Abstract
O-antigen variation due to the presence of different types of sugars and sugar linkages is important for the survival of bacteria threatened by host immune systems. The O antigens of Shigella dysenteriae type 7 and Escherichia coli O7 contain 4-(N-acetylglycyl)amino-4,6-dideoxy-d-glucose (d-Qui4NGlyAc) and 4-acetamido-4,6-dideoxy-d-glucose (d-Qui4NAc), respectively, which are sugars not often found in studied polysaccharides. In this study, we characterized the biosynthetic pathways for dTDP-d-Qui4N and dTDP-d-Qui4NAc (the nucleotide-activated precursors of d-Qui4NGlyAc and d-Qui4NAc in O antigens). Predicted genes involved in the synthesis of the two sugars were cloned, and the gene products were overexpressed and purified as His-tagged fusion proteins. In vitro enzymatic reactions were carried out using the purified proteins, and the reaction products were analyzed by capillary electrophoresis, electrospray ionization-mass spectrometry, and nuclear magnetic resonance spectroscopy. It is shown that in S. dysenteriae type 7 and E. coli O7, dTDP-d-Qui4N is synthesized from α-d-glucose-1-phosphate in three reaction steps catalyzed by glucose-1-phosphate thymidyltransferase (RmlA), dTDP-d-glucose 4,6-dehydratase (RmlB), and dTDP-4-keto-6-deoxy-d-glucose aminotransferase (VioA). An additional acetyltransferase (VioB) catalyzes the conversion of dTDP-d-Qui4N into dTDP-d-Qui4NAc in E. coli O7. Kinetic parameters and some other properties of VioA and VioB are described and differences between VioA proteins from S. dysenteriae type 7 (VioAD7) and E. coli O7 (VioAO7) discussed. To our knowledge, this is the first time that functions of VioA and VioB have been biochemically characterized. This study provides valuable enzyme sources for the production of dTDP-d-Qui4N and dTDP-d-Qui4NAc, which are potentially useful in the pharmaceutical industry for drug development.
Lipopolysaccharide, also known as endotoxin, is the major component of the outer membranes of gram-negative bacteria. It consists of three moieties: lipid A, the core oligosaccharide, and the O-specific polysaccharide (O antigen). The O antigen is composed of a number of repeats of an oligosaccharide unit (O unit). The O unit usually contains 2 to 8 residues from a broad range of monosaccharides and their derivatives (6, 9). The O antigen is one of the most varied cell constituents due to the variation in the types of sugars present, the arrangement of the sugars within the O unit, and the linkages between sugars. More than 180 O-antigen forms have been recognized in Escherichia coli (including Shigella) (33). The O antigen is a major target of the immune system and bacteriophages, and O-antigen variation is important for bacterial survival and pathogenicity (31). Several studies have indicated that the composition of the O antigen might be an indicator of virulence potential (18), and some O-antigen forms, including that in E. coli O7, are most commonly found in pathogenic E. coli strains (25). Shigella strains are important human pathogens causing diseases such as diarrhea and bacillary dysentery (26). Genes for the synthesis of the O antigen are normally located in a gene cluster which maps between galF and gnd on the chromosomes of E. coli and Shigella (32). Many O-antigen gene clusters have been sequenced, with gene functions proposed (http://www.mmb.usyd.edu.au/BPGD/default.htm). Biosynthetic pathways for several O-antigen sugar precursors, such as UDP-l-FucNAc, have been characterized biochemically (15, 16, 22, 27, 30, 36).
The O antigen of Shigella dysenteriae type 7 is composed of a tetrasaccharide O unit containing a residue of 4-(N-acetylglycyl)amino-4,6-dideoxy-d-glucose (d-Qui4NGlyAc) (17). The same O-antigen structure is also present in E. coli O121, which belongs to Shiga toxin-producing E. coli causing hemolytic uremic syndrome (28, 38). The E. coli O7 O antigen is composed of a pentasaccharide O unit containing a residue of 4-acetamido-4,6-dideoxy-d-glucose (d-Qui4NAc) (19). Both Qui4NGlyAc and Qui4NAc belong to dideoxy(acetylglycyl/acetyl)amino hexoses, which are sugars not often found in studied polysaccharides.
In our previous study, the O-antigen gene cluster of S. dysenteriae type 7 was sequenced and three genes from this organism, rmlAD7, rmlBD7, and vioAD7, were proposed to be responsible for the synthesis of dTDP-d-Qui4N (the nucleotide-activated precursor of d-Qui4NGlyAc) (10). Glucose-1-phosphate thymidyltransferase (RmlA) catalyzes the conversion of glucose-1-phosphate to dTDP-d-glucose (dTDP-d-Glc), which is then converted to dTDP-4-keto-6-deoxy-d-glucose (dTDP-d-Glc4O) by dTDP-d-glucose 4,6-dehydratase (RmlB). Both reaction steps have been biochemically verified in a number of bacterial strains (11, 30). VioA, a putative sugar aminotransferase (SAT) of the DegT/DnrJ/EryC1/StrS family, was proposed to catalyze the conversion of dTDP-d-Glc4O to dTDP-d-Qui4N. The E. coli O7 O-antigen gene cluster was also sequenced (20), and the biosynthetic pathway for dTDP-d-Qui4NAc was proposed, in which dTDP-d-Qui4NAc is derived from dTDP-d-Qui4N by a transacetylation reaction catalyzed by a putative acetyltransferase of the NodL-LacA family, VioBO7 (20). However, the functions of VioA and VioB have not yet been confirmed.
In this study, we characterized the biosynthetic pathways of dTDP-d-Qui4N and dTDP-d-Qui4NAc. Genes encoding RmlA, RmlB, VioA, and VioB were cloned from S. dysenteriae type 7 (rmlAD7, rmlBD7, and vioAD7) and/or E. coli O7 (vioAO7 and vioBO7), and the gene products were overexpressed and purified as His-tagged fusion proteins. In vitro enzymatic reactions were carried out, and the products were analyzed by spectroscopic methods. VioA was identified as a novel aminotransferase catalyzing the conversion of dTDP-d-Glc4O to dTDP-d-Qui4N in S. dysenteriae type 7 and E. coli O7, and VioB was identified as a novel acetyltransferase catalyzing the conversion of dTDP-d-Qui4N to dTDP-d-Qui4NAc in E. coli O7. Enzymatic properties of VioA and VioB were investigated, and differences between VioAD7 and VioAO7 are discussed.
MATERIALS AND METHODS
Materials.
α-d-Glucose-1-phosphate, dTTP, dTDP-d-Glc, pyridoxal-5-phosphate (PLP), inorganic pyrophosphatase, acetyl coenzyme A (acetyl-CoA), SH-CoA, and aminooxy acetic acid were purchased from Sigma-Aldrich (St. Louis, MO), methanol and acetonitrile were purchased from Fisher (Pittsburgh, PA), and acetic acid was purchased from Fluka (Buchs SG, Switzerland). All the chemicals were at the highest purity available. Restriction enzymes and recombinant Taq DNA polymerase were from TaKaRa (Japan), and T4 DNA ligase was from Promega (Madison, WI). Other chemicals and reagents were from Sangon Co., Ltd. (Shanghai, China). Bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Bacterial strain, plasmid, or primer | Description or sequencea | Sourceb |
|---|---|---|
| Strains | ||
| G1222 | S. dysenteriae serotype 7 type strain | IMVS |
| G1112 | E. coli O7 type strain | NICPBP |
| E. coli BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| E. coli DH5α | F− φ80lacZΔM15 endA recA1 hsdR(rK− mK−) supE44 thi-1 gyrA96 relA1 (lacZYA-argF)U169 | TBC |
| Plasmids | ||
| pET28a+ | T7 expression vector, Kanr | Novagen |
| pLW1048 | pET28a+ containing N-terminally six-histidine-tagged rmlAD7 at the EcoRI/XhoI site | This work |
| pLW1051 | pET28a+ containing C-terminally six-histidine-tagged rmlBD7 at the NcoI/XhoI site | This work |
| pLW1049 | pET28a+ containing N-terminally six-histidine-tagged vioAD7 at the EcoRI/XhoI site | This work |
| pLW1195 | pET28a+ containing N-terminally six-histidine-tagged vioAO7 at the BamHI/HindIII site | This work |
| pLW1155 | pET28a+ containing C-terminally six-histidine-tagged vioBO7 at the NcoI/SalI site | This work |
| Primers | AuGCT | |
| wl-4235 (rmlA-D7F) | 5′-TAGAATTCATGGCTTACTCAGCAGTATG-3′ | |
| wl-4236 (rmlA-D7R) | 5′-TTGGCTCTCGAGTTATTTGTCCTTAAC-3′ | |
| wl-4241 (rmlB-D7F) | 5′-GCAGCCATGGAAATCCTTATTACAG-3′ | |
| wl-4242 (rmlB-D7R) | 5′-GTCCTCGAGTTTCATTTCTCCATACTG-3′ | |
| wl-4243 (vioA-D7F) | 5′-GAGAATTCATGGAAAAGCCAATCTTTGTAAC-3′ | |
| wl-4244 (vioA-D7R) | 5′-GCACTTCTCGAGTCATTTAATCTCCCTAATC-3′ | |
| wl-5702 (vioA-O7F) | 5′-GCAGGATCCATGAACGATAAAACTATTCCAGTAAC-3′ | |
| wl-5703 (vioA-O7R) | 5′-GTCGAAGCTTTCACATCTTACCCAATAATAATTTG-3′ | |
| wl-4704 (vioB-O7F) | 5′-GGTCCCATGGCCTATTTAGATGAAATAC-3′ | |
| wl-4705 (vioB-O7R) | 5′-CGGAAGTCGACCAGATTATCTCCAATAG-3′ |
The underlined sequences indicate restriction sites.
IMVS, the Institute of Medical and Veterinary Science, Adelaide, Australia; NICPBP, the National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China; TBC, Tianjin Biochip Corporation, Tianjin, China; AuGCT, AuGCT Biotechnology Corporation, Beijing, China.
Cloning and plasmid construction.
Chromosomal DNA was prepared as previously described (3). The rmlAD7, rmlBD7, and vioAD7 genes were PCR amplified from strain G1222, and the vioAO7 and vioBO7 genes were amplified from strain G1112. Primers used are listed in Table 1. A total of 30 cycles (25 cycles for vioAO7) were performed using the following conditions: denaturation at 95°C for 30 s, annealing at 50°C (55°C for vioAO7) for 30 s, and extension at 72°C for 1 min in a final volume of 25 μl. The amplified genes were cloned into pET28a+ to construct plasmids pLW1048 (containing rmlAD7), pLW1051 (containing rmlBD7), pLW1049 (containing vioAD7), pLW1195 (containing vioAO7), and pLW1155 (containing vioBO7) (Table 1), and the inserts were sequenced by Tianjin Biochip Cooperation using an ABI 3730 sequencer.
Protein expression and purification.
E. coli BL21(DE3) carrying each of the recombinant plasmids was grown overnight at 37°C in LB medium containing 50 μg/ml kanamycin. The overnight culture (5 ml) was inoculated into 500 ml of fresh LB medium and grown until the A600 reached 0.6. The expression of RmlAD7, VioAD7, and VioBO7 was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 4 h, and expression of RmlBD7 and VioAO7 was induced with 0.1 mM IPTG at 12°C for 8 h and at 25°C for 4 h. After IPTG induction, cells were harvested by centrifugation, washed with 50 mM Tris-HCl (pH 8.0) containing 300 mM NaCl and 10 mM imidazole, resuspended into 5 ml of the same buffer, and sonicated. The cell debris was removed by centrifugation, and total soluble proteins in the supernatant were collected. The His6-tagged fusion proteins in the supernatant were purified by nickel ion affinity chromatography with a chelating Sepharose Fast Flow (GE Healthcare) column according to the manufacturer's instructions. Unbound proteins were washed out with 100 ml of wash buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 25 mM imidazole). The fusion proteins were eluted with 3 ml of elution buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, and 250 mM imidazole) and dialyzed overnight against 50 mM potassium phosphate buffer (pH 7.4) at 4°C. Protein concentration was determined by the Bradford method.
Enzyme activity assays.
The reaction mixture for RmlA contained 50 mM potassium phosphate buffer (pH 7.4), 5 mM MgCl2, 4 mM α-d-glucose-1-phosphate, 4 mM dTTP, 0.02 U inorganic pyrophosphatase, and a 0.13 μM concentration of purified RmlAD7 (39). The reaction mixture for RmlB contained 50 mM potassium phosphate buffer (pH 7.4), 5 mM MgCl2, 8 mM dTDP-d-Glc, and a 2.65 μM concentration of purified RmlBD7 (39). To assay the VioA activity, the RmlB mixture was supplemented with 50 mM l-glutamate (l-Glu), 0.2 mM PLP, and 2 μM VioAD7 or 0.66 μM VioAO7 after the RmlB reaction (30). To assay the VioB activity, the VioA mixture was supplemented with 2.5 mM acetyl-CoA and 1.93 μM VioBO7 after the VioA reaction (30). All reactions were carried out in a final volume of 50 μl at 37°C for 2 h unless otherwise indicated. Products from each of the reactions were analyzed by capillary electrophoresis (CE), electrospray ionization mass spectrometry (ESI-MS), and nuclear magnetic resonance spectroscopy (NMR). Enzyme activities were indicated by the conversion of substrates into products.
Kinetic-parameter measurements.
To measure Km and Vmax values of VioAD7 and VioAO7, reactions were carried out with various concentrations of dTDP-d-Glc4O (0.05 to 2 mM for VioAD7 and 0.02 to 0.1 mM for VioAO7) with 1 μM VioAD7 and 0.022 μM VioAO7. To measure Km and Vmax values of VioB, reactions were carried out with various concentrations of dTDP-d-Qui4N (0.08 to 0.48 mM), a constant concentration of acetyl-CoA (3 mM), various concentrations of acetyl-CoA (0.187 to 1.87 mM), and a constant concentration of dTDP-d-Qui4N (1.9 mM) with a 3.86 nM concentration of purified VioBO7. All reactions were performed in a final volume of 20 μl, and reactions were terminated by adding an equal volume of chloroform. Conversion from dTDP-d-Glc4O to dTDP-d-Qui4N and from dTDP-d-Qui4N to dTDP-d-Qui4NAc was examined by CE. Conversion of acetyl-CoA to SH-CoA was measured with Ellman's reagent according to the method of Pfoestl et al. (30). Km and Vmax values were calculated based on the Michaelis-Menten equation. The data are averages of results from three independent experiments.
Determination of temperature optimum, divalent-cation effects, and amino donor requirements for VioAD7, VioAO7, and VioBO7.
To determine the temperature optimum for each enzyme, reactions were carried out at 4, 15, 25, 37, 50, 65, and 75°C, respectively. To test the effects of different cations on enzyme activity, reactions were carried out in the presence of 5 mM MgCl2, MnCl2, FeSO4, CoCl2, and CaCl2 for 1 h (VioAD7), 30 min (VioAO7), or 20 min (VioBO7). To test the amino donors for the transamination reactions catalyzed by VioAD7 and VioAO7, reactions were carried out in the presence of 50 mM l-arginine (l-Arg), l-aspartic acid (l-Asp), l-asparagine (Asn), l-glycine (l-Gly), l-alanine (l-Ala), l-glutamine (l-Gln), or l-Glu. Enzyme activities were examined by CE, and the data are the averages of results from three independent experiments.
CE.
CE was performed using a Beckman Coulter P/ACE MDQ CE system with a photoelectricity diode array detector (Beckman Coulter, CA). The capillary was bare silica (inside diameter, 75 μm by 57 cm, with the detector at 50 cm) and conditioned before each run by washing it with 0.1 M NaOH first, with deionized water next, and with 25 mM borate-sodium hydroxide, pH 9.4 (used as the mobile phase) last for 2 min each time. Samples were loaded by pressure injection at 0.5 lb/in2 for 10 s, and separation was carried out at 20 kV. Peak integration and trace alignments were done with Beckman P/ACE Station software (32 Karat, version 5.0). Conversion ratios were calculated by comparing the peak areas of the substrate and product.
ESI-MS and tandem MS.
The reaction mixtures of RmlBD7, VioAD7, and VioBO7 were separated by reverse-phase high-performance liquid chromatography (RP-HPLC) using a BioCAD 700E perfusion chromatography workstation (Applied Biosystems, CA) with an Venusil MP-C18 column (5-μm particle size, 4.6 by 250 mm) (Agela Technologies, Inc.). The mobile phase used was composed of 10% acetonitrile and 90% 50 mM triethylamine-acetic acid (pH 6.0), and the flow rate was 0.6 ml/min. Fractions containing the expected products were collected, lyophilized, and redissolved in 50% methanol before they were injected into a Finnigan LCQ Advantage MAX ion trap mass spectrometer (Thermo Electron, CA) in the negative mode (4.5 kV, 250°C) for ESI-MS analysis. For second and third MS (MS2 and MS3) analyses, nitrogen was used as the collision gas and helium as the auxiliary gas and collision energies used were typically 20 to 30 eV.
NMR spectroscopy.
A sample of dTDP-d-Qui4NAc (0.2 mg) was deuterium exchanged by freeze-drying from D2O, dissolved in 99.96% D2O (150 μl), and examined using a Shigemi (Japan) microtube. NMR spectra were recorded on a Bruker DRX-500 spectrometer (Germany) at 30°C using internal sodium trimethylsilyl- [2,2,3,3-2H4]propanoate (δH 0.00) and external aqueous 85% H3PO4 (δP 0) as references. Two-dimensional NMR spectra were obtained using standard pulse sequences from the manufacturer, and the XWinNMR 2.6 program (Bruker) was used to acquire and process the NMR data. A mixing time of 200 ms was used in a total correlation spectroscopy (TOCSY) experiment, which was employed to correlate all coupled protons within each spin system (4).
RESULTS
Overexpression and purification of enzymes.
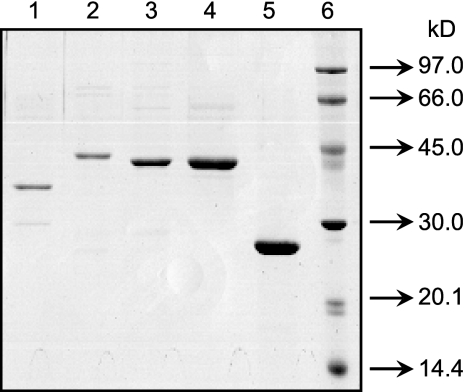
Plasmids pLW1048 (containing rmlAD7), pLW1051 (containing rmlBD7), pLW1049 (containing vioAD7), pLW1195 (containing vioAO7), and pLW1155 (containing vioBO7) were constructed, and the expression of each of the five genes in E. coli BL21(DE3) carrying the corresponding plasmids was induced by IPTG as described in Materials and Methods. The majority of each protein was found in the soluble fraction (data not shown), and the proteins were purified to near homogeneity by nickel ion affinity chromatography (Fig. 1). The apparent molecular masses of histidine-tagged RmlAD7, RmlBD7, VioAD7, VioAO7, and VioBO7 estimated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis were 38.3, 45.6, 43.7, 43.8, and 27.1 kDa, respectively, correlating well with the predicted molecular masses (36.1, 42.3, 42.5, 45.2, and 22.4 kDa).
FIG. 1.
SDS-polyacrylamide gel electrophoresis of purified RmlAD7 (lane 1), RmlBD7 (lane 2), VioAD7 (lane 3), VioAO7 (lane 4), and VioBO7 (lane 5). Proteins were denatured at 100°C for 5 min in 0.1% SDS and 1% 2-mercaptoehanol before being loaded in a 5% (wt/vol) stacking gel and separated in a 12% (wt/vol) separation gel. The gel was stained with Coomassie bright blue R250. The molecular weight markers from the LMW-SDS marker kit (GE Healthcare) are indicated at the right of the panel.
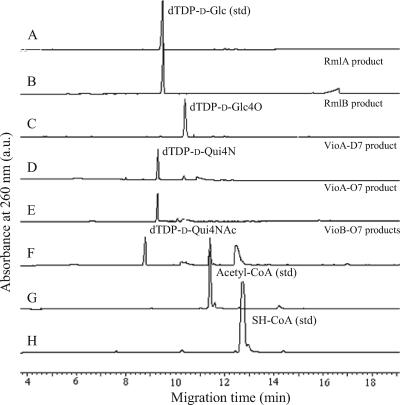
In vitro characterization of dTDP-d-Qui4N and dTDP-d-Qui4NAc biosynthetic pathways by CE.
Biosynthetic pathways of dTDP-d-Qui4N and dTDP-d-Qui4NAc were characterized by carrying out each of the enzymatic steps sequentially in a single reaction mixture containing α-d-glucose-1-phosphate as the initial substrate and by analyzing each of the reaction products by CE. When RmlAD7, the first enzyme of the pathways, was added, a peak eluted at the same time (9.5 min) that the standard dTDP-d-Glc (Fig. 2A and B) was produced, confirming the function of RmlAD7 as RmlA. When the second enzyme, RmlBD7, was added, the peak corresponding to dTDP-d-Glc was converted to a peak that eluted at 10.4 min, indicating that the emerging peak is the product of RmlB (Fig. 2C). The peak at 10.4 min was converted to a new peak that eluted at 9.3 min when l-glutamate, PLP, and purified VioAD7 were added next (Fig. 2D), and this peak had not appeared when VioAD7 was heat denatured before the addition or in the presence of the PLP-dependent aminotransferase inhibitor aminooxy acetic acid (12; data not shown). The same was found when purified VioAO7 was used instead of VioAD7 (Fig. 2E). These results indicated that the peak at 9.3 min was the product of VioAD7 and VioAO7 and that both enzymes are PLP-dependent aminotransferases catalyzing the transfer of an amino group onto the RmlB product. The peak at 9.3 min disappeared almost completely, and another new peak that migrated at 8.8 min appeared when acetyl-CoA and purified VioBO7 were added to the reaction mixture (Fig. 2F). Conversion of acetyl-CoA to SH-CoA was also detected by comparing the migration times of the peaks before and after the reaction with those of the standard chemicals (Fig. 2G and H), indicating that the acetyl group was transferred to the VioA product. No products were produced when VioBO7 was heat denatured before the addition (data not shown).
FIG. 2.
CE chromatographs of reaction products. Shown are chromatographs of standard (std) dTDP-d-Glc without addition (A) and after the addition of RmlAD7 (B); RmlAD7 and RmlBD7 (C); RmlAD7, RmlBD7, and VioAD7 (D); RmlAD7, RmlBD7, and VioAO7 (E); RmlAD7, RmlBD7, VioAO7, and VioBO7 (F); standard acetyl-CoA (G); and standard SH-CoA (H). a.u., arbitrary units.
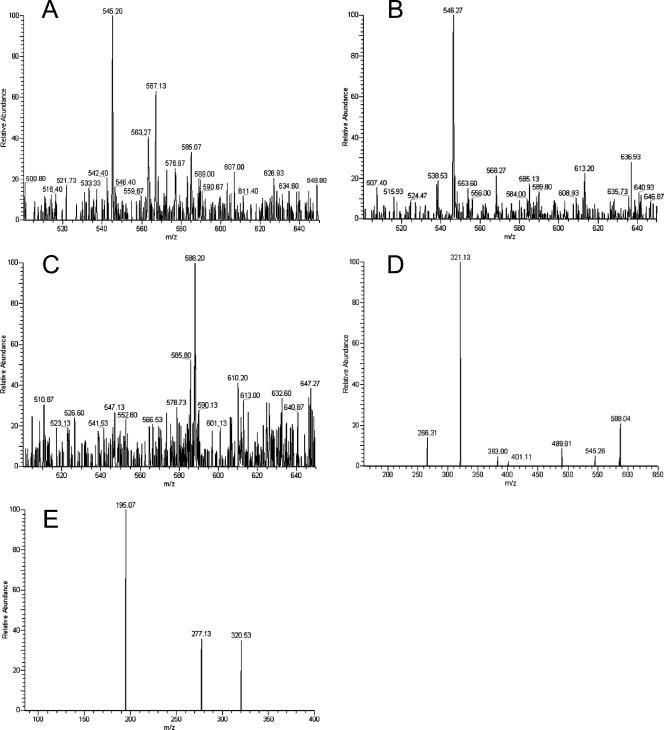
Structural identification of the reaction products by ESI-MS and tandem MS.
Products of RmlBD7, VioAD7, and VioBO7 were separated and purified by RP-HPLC (data not shown). Fractions containing each of the RmlB, VioA, and VioB products were collected and analyzed by ESI-MS (Fig. 3A to C). Ion peaks at m/z 545.20 (Fig. 3A), 546.27 (Fig. 3B), and 588.20 (Fig. 3C) were obtained, which are in agreement with the expected masses for dTDP-d-Glc4O (m/z 544.32), dTDP-d-Qui4N (m/z 547.34), and dTDP-d-Qui4NAc (m/z 589.38). Ion peaks representing each sugar plus one or two Na+ ions were also detected (Fig. 3A to C; Table 2).
FIG. 3.
ESI-mass spectra of dTDP-d-Glc4O (A), dTDP-d-Qui4N (B), and dTDP-d-Qui4NAc (C); MS2 analysis of the ion with m/z 588.20 in panel C (D); and MS3 analysis of the ion with m/z 321.13 in panel D (E).
TABLE 2.
Interpretations of the ion peaks shown in Fig. 3
| Composition of fragment | Molecular formula | Molecular wt | Mass (negative) |
|---|---|---|---|
| dTDP-d-Glc4O (full scan) | |||
| dTDP-d-Glc4O-Na2 | C16H22P2O15N2Na2 | 590.27 | 589.00 |
| dTDP-d-Glc4O-Na | C16H23P2O15N2Na | 568.29 | 567.13 |
| dTDP-d-Glc4O | C16H22P2O15N2 | 544.32 | 545.20 |
| dTDP-d-Qui4N (full scan) | |||
| dTDP-d-Qui4N-Na2 | C16H25P2O14N3Na2 | 591.31 | 589.80 |
| dTDP-d-Qui4N-Na | C16H25P2O14N3Na | 569.32 | 568.27 |
| dTDP-d-Qui4N | C16H27P2O14N3 | 547.34 | 546.27 |
| dTDP-d-Qui4NAc(full scan) | |||
| dTDP-d-Qui4NAc-Na2 | C18H27P2O15N3Na2 | 633.34 | 632.60 |
| dTDP-d-Qui4NAc-Na | C18H28P2O15N3Na | 611.35 | 610.20 |
| dTDP-d-Qui4NAc | C18H29P2O15N3 | 589.38 | 588.20a |
| dTDP-d-Qui4NAc (MS2, 588.20) | |||
| dTDP-d-Qui4NAc minus acetyl group | C16H26P2O14N3 | 546.32 | 545.26 |
| dTDP-d-Qui4NAc minus CH3-CO-NH-CH-CH-CH3 | C13H20P2O14N2 | 490.25 | 489.91 |
| dTDP plus H2O | C10H15P2O11N2 | 401.18 | 401.11 |
| dTDP | C10H13P2O10N2 | 383.16 | 383.00 |
| Qui4NAc-PO3-PO3 minus acetyl group | C6H14P2O10N | 322.12 | 321.13a |
| Qui4NAc-PO3-PO3 minus CH3-CO-NH-CH-CH-CH3 | C3H8P2O10 | 266.04 | 266.31 |
| dTDP-d-Qui4NAc (MS3, 321.13) | |||
| Qui4NAc-PO3-PO3 minus acetyl and CH3-CH-O | C4H10P2O9N | 278.07 | 277.13 |
| PO3-PO3-2H2O | P2O8H6 | 195.99 | 195.07 |
The ESI-mass peaks at m/z 588.20 and 321.13 were selected as parent ions for MS2 and MS3, respectively.
Further analyses by MS2 and MS3 were performed to confirm the molecular mass of the VioBO7 product, the end product of the Qui4NAc biosynthetic pathway. MS2 analysis of the product peak at m/z 588.20 produced peaks at m/z 545.26, 401.11, 383.00, and 321.13, matching the masses of the dTDP-d-Qui4NAc minus acetyl group, dTDP-H2O, dTDP, and the Qui4NAc-PO3-PO3 minus acetyl group, respectively. Peaks corresponding to dTDP-d-Qui4NAc minus CH3-CO-NH-CH-CH-CH3 (m/z 489.91) and Qui4NAc-PO3-PO3 minus CH3-CO-NH-CH-CH-CH3 (m/z 266.31) resulting from fragmentation of the hexose rings between C-3-C-4 and C-5-O were also detected (Fig. 3D; Table 2). When the ion peak at m/z 321.13 was selected as a parent peak for MS3 analysis, a peak at m/z 277.13, which matched the fragment of Qui4NAc-PO3-PO3 minus acetyl and CH3-CH-O, resulted from fragmentation of the hexose rings between C-4-C-5 and O-C-1 (Fig. 3E; Table 2). These results indicated that an acetamido group had been added to the C-4 position of the glucose moiety of the VioBO7 product, confirming the identity of Qui4NAc as the product of VioBO7. Fragments corresponding to each peak are depicted in Table 2.
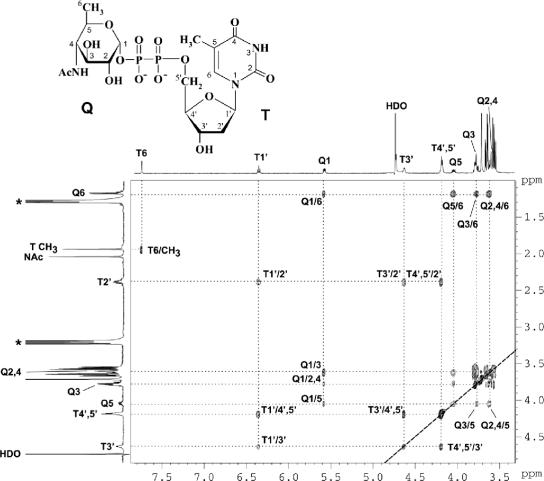
Determination of the configuration at C-4 of the hexose unit in the VioBO7 product by NMR spectroscopy.
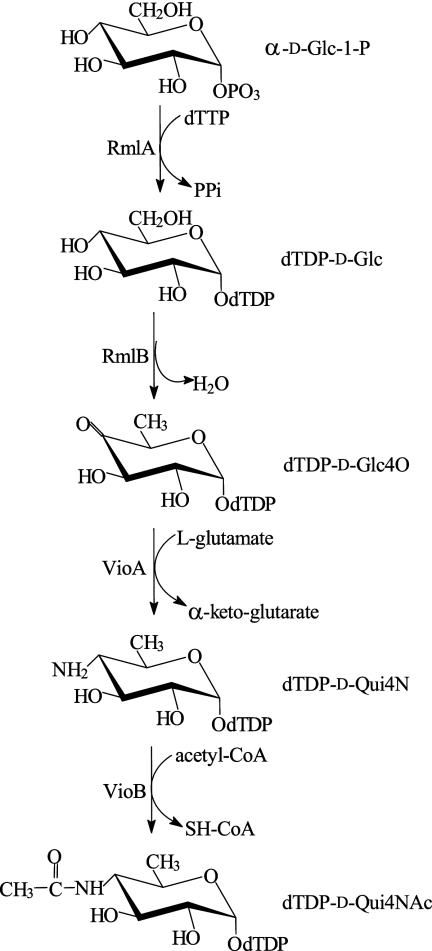
The 1H NMR spectrum of dTDP-d-Qui4NAc was assigned using two-dimensional 1H,1H COSY and TOCSY experiments. In the TOCSY spectrum (Fig. 4), cross-peaks were generated between all coupled protons within each spin system, including those between H-1 and H-2-H-6 of the QuiNAc moiety at δ 5.58 (H-1)/3.61 (H-2), 3.77 (H-3), 3.63 (H-4), 4.06 (H-5), and 1.18 (H-6); between H-1′ and H-2′-H-5′ of 2′-deoxyribose in the dTDP moiety at δ 6.35 (H-1)/2.38 (H-2), 4.63 (H-3), and 4.18 (H-4-H-5a and -H-5b); and between H-6 and CH3 of thymine at δ 7.74/1.93. The assigned chemical shifts were in good agreement with those published for dTDP (24) and α-Qui4NAc (29). Relatively large 3J2,3, 3J3,4, and 3J4,5 coupling constants (ca. 9 to 10 Hz) determined from the 1H NMR spectrum are characteristic for the all-axial orientation of the Qui4NAc ring protons; hence, the sugar has the gluco configuration and is thus 4-acetamido-4,6-dideoxy-α-d-glucose. The 31P NMR spectrum contained signals for a diphosphate group at δ −11.1 and −12.7, which, as expected, showed strong correlations with the H-5′ signal of dTDP at δ −11.1/4.18 and H-1 of α-Qui4NAc at δ −12.7/5.58 in the 1H,31P heteronuclear multiquantum coherence spectrum. These data proved finally that the enzymatic product of VioBO7 is dTDP-d-QuiNAc, whose structure is depicted in Fig. 4. The biosynthetic pathways for dTDP-d-Qui4N and dTDP-d-Qui4NAc are summarized in Fig. 5.
FIG. 4.
Part of a 1H,1H TOCSY spectrum of dTDP-d-Qui4NAc. The corresponding parts of the 1H NMR spectrum are displayed along the axes. Arabic numerals refer to protons in the Qui4NAc and dTDP moieties designated Q and T, respectively. Signals of contaminating triethylamine are marked with an asterisk. Connectivities between coupled protons in the spin systems of Qui4NAc (Q1-Q6), 2′-deoxyribose (T1′-T5′), and thymine (T6-CH3) are traced by dotted lines. The structure of dTDP-d-Qui4NAc, the enzymatic product of VioBO7, is shown.
FIG. 5.
Pathways for the biosynthesis of dTDP-d-Qui4N and dTDP-d-Qui4NAc. α-d-Glc-1-P, α-d-glucose-1-phosphate; PPi, inorganic pyrophosphate.
Kinetic parameters for VioAD7, VioAO7, and VioBO7.
Kinetic parameters for the three enzymes investigated are listed in Table 3. The kinetics of the reactions catalyzed by VioAD7, VioAO7, and VioBO7 fit reasonably well to the Michaelis-Menten model (Fig. S1 in the supplemental material). The Km values for dTDP-d-Glc4O of VioAD7 and VioAO7 are 980 μM and 45.8 μM, respectively, and the Km values of VioBO7 are 142 μM for dTDP-d-Qui4N and 554 μM for acetyl-CoA (Table 3).
TABLE 3.
Kinetic parameters
| Enzyme | Substrate | Km (μM) | Vmax (μM/s) | kcat/s |
|---|---|---|---|---|
| VioAD7 | dTDP-d-Glc4O | 980 ± 157 | 0.74 ± 0.42 | 0.74 ± 0.42 |
| VioAO7 | dTDP-d-Glc4O | 45.8 ± 7.3 | 0.125 ± 0.009 | 5.66 ± 0.41 |
| VioBO7 | SH-CoA | 554 ± 73 | 0.953 ± 0.059 | 21.6 ± 1.3 |
| VioBO7 | dTDP-d-Qui4N | 142 ± 15 | 0.601 ± 0.159 | 155.3 ± 41.4 |
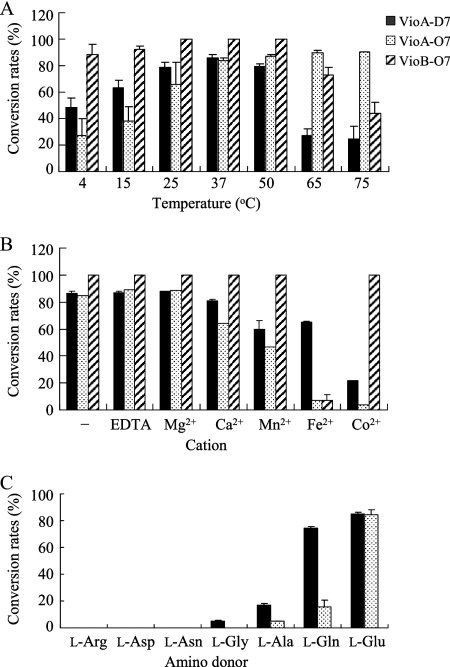
Determination of temperature optimum and shelf life for VioAD7, VioAO7, and VioBO7.
Activities of VioAD7, VioAO7, and VioBO7 at temperatures ranging from 4 to 75°C are shown in Fig. 6A. Different temperature effects were observed for VioAD7 and VioAO7. For VioAO7, the conversion rate increased along with a rise in temperature and reached to 90.3% at 75°C. For VioAD7, the conversion rate was highest (90.7%) at 37°C and greatly decreased at 65°C and above. At lower temperatures ranging from 4 to 25°C, the conversion rate for VioAO7 was much lower than for VioAD7. VioBO7 showed high activity over a wide temperature range from 4 to 65°C, with complete conversion at temperatures of 25, 37, and 50°C (Fig. 6A). Conversion rates at temperatures above 75°C were not tested due to the instability of nucleotide-activated sugars at high temperatures, as indicated by the detection of the breakdown products by CE (data not shown). After being preserved in 50% glycerol for 2 months at −80°C, no significant loss of activity was observed for VioAD7, VioAO7, and VioBO7.
FIG. 6.
Effects of temperature (A), cations (B), and amino donors (C) on the conversion rates of VioAD7, VioAO7, and VioBO7.
Effects of divalent cations on the completion of enzyme-substrate reactions.
The effects of divalent cations, including Mg2+, Ca2+, Mn2+, Fe2+, and Co2+, on the activities of VioAD7, VioAO7, and VioBO7 are shown in Fig. 6B. For VioAD7 and VioAO7, Mg2+ had no effects on the activities of both enzymes, but other cations tested showed different levels of inhibition of the activities, with Co2+ being the strongest inhibitor. VioAO7 was more prone to divalent-cation inhibition than VioAD7, with the rate of conversion in the presence of Co2+ being only 3.9%, compared with 22.2% for VioAD7. For VioBO7, complete conversion was obtained in the absence of cations and in the presence of Mg2+, Ca2+, Mn2+, and Co2+; however, the conversion rate obtained in the presence of Fe2+ was only 4.2%, indicating that all cations tested except for Fe2+ have no effects on the activity of VioBO7. The inhibition of all cations was released when EDTA as the cation-chelating agent was added (data not shown). All those indicate that all three enzymes are divalent cation independent. EDTA also had no effect on the activities of all three enzymes (Fig. 6B).
Analysis of amino donors for VioAD7 and VioAO7.
Seven amino acids, including l-Arg, l-Asp, l-Asn, l-Gly, l-Ala, l-Gln, and l-Glu, were tested as amino donors for the transamination reaction catalyzed by VioAD7 and VioAO7 (Fig. 6C). High conversion rates for both VioAD7 (84.9%) and VioAO7 (84.5%) were obtained when l-Glu was utilized, and this is consistent with other PLP-dependent SATs (12). l-Gln was also an effective amino donor for VioAD7, with a 74.1% conversion rate being detected, but less effective for VioAO7 (15.9%). In addition, l-Ala could be used by both VioA enzymes and l-Gly by only VioAD7 as poor amino donors.
DISCUSSION
This is the first report on the characterization of the biosynthetic pathways for dTDP-d-Qui4N and dTDP-d-Qui4NAc. Genes encoding enzymes for all four reaction steps were functionally confirmed by in vitro experiments. Until now, functions of VioA and VioB have not unambiguously been defined. An early study reported purification of a SAT from E. coli strain B for the conversion of TDP-d-Glc4O to TDP-d-Qui4N; however, no structural verification of the enzyme product was carried out, and a putative gene was not proposed (21). RmlA and RmlB catalyzing the first two steps of the pathways have been functionally identified in several other bacterial species (11, 30). In this study, we confirmed the functions of RmlA and RmlB from S. dyenteriae type 7 and utilized those two enzymes to produce the substrate (dTDP-d-Glc4O) for the assay of VioA. The product of the VioA reaction was then used as the substrate (dTDP-d-Qui4N) for the VioB reaction. Neither of the two substrates is commercially available. Accordingly, concentrations of those two chemicals were calculated based on conversion rates of the substrates from the previous reaction steps.
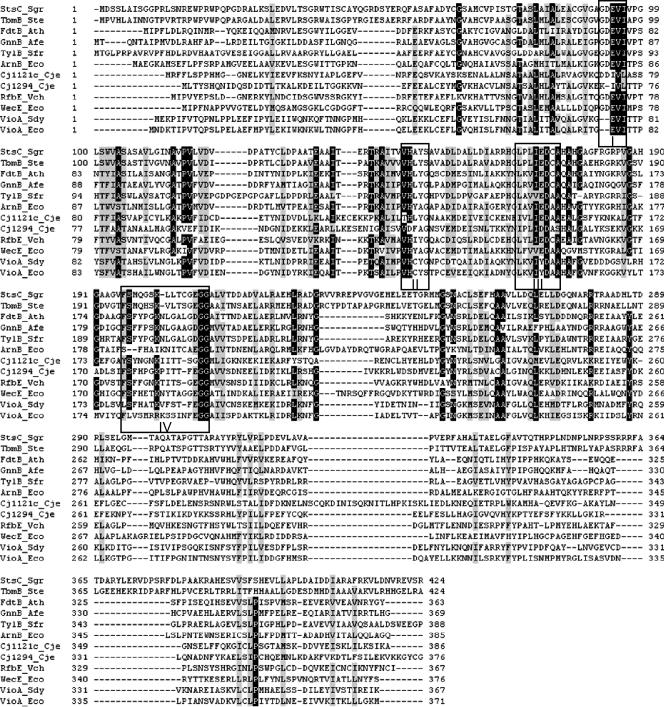
VioAD7 and VioAO7 catalyze the same transamination reaction. Sequence analysis reveals that the two SATs share 53% identity (67% similarity), which is much higher than the levels they share with other reported SATs (Table S1 in the supplemental material). Based on the position of the amino receptor, SATs can be divided into three subgroups acting on scyllo-inosose (1, 13, 14), NDP-3-keto sugars (7, 23, 30, 35), or NDP-4-keto sugars (2, 5, 12, 27, 34, 36). This is also supported by the phylogenetic analysis of SATs (Fig. S2 in the supplemental material). As expected, VioAD7 and VioAO7 fall into the NDP-4-keto sugar subgroup but form a separate branch within the subgroup in the phylogenetic tree.
Sequence alignment of VioAD7, VioAO7, and other reported SATs reveals the substitution of amino acids at the potential activity sites in motifs II and IV in VioAO7 (Fig. 7). A glycine residue in motif II is replaced by a serine residue in VioAO7, and the same was found in StsC and TbmB, the glutamine:scyllo-inosose aminotransferases from Streptomycetes (1, 14). A lysine residue in motif IV has been suggested to form a Schiff base with cofactor PLP (12). In VioAO7, we found two arginine residues and one lysine residue in tandem at the site. As in lysine, arginine also contains the ɛ-NH2 group and is expected to contribute to the binding of PLP. This may explain the higher activity of VioAO7, which has a Km value that is 20 times lower and a kcat/Km ratio that is about 150 times higher than those of VioAD7. Furthermore, the Km of VioAO7 is the lowest among all kinetically characterized SATs, including Cj1294 (27), Cj1121c (36), WecE (12), and RfbE (2). In future work, it is worthwhile to perform site-directed mutagenesis at the potential active sites to investigate any significance of amino acid substitution in relation to enzyme activity.
FIG. 7.
Sequence alignments of VioAD7, VioAO7, and other reported sugar aminotransferases. Multiple alignments were performed with the Clustal W program using the BioEdit sequence alignment software. The GenBank accession numbers for StsC_Sgr, TbmB_Ste, FdtB_Ath, GnnB_Afe, TylB_Sfr, ArnB_Eco, Cj1121c_Cje, Cj1294_Cje, RfbE_Vch, WecE_Eco, VioA_Sdy, and VioA_Eco are CAA70012, Q2MF17, AAS55722, AAS48422, S49052, AAM92146, CAL35238, AAT12282, CAA42137, AAC76796, AAR97958, and AAD44154, respectively. Four motifs are boxed. Sgr, Streptomyces griseus; Ste, Streptomyces tenebrarius; Ath, Aneurinibacillus thermoaerophilus; Afe, Acidithiobacillus ferrooxidans; Sfr, Streptomyces fradiae; Eco, Escherichia coli; Cje, Campylobacter jejuni; Vch, Vibrio cholerae; Sdy, Shigella dysenteriae.
Having the maximum activity at 37°C while losing most of the activity at higher temperatures (65°C and above) for VioAD7, but not for VioAO7, may reflect the different niches occupied by Shigella and E. coli. Shigella strains live only inside the human intestine, and E. coli strains can adapt to various environmental niches, either inside or outside the human body. The activity of VioBO7 is also maintained over a wide temperature range.
Rare sugars are potentially useful in the pharmaceutical and chemical industries for drug development. Amino sugars have been reported as components of many macrolide antibiotics, including tylosin, desosamine, and erythromycin (7, 8, 23, 37). Chemical synthesis of rare sugars is difficult due to the need for multistep reactions of protection and deprotection. This work provides genetic and biochemical means for the synthesis of Qui4N and Qui4NAc, which are not commonly found and are not yet commercially available.
Supplementary Material
Acknowledgments
We thank Dawei Zhou for technical assistance with RP-HPLC.
This work was supported by the NSFC General Program (grants 30370023, 30670038, and 30500024), the NSFC Key Program (grant 30530010), the Tianjin Municipal Special Fund for Science and Technology Innovation (grant 05FZZDSH00800), the National 863 Program of China (grant 2006AA020703), the RFBR (grants 05-04-48992 and 05-04-39015 to A.V.P. and Y.A.K.), the Council on Grants of the president's office of the Russian Federation for Support of Young Russian Scientists (project MK-157.2007.4) to A.V.P., and the Russian Science Support Foundation.
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahlert, J., J. Distler, K. Mansouri, and W. Piepersberg. 1997. Identification of stsC, the gene encoding the l-glutamine:scyllo-inosose aminotransferase from streptomycin-producing Streptomycetes. Arch. Microbiol. 168:102-113. [DOI] [PubMed] [Google Scholar]
- 2.Albermann, C., and W. Piepersberg. 2001. Expression and identification of the RfbE protein from Vibrio cholerae O1 and its use for the enzymatic synthesis of GDP-d-perosamine. Glycobiology 11:655-661. [DOI] [PubMed] [Google Scholar]
- 3.Bastin, D. A., and P. R. Reeves. 1995. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 164:17-23. [DOI] [PubMed] [Google Scholar]
- 4.Bax, A., and D. G. Davis 1985. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65:355-360. [Google Scholar]
- 5.Breazeale, S. D., A. A. Ribeiro, and C. R. Raetz. 2003. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-l-arabinose. J. Biol. Chem. 278:24731-24739. [DOI] [PubMed] [Google Scholar]
- 6.Caroff, M., D. Karibian, J. M. Cavaillon, and N. Haeffner-Cavaillon. 2002. Structural and functional analyses of bacterial lipopolysaccharides. Microbes Infect. 4:915-926. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F., G. M. Evins, W. L. Cook, R. Almeida, N. Hargrett-Bean, and I. K. Wachsmuth. 1991. Genetic diversity among toxigenic and nontoxigenic Vibrio cholerae O1 isolated from the western hemisphere. Epidemiol. Infect. 107:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhillon, N., R. S. Hale, J. Cortes, and P. F. Leadlay. 1989. Molecular characterization of a gene from Saccharopolyspora erythraea (Streptomyces erythraeus) which is involved in erythromycin biosynthesis. Mol. Microbiol. 3:1405-1414. [DOI] [PubMed] [Google Scholar]
- 9.Erridge, C., E. Bennett-Guerrero, and I. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4:837-851. [DOI] [PubMed] [Google Scholar]
- 10.Feng, L., J. Tao, H. Guo, J. Xu, Y. Li, F. Rezwan, P. Reeves, and L. Wang. 2004. Structure of the Shigella dysenteriae 7 O antigen gene cluster and identification of its antigen specific genes. Microb. Pathog. 36:109-115. [DOI] [PubMed] [Google Scholar]
- 11.Graninger, M., B. Kneidinger, K. Bruno, A. Scheberl, and P. Messner. 2002. Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-beta-l-rhamnose biosynthesis in the gram-positive thermophile Aneurinibacillus thermoaerophilus DSM 10155. Appl. Environ. Microbiol. 68:3708-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang, B. Y., H. J. Lee, Y. H. Yang, H. S. Joo, and B. G. Kim. 2004. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem. Biol. 11:915-925. [DOI] [PubMed] [Google Scholar]
- 13.Kharel, M. K., D. B. Basnet, H. C. Lee, K. Liou, J. S. Woo, B. G. Kim, and J. K. Sohng. 2004. Isolation and characterization of the tobramycin biosynthetic gene cluster from Streptomyces tenebrarius. FEMS Microbiol. Lett. 230:185-190. [DOI] [PubMed] [Google Scholar]
- 14.Kharel, M. K., B. Subba, H. C. Lee, K. Liou, and J. K. Sohng. 2005. Characterization of l-glutamine:2-deoxy-scyllo-inosose aminotransferase (tbmB) from Streptomyces tenebrarius. Bioorg. Med. Chem. Lett. 15:89-92. [DOI] [PubMed] [Google Scholar]
- 15.Kneidinger, B., S. Larocque, J. Brisson, N. Cadotte, and J. Lam. 2003. Biosynthesis of 2-acetamido-2,6-dideoxy-l-hexoses in bacteria follows a pattern distinct from those of the pathways of 6-deoxy-l-hexoses. Biochem. J. 371:989-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kneidinger, B., K. O'Riordan, J. Li, J. Brisson, J. Lee, and J. Lam. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. J. Biol. Chem. 278:3615-3627. [DOI] [PubMed] [Google Scholar]
- 17.Knirel, Y. A., V. V. Dashunin, A. S. Shashkov, N. K. Kochetkov, B. A. Dmitriev, and I. L. Hofman. 1988. Somatic antigens of Shigella: structure of the O-specific polysaccharide chain of the Shigella dysenteriae type 7 lipopolysaccharide. Carbohydr. Res. 179:51-60. [DOI] [PubMed] [Google Scholar]
- 18.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 19.L'vov, V. L., A. S. Shashkov, B. A. Dmitriev, N. K. Kochetkov, B. Jann, and K. Jann. 1984. Structural studies of the O-specific side chain of the lipopolysaccharide from Escherichia coli O7. Carbohydr. Res. 126:249-259. [DOI] [PubMed] [Google Scholar]
- 20.Marolda, C. L., M. F. Feldman, and M. A. Valvano. 1999. Genetic organization of the O7-specific lipopolysaccharide biosynthesis cluster of Escherichia coli VW187 (O7:K1). Microbiology 145:2485-2495. [DOI] [PubMed] [Google Scholar]
- 21.Matsuhashi, M., and J. L. Strominger. 1966. Thymidine diphosphate 4-acetamido-4,6-dideoxyhexoses III. Purification and properties of thymidine diphosphate 4-keto-6-deoxy-d-glucose transaminase from Escherichia coli strain B. J. Biol. Chem. 241:4738-4744. [PubMed] [Google Scholar]
- 22.McNally, D., I. Schoenhofen, E. Mulrooney, D. Whitfield, E. Vinogradov, J. Lam, S. Logan, and J. Brisson. 2006. Identification of labile UDP-ketosugars in Helicobacter pylori, Campylobacter jejuni and Pseudomonas aeruginosa: key metabolites used to make glycan virulence factors. Chem. Biol. Chem. 7:1865-1868. [DOI] [PubMed] [Google Scholar]
- 23.Merson-Davies, L. A., and E. Cundliffe. 1994. Analysis of five tylosin biosynthetic genes from the tylIBA region of the Streptomyces fradiae genome. Mol. Microbiol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 24.Nakano, Y., N. Suzuki, Y. Yoshida, T. Nezu, Y. Yamashita, and T. Koga. 2000. Thymidine diphosphate-6-deoxy-l-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J. Biol. Chem. 275:6806-6812. [DOI] [PubMed] [Google Scholar]
- 25.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niyogi, S. K. 2005. Shigellosis. J. Microbiol. 43:133-143. [PubMed] [Google Scholar]
- 27.Obhi, R. K., and C. Creuzenet. 2005. Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aminotransferase that generates UDP-4-amino-4,6-dideoxy-GalNAc. J. Biol. Chem. 280:20902-20908. [DOI] [PubMed] [Google Scholar]
- 28.Parolis, H., L. A. Parolis, and G. Olivieri. 1997. Structural studies on the Shigella-like Escherichia coli O121 O-specific polysaccharide. Carbohydr. Res. 303:319-325. [DOI] [PubMed] [Google Scholar]
- 29.Perepelov, A. V., D. Babicka, S. N. Senchenkova, A. S. Shashkov, H. Moll, A. Rozalski, U. Zahringer, and Y. A. Knirel. 2001. Structure of the O-specific polysaccharide of Proteus vulgaris O4 containing a new component of bacterial polysaccharides, 4,6-dideoxy-4-{N-[(R)-3-hydroxybutyryl]-l-alanyl}amino-d-glucose. Carbohydr. Res. 331:195-202. [DOI] [PubMed] [Google Scholar]
- 30.Pfoestl, A., A. Hofinger, P. Kosma, and P. Messner. 2003. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 278:26410-26417. [DOI] [PubMed] [Google Scholar]
- 31.Reeves, P. R. 1995. Role of O-antigen variation in the immune response. Trends Microbiol. 3:381-386. [DOI] [PubMed] [Google Scholar]
- 32.Reeves, P. R., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 33.Stenutz, R., A. Weintraub, and G. Widmalm. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 30:382-403. [DOI] [PubMed] [Google Scholar]
- 34.Stroeher, U. H., L. E. Karageorgos, M. H. Brown, R. Morona, and P. A. Manning. 1995. A putative pathway for perosamine biosynthesis is the first function encoded within the rfb region of Vibrio cholerae O1. Gene 166:33-42. [DOI] [PubMed] [Google Scholar]
- 35.Sweet, C. R., A. A. Ribeiro, and C. R. Raetz. 2004. Oxidation and transamination of the 3"-position of UDP-N-acetylglucosamine by enzymes from Acidithiobacillus ferrooxidans. Role in the formation of lipid a molecules with four amide-linked acyl chains. J. Biol. Chem. 279:25400-25410. [DOI] [PubMed] [Google Scholar]
- 36.Vijayakumar, S., A. Merkx-Jacques, D. B. Ratnayake, I. Gryski, R. K. Obhi, S. Houle, C. M. Dozois, and C. Creuzenet. 2006. Cj1121c, a novel UDP-4-keto-6-deoxy-GlcNAc C-4 aminotransferase essential for protein glycosylation and virulence in Campylobacter jejuni. J. Biol. Chem. 281:27733-27743. [DOI] [PubMed] [Google Scholar]
- 37.Volchegursky, Y., Z. Hu, L. Katz, and R. McDaniel. 2000. Biosynthesis of the anti-parasitic agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. Mol. Microbiol. 37:752-762. [DOI] [PubMed] [Google Scholar]
- 38.Yatsuyanagi, J., S. Saito, and I. Ito. 2002. A case of hemolytic-uremic syndrome associated with Shiga toxin 2-producing Escherichia coli O121 infection caused by drinking water contaminated with bovine feces. Jpn. J. Infect. Dis. 55:174-176. [PubMed] [Google Scholar]
- 39.Yoshida, Y., Y. Nakano, T. Nezu, Y. Yamashita, and T. Koga. 1999. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-d-fucose. J. Biol. Chem. 274:16933-16939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.