Abstract
Adeno-associated viruses (AAVs) are being developed as gene therapy vectors, and their efficacy could be improved by a detailed understanding of their viral capsid structures. AAV serotype 8 (AAV8) shows a significantly greater liver transduction efficiency than those of other serotypes, which has resulted in efforts to develop this virus as a gene therapy vector for hemophilia A and familial hypercholesterolemia. Pseudotyping studies show that the differential tissue tropism and transduction efficiencies exhibited by the AAVs result from differences in their capsid viral protein (VP) amino acids. Towards identifying the structural features underpinning these disparities, we report the crystal structure of the AAV8 viral capsid determined to 2.6-Å resolution. The overall topology of its common overlapping VP is similar to that previously reported for the crystal structures of AAV2 and AAV4, with an eight-stranded β-barrel and long loops between the β-strands. The most significant structural differences between AAV8 and AAV2 (the best-characterized serotype) are located on the capsid surface at protrusions surrounding the two-, three-, and fivefold axes at residues reported to control transduction efficiency and antibody recognition for AAV2. In addition, a comparison of the AAV8 and AAV2 capsid surface amino acids showed a reduced distribution of basic charge for AAV8 at the mapped AAV2 heparin sulfate receptor binding region, consistent with an observed non-heparin-binding phenotype for AAV8. Thus, this AAV8 structure provides an additional platform for mutagenesis efforts to characterize AAV capsid regions responsible for differential cellular tropism, transduction, and antigenicity for these promising gene therapy vectors.
Gene therapy has shown great promise for the treatment of diseases caused by defective genes. However, efficiency in the delivery of the corrective genes into the target cells and long-term expression of the genes have been two major challenges of this treatment strategy. Over the past 2 decades, adeno-associated viruses (AAVs) have been investigated extensively as potential gene therapy vectors which can deliver corrective genes into the cells or tissues of a target organism. The AAVs belong to the Dependovirus genus of the Parvoviridae and require helper viruses such as an adenovirus or herpesvirus to cause productive infection (36). Their nonpathogenicity and the ability of the AAVs to package and deliver foreign DNA to target cells have made them ideal viral vehicles for gene delivery (14). To date, 11 distinct serotypes, AAV serotype 1 (AAV1) to AAV11, have been identified in primates, with many more recombinant isolates discovered in different tissues of human and nonhuman primates (8, 15, 16, 17, 35). The capsid amino acid sequence homologies between the AAV serotypes range from ∼55 to 99% (17, 35), and pseudotyping experiments demonstrate that their various cell tropisms and transduction efficiencies are dictated by their capsid sequences (8, 15, 16, 17, 23, 42, 51).
AAV8 is an isolate from rhesus monkey tissue with high homology to the other AAVs, but it has a liver cell transduction efficiency reported to be far greater than those of all others AAVs tested to date. For example, AAV8 has been reported to have an ∼50-fold greater liver transduction efficiency than AAV2 in a mouse model (17). This observation, combined with low reactivity to antibodies against human AAVs, has resulted in efforts to develop AAV8 as a viral vector for liver-directed gene therapy applications. In mouse models, long-term correction of hemophilia A, familial hypercholesterolemia, and glycogen storage disease type II by using AAV8 as a gene therapy vector has been reported (32, 45, 48). Also, in a canine model, AAV8 has been used successfully for liver-directed gene therapy (53), and it has been shown to efficiently cross the blood vessel barrier to attain systemic gene transfer in both skeletal and cardiac muscles of mice and hamsters (54). Successful targeting to the pancreas has also been reported for mice (55).
The tissue tropism and transduction efficiency of AAVs are reported to depend on capsid amino acids which must control initial receptor attachment and/or entry mechanisms. Specific glycan motifs have been identified as primary receptors for some of the AAV serotypes. AAV2, AAV3, and AAV6 show affinity for heparin sulfate and utilize this glycan for cell recognition. However, the interaction of AAV6 with heparin sulfate is not necessary for entry and transduction (20, 59). AAV1, AAV4, and AAV5, on the other hand, bind to glycans with a terminal sialic acid. AAV1 recognizes sialic acids that are bonded α2,3 and α2,6 to the adjacent carbohydrate in an N-linked oligosaccharide (59); AAV4 recognizes sialic acids that are bonded α2,3 to the adjacent carbohydrate in an O-linked oligosaccharide; and AAV5 binds to N-linked oligosaccharides containing sialic acid bonded α2,3 to the adjacent carbohydrate. AAV6 has also been shown to utilize its interaction with sialic acid for cellular transduction (59). The specific glycan motifs recognized for cell entry and/or transduction by the more recently identified serotypes, such as AAV7-11, are not known. However, AAV8 has been reported to have no affinity for heparin sulfate, despite its high amino acid sequence homology with AAV2 (83%) and AAV3 (85%) (33).
Recently, the 37/67-kDa laminin receptor (LamR) was identified as the host cell receptor for AAV8 (3). In cells, the 37/67-kDa LamR protein is reported to have multiple functions, including serving as a ribosomal structural protein as well as a receptor for laminin (4, 5). The interaction of laminin and LamR facilitates tumor metastasis in cancer cells, with the 37/67-kDa LamR protein showing increased expression in cancer cell lines, such as colon and breast cancer lines (34, 44). LamR was also shown to function as a receptor for prion proteins in neuronal cells and for selective serotypes of dengue virus (21, 50). AAV8 was shown to have high selectivity toward LamR compared to other serotypes (3), suggesting a possible new avenue for the utilization of AAV8 capsids in cancer cell-targeted gene therapy. Interestingly, while laminin is glycosylated, LamR is not modified by carbohydrates. Thus, given the reports of the involvement of glycans in cellular infection/transduction by the other AAV serotypes, it is likely that other cellular molecules which have yet to be identified also play a role in AAV8 cell recognition and transduction.
Efforts to understand the basic biology of the AAVs in terms of capsid amino acids responsible for receptor attachment, cellular transduction, and antibody recognition have generated a need to determine the three-dimensional structures of their capsids. The AAV capsids contain 60 copies (in total) of three viral proteins (VPs), VP1, VP2, and VP3, in a predicted ratio of 1:1:10, arranged with T=1 icosahedral symmetry. The three VPs are translated from the same mRNA, with VP1 containing a unique N-terminal domain in addition to the entire VP2 sequence at its C-terminal region. VP2 contains an extra N-terminal sequence in addition to VP3 at its C terminus. In X-ray crystal structures of the AAV2 (61) and AAV4 (18) capsids and all other structures determined for parvovirus capsids, only the C-terminal polypeptide sequence common to all the capsid proteins (∼530 amino acids) is observed. The N-terminal unique region of VP1, the VP1-VP2 overlapping region, and the first 14 to 16 N-terminal residues of VP3 are disordered (18, 61). Cryo-electron microscopy and image reconstruction data suggest that in intact AAV capsids, the N-terminal regions of the VP1 and VP2 proteins are located inside the capsid (40, 61) and are inaccessible for receptor and antibody binding (28). Thus, receptor attachment and transduction phenotypes are determined by the amino acid sequences within the common C-terminal domain of VP1-VP3. The VP1 unique region has been shown to harbor a functional phospholipase A2 enzyme required for endosomal escape during trafficking to the nucleus after receptor attachment, in addition to containing nuclear localization sequences for nuclear targeting (19, 47).
Here we report the crystal structure of the AAV8 viral capsid determined to 2.6-Å resolution. The VP topology is very similar to those of previously reported AAV structures, but with variations in regions previously shown to be important for receptor attachment, transduction, and antibody recognition by AAV2, the best-characterized serotype. The observed structural differences between the highly homologous AAV8 and AAV2 capsids provide a basis for future molecular studies to decipher the mechanism of AAV8's enhanced tropism for liver cells towards the goal of developing optimal cell/tissue-targeted AAV gene therapy vectors. Ordered electron density within the AAV8 capsid that could be interpreted as a single nucleotide was observed in the same interior pocket as that reported for the structurally diverse AAV4 (18), suggesting a common AAV DNA recognition site. The role of this interaction in the AAV life cycle is currently unknown.
MATERIALS AND METHODS
Capsid production, purification, crystallization, data collection, and reduction.
The AAV8 capsid VPs were expressed and assembled using a baculovirus system. Construction of a recombinant baculovirus encoding AAV8 VP1, VP2, and VP3 and purification of the viral capsid were previously described (30). A detailed description of the crystallization processes, in addition to the preliminary characterization of the AAV8 capsid crystals, including data collection, data indexing, processing, scaling, and reduction, has been reported previously (30). Briefly, hexagonal crystals belonging to the space group P6322 were obtained from a crystallization condition which initially contained 20 mM Tris-HCl, pH 7.5, 1.5% polyethylene glycol 8000 (PEG 8000), 25 mM Li2SO4, and 6% glycerol and later became severely dehydrated. These crystals were later reproduced from a condition containing 50 mM Tris-HCl, pH 7.5, 4% PEG 8000, 1 M NaCl, and 6% glycerol.
The original crystals were flash-cooled with Paratone N (Hampton Research, Laguna Niguel, CA), and the X-ray diffraction data were collected at the 22-ID beamline of the South East Regional Collaborative Access Team facilities at the Advanced Photon Source, Argonne National Laboratory (30). The latter crystals, grown in 1 M NaCl, were cryo-protected in a solution containing 50 mM Tris, pH 7.5, 4% PEG 8000, 1 M NaCl, and 24% glycerol prior to X-ray diffraction data collection. New data sets were collected at the A1 and F1 beamlines of the Cornell High Energy Synchrotron Source (Cornell University, Ithaca, NY). The A1 and F1 data were collected with ADSC Quantum 210 and Quantum 4 charge-coupled device detectors, respectively, with crystal-to-detector distances of 200 mm and 300 mm, an oscillation angle of 0.3°, and exposure times of 40 to 60 seconds per image. To obtain information regarding divalent cation binding sites on the AAV8 capsid, crystals were equilibrated in cryo-solution containing 24% glycerol and 20 mM gadolinium chloride (GdCl3) or 10 mM EDTA for ∼24 h prior to data collection at the F1 beamline at the Cornell High Energy Synchrotron Source. The diffraction data were collected using an ADSC Quantum 4 charge-coupled device detector with a crystal-to-detector distance of 300 mm, an oscillation angle of 0.3°, and an exposure time of 60 seconds per image. The data were indexed and processed with DENZO and scaled and reduced with SCALEPACK (39).
Structure determination and refinement.
The phasing of the original X-ray diffraction data with the AAV2 structure (61), using the molecular replacement method, has been described previously (30). The later data sets were phased using the same method. The structure refinement for all data sets followed the same protocol as that described below.
The orientations and positions of the 10 VP3 monomers in the AAV8 capsid asymmetric unit were refined using the rigid-body option of the CNS program (7). The AAV2 amino acid residues for a reference VP3 monomer were mutated to those of the AAV8 sequence by using the SWISS-MODEL program (46). This VP3 monomer structure was refined by several iterative cycles of the rigid-body refinement, simulated annealing, energy minimization, individual B-factor (temperature factor) refinement, and anisotropic temperature factor refinement options of the CNS program (7), constrained by 10-fold noncrystallographic symmetry (NCS). Each refinement step was followed by manual model rebuilding into averaged Fourier 2Fo-Fc and Fo-Fc (where Fo represents observed structure factors and Fc represents the calculated structure factors) electron density maps (generated in CNS by using a molecular mask while applying 10-fold NCS), using the O program (22). The 10 NCS operators used throughout the refinement and averaging procedures were generated from the superimposition of the reference VP3 monomer onto the nine other VP3s in the phasing model and were improved using the program IMP, which is part of the Uppsala Software package (25). A test data set of 5% was partitioned for monitoring of the refinement process (7).
Following the refinement of AAV8 VP3 residues 220 to 738 (the last C-terminal residue [VP1 numbering]), water molecules were assigned in Fo-Fc electron density (at 3σ or above) constrained by hydrogen-bonding-distance geometry. A significantly large peak (8σ) observed in the Fo-Fc map was assigned as a putative ion binding site. Using comparison of the Fo-Fc map generated for the AAV8 capsid data alone to that for data collected for AAV8 capsid crystals soaked with 20 mM GdCl2 or 10 mM EDTA, this ion was interpreted as not being a divalent cation. This density was tentatively assigned as a Na+ ion. The refinement process (for all structures) was complete when there was no longer any improvement in the agreement between the observed (Fo) and calculated (Fc) structure factors (Rfactor) (Table 1). The refined parameters for the final VP3 model, water molecules, and a cation, in terms of average temperature factors and root mean square deviations (RMSD) from ideal bond lengths and angles, were calculated using CNS (7), and the RMSD for main-chain torsion angles were calculated using the PROCHECK program (31). Refinement statistics are given in Table 1. The figures were generated using the programs Chimera (41), GRASP (37), PyMOL (11), and “Roadmap” (60).
TABLE 1.
Refinement statistics for the AAV8 capsid
| Parameter | Valued |
|---|---|
| Resolution (Å) | 40-2.6 (2.69-2.60) |
| Space group/crystal system | P6322 |
| Unit cell (Å) | a = 254.8, b = 445.4 |
| Rsyma | 0.110 (0.338) |
| Completeness (%) | 78.2 (62.2) |
| I/σ | 11.7 (2.4) |
| Rfactorb | 0.255 |
| Rfreec | 0.259 |
| No. of observed reflections | 437,012 |
| No. of unique reflections | 202,494 |
| No. of protein atoms | 4,138 |
| No. of metal atoms | 1 |
| No. of solvent atoms | 89 |
| RMSD for bond lengths (Å) | 0.007 |
| RMSD for bond angles (°) | 1.54 |
| Avg B factor, main chain (Å2) | 34.3 |
| Avg B factor, side chain and solvent | |
| atoms (Å2) | 34.4 |
| Residues in the most/additional/generously | |
| allowed regions (%) | 85.3/14.5/0.2 |
Calculated as Σ|Ihkl − <Ihkl>|/Σ<Ihkl>, where Ihkl is a single value of the measured intensity of the hkl reflection and <Ihkl> is the mean of all measured values of the intensity of the hkl reflections.
Calculated as Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Same as Rfactor, but calculated with a 5% randomly selected fraction of the reflection data not included in the refinement.
Numbers in parentheses are for the highest-resolution shell.
Structural alignment of the AAV8, AAV2, and AAV4 VP3 structures.
To enable comparison of the available AAV viral capsid crystal structures, the VP3 coordinates of AAV8, AAV2 (Protein Data Bank [PDB] accession no. 1LP3), and AAV4 (PDB accession no. 2G8G) were aligned using the secondary structure matching (SSM) program (27). The program also generates a list of atomic distances (in Å) between each C-α position aligned. This list was used to identify amino acid residues that are structurally equivalent and regions of amino acid/structural insertions/difference (defined as C-α atoms that are >1.0 Å apart).
Heparin sulfate affinity column chromatography of AAV capsids.
The binding of AAV2, AAV5, and AAV8 to heparin sulfate was analyzed using a heparin sulfate-agarose column as described previously (38). Briefly, silicon dioxide-pretreated Bio-Rad microspin columns were utilized to pack the heparin-agarose. A 0.5-ml heparin-agarose gravity column (Sigma H-6508) was prepared for the binding assay by being washed with 3 column volumes each of TD buffer (137 mM NaCl, 15 mM KCl, 10 mM Na2PO4, 5 mM MgCl2, 2 mM KH2PO4 [pH 7.4]), TD buffer containing 2 M NaCl, and TD buffer again. Approximately equal amounts of AAV2, AAV5, and AAV8 capsids were added to 500 μl of TD buffer and loaded onto the column. The column was washed with 10 column volumes of TD buffer. Bound virus was eluted with TD buffer containing 2 M NaCl. The entire volume of the flowthrough, wash, and eluate fractions was pooled separately, denatured by boiling in sodium dodecyl sulfate, and slot blotted onto a polyvinylidene difluoride membrane for immunoblot analysis. The membrane was blocked with phosphate-buffered saline-0.05% Tween 20 with 10% dry milk and incubated with the B1 antibody (which recognizes denatured AAV VPs) (38) at a 1:3,000 dilution for 18 h at 4°C. Anti-mouse immunoglobulin G-horseradish peroxidase was used to detect bands by enhanced chemiluminescence (Amersham Pharmacia).
Protein structure accession numbers.
The refined coordinates for the AAV8 VP3 structure at 2.6-Å resolution have been deposited in the PDB with accession number 2QA0.
RESULTS AND DISCUSSION
The AAV8 viral capsid structure has been determined from two independent data sets, to 3.0-Å (data set 1) and 2.6-Å (data set 2) resolution, respectively. The data sets differ in that the 3.0-Å-resolution structure was determined from crystals cryo-protected using Paratone N prior to data collection, while that at 2.6-Å resolution was collected from crystals cryo-protected with glycerol. The two data sets could not be merged due to unit cell differences. The two structures were determined independently and compared. The structural features of the VP region ordered in the electron density were identical except at a capsid surface loop comprising residues 266 to 269, which were disordered in the 3.0-Å-resolution structure. The final refinement statistics for the 2.6-Å-resolution structure are given in Table 1. The final Rfactor and Rfree values, 0.256 and 0.260, respectively, are comparable to or better than those reported for other parvovirus structures (and structures for members of other virus families) at comparable resolutions (see the VIPER database [43]). The similarity of Rfactor and Rfree stems from the 10-fold NCS used during structure determination. The stereochemical parameters and geometries for the final AAV8 capsid VP3 models are also consistent with those for VP models built into comparable-resolution electron densities for other viruses. A representative piece of the 2Fo-Fc density map is shown in Fig. 1A. Our discussion of the AAV8 viral capsid in the remainder of this report is based on the structure determined at 2.6-Å resolution.
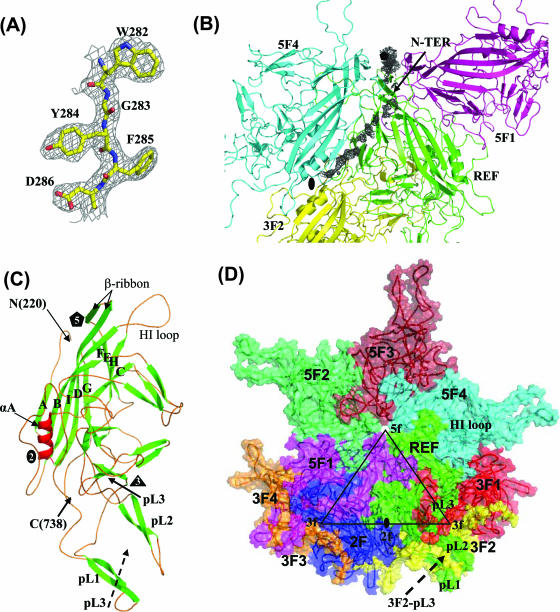
FIG. 1.
Structure of AAV8. (A) Part of the 2Fo-Fc electron density map (gray mesh) for residues 282 to 286, contoured at 1.5σ. The map was calculated using data at a 2.6-Å resolution. The atoms are colored according to atom type. (B) Portion of Fo-Fc electron density map (gray) showing extra density at the fivefold channel and the inner surface of the capsid. Three of the fivefold symmetry-related VP monomers are shown, in green (REF = reference), magenta (5F1), and cyan (5F4), and a threefold monomer is shown in yellow (3F2). (C) Ribbon diagram of AAV8 VP3 showing the core eight-stranded β-barrel strands (green), stretches of small antiparallel β-strands (green), loops (orange), and a conserved parvoviral helical region (red). The first N-terminal residue observed (220), the C-terminal residue (738), the eight strands (βB to βI) that make up the core β-barrel, the loop between βH and βI (the HI loop), αA, and the loop regions (pL1 to pL3) that make up the protrusions that surround the icosahedral threefold axes are labeled. The locations of the five-, three-, and twofold axes are also labeled as black polygons. (D) AAV8 VP3s related to the reference monomer (REF) by five (5F1 to 5F4)-, three (3F)- and twofold (2F) symmetry relationships. The black triangle depicts a viral asymmetric unit bounded by five-, three-, and twofold icosahedral symmetry axes. The HI loop and pL1 to pL3 are labeled.
Overview of the AAV8 capsid structure.
Although all three AAV viral capsid VPs were present in the virus sample used to obtain the AAV8 crystals (30), only the C-terminal 519 amino acid residues could be assigned into the averaged electron density map at ≥1σ. This corresponds to residues 220 to 738 of the common overlapping capsid VPs (Fig. 1C). The VP1 unique region (residues 1 to 137 [VP1 numbering]), the region of VP2 that overlaps with VP1 (residues 138 to 203), and the first N-terminal 16 residues of VP3 are not ordered. However, density at ∼0.5σ extending from residue 220 could be modeled as additional N-terminal residues extending either through the channel at the icosahedral fivefold axis of the capsid (residues 210 to 217) or towards the twofold axis, running between the interfaces of fivefold symmetry-related VP monomers (residues 207 to 219) (Fig. 1B). The lack of ordering of the N-terminal regions of VP1-VP3 is consistent with observations for all parvovirus structures determined to date (reviewed in reference 9). The low copy numbers of VP1 and VP2 and a possible multiple conformation of the N termini in all three capsid VPs, which is incompatible with the imposed icosahedral symmetry used during the structure determination, are likely responsible for this disorder. The observation of weaker density close to the last ordered N-terminal residue within the fivefold channel and extending into the interior of the capsid has also been reported for the autonomous parvoviruses canine parvovirus (62) and minute virus of mice (2). Density within the fivefold channel is consistent with the postulation that this capsid region is the site for externalization of the unique N-terminal region of VP1 for its phospholipase A2 activity and for the VP1 and VP2 nuclear localization signal sequences (19, 47).
The structural topology of the ordered region of the AAV8 VP monomer (residues 220 to 738) consists of a core eight-stranded (labeled βB to βI) β-barrel motif (common in most virus structures) and long loops between the strands (Fig. 1C), as observed for AAV2 (61), AAV4 (18), and all other parvovirus structures determined to date (reviewed in reference 9). Small regions of conserved β-strands are observed within the large loops. The β-barrel represents ∼17% of the amino acid sequence of the ordered VP region. The elaborate loops between the β-barrel strands form the surface of the capsid.
Residues 654 to 673, located between βH and βI (the HI loop) from each VP monomer, are positioned on the surface of the capsid above residues 251 to 253, 362 to 374, and 676 to 680 of an adjacent monomer in a depression that surrounds each fivefold axis of the parvovirus capsid (Fig. 1C and D). The above amino acids, in addition to residues 223 to 233 in βA and 400 to 408, form the interactions between fivefold symmetry-related monomers. A cylindrical channel at the fivefold axis, which is the only pore in the assembled parvovirus capsid, is formed by a conserved β-ribbon between βD and βE (Fig. 1C and D). Residues from two threefold symmetry-related VP monomers (residues 443 to 468 and 580 to 597 from one VP and 486 to 511 from the other VP) interact to form the characteristic protrusions that are offset (by ∼34 Å in AAV8) from the icosahedral threefold axis of the AAV capsids (18, 29, 40, 52, 61). Residues 443 to 468 and 580 to 597 (labeled pL1 and pL2, respectively, in Fig. 1C and D) form finger-like loops in one VP monomer, stabilized by small stretches of β-strand, between which are located residues 486 to 511 (labeled pL3 in Fig. 1C and D) from the threefold symmetry-related monomer in an assembled capsid. These threefold interactions are the most extensive in AAV8 (Fig. 1D), as also reported for AAV2 and AAV4. The icosahedral twofold axis, located in a surface depression, contains the smallest number of intermonomer interacting residues between VP monomers (residues 695 to 709 from each monomer) in the capsid and is the thinnest region, being only one polypeptide chain thick (Fig. 1C). An α-helix (residues 295 to 304) that forms the wall of the twofold depression in all parvovirus structures determined to date is also conserved in AAV8 (Fig. 1C).
Variable AAV loop regions.
A structure alignment of the ordered VP region of AAV8 with those for AAV2 and AAV4 shows that the β-strands forming the core β-barrel are highly conserved (Fig. 2A). A superimposition of the coil representations of the VP monomers from these three AAVs shows them to have very similar topologies (Fig. 2B). In particular, the structure of AAV8 is closely related to that of AAV2, to which it is ∼83% identical in amino acid sequence, with an RMSD of only 0.79 Å between the equivalent C-α atoms for residues 220 to 738 of AAV8 superimposed onto residues 219 to 735 of AAV2. The RMSD for a similar comparison of AAV8 and AAV4, which are ∼59% identical, is 1.6 Å for residues 220 to 738 of AAV8 and 213 to 734 of AAV4.
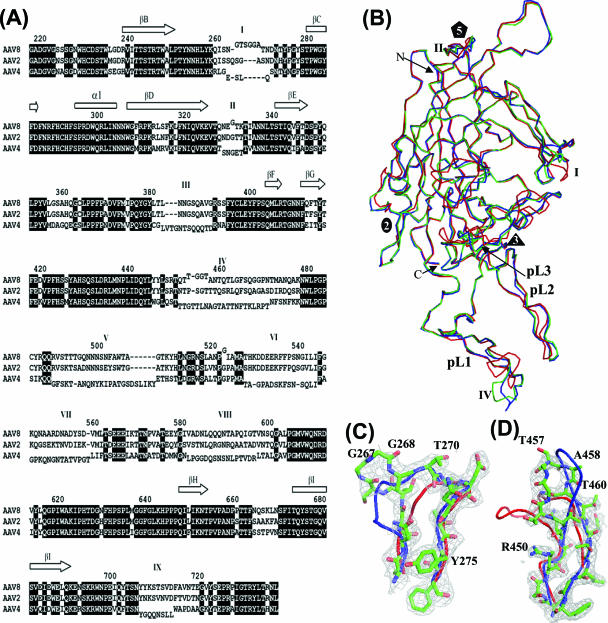
FIG. 2.
Superimposition of AAV structures. (A) Sequence/structure alignment of the AAV2, AAV4, and AAV8 VP3 amino acids. The core β-strands forming the β-barrel are represented as arrows at the top of the alignment, and variable regions I to IX (as defined in reference 18) are indicated above the sequence. VP regions that differ structurally between the viruses are offset above (for AAV8) and below (for AAV4) the alignment. The residue numbering above the alignment is based on AAV8 VP1 and refers to the center residue. (B) Superimposition of C-α atoms of AAV2 (blue), AAV4 (red), and AAV8 (green). The three loops at which AAV8 differs from AAV2 are located at variable regions I (residues 263 to 271), II (residues 329 to 333), and IV (residues 452 to 471). (C) Superimposition of the variable region I surface loops of AAV2 (blue), AAV4 (red), and AAV8 (green) and the 2Fo-Fc map of AAV8 (gray mesh). (D) Superimposition of the variable region IV loops and the 2Fo-Fc map of AAV8. The color scheme follows that of panel C.
Structurally variable regions between the AAV VPs (I to IX) (defined in reference 18) lie within the loops between the β-strands. However, while the structural comparison of AAV2 and AAV4 identified nine regions with C-α positions that are >1.0 Å apart (18), the most prominent differences between AAV8 and AAV2 occur only at variable regions I, II, and IV (Fig. 2). Significantly, AAV2 residues within these regions have been shown to play a role in tissue transduction and antibody recognition (24, 33, 38, 56, 57). Structurally analogous regions are also implicated in tissue tropism and pathogenicity determination for members of the autonomous parvoviruses (reviewed in reference 1).
Variable region I in AAV8 (residues 263 to 271) contains two and five additional amino acids compared to those in AAV2 (residues 262 to 268) and AAV4 (residues 256 to 259), respectively, and adopts a different configuration from those observed in the latter viruses (Fig. 2B and C). The B factors for the residues in this AAV8 loop are higher (∼68 to 80 Å2) than those for the rest of the structure (average, ∼34 Å2) (Table 1), indicating the flexible nature of this surface loop. The B factors for residues in the analogous loop in AAV2 and AAV4 (PDB accession no. 1LP3 and 2G8G, respectively) are also high, consistent with structural flexibility. Mutations of residues 265 to 271 (AAV2 VP1 numbering) within or close to this loop in AAV2 were reported to significantly affect tissue transduction (101- to 10−5-fold changes) (33). This region was proposed as part of a non-heparin sulfate binding-related transduction “dead zone” mapped onto the AAV2 capsid by mutagenesis of all surface-exposed residues (33). This observation suggests that the region I residues are involved in cellular interactions required for infection and that the variation in the AAVs may control differential tissue recognition and/or transduction efficiency. Residues within variable region I also form a minor component of the AAV2 A20 capsid antibody epitope (56) located in the raised capsid region between the depressions at the twofold and surrounding the fivefold axis (Fig. 3B). This observation is consistent with the lack of recognition of the AAV8 capsid by the A20 antibody (unpublished data). The A20 antibody is reported to recognize only AAV2 and AAV3 (56), which contain the same amino acids in this region and are predicted to be structurally similar (40).
FIG. 3.
Comparison of the AAV8, AAV2, and AAV4 capsids. Depth-cued surface representations of the AAV8 (green) (A), AAV2 (blue) (B), and AAV4 (red) (C) capsids are shown. The whole-capsid surface topologies are shown at the top, and close-up views of the threefold symmetry axis region are shown at the bottom. Locations of variable regions I, II, and IV are indicated by circles on the capsid surface of AAV8 and the close-up views.
Variable region II (residues 329 to 333) is located in the turn between the β-ribbon that forms the channels at the fivefold axis (Fig. 1C and 2A and B), resulting in different surface topologies of this capsid region in AAV8, AAV2, and AAV4 (Fig. 3A to C). This VP region has also been observed to be variable between the autonomous parvovirus capsids (26). Flexibility in this VP region is expected and consistent with transitions that would be necessary to enable the postulated pH-induced externalization of the N termini of VP1 and VP2 through the fivefold channel while the capsid remains intact during endosomal trafficking (12, 13, 28, 47). Mutations of AAV2 residues 325, 327, 328, and 330 within this loop were reported to affect transduction (33), although their role is unknown.
The third VP variable region (IV) between AAV8 and AAV2 is at residues 454 to 457 (AAV8 VP1 numbering), which are located in the large loop region between β-strands βG and βH (referred to as the GH loop) (Fig. 2A). Residues 454 to 457 form the top of the outermost finger-like projection in each VP monomer (pL1 in Fig. 1C and D and 2B), contributing to the protrusions that surround the threefold axis of the capsid (Fig. 3A and B). Variable region IV (Fig. 2B and D) and AAV variable region V (equivalent to AAV8 pL3 in Fig. 2B) of the AAV4 VP are dramatically different in conformation from those of both AAV8 and AAV2, resulting in a “rounder” appearance of the AAV4 protrusions (Fig. 3C) (18). Mutation of AAV2 residue R459 (AAV2 VP1 numbering), located in variable region IV, was reported to increase cellular transduction threefold (33).
Role of the AAV VP regions surrounding the threefold axis in receptor binding.
The representative members of the primate AAV clades (16) for which the cell surface glycans recognized for cellular infection have been characterized fall into two broad groups, those that bind heparin sulfate (AAV2, AAV3, and AAV6) and those that bind sialic acid (AAV1, AAV4, AAV5, and AAV6). For the remaining members, AAV7, AAV8, and AAV9, the glycans recognized are not known, although AAV8, which is highly homologous to AAV2 in amino acid sequence, has been reported to not have an affinity for heparin sulfate-agarose (33). This observation was further confirmed, as shown in Fig. 4. Chromatographic analysis of AAV2 (as a positive control), AAV5 (as a negative control), and AAV8 on a heparin-agarose column showed binding of AAV2 and no interaction for AAV5 and AAV8. The non-heparin-binding phenotype observed for AAV5 is consistent with previous reports (38).
FIG. 4.
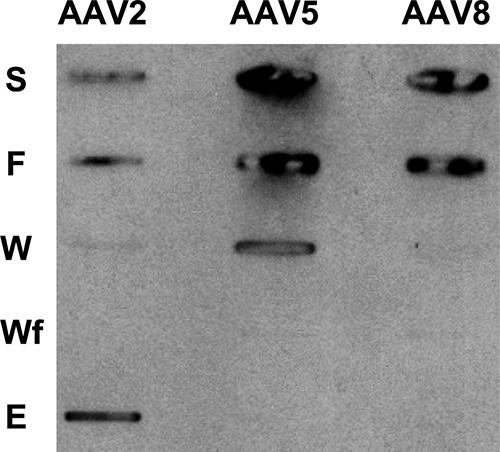
Heparin sulfate binding phenotypes of AAV capsids. Slot blot analysis was performed with flowthrough (F), wash 1 (W), wash 2 (Wf), and elution (E) fractions of AAV2, AAV5, and AAV8 capsids following loading onto a heparin-agarose column. The loaded sample is shown in the sample (S) slot. AAV2, positive control; AAV5, negative control.
Mutational and structural analyses have mapped the AAV2 heparin sulfate binding region to VP residues R484, R487, K532, R585, and R588 (AAV2 VP1 numbering) (24, 38, 61). Residue K527 was also reported to play a minor role in this interaction (24, 38, 61). Residues R585 and R588 are located on the wall of the protrusions surrounding the icosahedral threefold axis, and the other residues are located on the floor of the valley that runs between the twofold and threefold axes (24, 38, 61). Table 2 shows an amino acid alignment of AAV1 to AAV9 at residues structurally predicted to be equivalent or proximate to the AAV2 heparin binding residues in the other serotypes. The two AAV2 residues reported to be the most critical for the heparin sulfate binding phenotype, R585 and R588, are not present in the other serotypes compared. These positions align with Q588 and T591, respectively, in AAV8. The other four amino acids implicated in AAV2 heparin sulfate recognition, R484, R487, K527, and K532, are conserved in AAV3 and AAV6, which also bind heparin sulfate, and in AAV8, as R487, R490, K530, and R535 (AAV8 VP1 numbering) (Table 2), respectively, although this virus does not bind this glycan (Fig. 4) (33). The residues are also conserved in AAV1, which utilizes sialic acid recognition rather than heparin sulfate for cellular infection (10, 58, 59) (Table 2).
TABLE 2.
Comparison of residue types at the AAV2 heparin sulfate binding amino acid positions among AAV serotypes 1 to 9
| Glycan targeta | AAV serotype | Residue at positionb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 487 | 490 | 530 | 533 | 535 | 578 | 588 | 591 | 596 | ||
| ? | 8 | R | R | K | E | R | E | Q | T | G |
| S | 1 | R | R | K | E | K | R | S | T | G |
| H | 2 | R | R | K | E | K | Q | R | R | A |
| H | 3 | R | R | K | E | K | Q | S | T | R |
| S | 4 | K | G | G | A | S | M | S | N | D |
| S | 5 | R | G | K | S | N | V | S | T | G |
| H/S | 6 | R | R | K | K | K | R | S | T | G |
| ? | 7 | R | R | K | E | R | E | A | T | Q |
| ? | 9 | R | R | K | E | R | S | S | A | G |
H, heparin; S, sialic acid; ?, unknown.
Numbering is based on the AAV8 sequence. Basic residues are shown in bold, and acidic residues are italicized.
The non-heparin-sulfate-binding phenotype of AAV8 and the limited binding by AAV1 (58) compared to that by AAV3 and AAV6, despite the conservation of the same AAV2 heparin binding residues in the former viruses, may be due to either the fact that additional amino acids contribute to this phenotype in AAV3 and AAV6 or a difference in the side chain conformations of the conserved basic residues in the AAV capsid structures. In addition to the four conserved basic amino acids, AAV6 uniquely contains an additional basic amino acid, K531 (AAV6 VP1 numbering), which aligns with glutamic acid in AAV2 (E530), AAV8 (E533), and most of the other serotypes, with the exception of AAV4 and AAV5 (Table 2). AAV6 K531 is one of the six AAV1/AAV6 differing residues and is predicted to be located on the capsid surface, forming a continuous basic patch with conserved basic residues R488, K528, and K533 (AAV6 VP1 numbering), equivalent to AAV2 heparin binding residues 487, 527, and 532 (58). AAV6 K531 has been shown to be involved in heparin sulfate binding by AAV6, suggesting that the absence of this residue in AAV1 limits its heparin sulfate recognition (58). In AAV3, there is an additional basic residue, R594 (AAV3 VP1 numbering), which aligns with an alanine in AAV2 (A593) and a glycine in AAV8 (G596) and the other serotypes compared, except AAV4, which contains an aspartic acid, and AAV7, which contains a glutamine (Table 2). A three-dimensional model of AAV3, built based on the AAV2 VP coordinates (PDB accession no. 1LP3) (40; data not shown), predicts that R594 will be located on the capsid surface close to the icosahedral threefold axis (equivalent to AAV2 A593 in Fig. 5B). However, this residue does not form a continuous basic patch with the other residues implicated in AAV2 heparin sulfate binding (Fig. 5B). The possible contribution of this AAV3 unique basic residue to its heparin binding phenotype remains to be determined.
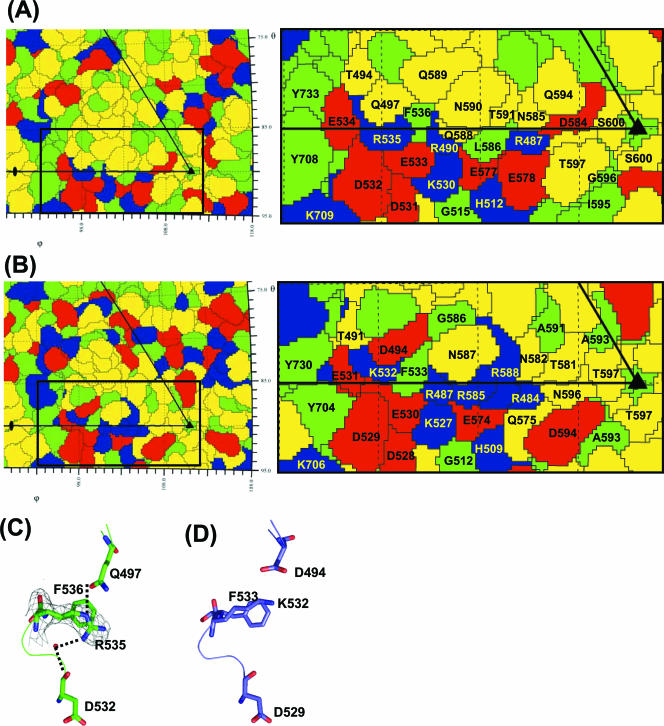
FIG. 5.
Comparison of AAV2 and AAV8 capsid surface residues at the mapped AAV2 heparin sulfate region. (A and B) Schematic “Roadmap” projections (60) of a section of the asymmetric unit surface residues on the capsid crystal structures of AAV8 and AAV2, respectively, with close-up views on the right. Red, acidic residues; blue, basic residues; yellow, polar residues; green, hydrophobic residues. The boundary for each residue is shown in black. The residues are labeled. (C) 2Fo-Fc electron density map (gray) of R535 contoured at 1σ and stick representation of residues that interact with this residue in AAV8. C-α traces of the residues are shown as coils. The side chain for this residue is highly ordered in the electron density map. Hydrogen bonds positioning the R535 side chain are shown as dotted lines. (D) Equivalent region of AAV2 to that shown for AAV8 in panel C. The side chain of K532 in AAV2 (C-α equivalent to AAV8 R535) is not involved in any VP interactions.
A comparison of the capsid surface amino acids within one viral asymmetric unit for the AAV2 and AAV8 capsid crystal structures shows a clear difference in the distribution of acidic and basic residues close to the AAV2 heparin binding residues (Fig. 5A and B). The two critical AAV2 heparin sulfate binding residues, R585 and R588, significantly increase the basic surface area for AAV2 compared to AAV8. In addition to this reduced basic surface area on the AAV8 capsid, a difference in side chain orientation is observed for one of the residues when comparing the two structures at the conserved basic amino acid positions (AAV8 residues 487, 490, 530 and 535 to AAV2 residues 484, 487, 527 and 532). The terminal amino groups of AAV8 R535 projects downwards rather than up towards the capsid surface as observed for terminal amino group of the equivalent K532 in AAV2 (Table 2 and Fig. 5C and D). The Nɛ atom of AAV8 R535 makes a hydrogen bond with the side chain of Q497; the terminal amide group makes a water mediated hydrogen bond with D532 (Fig. 5C). The side chain of K532 in AAV2 is not able to form a hydrogen bond with the shorter side chain of the adjacent D494 (Fig. 5D). Compared to the AAV2 K532 side chain, AAV8 R535 is less flexible due to the hydrogen bonding interactions. The altered orientation of the AAV8 R535 terminal amino groups could make this residue less available for receptor binding. This would suggest that a minimum basic surface required for heparin sulfate binding by the AAV capsids, as suggested by mutational analysis of AAV1/AAV6 studies (58), is missing in AAV8, providing a potential explanation for its non-heparin sulfate binding phenotype. These observations also suggest that if the arginines in AAV7 and AAV9 that align to AAV8 R535 (Table 2) adopt a similar conformation as in AAV8; they should also have a non-heparin sulfate binding phenotype. Work is in progress to identify potential cell surface glycan(s) that mediate the binding of AAV7, AAV8 and AAV9 to host cells.
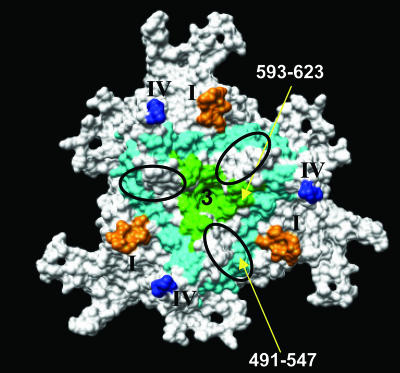
AAV8 cellular transduction utilizing the 37/67-kDa LamR was reported to be mediated by two stretches of amino acids in AAV8, namely, residues 491 to 557 and 593 to 623 (3). The structure of AAV8 shows that these residues are located at the threefold region of the capsid, with residues 491 to 557 within the protrusions that surround this axis and residues 593 to 623 at or near the threefold axis (Fig. 6). The regions contact the heparin sulfate binding region of AAV2 discussed above and contain conserved basic positions K530 and R535 (Fig. 5). Both regions are located within the large loop between βG and βH, which contain AAV variable regions V, VI, VII, and VIII defined during the comparison of AAV2 to AAV4 but are structurally similar in AAV2 and AAV8 (Fig. 2A). The report that LamR also mediates cellular transduction by AAV2, AAV3, and AAV9 suggests that AAV3 and AAV9 have similar structural topologies to those of AAV2 and AAV8 in these VP regions. However, variable regions I and IV discussed above are structurally adjacent to the two residue stretches (Fig. 6). Thus, the report that AAV8's utilization of LamR is different from that by AAV2, with AAV8 absolutely requiring LamR for transduction while AAV2 can transduce cells independently of LamR interaction (3), may be due to these structural variations. It is also possible that differences in amino acids between the two serotypes (Fig. 2A) in the two mapped amino stretches also play a role in their different LamR binding phenotypes.
FIG. 6.
AAV8 binding site for 37/67-kDa LamR. An icosahedral threefold region is shown with stretches of amino acids reported to be involved in 37/67-kDa LamR binding by AAV8, colored in cyan (residues 491 to 547) and green (residues 593 to 623). The locations of variable loops I and IV are shown in orange and blue, respectively, and the heparin sulfate binding region is depicted with an oval.
Putative ion binding site on the AAV8 capsid.
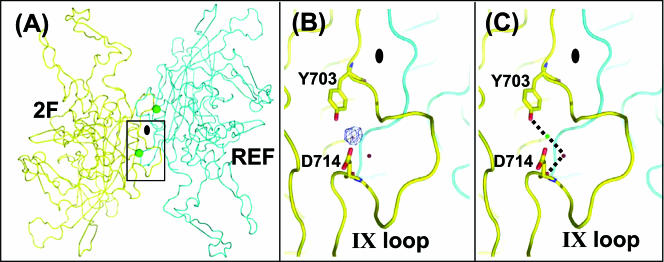
During the course of structural refinement, a strong Fo-Fc electron density peak (8σ) was observed near the icosahedral twofold symmetry axis of the capsid (Fig. 7A and B). In order to verify if this density was due to a divalent cation, an X-ray diffraction data set was collected from crystals soaked with 20 mM GdCl3 and 10 mM EDTA. The electron density map calculated for the GdCl3-soaked crystal did not show a significant increase in the density level at the twofold axis, nor did the data for the EDTA-soaked crystal show a decrease in the density (data not shown), suggesting that the observed density is probably not due to a divalent cation. Thus, this density was interpreted as either a highly ordered water molecule or a Na+ ion, since the crystallization condition contained a high concentration of NaCl (1 M). A solvent molecule could be refined with a low B-factor value of 9 Å2 at this position, while a Na+ ion could also be refined with an acceptable B factor of 23 Å2. Efforts to model other ions, for example, Ca2+ and Cl−, into this density resulted in high B factors, of 50 and 42 Å2, respectively, and thus the density was modeled as being due to a Na+ ion. Despite its location close to the twofold axis, this putative ion is not involved in any intersubunit interactions. It is, however, engaged in a solvent-mediated interaction between residues Y703 and D714 from one monomer, stabilizing the loop formed between these residues (defined as variable loop IX in reference 18) (Fig. 7A and C). This loop forms the majority of the twofold symmetry-related interactions in all AAV capsid structures (18). The location of this putative ion at the twofold axis, the thinnest region of the capsid, holding an AAV variable loop in place, suggests a role in capsid stability, although its real functional role remains to be verified.
FIG. 7.
Putative AAV8 ion binding site. (A) Overall view of the cation binding site close to the icosahedral twofold axis region. The cation binding site is depicted as a green sphere. The reference VP monomer is shown in cyan, and the twofold symmetry-related monomer is shown in yellow. The approximate twofold axis is shown as a filled oval. (B) Fo-Fc electron density map of native crystals (blue mesh). The view is a zoomed-in version of the boxed region in panel A. The map was generated using the CNS program (7) and is shown at a 4σ contour level. The typical density for a solvent molecule (in red) contoured at 4σ is shown for comparison. (C) The environment of the cation binding site. The view is the same as that for panel B. Residues making hydrogen bonds (dotted lines) with the cation (green sphere), i.e., Y703 and D714, and a solvent molecule (red sphere) are shown.
A DNA nucleotide binding site inside the AAV8 capsid.
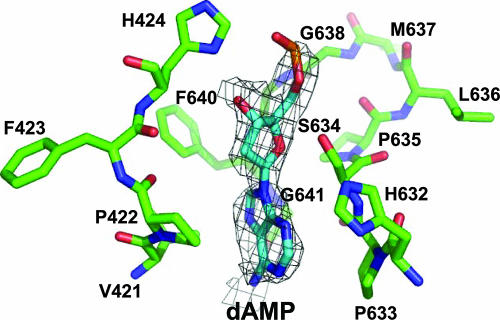
In the AAV4 crystal structure, electron density for a single dAMP nucleotide was observed in the capsid interior near the threefold axis (18). A similar piece of density was ordered in the AAV8 structure observed at a lower σ level (0.5) in the 2Fo-Fc map (Fig. 8). The identity of the ordered purine base could not be determined from the density. As discussed in the AAV4 structure report (18), this ordering of DNA density was unexpected because the capsid packages just one copy of the genome, which is inconsistent with the icosahedral symmetry assumed during the structure determination procedures. The binding site, containing AAV8 residues 421 to 424, 633 to 635, and 638 to 641, is configured from residues that are completely conserved in all AAV serotypes. The location of the purine base is similar to that observed in AAV4, sandwiched between P422 and P633, which are equivalent to P413 and P629 of AAV4. The densities for the deoxyribose sugar and the phosphate group are weaker (∼2σ in the Fo-Fc map) than that for the purine base (∼3σ in the Fo-Fc map) and are oriented differently from that of the AAV4 nucleotide (18). Residues 424, 634, 635, and 638 to 641 are within acceptable distances for forming polar interactions with the DNA. Observation of the base density in an analogous region of the AAV4 and AAV8 structures suggests that this is a common AAV nucleotide binding site.
FIG. 8.
An ordered DNA nucleotide in the AAV8 capsid. A stick representation of the residues in the DNA binding pocket and the DNA molecule is shown within a piece of the Fo-Fc density map (gray mesh; contoured at 1.8σ). The amino acids are labeled and colored according to atom type, except for carbon, which is colored green. The model (energy minimized) shown is that of dAMP, although it is possible that the ordered base is a guanine.
In contrast to the wild-type AAV4 capsids used in the study by Govindasamy et al. (18), which contained a full-length genome, the AAV8 capsids were expressed using a baculovirus construct containing only the cap gene and no rep gene and, as such, were devoid of genomic DNA. However, the ordering of the nucleotide density inside the AAV8 capsid suggests that either the cap gene or random cellular DNA must be being packaged. Preliminary DNA extraction data showed that baculovirus- and adenovirus-expressed AAV capsids package small amounts of cellular DNA (data not shown). This observation was unexpected because AAV packaging is reported to require the functions of the Rep proteins (reviewed in reference 49). The AAV8 dAMP/dGMP must become ordered from the cellular DNA inside a common AAV binding site, but at a lower occupancy in AAV8 than that observed for the AAV4 structure. The lower occupancy is likely due to all 60 sites within the capsid not being occupied in the AAV8 capsid. Work is in progress to identify the cellular DNA sequences being packaged in the expressed AAV capsids and to determine if specific DNA elements are involved in packaging and interaction with this threefold region. This information will provide valuable insights into DNA packaging and interaction requirements for AAVs that could aid efforts to improve vector packaging efficiency.
Summary.
The 2.6-Å-resolution structure presented for AAV8 further highlights the high overall homology between the VP structures of the AAVs, which likely dictates successful assembly. Structural differences between AAV8 and AAV2, the best-characterized AAV serotype, reside in the protrusions that surround the threefold axis, which is utilized for receptor recognition by the two viruses, and also in the raised capsid region between the two- and fivefold axes, the A20 antibody epitope for AAV2. The high transduction rate for liver cells observed for AAV8 compared to AAV2 and other serotypes is likely mediated, in part, by receptor binding phenotypes. Thus, the structural variations between AAV8 and AAV2, which were also observed in comparing AAV2 and AAV4, are consistent with evolutionary diversity in surface regions that confers tissue tropism and transduction efficiency. These differences also confer antigenic specificity. Future research efforts are directed to identifying the specific glycans utilized by AAV8 for cellular transduction and toward further characterization of the DNA binding pocket in the AAV capsid. The data presented here and those attainable in the future should facilitate engineering of AAV vector capsids for improved gene therapy applications in general.
Acknowledgments
We thank the staff at the SER-CAT 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, and the staff at the Cornell High Energy Synchrotron Source for assistance during X-ray diffraction data collection. We thank Robbie Reutzel and Lakshmanan Govindasamy for help with X-ray diffraction data collection.
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. This project was funded by a UF University Scholars Program award (to M.D.L.) and by NIH projects P01 HL59412 and P01 HL51811 (to N.M., S.Z., and M.A.-M.).
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Agbandje-McKenna, M., and M. S. Chapman. 2006. Correlating structure with function in the viral capsid, p. 125-139. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, NY.
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Akache, B., D. Grimm, K. Pandey, S. R. Yant, H. Xu, and M. A. Kay. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 80:9831-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardini, E., B. Sporchia, L. Pollegioni, M. Modugno, C. Ghirelli, F. Castiglioni, E. Tagliabue, and S. Menard. 2002. Identification of a novel function for 67-kDa laminin receptor: increase in laminin degradation rate and release of motility fragments. Cancer Res. 62:1321-1325. [PubMed] [Google Scholar]
- 5.Auth, D., and G. Brawerman. 1992. A 33-kDa polypeptide with homology to the laminin receptor: component of translation machinery. Proc. Natl. Acad. Sci. USA 89:4368-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, B., B. Bruccoleri, D. Olafson, D. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization and dynamics calculation. J. Comp. Chem. 4:187-217. [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 8.Burger, C., O. S. Gorbatyuk, M. J. Velardo, C. S. Peden, P. Williams, S. Zolotukhin, P. J. Reier, R. J. Mandel, and N. Muzyczka. 2004. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 10:302-317. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, M. S., and M. Agbandje-McKenna. 2006. Atomic structure of viral particles, p. 107-123. In J. R. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), Parvoviruses. Edward Arnold, Ltd., New York, NY.
- 10.Chen, C. L., R. L. Jensen, B. C. Schnepp, M. J. Connell, R. Shell, T. J. Sferra, J. S. Bartlett, K. R. Clark, and P. R. Johnson. 2005. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 79:14781-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 12.Farr, G. A., S. F. Cotmore, and P. Tattersall. 2006. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 80:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farr, G. A., L. G. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flotte, T. R., and B. J. Carter. 1995. Adeno-associated virus vectors for gene therapy. Gene Ther. 2:357-362. [PubMed] [Google Scholar]
- 15.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 100:6081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., L. H. Vandenberghe, R. Alvarez, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govindasamy, L., E. Padron, R. McKenna, N. Muzyczka, N. Kaludov, J. A. Chiorini, and M. Agbandje-McKenna. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J. Virol. 80:11556-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grieger, J. C., S. Snowdy, and R. J. Samulski. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 80:5199-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hundt, C., J. M. Peyrin, S. Haik, S. Gauczynski, C. Leucht, R. Rieger, M. L. Riley, J. P. Deslys, D. Dormont, C. I. Lasmezas, and S. Weiss. 2001. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 20:5876-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 23.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleywegt, G. J., and R. J. Read. 1997. Not your average density. Structure 5:1557-1569. [DOI] [PubMed] [Google Scholar]
- 26.Kontou, M., L. Govindasamy, H. J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 79:10931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 28.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kronenberg, S., J. A. Kleinschmidt, and B. Bottcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, M. D., H.-J. Nam, E. Padron, B. Gurda-Whitaker, E. Kohlbrenner, G. Aslanidi, B. Byrne, R. McKenna, N. Muzyczka, S. Zolotukhin, and M. Agbandje-McKennaa. 2005. Production, purification, crystallization and preliminary X-ray analysis of adeno-associated virus serotype 8. Acta Crystallogr. F 61:558-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 32.Lebherz, C., G. Gao, J. P. Louboutin, J. Millar, D. Rader, and J. M. Wilson. 2004. Gene therapy with novel adeno-associated virus vectors substantially diminishes atherosclerosis in a murine model of familial hypercholesterolemia. J. Gene Med. 6:663-672. [DOI] [PubMed] [Google Scholar]
- 33.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martignone, S., S. Menard, R. Bufalino, N. Cascinelli, R. Pellegrini, E. Tagliabue, S. Andreola, F. Rilke, and M. I. Colnaghi. 1993. Prognostic significance of the 67-kilodalton laminin receptor expression in human breast carcinomas. J. Natl. Cancer Inst. 85:398-402. [DOI] [PubMed] [Google Scholar]
- 35.Mori, S., L. Wang, T. Takeuchi, and T. Kanda. 2004. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology 330:375-383. [DOI] [PubMed] [Google Scholar]
- 36.Muzyczka, N., and K. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2360. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, New York, NY.
- 37.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 38.Opie, S. R., K. H. J. Warrington, M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 296:307-326. [DOI] [PubMed] [Google Scholar]
- 40.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 42.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddy, V. S., P. Natarajan, B. Okerberg, K. Li, K. V. Damodaran, R. T. Morton, C. L. Brooks III, and J. E. Johnson. 2001. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 75:11943-11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanjuan, X., P. L. Fernandez, R. Miquel, J. Munoz, V. Castronovo, S. Menard, A. Palacin, A. Cardesa, and E. Campo. 1996. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. J. Pathol. 179:376-380. [DOI] [PubMed] [Google Scholar]
- 45.Sarkar, R., R. Tetreault, G. Gao, L. Wang, P. Bell, R. Chandler, J. M. Wilson, and H. H. J. Kazazian. 2004. Total correction of hemophilia A mice with canine FVIII using an AAV 8 serotype. Blood 103:1253-1260. [DOI] [PubMed] [Google Scholar]
- 46.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80:11040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, B., H. Zhang, L. M. Franco, S. P. Young, A. Schneider, A. Bird, A. Amalfitano, Y. T. Chen, and D. D. Koeberl. 2005. Efficacy of an adeno-associated virus 8-pseudotyped vector in glycogen storage disease type II. Mol. Ther. 11:57-65. [DOI] [PubMed] [Google Scholar]
- 49.Tal, J. 2000. Adeno-associated virus-based vectors in gene therapy. J. Biomed. Sci. 7:279-291. [DOI] [PubMed] [Google Scholar]
- 50.Thepparit, C., and D. R. Smith. 2004. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J. Virol. 78:12647-12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walters, R., S. M. Yi, S. Keshavjee, K. Brown, M. Welsh, J. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 52.Walters, R. W., M. Agbandje-McKenna, V. D. Bowman, T. O. Moninger, N. H. Olson, M. Seiler, J. A. Chiorini, T. S. Baker, and J. Zabner. 2004. Structure of adeno-associated virus serotype 5. J. Virol. 78:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, L., R. Calcedo, T. C. Nichols, D. A. Bellinger, A. Dillow, I. M. Verma, and J. M. Wilson. 2005. Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8-mediated, liver-directed gene therapy. Blood 105:3079-3086. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Z., T. Zhu, C. Qiao, L. Zhou, B. Wang, J. Zhang, C. Chen, J. Li, and X. Xiao. 2005. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 23:321-328. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Z., T. Zhu, K. K. Rehman, S. Bertera, J. Zhang, C. Chen, G. Papworth, S. Watkins, M. Trucco, P. D. Robbins, J. Li, and X. Xiao. 2006. Widespread and stable pancreatic gene transfer by adeno-associated virus vectors via different routes. Diabetes 55:875-884. [DOI] [PubMed] [Google Scholar]
- 56.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, Z., A. Asokan, J. C. Grieger, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 80:11393-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, Z., E. Miller, M. Agbandje-McKenna, and R. J. Samulski. 2006. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J. Virol. 80:9093-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao, C., and M. G. Rossmann. 2007. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]