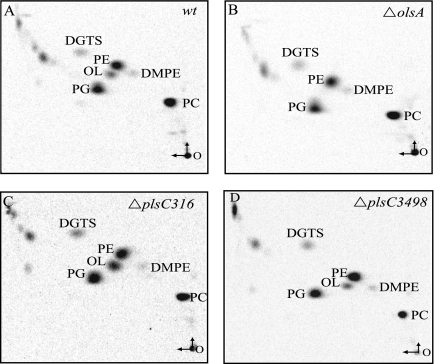
Total lipid compositions of the PlsC316− and PlsC3498− mutants were next examined after labeling with [1-14C]acetate followed by extraction and 2D-TLC separation, as described in Materials and Methods. The data showed no qualitative differences between the PlsC316− and PlsC3498− mutants and the wild-type parental strain MT1131 (Fig. 4). Quantitation of polar lipids was performed using ImageQuant software (Typhoon 9410) (Table 2). Compared with a wild-type strain, inactivation of plsC316 decreased the relative amounts of PE and increased those of PG and OL, whereas inactivation of olsA mainly abolished OL production. Overproduction of OL in the absence of plsC316 (about 10% versus 4% of total lipids in its presence) suggested that in this mutant OlsA activity might have increased to sustain sufficient PA production, concomitantly leading to higher OL production. On the other hand, absence of plsC3498 had no affect on the total lipid composition of R. capsulatus (data not shown), again suggesting that it was unrelated to membrane lipid biosynthesis.
FIG. 4.
Total lipid composition of plsC316 and plsC3498 null mutants of R. capsulatus. In all cases, total polar lipids were extracted from [1-14C]acetate-labeled cells, similar amounts (60,000 cpm) were deposited on TLC plates, and 2D-TLC analyses were carried out as described in Materials and Methods. DGTS, diacylglyceryl trimethyl-homoserine; DMPE, phosphatidyl-N,N dimethylethanolamine. The vertical and horizontal arrows at the origin O refer to the first and second dimension of solvent migrations, respectively. The radioactivity associated with each spot was determined and is given in Table 2.
TABLE 2.
Comparison of polar membrane lipid composition and fatty acid profiles of R. capsulatus wild-type, OlsA−, and PlsC316− mutant strains
| Strain | Lipid (%)b
|
Fatty acid (%)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PE | PG | PC | OL | Other | C10:0 3OH | C16:0 | C16:1ω7c+C16:1 ω6c | C18:0 | C18:0 3OH | C18:1 ω7c | C18:1 ω5c | |
| Wild type | 28.5 | 18.2 | 37.7 | 4.0 | 11.6 | 2.60 | 2.36 | 6.02 | 2.09 | 2.12 | 79.11 | 2.98 |
| ΔolsA strain | 25.7 | 21.0 | 44.5 | ND | 8.9 | 2.17 | 5.74 | 4.14 | 3.42 | 1.92 | 76.91 | 2.44 |
| ΔplsC316 strain | 16.5 | 27.9 | 35.2 | 10.6 | 9.7 | 2.96 | 1.19 | 0.59 | 4.73 | 2.87 | 82.39 | 2.53 |
a All strains were grown on enriched MPYE medium by respiration in the presence of [14C]acetate, and their polar membrane lipids and total fatty acids were analyzed as described in Materials and Methods.
Data are the percentages relative to total 14C. ND, not detected.
Total fatty acid profiles of olsA or plsC316 mutants were also compared with the R. capsulatus wild-type strain MT1131 by using fatty acid methyl ester analysis, as described in Materials and Methods. The data showed that the fatty acid composition of the membrane lipids was altered in the olsA and plsC316 null mutants (Table 2). In comparison with a wild-type strain, inactivation of plsC316 decreased and increased modestly the relative amounts of saturated C16 and C18 fatty acids, respectively. Moreover, it drastically decreased the amount of unsaturated C16 but not unsaturated C18 fatty acids. On the other hand, inactivation of olsA somewhat increased the amounts of saturated, but not unsaturated, C16 and C18 fatty acids compared to a wild-type strain.
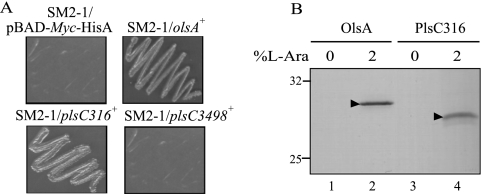
Both R. capsulatus olsA and plsC316 can complement an E. coli plsC mutant in vivo.
Pronounced similarities observed between various PlsC homologues (Fig. 2) led us to probe whether any of the R. capsulatus plsC homologues could complement the E. coli plsC(Ts) mutant, SM2-1, producing a temperature-sensitive AGPAT (12). Plasmid pBAD derivatives, expressing upon induction by l-arabinose either olsA, plsC316, or plsC3498, were constructed as described in Materials and Methods and transformed into the strain SM2-1 at 30°C. Appropriate transformants were tested for their ability to grow at 42°C in the presence of 2% l-arabinose. The plasmid pSEM17 or pSEM25 carrying either olsA or plsC316 was able to complement the E. coli plsC(Ts) mutant, SM2-1, for growth at 42°C but only upon induction with l-arabinose (Fig. 5A). Under similar conditions, no complementation was observed with the plasmid pDML1 carrying plsC3498. Thus, both OlsA and PlsC316, but not PlsC3498, acted as functional homologues of E. coli PlsC and produced apparently temperature-resistant AGPAT activity. Furthermore, it was also noted that plsC316 provided a more vigorous growth than olsA. Immunoblot analyses were carried out to confirm that genetic complementation was due to the production in E. coli of R. capsulatus OlsA or PlsC316. As expected, upon induction by l-arabinose, α-Myc epitope-tagged proteins with molecular masses of approximately 31 and 29.5 kDa were detected by using anti α-Myc antibodies in the E. coli SM2-1 derivatives harboring OlsA (SM2-1/pSEM17) and PlsC316 (SM2-1/pSEM25), respectively (Fig. 5B, lanes 2 and 4).
FIG. 5.
Expression of R. capsulatus olsA and plsC316 in E. coli. (A) The E. coli plsC(Ts) strain SM2-1 harboring plasmids carrying olsA, plsC316, or plsC3498 of R. capsulatus was grown on 2% l-arabinose-containing LB plates at 42°C to score heterologous complementation. SM2-1 cells carrying the cloning vector pBAD/Myc-His A were used as a control. (B) Expression of R. capsulatus olsA and plsC316 in E. coli plsC(Ts) mutant SM2-1 cells before (0) and after (2) induction with 2% l-arabinose for 4 h at 30°C. Following induction cells were resuspended in 2× SDS loading buffer, and expressed proteins were detected by SDS-PAGE and immunoblotting using anti-Myc antibody as described in Materials and Methods. The triangles point out the R. capsulatus OlsA and PlsC316 proteins (31 and 29.5 kDa, respectively) together with the 32.5- and 25-kDa molecular mass markers.
Availability of plasmids carrying α-Myc epitope-tagged alleles of OlsA and PlsC316 allowed us to probe whether these proteins were produced in active forms in R. capsulatus. The plasmids pSEM18 and pSEM26 carrying olsA and plsC316, respectively, were crossed into SA4 [Δ(olsA::spe)] and SA13 [Δ(plsC316::kan)]. Transconjugants SA4/pSEM18 and SA13/pSEM26 thus obtained were grown in MPYE medium with or without 2% l-arabinose. Immunoblot analyses revealed that they contained proteins of approximately 31 kDa and 29.5 kDa that reacted with anti-Myc antibodies (data not shown). The levels of the proteins produced in R. capsulatus were lower than those seen in E. coli, but the wild-type phenotypes of the transconjugants in respect to OL, c-type cyt production, and better growth indicated that the epitope-tagged versions of OlsA and PlsC316 were functional.
The E. coli plsB and plsC gene products, conferring GPAT and AGPAT activities, share partial amino acid sequence homologies and are thought to function coordinately (Fig. 1A) (13). Considering that some acyltransferases, like the Clostridium butyricum plsD exhibiting functional GPAT activity, can complement an E. coli PlsB− mutant (23) and that plsC3498 showed similarity to plsD (20% identity and 34% similarity), we used the E. coli mutant SJ22 to investigate whether olsA, plsC316, or plsC3498 exhibited functional GPAT activity. This mutant carries both the plsB26 and plsX50 mutations and requires supplementation with G3P for growth (39). Upon transformation of the plasmids pSEM17 (olsA), pSEM25 (plsC316), and pDML1 (plsC3498) into SJ22, no complementation for G3P auxotrophy was observed, indicating that none of these genes produced GPAT activity and especially that plsC3498 was not a homologue of plsB in R. capsulatus.
R. capsulatus OlsA and PlsC316 exhibit AGPAT activities and synthesize PA in vitro.
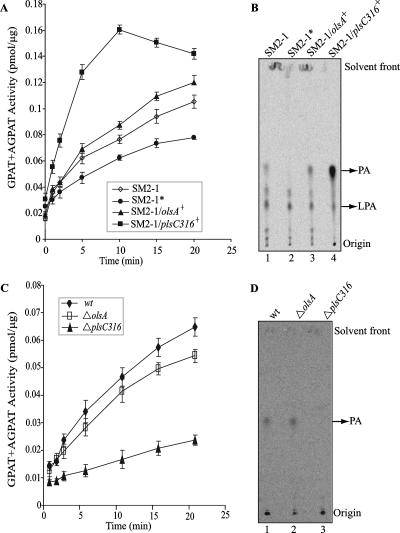
In an attempt to define the biochemical function(s) of OlsA and PlsC316, combined GPAT-AGPAT activities were assayed in vitro by using radiolabeled G3P as the acyl acceptor and acyl-ACP as the acyl donor, as described in Materials and Methods. Unlike the E. coli GPAT and AGPAT enzymes, which can use either acyl-CoA or acyl-ACP as acyl donors (47), Rhodobacter sphaeroides enzymes exhibit high specificity for acyl-ACP compared to acyl-CoA (34). No significant enzyme activity was observed with the acyl-CoA substrate in R. capsulatus (data not shown) as in R. sphaeroides. Considering that R. capsulatus lipids contain predominantly cis-vaccenic acid (cis-11-18:1) fatty acid, cis-vaccenyl-ACP was prepared as the acyl donor. Purified membrane particles (see the supplemental material) from E. coli plsC(Ts) mutant SM2-1 derivatives harboring olsA or plsC316 and grown at 42°C in the presence of l-arabinose were assayed. Time course assays monitoring the production of radiolabeled LPA and PA were carried out as described in Materials and Methods. Control experiments established that the activities measured were vaccenyl-ACP and membrane particle dependent (data not shown), and the endogenous activity detected using membranes from SM2-1 cells grown at 30°C and subsequently incubated at 42°C was very low. The data obtained revealed that membranes from SM2-1 derivatives producing either OlsA or PlsC316 exhibited measurable amounts of combined GPAT-AGPAT activity (Fig. 6A). Moreover, PlsC316-containing membrane particles displayed much higher specific activities than either those containing OlsA or those from SM2-1 cells grown at 30°C.
FIG. 6.
GPAT-AGPAT activities exhibited by appropriate E. coli plsC(Ts) mutants harboring R. capsulatus plsC homologues as well as R. capsulatus wild-type, olsA, and plsC316 mutants. (A) Time course assays of GPAT-AGPAT activities in E. coli plsC mutant harboring olsA or plsC316 were performed using radioactive G3P, vaccenyl-ACP, and membrane particles (prepared as described in the supplemental material) from SM2-1 cells grown at 30°C (SM2-1), SM2-1 cells grown at 30°C with a subsequent 30-min incubation at 42°C (SM2-1*), SM2-1 cells harboring olsA, and SM2-1 cells harboring plsC316, as described in Materials and Methods. The data shown are the means of two independent experiments with the standard errors, as indicated. (B) Assays similar to those shown in panel A were performed at 35°C for 5 min, and labeled lipids (approximately 7,000 cpm total) were extracted and separated by 1D-TLC, as described in Materials and Methods. LPA and PA produced using membranes from SM2-1 cells grown at 30°C (lane 1), SM2-1 grown at 30°C with a subsequent 30-min incubation at 42°C (lane 2), SM2-1 cells harboring olsA (lane 3), and SM2-1 cells harboring plsC316 (lane 4) are shown. Note the absence of PA production in lane 2 and PA overproduction in lane 4. (C) Time course assays of GPAT-AGPAT activities in wild-type (wt), ΔolsA (SA4), and ΔplsC316 (SA13) strains were performed as described for panel A. The data shown are the means of two independent experiments with the standard errors as indicated. (D) Labeled lipids (approximately 2,000 cpm total) were prepared and separated by 1D-TLC, as described for panel B. Note that the PA produced using membranes from the wild-type strain MT1131 and the ΔolsA mutant are readily seen while that produced by the ΔplsC316 mutant is barely detectable.
As the combined GPAT-AGPAT assay using radioactive G3P reflects the production of both LPA and PA, separate formation of LPA via GPAT and of PA via AGPAT activities was also determined. Products of a similar enzymatic reaction were analyzed by 1D-TLC, and LPA and PA were identified by comparison of their Rf values with those of standard markers (Fig. 6B). As expected, all membrane particles produced some amounts of LPA, which reflected the intact GPAT activity of the E. coli host SM2-1. On the other hand, membrane particles from heat-treated (42°C for 30 min) SM2-1 cells (grown at 30°C) did not produce any (Fig. 6B, lane 2), whereas those from non-heat-treated cells produced detectable amounts of PA. Similarly, E. coli SM2-1 derivatives harboring the R. capsulatus olsA or plsC316 contained AGPAT activity even when grown at 42°C. Moreover, membrane particles harboring PlsC316 or OlsA produced visibly more PA than their parent SM2-1 grown at 30°C (Fig. 6B, lanes 1, 3, and 4). Quantitative estimations using ImageQuant software indicated that the PA production rate was highest (approximately 10 pmol/min/μg of membrane protein) in SM2-1 cells with PlsC316, followed by cells with OlsA (0.875 pmol/min/μg of membrane protein), and lowest in SM2-1 cells grown at 30°C (0.34 pmol/min/μg of membrane protein). Apparently, expression of OlsA or PlsC316 yielded, respectively, approximately 2.5- or 11-fold more PA production than the endogenous activity present in the E. coli plsC(Ts) mutant SM2-1 grown at 30°C. We therefore concluded that both R. capsulatus olsA and plsC316 gene products have AGPAT activities, which explained why the presence of either gene was sufficient for membrane glycerophospholipid production and growth of this species. In addition, the vigorous AGPAT activity and the inability to produce OL distinguished PlsC316 from the bifunctional OlsA involved in both PA and OL synthesis and suggested that PlsC316 might be the major enzyme responsible for PA biosynthesis in R. capsulatus.
AGPAT activities of R. capsulatus PlsC316− or OlsA− mutants.
Combined GPAT-AGPAT activities in vitro were also determined using membrane preparations from R. capsulatus OlsA− (SA4) or PlsC316− (SA13) mutants to further establish that PlsC316 is the main enzyme carrying out PA biosynthesis in this species. As expected, the OlsA− mutant exhibited a combined GPAT-AGPAT activity that was approximately the same as that seen with the wild-type strain MT1131, and the PlsC316− mutant exhibited much lower (four- to fivefold) GPAT-AGPAT activity relative to both the wild-type strain MT1131 and the OlsA− mutant SA4 (Fig. 6C). Moreover, TLC with quantitative estimations using ImageQuant software showed that the PA production rate in the OlsA− mutant was almost identical (approximately 0.8 pmol/min/μg of membrane protein) to that seen with the wild-type strain MT1131 (Fig. 6D, lanes 1 and 2), whereas the PlsC316− mutant produced barely detectable amounts of PA in vitro (Fig. 6D, lane 3), in agreement with the GPAT-AGPAT activities measured. Therefore, in R. capsulatus PlsC316 is apparently the main AGPAT enzyme producing PA for membrane glycerophospholipid synthesis.
DISCUSSION
At the outset of this work, the genes encoding GPAT and AGPAT enzymes were unidentified experimentally in Rhodobacter species. Our previous studies on c-type cyt biogenesis led us to the identification of the OL biosynthesis genes, olsA and olsB, of R. capsulatus (3) and indicated that the identity of the gene carrying out PA biosynthesis was unclear. The evidence that OlsA− mutants still produced quasi-normal amounts of PA and glycerophospholipids and the occurrence of at least two additional PlsC homologues on the R. capsulatus genome led us to investigate the gene responsible for the AGPAT activity dedicated to PA biosynthesis.
The data obtained in this work indicated that R. capsulatus plsC3498 is not involved in either PA or OL synthesis. PlsC3498 shares similarity with both NlaA (15% identity and ∼25% similarity) and NlaB (∼13% identity and ∼28% similarity) from N. meningitidis. It possesses the HX4D sequence thought to correspond to the catalytic motif of GPATs and AGPATs, but compared to OlsA and PlsC316, the substrate binding motif (PEGTR) of AGPATs is not conserved (Fig. 2). It has homology to the C. butyricum PlsD (20% identity and 34% similarity), but, unlike PlsD (23), it cannot complement a GPAT-less E. coli mutant and does not appear to be a functional homologue of PlsB. Thus, the role of plsC3498 in R. capsulatus remains unknown. Moreover, whether R. capsulatus has a true PlsB homologue or whether it utilizes exclusively the PlsX/PlsY pathway for LPA biosynthesis (33) awaits the study of R. capsulatus ORFs RRC01510 and RRC02960, which exhibit significant homologies to PlsX (pfam02504/COG0416) (10) and PlsY (pfam02660/COG0344), respectively.
A major outcome of this work were the findings that the gene products of both olsA and plsC316 have AGPAT activities and that R. capsulatus, unlike E. coli, possesses two AGPAT isozymes capable of producing PA. The AGPAT activities of OlsA and PlsC316 were demonstrated by their ability to complement an E. coli mutant that has a temperature-sensitive PlsC and by GPAT-AGPAT activity assays in vitro using membrane particles prepared from appropriate E. coli and R. capsulatus strains. It was noted that PlsC316 conferred higher AGPAT activities than OlsA but displayed no OL synthesis activity at least in vivo, as OlsA− mutants are devoid of OL. Moreover, PlsC316− mutation had no effect on the steady-state amounts of c-type cyt, consistent with their OL contents. Thus, our overall findings suggested that PlsC316 is the major AGPAT enzyme, dedicated to PA biosynthesis only. This finding was further supported by the fact that R. capsulatus PlsC316− mutants have much lower AGPAT activities than OlsA− mutants. On the other hand, OlsA is primarily responsible for OL biosynthesis and also produces some PA to sustain slower growth of R. capsulatus. Although OlsA and PlsC316 share homologies with E. coli PlsC and act as AGPAT isozymes, they have distinct but overlapping cellular functions. Finally, as double mutants lacking both of these enzymes are lethal, no other gene encoding another functional AGPAT enzyme appears to be present in the R. capsulatus genome.
The O-acyltransferase OlsA is able to recognize both lyso-ornithine lipid ([LOL] a long-chain acyl amide of ornithine) and LPA (esterified sn-G3P) as substrates to which it transfers an acyl group from an acyl-ACP to yield OL and PA, respectively. In both cases, the reaction catalyzed is esterification of an α-CHOH moiety, suggesting broad substrate specificity for this enzyme beyond the accepting group. However, this relaxed substrate recognition does not seem to be a general property of all OlsA enzymes. Apparently, homologues of OlsA from some other bacteria, e.g., Sinorhizobium meliloti (48) and P. fluorescens (14), do not display any AGPAT activity, as indicated by their inability to complement an E. coli plsC(Ts) mutant, unlike the R. capsulatus OlsA. Although OlsA enzymes from different species show pronounced similarities to AGPATs of prokaryotes and eukaryotes and contain two conserved domains and the consensus (HX4D) catalytic motif, it is unclear why some of them are bifunctional and can produce both OL and PA while others can synthesize only OL. A possibility is that different OlsA enzymes might have differing specificities for their acyl donor substrates (acyl-ACP) rather than acyl acceptor substrates (LOL and LPA). If this is the case, then the R. capsulatus but not the S. meliloti or P. fluorescens OlsA seems to recognize E. coli ACP efficiently. Also consistent with the more selective behavior of S. meliloti OlsA is our earlier observation that S. meliloti OlsA− mutants can be complemented with R. capsulatus OlsA but not vice versa (3), suggesting that the latter enzyme has a more relaxed ACP specificity to recognize S. meliloti ACP for OL synthesis.
Why some organisms have multiple AGPAT isozymes is interesting. In eukaryotes, the fact that AGPATs are involved in different regulatory circuits with different substrate preferences, like cellular responses to cytokines and growth factors, has been suggested as an explanation the occurrence of multiple AGPATs expressed in different tissues (9, 20, 32). Similarly, some bacterial species including N. meningitidis, N. gonorrhoeae, and P. fluorescens have multiple AGPATs, whereas others, like E. coli, appear to have only one such enzyme. It has been suggested that the different isozymes might play different roles, such as fine-tuning the membrane lipid and fatty acid profiles in diverse environments (14, 42, 45). Indeed, while P. fluorescens OlsA− mutants exhibited no major changes in the membrane phospholipid and fatty acid profiles, inactivation of P. fluorescens AGPAT isozymes PatB and HdtS did alter the fatty acid profile of phospholipids and some membrane properties (14), as seen here with R. capsulatus OlsA− and PlsC− mutants.
In the case of N. meningitidis, apparently both NlaA and NlaB proteins displayed AGPAT activity in vitro as they complemented a temperature-sensitive E. coli plsC(Ts) mutant. Furthermore, this species might have at least an additional enzyme with AGPAT activity as an NlaA− NlaB− double mutant is viable and has AGPAT activity (42, 45). Indeed, R. capsulatus OlsA and PlsC316 show noteworthy similarities to NlaA (OlsA, ∼21% identity and ∼32% similarity; PlsC316, ∼16% identity and ∼30% similarity) and NlaB (OlsA, ∼16% identity and 27% similarity; PlsC316, ∼26% identity and ∼32% similarity), as depicted in Fig. 2. But a closer examination suggests that NlaB seems to be more homologous to PlsC316 and NlaA to OlsA, especially based on the pfam01553/COG0204 motif, suggesting that N. meningitidis might contain OL.
In summary, this work demonstrated that of the three plsC homologues encountered in the R. capsulatus genome, plsC3498 is not involved in membrane phospholipids or OL biosynthesis. On the other hand, olsA and plsC316 encode efficient AGPAT enzymes able to sustain membrane glycerophospholipid synthesis and growth of R. capsulatus; of the two isozymes, PlsC316 seems to be the major enzyme responsible for PA biosynthesis. Finally, the finding that R. capsulatus OlsA produces both OL and PA demonstrated for the first time that some OlsA homologues are bifunctional enzymes with overlapping activities. Future studies may shed light on why nature has evolved and conserved multifunctional AGPAT enzymes and how organisms use the specificity and control the promiscuity of these isoenzymes in response to their changing environments.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM38237 (to F.D.) and AI45153 (to H.G.) and Department of Energy grant ER20052 (to F.D.).
We thank Damla Erdogan for help with the constructions of plasmids pDML1, pDML3, and pDML4 and Dong-Woo Lee for assistance with purification of cis-vaccenyl-ACP.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguado, B., and R. D. Campbell. 1998. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J. Biol. Chem. 273:4096-4105. [DOI] [PubMed] [Google Scholar]
- 2.Athenstaedt, K., and G. Daum. 1999. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266:1-16. [DOI] [PubMed] [Google Scholar]
- 3.Aygun-Sunar, S., S. Mandaci, H.-G. Koch, I. V. J. Murray, H. Goldfine, and F. Daldal. 2006. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol. Microbiol. 61:418-435. [DOI] [PubMed] [Google Scholar]
- 4.Baysse, C., M. Cullinane, V. Denervaud, E. Burrowes, J. M. Dow, J. P. Morrissey, L. Tam, J. T. Trevors, and F. O'Gara. 2005. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology 151:2529-2542. [DOI] [PubMed] [Google Scholar]
- 5.Bonham, L., D. W. Leung, T. White, D. Hollenback, P. Klein, J. Tulinsky, M. Coon, P. De Vries, and J. W. Singer. 2003. Lysophosphatidic acid acyltransferase-beta: a novel target for induction of tumour cell apoptosis. Expert Opin. Ther. Targets 7:643-661. [DOI] [PubMed] [Google Scholar]
- 6.Bourgis, F., J.-C. Kader, P. Barret, M. Renard, D. Robinson, C. Robinson, M. Delseny, and T. J. Roscoe. 1999. A plastidial lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol. 120:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, A. P., J. Coleman, A. M. Tommey, M. D. Watson, and R. Slabas. 1994. Isolation and characterisation of a maize cDNA that complements a 1-acyl-sn-glycerol-3-phosphate acyltransferase mutant of Escherichia coli and encodes a protein which has similarities to other acyltransferases. Plant Mol. Biol. 26:211-223. [DOI] [PubMed] [Google Scholar]
- 8.Brown, A. P., C. L. Brough, J. T. Kroon, and J. R. Slabas. 1995. Identification of a cDNA that encodes a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii. Plant Mol. Biol. 29:267-278. [DOI] [PubMed] [Google Scholar]
- 9.Bursten, S. L., W. E. Harris, K. Bomsztyk, and D. Lovett. 1991. Interleukin-1 rapidly stimulates lysophosphatidate acyltransferase and phosphatidate phosphohydrolase activities in human mesangial cells. J. Biol. Chem. 266:20732-20743. [PubMed] [Google Scholar]
- 10.Carty, S. M., A. Colbeau, P. M. Vignais, and T. J. Larson. 1994. Identification of the rpmF-plsX-fabH genes of Rhodobacter capsulatus. FEMS Microbiol. Lett. 118:227-231. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, J. 1990. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3 phosphate acyltransferase activity. J. Biol. Chem. 265:17215-17221. [PubMed] [Google Scholar]
- 12.Coleman, J. 1992. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC). Mol. Gen. Genet. 232:295-303. [DOI] [PubMed] [Google Scholar]
- 13.Cronan, J. E., Jr., and C. O. Rock. 1987. Biosynthesis of membrane lipids, p. 474-497. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 14.Cullinane, M., C. Baysse, J. P. Morrissey, and F. O'Gara. 2005. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology 151:3071-3080. [DOI] [PubMed] [Google Scholar]
- 15.Daldal, F., S. Cheng, J. Applebaum, E. Davidson, and R. C. Prince. 1986. Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 83:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Rudder, K. E. E., J. E. Thomas-Oates, and O. Geiger. 1997. Rhizobium meliloti mutants deficient in phospholipids N-methyltransferase still contain phosphatidylcholine. J. Bacteriol. 179:6921-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dircks, L., and H. S. Sul. 1999. Acyltransferases of de novo glycerophospholipid biosynthesis. Prog. Lipid Res. 38:461-479. [DOI] [PubMed] [Google Scholar]
- 18.Ditta, G., S. Stanfield, D. Corbin, and D. Helinski. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditta, G., T. Schmidhauser, E. Yacobson, P. Lu, X. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 20.Eberhardt, C., P. W. Gray, and L. W. Tjoelker. 1997. Human lysophosphatidic acid acyltransferase. cDNA cloning, expression, and localization to chromosome 9q34.3. J. Biol. Chem. 272:20299-20305. [DOI] [PubMed] [Google Scholar]
- 21.Goldfine, H. 1969. Filter paper disk assay for lipid synthesis. Methods Enzymol. 14:649-651. [Google Scholar]
- 22.Hanke, C., F. P. Wolter, J. Coleman, G. Peterek, and M. Frentzen. 1995. A plant acyltransferase involved in triacylglycerol biosynthesis complements an Escherichia coli sn-1-acylglycerol-3-phosphate acyltransferase mutant. Eur. J. Biochem. 232:806-810. [PubMed] [Google Scholar]
- 23.Heath, R. J., H. Goldfine, and C. O. Rock. 1997. A gene (plsD) from Clostridium butyricum that functionally substitutes for the sn-glycerol-3-phosphate acyltransferase gene (plsB) of Escherichia coli. J. Bacteriol. 179:7257-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath, R. J., and C. O. Rock. 1998. A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 180:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu, S. M., and E. Soban. 1982. Color modification of diaminobenzidine (DAB) precipitation by metallic ions and its application for double immunohistochemistry. J. Histochem. Cytochem. 30:1079-1082. [DOI] [PubMed] [Google Scholar]
- 26.Kim, H. U., Y. Li, and A. H. C. Huang. 2005. Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17:1073-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutzon, D. S., K. D. Lardizabal, J. S. Nelsen, J. L. Bleibaum, H. M. Davies, and J. G. Metz. 1995. Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol. 109:999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch, H.-G., O. Hwang, and F. Daldal. 1998. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kume, K., and T. Shimizu. 1997. cDNA cloning and expression of murine 1-acyl-sn-glycerol-3-phosphate acyltransferase. Biochem. Biophys. Res. Commun. 237:663-666. [DOI] [PubMed] [Google Scholar]
- 30.Läemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Li, D., L. Yu, H. Wu, Y. Shan, J. Guo, Y. Dang, Y. Wei, and S. Zhao. 2003. Cloning and identification of the human LPAAT-zeta gene, a novel member of the lysophosphatidic acid acyltransferase family. J. Hum. Genet. 48:438-442. [DOI] [PubMed] [Google Scholar]
- 32.Lu, B., Y. J. Jiang, Y. Zhou, F. Y. Xu, G. M. Hatch, and P. C. Choy. 2005. Cloning and characterization of murine 1-acyl-sn-glycerol-3-phosphate acyltransferases and their regulation by PPARα in murine heart. Biochem. J. 385:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, Y., Y. Zhang, K. Grimes, J. Qi, R. Lee, and C. Rock. 2006. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Mol. Cell 23:765-772. [DOI] [PubMed] [Google Scholar]
- 34.Lueking, D. R., and H. Goldfine. 1975. sn-Glycerol-3-phosphate acyltransferase activity in particulate preparations from anaerobic, light-grown cells of Rhodopseudomonas sphaeroides. J. Biol. Chem. 250:8530-8535. [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Myllykallio, H., F. E. Jenney, Jr., C. R. Moomaw, C. A. Slaughter, and F. Daldal. 1997. Cytochrome cy of Rhodobacter capsulatus is attached to the cytoplasmic membrane by an uncleaved signal sequence-like anchor. J. Bacteriol. 179:2623-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagiec, M. M., G. B. Wells, R. L. Lester, and R. C. Dickson. 1993. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J. Biol. Chem. 268:22156-22163. [PubMed] [Google Scholar]
- 38.Rock, C. O., S. E. Goelz, and J. E. Jr. Cronan. 1981. Phospholipid synthesis in Escherichia coli. Characteristics of fatty acid transfer from acyl-acyl carrier protein to sn-glycerol 3-phosphate. J. Biol. Chem. 256:736-742. [PubMed] [Google Scholar]
- 39.Rock, C. O., and S. Jackowski. 1982. Regulation of phospholipid synthesis in Escherichia coli. Composition of the acyl-acyl carrier protein pool in vivo. J. Biol. Chem. 257:10759-10765. [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schägger, H., and G. Von Jagow. 1987. Tricine-sodium dodecyl sulphate sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Shih, G. C., C. M. Kahler, J. S. Swartley, M. M. Rahman, J. Coleman, R. W. Carlson, and D. S. Stevens. 1999. Multiple lysophosphatidic acid acyltransferases in Neisseria meningitidis. Mol. Microbiol. 32:942-952. [DOI] [PubMed] [Google Scholar]
- 43.Sistrom, W. R. 1960. A requirement for sodium in the growth medium of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22:77-85. [DOI] [PubMed] [Google Scholar]
- 44.Stamps, A. C., M. A. Elmore, M. E. Hill, A. A. Makda, and M. J. Finnen. 1997. A human cDNA sequence with homology to non-mammalian lysophosphatidic acid acyltransferases. Biochem. J. 326:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swartley, J. S., J. T. Balthazar, J. Coleman, W. M. Shafer, and D. S. Stephens. 1995. Membrane glycerophospholipid biosynthesis in Neisseria meningitidis and Neisseria gonorrhoeae: identification, characterization, and mutagenesis of a lysophosphatidic acid acyltransferase. Mol. Microbiol. 18:401-412. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 47.Van den Bosch, H., and P. R. Vagelos. 1970. Fatty acyl-CoA and fatty acyl-acyl carrier protein as acyl donors in the synthesis of lysophosphatidate and phosphatide in Escherichia coli. Biochim. Biophys. Acta 218:233-248. [Google Scholar]
- 48.Weissenmayer, B., J.-L. Gao, I. M. López-Lara, and O. Geiger. 2002. Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 45:721-733. [DOI] [PubMed] [Google Scholar]
- 49.West, J., C. K. Tompkins, N. Balantac, E. Nudelman, B. Meengs, T. White, S. Bursten, J. Coleman, A. Kumar, J. W. Singer, and D. W. Leung. 1997. Cloning and expression of two human lysophosphatidic acid acyltransferase cDNAs that enhance cytokine-induced signaling responses in cells. DNA Cell Biol. 16:691-701. [DOI] [PubMed] [Google Scholar]
- 50.Ye, G. M., C. Chen, S. Huang, D. D. Han, J. H. Guo, B. Wan, and L. Yu. 2005. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7. DNA Seq. 16:386-390. [DOI] [PubMed] [Google Scholar]
- 51.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.