Abstract
A new highly pathogenic clone of Escherichia coli meningitis strains harboring the unusual serogroup O45 has recently emerged in France. To gain insight into the pathogenicity of this new clone, we investigated the possible role of antigen O45 in the virulence of strain S88 (O45:K1:H7), representative of this emerging clone. We first showed that the S88 O-antigen gene cluster sequence differs from that of O45 in the reference strain E. coli 96-3285, suggesting that the two O45 polysaccharides, while probably sharing a community of epitopes, represent two different antigens. The unique functional organization of the two O-antigen gene clusters and the low DNA sequence homology of the orthologous genes suggest that the two loci originated from a common ancestor and have since undergone multiple recombination events. Phylogenetic analysis based on the flanking gene gnd sequences indicates that the S88 antigen O45 (O45S88) gene cluster may have been acquired, at least in part, from another member of the Enterobacteriaceae. Mutagenesis of the O45S88 antigen gene cluster was used for functional analysis of the loci and revealed the crucial role of the O polysaccharide in S88 virulence in a neonatal rat meningitis model. We also developed a PCR method to specifically identify the O45S88 antigen gene cluster. Together, our findings suggest that horizontal acquisition of a new O-antigen gene cluster, at least partly from another species, may have been a key event in the emergence and virulence of the E. coli O45:K1:H7 clone in France.
O antigen, the polysaccharide constituent of lipopolysaccharide (LPS), is a major focus of studies of infections caused by extraintestinal pathogenic Escherichia coli (ExPEC) strains for two main reasons. First, it can be used for typing studies, which have shown the highly clonal organization of some ExPEC strains, such as neonatal meningitis strains (39). Indeed, the most important globally distributed neonatal meningitis clone is the so-called archetypal clone O18:K1:H7 (1). Second, O antigens may play a major role in the virulence of ExPEC and notably in resistance to serum bactericidal activity; their characterization may thus help to understand the pathophysiology of ExPEC infections (22).
Serotyping combined with molecular methods previously allowed us to detect the emergence in France of a highly virulent meningitis-causing clone closely related to the O18:K1:H7 clone but harboring the unusual serogroup O45, as well as capsular antigen K1 and flagellar antigen H7 (12). O45 antigen has only sporadically been described in ExPEC strains (34, 46) and is absent from most E. coli meningitis strains in American and European collections (1, 23), except in Hungary (14).
In contrast, several intestinal pathogenic E. coli (InPEC) strains have been identified as belonging to serogroup O45, such as enterotoxigenic E. coli and Shiga toxin-producing E. coli (STEC) (9, 41). In addition, E. coli O45 strains producing cytotoxic necrotizing factor or expressing the “attaching and effacing” phenotype have been isolated from several diarrheic animals (4, 32). The importance of O45 antigen in InPEC strains led DebRoy et al. to sequence the genomic region involved in O-antigen synthesis in the CDC reference strain 96-3285 (O45:H2) and to develop a specific PCR method to detect E. coli O45 (16).
Many PCR tests for E. coli serogroups have recently been developed by targeting the genes involved in O-antigen synthesis and clustered in the so-called O-antigen gene cluster. Among these genes, those responsible for O-unit processing—wzx and wzy, encoding the O-antigen flippase and O-antigen polymerase, respectively—are specific for the O-unit composing the polysaccharide (38). These genes may therefore be ideal targets for specific PCR. Indeed, the classical agglutination reaction with specific antiserum is laborious and expensive, and cross-reactions between serogroups can occur (33).
The emergence of the unusual O45 antigen in ExPEC strains prompted us to analyze the O-antigen gene cluster sequence of S88, a strain representative of the O45:K1:H7 clone causing neonatal meningitis, by comparison with the published sequence of strain 96-3285 (O45:H2) (16). We show that the two O-antigen gene clusters are genetically related but not identical, suggesting that S88 expresses a new O antigen. We also performed a functional analysis of the S88 O-antigen gene cluster and developed a specific PCR to detect strains harboring the S88 somatic antigen.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The E. coli meningitis strain S88 is representative of the French virulent clone O45:K1:H7 (12). This serotype was kindly determined by three different National Reference Centers in Denmark (Flemming Schultz, Serum Staten Institut, Copenhagen, Denmark), Germany (Lothar Beutin, Robert Koch Institute, Berlin, Germany), and Spain (Jorge Blanco, Lugo, Spain). Strain S88 belongs to phylogenetic group B2 and harbors ribotype B21 (12). The CDC reference strain for antigen O45, E. coli strain 96-3285 (O45:H2), the O-antigen gene cluster of which was recently sequenced, was kindly provided by Chitrita DebRoy (E. coli Reference Center, The Pennsylvania State University). This strain was shown in a previously described PCR grouping method to belong to the nonvirulent phylogenetic group B1 (8) and harbors none of the virulence factors usually encountered among meningitis strains (12). The Danish O45 reference strain, strain H61 (O45:K1:H10), which belongs to phylogenetic group A, was purchased from the Staten Serum Institut (Copenhagen, Denmark). Thirty-six meningitis strains and nine urosepsis strains, all belonging to serotype O45:K1:H7 (determined by the Spanish National Reference Center) and to ribotype B21 (like S88) and sharing the same virulence genotype, were also used (7, 12). A total of 130 reference strains for the different somatic antigens (O antigens 1, 2, 4 to 24, 26 to 30, 32 to 45, 48 to 53, 55, 57, 59 to 61, 63, 64, 68 to 70, 73 to 77, 79, 81, 83 to 86, 89 to 91, 93, 95, 98, 101 to 121, 123, 125 to 129, 134 to 136, 138 to 142, 144 to 152, 154 to 162, and 164) were kindly provided by Patrick Grimont (Institut Pasteur, Paris, France). The 72 E. coli reference (ECOR) strains (31) were also used.
The plasmids pKD46, pKD3, and pCP20 were kindly provided by Lionello Bossi (Centre de Genetique Moleculaire, CNRS, Gif sur Yvette, France).
Luria-Bertani (LB) broth and agar were routinely used and were supplemented with chloramphenicol (12 μg/ml) or ampicillin (50 μg/ml) as necessary.
Sequencing of the S88 O-antigen gene cluster.
The O-antigen gene cluster in strain S88 was sequenced as part of a whole-genome sequencing project (ColiScope; www.genoscope.cns.fr) at the Genoscope sequencing center (Evry, France). Sequencing and assembly of the S88 genome were performed as previously described (3). Briefly, the complete genome sequence of E. coli S88 was determined using the whole-genome shotgun method (10× coverage). Three libraries were made as follows: two plasmid libraries of 3 kb and 10 kb, obtained by mechanical shearing, were constructed in plasmid pcDNA2.1 (Invitrogen) and in home vector pCNS (pSU18 modified), respectively. One bacterial artificial chromosome library of average insert size of 30 kb was constructed by enzymatic digestion (HindIII) into pBeloBacII (CalTech). The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment. To resolve contigs, sequence finishing was performed by PCR amplification, primer walking, and/or transposition.
The MaGe (magnifying genomes) software program was used for gene annotation and comparative analysis of the S88 genome (43). Using the AMIGene (annotation of microbial genes) (10) program, a total of 4,859 coding sequences were predicted (and assigned a unique identifier prefixed with ECOS88_) and submitted to automatic functional annotation, including synteny computation (that is, conservation of the chromosomal colocalization between pairs of orthologous genes from different genomes). Manual validation of the automatic annotation was performed using the MaGe interface, which allows graphic visualization of the E. coli S88 annotations enhanced by a synchronized representation of synteny groups in other genomes chosen for comparisons (43). Protein motifs and domains were identified by using the InterPro databank (2). TMHMM, version 2.0, was used to identify transmembrane domains (26). Sequence data for comparative analyses were obtained from NCBI databases (ftp://ftp.ncbi.nlm.nih.gov). Annotations of the O-antigen gene cluster described in this paper range from ECOS88_2129 (gnd) to ECOS88_2139 (galF).
Phylogenetic analysis.
To gain insight into the evolutionary history of the O45 antigen gene cluster in strains S88 and 96-3285, nucleotide sequences of the internal genes rmlABC and the external gene gnd from different E. coli strains and several other representative gram-negative bacteria were extracted from the GenBank database. The gnd sequence of strain 96-3285 was not available and was determined here from a PCR product obtained with the primers indicated in Table 1. The ClustalW program was used to align the sequences (40). Phylogenetic and molecular evolutionary relationships were examined by using the neighbor-joining method implemented with MEGA, version 3.1, software (27). Bootstrap confidence values for each node of the trees were calculated over 100 replicate trees. Phylogenetic analysis was also performed using the maximum parsimony method, also implemented in MEGA, version 3.1, software.
TABLE 1.
Oligonucleotide primers for PCR sequencing, mutant construction, and O45 PCR assay
| Namea | Target | Sequence (5′ to 3′)b | Amplicon size (bp) |
|---|---|---|---|
| gndF | gnd | ATGTCCAAGCAACAGATCGGCGT | 1,407 |
| gndR | gnd | TTAATCCAGCCATTCGGTATGGA | |
| rfb405S88.P0 | ORF 2138 | GATTCCCTTGGTTATTCTCAATGCTCTCGAAGGGAAATCGTGTGTAGGCTGGAGCTGCTT | |
| rfb397S88.P2 | ORF 2130 | CACCTGATTATCATAGAAATAGAGTCCAGTTATGGCCCAACATATGAATATCCTCCTTAG | |
| rfb399S88.P0 | ORF 2132 | TCTGGTGTTTATACAGGGTAAATGTAAGGTTTCATCGCAATGTGTAGGCTGGAGCTGCTT | |
| rfb399S88.P2 | ORF 2132 | GGAAGGTTAGTTTAAGGCAGGGGAGCAGGCATAATAACATCATATGAATATCCTCCTTAG | |
| C1 | cat | TTATACGCAAGGCGACAAGG | |
| C2 | cat | GATCTTCCGTCACAGGTAGG | |
| rfb405S88.FR1b | ORF 2138 | CGTACATACGGTCTTCCAAC | |
| rfb397S88.FR2b | ORF 2130 | CATTAAGCATATCCCGCTCA | |
| rfb400S88.FR1 | ORF 2133 | GAGTAGCTTTGTCTTGCGC | |
| rfb399S88.FR2 | ORF 2132 | TATTCCTGCGTATCCTGCTA | |
| wzxS88b.F | ORF 2134 | GTTGCGATAGTCATGTACTG | 119 |
| wzxS88b.R | ORF 2134 | GCTACAACCCCTCCCCAGAT | |
| wzyS88b.F | ORF 2132 | GGTATCGTTCACATCGCTTA | 330 |
| wzyS88b.R | ORF 2132 | GAGAAAATACTCGGTTCGGC | |
| ColiBglu.1 | uidA | TATGAACTGTGCGTCACAGCC | 186 |
| ColiBglu.2 | uidA | CATCAGCACGTTATCGAATCC |
The oligonucleotide primers used for gene recombination are designated by the suffixes P0 and P2; the oligonucleotide primers used to control correct introduction and excision of the cat gene are designated by the suffixes FR1 and FR2 and flank the DNA target segment. Oligonucleotide primers used for O45 triplex PCR are shown in bold characters.
Boldface characters indicate the 20 nucleotides homologous to the cat gene sequence.
Construction of mutants.
S88 mutants were obtained by the method of Datsenko and Wanner (15) as previously described (29). Briefly, this PCR-based method uses the plasmid pKD46, which allows homologous recombination to occur directly with PCR products. This plasmid is a temperature-sensitive replicon that carries the bacteriophage λ Red system under the control of an arabinose-inducible promoter. Once introduced into the bacterium by electroporation, it renders S88 transformable with linear DNA obtained by PCR. The chloramphenicol acetyltransferase (cat) gene, carried by plasmid pKD3, was amplified by PCR with primers bearing extensions of 40 nucleotides homologous to the initial and final portions of the DNA target segment. All the primers used in this study are listed in Table 1. Transformation by electroporation of strain S88 expressing bacteriophage λ Red functions with the PCR product yielded recombinants carrying the DNA target fused to the cat gene. Correct introduction of the cat gene was controlled by PCR with primers (Table 1) flanking the initial and final portions of the DNA target segment and primers homologous to the cat gene, as previously described (29). Conservation of the main virulence determinants in the mutants was controlled by multiplex PCR with primers located in the main extraintestinal virulence genes of S88 (fyuA, papC, papGII, iucC, and iroN) as previously described (11). Antigens O45 and K1 were detected with specific antisera and phage from the Staten Serum Institute (Copenhagen, Denmark).
Analysis of the LPS.
The LPS was extracted with denaturing buffer composed as follows (final concentrations): 0.2% sodium dodecyl sulfate (SDS), 1% β-mercaptoethanol, 36% glycerol, 30 mM Tris-HCl (pH 7.4), and 0.001% bromophenol blue. Strains suspended in 1 ml of phosphate-buffered saline at an optical density at 600 nm of 2 were centrifuged at 3,500 × g for 10 min at 25°C, and the pellet was resuspended with 500 μl of denaturing buffer. The sample was denatured for 5 min at 95°C, and 3 μl of proteinase K was added after cooling to room temperature. The sample was incubated for 1 h at 55°C with proteinase K and then centrifuged for 30 min at 13,000 × g and 25°C. The LPS-containing supernatants were stored at −20°C. The LPS preparations were separated on 16% SDS-tricine-polyacrylamide gels at 30 V for 30 min and 100 V for 3 h. LPS was then visualized by silver staining as previously described (42).
Animal meningitis model.
E. coli bacteremia and meningitis were induced in newborn rats as described by Houdouin et al. (21). Briefly, pathogen-free Sprague-Dawley rats were obtained from Charles River Laboratories (L'Abresle, France) at 4 days of age, together with their mothers. At 5 days of age all the pups were inoculated intraperitoneally with ∼500 CFU of the tested strain in physiological saline. Eighteen hours later, 5 μl of blood was obtained by tail incision. The animals were then killed, and 5 μl of cerebrospinal fluid (CSF) was immediately obtained by cisternal puncture. All samples were quantitatively cultured by plating dilutions of blood and cerebrospinal fluid on LB agar.
O-serogroup-specific PCR assay.
Genes encoding O-antigen flippase (wzx) and O-antigen polymerase (wzy) in the O-antigen gene cluster are specific for each O antigen and are suitable targets for serogroup-specific PCR (36). In order to develop a specific PCR assay for identifying E. coli strains harboring a somatic antigen identical to that expressed by strains belonging to the O45:K1:H7 meningitis clone, we designed specific primers for wzx and wzy. Template DNA for the PCR assays was prepared by mixing 2 μl of bacterial colony formed on LB agar in 500 μl of sterile water and heating at 100°C for 10 min. Then, the suspension was centrifuged at 11,000 × g for 3 min at 4°C, and the supernatant containing DNA was used for PCR. PCR was performed in a final volume of 50 μl using a Qiagen Multiplex PCR kit with 5 μl of template DNA, 0.3 μM wzx primers, and a 0.2 μM concentration (each) of wzy and uidA primers. PCR was performed in an Icycler (Bio-Rad, Marnes la Coquette, France) as follows: denaturation at 95°C for 15 min; 30 cycles of 94°C for 30 s, 55°C for 90 s, and 72°C for 90 s; and final extension at 72°C for 10 min. The PCR products were electrophoresed in 2% standard agarose gels. The gels were stained with ethidium bromide and visualized under UV light. Positive samples were identified from the presence of bands of the expected sizes compared to results obtained with strain S88.
Nucleotide sequence accession number.
The DNA sequences of the S88 E. coli O-antigen gene cluster have been deposited in the GenBank database under the accession number CU463050.
RESULTS
Functional annotation of the E. coli S88 O-antigen gene cluster.
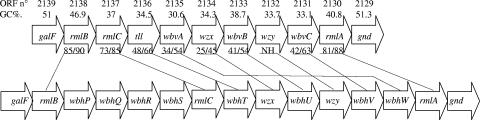
Analysis of the DNA sequence of the putative O-antigen gene cluster located between galF and gnd in strain S88 showed that it contains nine open reading frames (ORFs) with a total length of 8,379 bp, the same transcriptional direction from galF to gnd, and a low G+C content (30.6 to 46.9%) compared to the E. coli core genome (51%). The nine ORFs of the locus were assigned putative functions based on a protein database similarity search and were named using the system proposed by Reeves et al. (36) (Table 2). The most homologous proteins were almost always found in the products of the O-antigen gene cluster of E. coli 96-3285, the CDC reference strain for the O45 somatic antigen (Table 2).
TABLE 2.
Genes located in the O-antigen gene cluster of E. coli S88 serotype O45:K1:H7
| ORF | Proposed gene name | Location (nucleotides) | % G+C content | No. of amino acids in gene product | Putative function | Most significant homolog (strain) | % Amino acid identity | % Amino acid similarity |
|---|---|---|---|---|---|---|---|---|
| 2138 | rmlB | 2108855-2109937 | 46.9 | 1,083 | dTDP-glucose-4,6-dehydratase | dTDP-glucose-4,6-dehydratase (E. coli 96-3285) | 90 | 95 |
| 2137 | rmlC | 2108241-2108783 | 37 | 543 | dTDP-6-deoxy-d-glucose-3,5-epimerase | dTDP-4-dehydrorhamnose-3,5-epimerase (E. coli 96-3285) | 73 | 85 |
| 2136 | tll | 2107363-2108178 | 34.5 | 816 | dTDP-6-deoxy-l-xylo-4-hexulose reductase | dTDP-6-deoxy-l-xylo-4-hexulose reductase(E. coli O66) | 52 | 68 |
| 2135 | wbvA | 2106363-2107361 | 30.6 | 999 | Unknown | Hypothetical protein (E. coli 96-3285) | 34 | 51 |
| 2134 | wzx | 2105114-2106376 | 34.3 | 1,263 | O-antigen flippase | O-antigen flippase (E. coli 96-3285) | 23 | 42 |
| 2133 | wbvB | 2104150-2105127 | 38.7 | 978 | Glycosyl transferase | Glycosyl transferase (E. coli 96-3285) | 38 | 52 |
| 2132 | wzy | 2103066-2104157 | 33.7 | 1,092 | O-antigen polymerase | Putative membranous protein (Photobacterium profundum) | 34 | 54 |
| 2131 | wbvC | 2102570-2103100 | 31.1 | 531 | O-Acetyltransferase | Serine acetyltransferase (E. coli 96-3285) | 42 | 63 |
| 2130 | rmlA | 2101559-2102425 | 40.8 | 867 | Glucose-1-phosphate thymidyl transferase | Glucose-1-phosphate thymidyl transferase (E. coli 96-3285) | 81 | 88 |
Products of ORFs 2130, 2138, and 2137 shared a high level of identity (>70%) with RmlA, RmlB, and RmlC, respectively. ORF 2136, immediately downstream of rmlC, putatively encodes a protein sharing 52% identity with the tll product in the recently sequenced O66-antigen gene cluster (13). This gene and rmlABC are known to be involved in the biosynthesis pathway of 6-deoxy-l-talose, which may therefore compose the oligosaccharide unit of S88 O antigen. The other putative proteins encoded by the five remaining ORFs had identities lower than 50% with available proteins (Table 2). Two genes for S88 O-antigen unit processing, potentially encoding the O-antigen flippase (wzx) and the O-antigen polymerase (wzy), were presumptively identified. The putative Wzx (ORF 2134) had 12 predicted transmembrane segments, which is the typical number for O-antigen flippase (36). Moreover, it also showed 23% identity and 42% similarity to the putative Wzx of E. coli 96-3285. The ORF 2132 (putative wzy) had a predicted amino acid sequence corresponding to eight transmembrane segments and a large cytoplasmic loop of 60 amino acids compatible with an O-antigen polymerase function (38). However, this protein had no homology with previously published E. coli O-antigen polymerase sequences, and its function is therefore highly hypothetical. Among the three remaining genes, two putative transferases (ORFs 2131 and 2133) were identified; the putative function of the third gene (ORF 2135) could not be assigned (Table 2).
The maps of the O-antigen gene clusters of S88 and E. coli 96-3285, based on protein homologies, were highly similar, except for ORF 2135 (Fig. 1). For example, the rmlABC and tll genes (assuming that wbhT corresponds to tll) were in the same order. Moreover, rmlB and rmlA were identically situated at both extremities of each locus, a very unusual disposition for these two genes (38). The only important difference was the presence of four genes located between rmlB and rmlC in the 96-3285 O-antigen gene cluster but not in S88 (Fig. 1).
FIG. 1.
Genetic organization and comparison of the O-antigen gene clusters of E. coli S88 (upper row) and 96-3285 (lower rower). Putative orthologous genes of the O-antigen gene clusters are connected. Values below the ORF numbers are % G+C content; values below the S88 genes are percent amino acid identity/percent amino acid similarity. NH, no homology.
Phylogenetic analysis of genes lying within or flanking the O-antigen gene cluster.
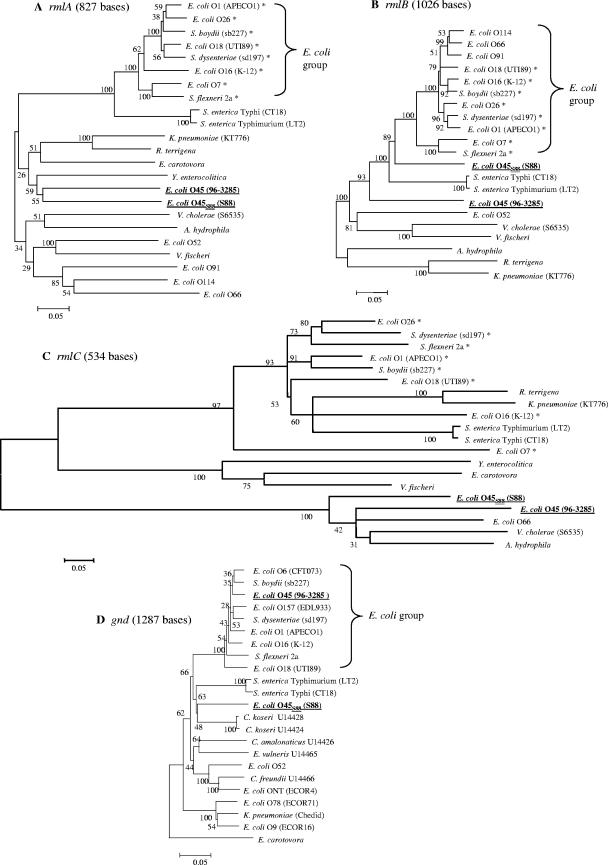
In order to determine the genetic relatedness of the S88 and 96-3285 O-antigen gene clusters, phylogenetic trees were constructed with GenBank DNA sequences of the rmlABC genes from E. coli and other genera. These three genes were chosen for their strong similarities at the protein level between E. coli strains S88 and 96-3285. Trees obtained with the neighbor-joining method are shown in Fig. 2. Similar topologies were obtained with the maximum parsimony method (data not shown). In the rmlA tree (Fig. 2A), a cluster containing only E. coli and Shigella strains clearly emerged. Of note, all these strains harbored the four rmlABCD genes required for l-rhamnose synthesis. All the other E. coli rmlA genes, including rmlA in strains S88 and 96-3285, which are not associated with a complete rmlABCD cluster, were distantly related to each other, with low DNA homologies. Indeed, the rmlA genes of S88 and 96-3285 were not significantly closer to each other than to the rmlA genes of different species such as Yersinia spp. (Fig. 2A). In the rmlB tree, S88 was again distantly related to the main group of E. coli strains, despite a mean identity of 81% with this group. Finally, in the rmlC tree, three major groups of genes with very low DNA homologies were distinguished. One of them contained E. coli S88, 96-3285, and the O66 reference strain, in which Vibrio cholerae and Aeromonas hydrophila are nearly equidistant.
FIG. 2.
Phylogenetic trees generated by the neighbor-joining method for rmlA, rmlB, rmlC, and gnd sequences of several representative gram-negative species. Numbers at the branches are bootstrap proportions obtained from 100 replicates. *, E. coli or Shigella strain harboring a complete rmlABCD cluster. Sequences were extracted from the GenBank database, except for the gnd sequences of strains 96-3285 and S88. The following E. coli serogroups are represented: O1 (strain APEC01; accession number CP000468), O6 (strain CFT073; accession number NC_004431), O7 (strain VW187; accession number AF125322), O9 (strain ECOR-16; accession number M64325), O16 (strain K-12, MG1655; accession number U00096), O18 (strain UTI89; accession number CP000243), O26 (accession number AF529080), O45 (strain 96-3285; accession number AY771223), O52 (strain G1066; accession number AY528413), O66 (accession number DQ069297), O78 (strain ECOR-71; accession number U14461), O91 (strain ECA95; accession number AY035396), O114 (strain G1088; accession number AY573377), and O157 (strain EDL933; accession number NC_002655), and O nontypeable (strain ECOR-4; accession number M64324). Other strains include the following: Escherichia vulneris (strain ATCC 33821; accession number U14465), Shigella boydii (strain sb227; accession number CP000036), Shigella dysenteriae (strain sd197; accession number CP000034), Shigella flexneri 2a (strain 301; accession number AE005674), Salmonella enterica serovar Typhi (strain CT18; accession number AL627273), S. enterica serovar Typhimurium (strain LT2; accession number NC_003197), Citrobacter koseri (strain CT19; accession number U14424 and strain CT42; acc. number U14428), Citrobacter freundii (strain ATCC 8090; accession number U14466), Citrobacter amalonaticus (strain CT28; accession number U14426), Klebsiella pneumoniae (strain Chedid gnd gene [accession number D21242] and strain KT776 rml genes [accession number AF097519]), Raoultella terrigena (strain ATCC 33257; accession number AY376146), Erwinia carotovora (strain SCRI1043; accession number NC_004547), Yersinia enterocolitica serogroup O:3 (strain 6471/76; accession number Z18920), Vibrio cholerae (strain S6535; accession number AY239000), Vibrio fischeri (strain ES114; accession number CP000020), and Aeromonas hydrophila (strain PPD134/91; accession number AF148126).
As the immediate flanking gene gnd is potentially a target for horizontal transfer of the O-antigen gene cluster (30), we investigated the genetic relationship of this gene between several E. coli strains and other Enterobacteriaceae (Fig. 2D). Most of the E. coli gnd sequences, including the gnd sequence of strain 96-3285, clustered together, with an average identity of 95%. In contrast, S88 gnd was only distantly related to this cluster (mean identity, 85%). As previously described, gnd sequences from E. coli ECOR-16, ECOR-4, and ECOR-71 were more closely related to Klebsiella spp. or Citrobacter spp., from which they are thought to have been acquired (30). However, S88 gnd could not be linked to any of these species or to other Enterobacteriaceae gnd sequences available in the GenBank database.
Functional analysis of the O-antigen gene cluster.
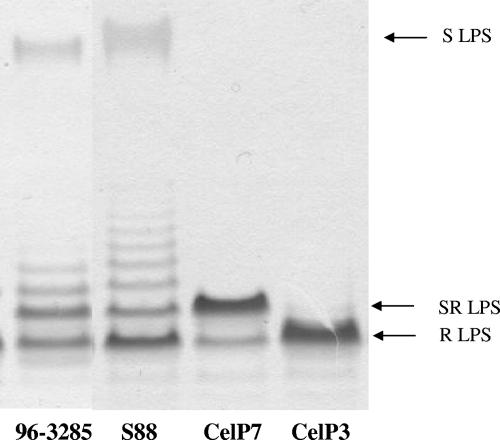
In order to determine the functional role of the putative O-antigen gene cluster and the putative O-antigen polymerase gene (wzy) in E. coli strain S88, two mutants were prepared according to the method described by Datsenko and Wanner (29). One mutant, named CelP3, had a deletion of the entire O-antigen gene cluster from the first gene (ORF 2138) to the last (ORF 2130), and the other mutant, named CelP7, had a deletion of the putative wzy gene. With both mutants, PCR using primers flanking the initial and final portions of the DNA target segment and primers homologous to the cat gene (29) demonstrated the introduction of the cat gene instead of the target genes (data not shown). Moreover, multiplex PCR amplification of the main extraintestinal virulence genes and capsular antigen K1 were positive for each mutant, as for the wild-type strain (data not shown). Mutants CelP3 and CelP7 did not agglutinate with O45-specific antiserum, in contrast to strain S88, suggesting the involvement of the two deleted loci in the biosynthesis of the polysaccharide somatic O antigen. We then analyzed the polysaccharide somatic O antigen in the two mutants and the wild-type strain by SDS-polyacrylamide gel electrophoresis (Fig. 3). The S88 polysaccharide somatic O antigen showed a wild-type bimodal distribution of LPS, characterized by a first band of lipid A-core and several more bands that corresponded to O-antigen chain subunits. The CelP3 mutant had a rough phenotype characterized by only one band of lipid A-core and no attached O-antigen chain, demonstrating that the DNA segment located between galF and gnd is effectively involved in the biosynthesis of the somatic O antigen in strain S88 and therefore corresponds to the O-antigen gene cluster. The CelP7 mutant had a semirough phenotype with only one O-antigen subunit substitution on the core oligosaccharide (Fig. 3). This result indicates that the gene that we presumptively identified as O-antigen polymerase (wzy) effectively encodes a polymerase responsible for the assembly of the oligosaccharide subunits.
FIG. 3.
Visualization of LPS from strains 96-3285 and S88 and mutants by SDS-polyacrylamide gel electrophoresis. Lane 1, 96-3285 (CDC reference strain O45); lane 2, S88 (wild type with long-chain LPS [S LPS]); lane 3, CelP7 (S88 with a deletion of wzy and the core replaced by one O unit [SR LPS]); and lane 4, CelP3 (S88 with a deletion of the O-antigen gene cluster and complete loss of O chains [R LPS]).
O somatic antigen has a major role in the virulence of S88.
O antigen has been recognized as a potential virulence factor in ExPEC infection. Indeed, somatic antigen O18 is known to play a key role in the virulence of the archetypal O18:K1:H7 meningitis clone. Thus, in order to investigate the pathogenic role of the S88 somatic antigen, we assessed the virulence of the CelP3 and CelP7 mutants, relative to S88, in two different experiments with a neonatal rat meningitis model. For comparison, strain 96-3285 was also assessed in our model. In the first experiment the average bacteremia observed with mutant CelP3 (2.94 ± 0.04 log CFU/ml) was far lower (P < 0.0001) than that observed with S88 (6.15 ± 0.89 log CFU/ml) (Table 3). No cases of meningitis were observed with the mutant, whereas five cases occurred with the wild-type strain. Similar results were obtained in the second experiment, in which the average bacteremia observed with mutant CelP7 (2.91 ± 0.04 log CFU/ml) was lower (P < 0.0001) than that observed with S88 (6.23 ± 1.68 log CFU/ml). Again, the only three cases of meningitis occurred with S88. Therefore, each mutant produced a level of bacteremia at least 3 logs lower than the wild-type strain S88. These results suggest that, in this model, the O antigen plays a major role in producing sustained high-level bacteremia. As expected 96-3285 was unable to induce any bacteremia in our model even when the inoculum was increased 100-fold (data not shown).
TABLE 3.
Mean bacterial counts in blood and CSF culture positivity in a neonatal rat model of meningitis with S88 and its mutants
| Expt. and strain | Description | No. of infected animals | Mean bacteremia (log CFU/ml [SD]) | No. of culture-positive CSF specimens |
|---|---|---|---|---|
| Expt. no. 1 | ||||
| S88 | S88 wild type | 15 | 6.15 (0.89) | 5 |
| CelP3 | S88 (O-antigen gene cluster with cat gene insertion) | 15 | 2.94 (0.04)a | 0 |
| Expt. no. 2 | ||||
| S88 | S88 wild type | 10 | 6.23 (1.68) | 3 |
| CelP7 | S88 (wzy::cat) | 10 | 2.91 (0.01)b | 0 |
P = 10−9 versus S88 bacterial count in experiment 1.
P = 10−4 versus S88 bacterial count in experiment 2.
O-serogroup PCR assay.
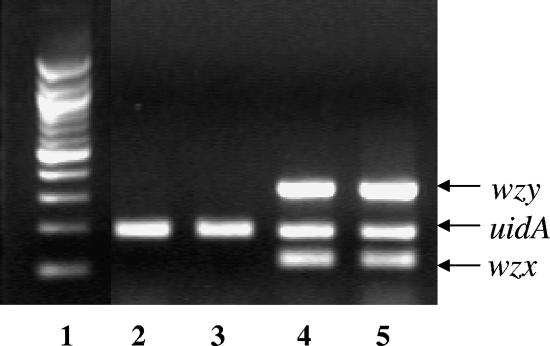
In order to develop an accurate typing system able to specifically recognize strains harboring the somatic antigen identical to that of strain S88, we designed primers to specifically amplify the wzx and wzy genes, which are usually O antigen specific. The two genes were amplified simultaneously with the uidA gene, which serves as an internal control, resulting in a triplex PCR assay (Fig. 4). First, we assessed the efficiency of our PCR assay with 44 clinical strains of E. coli O45:K1:H7 closely related to S88 (12). The 44 strains were PCR positive with both primer pairs. Specificity was then evaluated using as a template DNA extracted from 130 O reference strains, including the reference strains 96-3285 and H61 for O45 somatic antigen, and DNA extracted from the 72 ECOR reference strains. None of the 204 PCR assays was positive. These results showed the efficiency and specificity of our PCR assay.
FIG. 4.
O-serogroup PCR assay using wzx (119 bp), wzy (330 bp), and uidA (186 bp) primers. Lane 1, 100-bp molecular weight ladder; lane 2, 96-3285 (CDC reference strain O45); lane 3, H61 (Staten Serum Institute strain O45); lane 4, S88 (wild type); lane 5, clinical strain of E. coli O45:K1:H7, closely related to S88.
DISCUSSION
We recently described the emergence in France of a new highly virulent group B2 E. coli neonatal meningitis clone of serotype O45:K1:H7. While capsular antigen K1 and flagellar antigen H7 are common among E. coli strains causing neonatal meningitis (1), antigen O45 is rare and, intriguingly, is mainly shared by InPEC strains such as enterotoxigenic E. coli and STEC strains (4, 9, 32, 41) which do not usually belong to group B2 (18). One possible explanation for the expression of the same antigen by such different E. coli pathotypes belonging to different phylogenetic groups is horizontal transfer of the O-antigen gene cluster between these clones. The transferability of the O-antigen gene cluster has been demonstrated, particularly for the locus encoding the O157 antigen of the worldwide STEC O157:H7 clone (20). Moreover, older and recent phylogenetic analyses (1, 7, 24) showing the existence of common serogroups among distant clonal groups of ExPEC strains suggest that this transferability is shared by many other O-antigen gene clusters. In our study, comparing the published O-antigen gene cluster sequence of the O45 CDC reference strain and the O45-antigen gene cluster sequence obtained through the ongoing S88 sequencing project (www.genoscope.fr), we found that although the two loci shared some similarities, the serogroup identity cannot be explained by simple horizontal transfer of the locus between these two strains.
It is therefore likely that S88 expresses an O polysaccharide related but not identical to O45. The cross-reaction could be due in part to the probable presence of 6-deoxy-l-talose. This sugar is the product of the four genes rmlABC and tll (38), which we presumptively identified in the O-antigen gene cluster of strain S88. Moreover, 6-deoxy-l-talose is known to be present in O45 and O66 polysaccharides, for which antisera may cross-react (33). Until the structure of the S88 O polysaccharide is elucidated and specific antibodies are available, we propose to name its O serogroup O45S88.
Although not identical, the S88 and 96-3285 O-antigen gene clusters shared a high degree of similarity with respect to the physical map of the loci. rmlA and rmlB are important in the nucleotide sugar biosynthesis pathways converting glucose-1-phosphate to the dTDP-6-deoxy-d-xylo-4-hexulose intermediate, which is a branch point for several pathways (38). These two genes mainly have two types of disposition. In several E. coli strains and also in Salmonella spp. and Shigella spp., they are part of the four genes rmlABCD clustering at the 5′ end of the O-antigen cluster in the order rmlBDAC. In other E. coli strains and in several Enterobacteriaceae and non-Enterobacteriaceae, the rmlAB genes are present while rmlCD genes are absent, and rmlAB genes cluster together at the 5′ end of the O-antigen gene cluster in the order rmlBA. We exhaustively inspected all rmlAB genes available in the GenBank database and found no such scattered disposition. Therefore, the separate positions of rmlA and rmlB, each at one extremity of the locus in strains S88 and 96-3285, is unique. This result and the similar global disposition of the orthologous genes between the two loci, point to a common ancestor for the two O-antigen gene clusters. This common ancestor would have given rise to two different O-antigen gene clusters via several genetic events, such as deletion or acquisition of the four genes lying between rmlB and rmlC (Fig. 1).
In order to gain insight into the genetic evolution of the O45S88 gene cluster, we constructed phylogenetic trees of rmlABC DNA sequences. In the three trees, S88 and 96-3285 were distantly related to the main E. coli and Shigella group. Moreover, the two strains shared a low level of genetic identity, and their rmlABC genes did not appear to be more closely related to each other than to the rmlABC genes of other species. All these results suggest that although the global organization of the two loci encoding the O-antigen process is similar in S88 and 96-3285, none of the genes was recently exchanged between these strains. In line with the hypothesis postulating a common ancestor for the two loci, the large genetic distance observed between the orthologous genes may be explained by multiple horizontal gene transfers from different species and/or by multiple mutations during a long period of evolution.
Horizontal transfer of O-antigen gene clusters generally involves the flanking conserved genes and, notably, gnd, which serves as a target for recombination. Nelson and Selander sequenced gnd in several E. coli strains and other Enterobacteriaceae and found that gnd in certain E. coli strains had been imported from Klebsiella spp. and Citrobacter spp. (30). In order to determine whether the O45S88 gene cluster was acquired in part from other species, we compared the gnd sequences of E. coli strains S88 and 96-3285 and of several other Enterobacteriaceae. gnd in S88 was distantly related to gnd in other E. coli species but unrelated to the gene in previously identified donor genera such as Citrobacter and Klebsiella. These results indicate that the O45S88 gene cluster may, at least in part (including the rmlA gene), have been transferred from another, unidentified species. To confirm this it would be necessary to find a common donor for gnd and rmlA. In order to obtain more evidence of horizontal transfer, we also analyzed the genetic relationship of galF, the opposite flanking gene, and the JUMPStart sequence, a conserved sequence just upstream of the operon encoding many polysaccharides. However, galF and the JUMPStart sequences of S88 were strongly homologous to the sequences of other E. coli strains (data not shown) and could not, therefore, be used to support our hypothesis.
O polysaccharides contribute to the pathogenicity of ExPEC strains. This virulence factor belongs to the so-called protectin class, which plays an important role in protection against complement-mediated lysis and phagocytosis. Indeed, the O18 antigen plays a key role in virulence of the global meningitis clonal group O18:K1:H7. Pluschke et al., using E. coli mutants lacking somatic antigen, showed that polysaccharide O18 is involved in resistance to the classical complement pathway in guinea pig serum (35). In vivo, the same authors as well as Kim et al. demonstrated the crucial role of O18 in sustained high-level bacteremia, which is necessary for blood-brain barrier penetration (25, 35). However, several reports suggest that not all O polysaccharides have identical virulence properties. When exploring the virulence of the avian pathogenic E. coli O78:K80:H9, Mellata et al. constructed a mutant lacking antigen O78 and two derivative strains supplemented with antigen O1 or O26 (28). They found that the loss of O78 was associated with lower pathogenicity but that substitution by O1 or O26 did not fully restore the initial virulence. More strikingly, Russo et al. prepared an isogenic O-antigen-deficient mutant from a human blood isolate named CP9 (O4:K54:H5) and unexpectedly observed a slight increase in virulence compared with the parental strain (37). All these results underline the unpredictable nature of O-antigen involvement in ExPEC virulence. We evaluated the role of the somatic antigen in the virulence of the O45:K1:H7 clone in an experimental meningitis model and found that antigen O45 was crucial for the bacteremic step. Although we did not restore O45 polysaccharide expression in our mutants and thereby confirm the involvement of this factor in extraintestinal pathogenicity, we obtained the same results, namely, a complete loss of virulence in our experimental model, with two different mutants in two separate experiments. Thus, the implication of other, uncontrolled genetic events in the loss of virulence of our mutants is very unlikely. Therefore, the new somatic antigen O45S88 may in part account for the emergence of the French meningitis clone.
Our work underlined the limits of O serotyping by agglutination, owing to the risk of cross-reactions. Numerous O-antigen gene clusters have been sequenced in recent years, and PCR assays targeting the O-antigen unit processing genes, the O-antigen flippase (wzx) and the O-antigen polymerase (wzy), have been developed (5, 6, 13, 16, 17, 19, 44, 45). This O-genotyping approach has several advantages, including low cost, rapidity, and detection of strains with the O antigen of interest in a complex microflora. The PCR assay we developed here will allow strains harboring the O45S88 antigen to be distinguished from those expressing the reference O45 antigen. This method may also facilitate the search for the ecological niche and the mode of human acquisition of this antigen. Further studies will be necessary to understand the origin of this new somatic antigen, which could serve as a vaccine target for neonatal meningitis prevention. Meanwhile, our PCR test could be used to detect the highly virulent O45:K1:H7 clone in the microflora of neonates or even in the mother's vaginal flora, as is already the case for group B streptococci.
Acknowledgments
We thank Vanida Vongsouthi for technical assistance with LPS analysis and Olivier Clermont for finishing the sequencing of S88.
This work was supported by Fondation pour la Recherche Médicale grant 20050904666.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batisson, I., M. P. Guimond, F. Girard, H. An, C. Zhu, E. Oswald, J. M. Fairbrother, M. Jacques, and J. Harel. 2003. Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 71:4516-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., Q. Kong, L. Feng, Q. Wang, G. Krause, L. Leomil, Q. Jin, and L. Wang. 2005. Development of PCR assays targeting the genes involved in synthesis and assembly of the new Escherichia coli O174 and O177 O antigens. J. Clin. Microbiol. 43:5143-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., J. Tao, L. Feng, G. Krause, S. Zimmermann, K. Gleier, Q. Xia, and L. Wang. 2005. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J. Clin. Microbiol. 43:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidet, P., F. Mahjoub-Messai, J. Blanco, J. Blanco, M. Dehem, Y. Aujard, E. Bingen, and S. Bonacorsi. 2007. Combined multilocus sequence typing and O serogrouping distinguishes Escherichia coli subtypes associated with infant urosepsis and/or meningitis. J. Infect. Dis. 196:297-303. [DOI] [PubMed] [Google Scholar]
- 8.Bidet, P., A. Metais, F. Mahjoub-Messai, L. Durand, M. Dehem, Y. Aujard, E. Bingen, X. Nassif, and S. Bonacorsi. 2007. Detection and identification by PCR of a highly virulent phylogenetic subgroup among extraintestinal pathogenic Escherichia coli B2 strains. Appl. Environ. Microbiol. 73:2373-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanco, J., M. Blanco, J. I. Garabal, and E. A. Gonzalez. 1991. Enterotoxins, colonization factors and serotypes of enterotoxigenic Escherichia coli from humans and animals. Microbiologia 7:57-73. [PubMed] [Google Scholar]
- 10.Bocs, S., S. Cruveiller, D. Vallenet, G. Nuel, and C. Medigue. 2003. AMIGene: annotation of microbial genes. Nucleic Acids Res. 31:3723-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonacorsi, S., V. Houdouin, P. Mariani-Kurkdjian, F. Mahjoub-Messai, and E. Bingen. 2006. Comparative prevalence of virulence factors in Escherichia coli causing urinary tract infection in male infants with and without bacteremia. J. Clin. Microbiol. 44:1156-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonacorsi, S. P., O. Clermont, V. Houduoin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange, and E. Bingen. 2003. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J Infect. Dis. 187:1895-1906. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, J., B. Liu, D. A. Bastin, W. Han, L. Wang, and L. Feng. 2007. Genetic characterization of the Escherichia coli O66 antigen and functional identification of its wzy gene J. Microbiol. 45:69-74. [PubMed] [Google Scholar]
- 14.Czirok, E., M. Herpay, and H. Milch. 1993. Computerized complex typing of Escherichia coli strains from different clinical materials. Acta Microbiol. Hung. 40:217-237. [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DebRoy, C., P. M. Fratamico, E. Roberts, M. A. Davis, and Y. Liu. 2005. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl. Environ. Microbiol. 71:4919-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DebRoy, C., E. Roberts, J. Kundrat, M. A. Davis, C. E. Briggs, and P. M. Fratamico. 2004. Detection of Escherichia coli serogroups O26 and O113 by PCR amplification of the wzx and wzy genes. Appl. Environ. Microbiol. 70:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escobar-Paramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 19.Feng, L., W. Wang, J. Tao, H. Guo, G. Krause, L. Beutin, and L. Wang. 2004. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J. Clin. Microbiol. 42:3799-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. [DOI] [PubMed] [Google Scholar]
- 21.Houdouin, V., S. Bonacorsi, N. Brahimi, O. Clermont, X. Nassif, and E. Bingen. 2002. A uropathogenicity island contributes to the pathogenicity of Escherichia coli strains that cause neonatal meningitis. Infect. Immun. 70:5865-5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. R. 2003. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect. Dis. Clin. N. Am. 17:261-278, viii. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, J. R., K. L. Owens, C. R. Clabots, S. J. Weissman, and S. B. Cannon. 2006. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 8:1702-1713. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Investig. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 28.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss, 3rd, P. K. Brown, P. Arne, A. Bree, C. Desautels, and J. M. Fairbrother. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Negre, V. L., S. Bonacorsi, S. Schubert, P. Bidet, X. Nassif, and E. Bingen. 2004. The siderophore receptor IroN, but not the high-pathogenicity island or the hemin receptor ChuA, contributes to the bacteremic step of Escherichia coli neonatal meningitis. Infect. Immun. 72:1216-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson, K., and R. K. Selander. 1994. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc. Natl. Acad. Sci. USA 91:10227-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orden, J. A., J. A. Ruiz-Santa-Quiteria, D. Cid, S. Garcia, and R. de la Fuente. 1999. Prevalence and characteristics of necrotoxigenic Escherichia coli (NTEC) strains isolated from diarrhoeic dairy calves. Vet. Microbiol. 66:265-273. [DOI] [PubMed] [Google Scholar]
- 33.Orskov, I., F. Orskov, B. Jann, and K. Jann. 1977. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 41:667-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, M., L. Bender, G. Blum, M. Schmittroth, M. Achtman, H. Tschape, and J. Hacker. 1991. Virulence patterns and long-range genetic mapping of extraintestinal Escherichia coli K1, K5, and K100 isolates: use of pulsed-field gel electrophoresis. Infect. Immun. 59:2664-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluschke, G., J. Mayden, M. Achtman, and R. P. Levine. 1983. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect. Immun. 42:907-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 37.Russo, T. A., G. Sharma, C. R. Brown, and A. A. Campagnari. 1995. Loss of the O4 antigen moiety from the lipopolysaccharide of an extraintestinal isolate of Escherichia coli has only minor effects on serum sensitivity and virulence in vivo. Infect. Immun. 63:1263-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 39.Sarff, L. D., G. H. McCracken, M. S. Schiffer, M. P. Glode, J. B. Robbins, I. Orskov, and F. Orskov. 1975. Epidemiology of Escherichia coli K1 in healthy and diseased newborns. Lancet 1:1099-1104. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth, I., V. Karcagi, B. Nagy, I. Gado, and H. Milch. 1994. Examination of verocytotoxin producing capacity and determination of the presence of Shiga-like toxin genes in human Escherichia coli isolates. Acta Microbiol. Immunol. Hung. 41:259-264. [PubMed] [Google Scholar]
- 42.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 43.Vallenet, D., L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli, and C. Medigue. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34:53-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wullenweber, M., L. Beutin, S. Zimmermann, and C. Jonas. 1993. Influence of some bacterial and host factors on colonization and invasiveness of Escherichia coli K1 in neonatal rats. Infect. Immun 61:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]