Abstract
Assembly of infectious adenovirus particles requires seven functionally redundant elements at the left end of the genome, termed A repeats, that direct packaging of the DNA. Previous studies revealed that the viral IVa2 protein alone interacts with specific sequences in the A repeats but that additional IVa2-containing complexes observed during infection require the viral L4 22-kDa protein. In this report, we purified a recombinant form of the 22-kDa protein to characterize its DNA binding properties. In electrophoretic mobility shift assay analyses, the 22-kDa protein alone did not interact with the A repeats but it did form complexes on them in the presence of the IVa2 protein. These complexes were identical to those seen in extracts from infected cells and had the same DNA sequence dependence. Furthermore, we provide data that the 22-kDa protein enhances binding of the IVa2 protein to the A repeats and that multiple binding sites in the packaging sequence augment this activity. These data support a cooperative role of the IVa2 and 22-kDa proteins in packaging and assembly.
Adenovirus assembly proceeds through a series of ordered events: formation of an empty capsid comprised of scaffolding and major structural proteins; selective, polar packaging from the left end to the right end of the linear double-stranded DNA genome with its associated core proteins; and final maturation by the activity of the adenoviral encoded protease (25). Although viral proteins have been identified that specifically recognize portions of the genome required for packaging, the molecular mechanism responsible for completing encapsidation requires further investigation.
Early studies identified a cis-acting packaging sequence at the left end of the adenovirus genome that directs encapsidation of DNA (15). Genetic analysis of the human adenovirus type 5 (Ad5) genome narrowed the sequence important for packaging to nucleotides 194 to 358 (17). Sequence analysis of this region revealed seven repeated A/T-rich motifs, termed A repeats 1 to 7, from left to right on the genome. Successful propagation of Ad5 with truncated and deleted packaging sequences demonstrated that the A repeats are functionally redundant for packaging but ascribed functional dominance to the A1, A2, A5, and A6 repeats (9, 10, 30). Mutations introduced into homologous sequences of the dominant A repeats produced nonviable viruses, defining the crucial packaging repeat as a bipartite consensus: TTTGN8CG (30). This sequence became the target of further studies to identify protein-DNA interactions involved in packaging.
Subsequent studies using electrophoretic mobility shift assays (EMSAs) correlated the functional hierarchy of the A repeats in packaging to the relative binding affinities of specific viral factors. EMSA analyses using nuclear extracts from Ad5-infected cells and a probe encompassing the A1 and A2 repeats uncovered two virally induced complexes, named x and y in order of increasing mobility (32). Both the x and y complexes contained the adenovirus IVa2 protein, which also interacts with the packaging sequence in vivo as determined by chromatin immunoprecipitation assays (26, 28). The sequence-specific interaction of the IVa2 protein with the A repeats readily correlated with packaging function (26, 32). In subsequent experiments, the IVa2 protein purified from a prokaryotic expression system was found to reproduce the y complex, dependent on the CG motif in the A-repeat consensus sequence, with a half-maximal binding value of 15 nM IVa2 (31). The purified IVa2 protein also exhibited highest affinity to the functionally dominant A1 and A2 repeats compared to probes encompassing the other repeats and formed multiple complexes on a probe containing all seven repeats. However, generation of additional complexes on the A1-A2 probe with nuclear extracts from infected cells required at least concomitant expression of the viral L4 22-kDa protein with the IVa2 protein, as well as the TTTG motif (24).
Viruses containing early nonsense mutations in the coding sequences specific for the IVa2 or 22-kDa proteins do not produce infectious particles or, in the case of the IVa2 null virus, empty capsids; these viruses infect cells and express late structural proteins, suggesting the defect occurs during assembly (24, 33). Introduction of a stop codon in the sequence of the L4 transcripts encoding the 22-kDa and 33-kDa proteins, which possess a common amino terminus but different C termini, also results in a similar phenotype (6, 7). However, for the 22-kDa null virus, expression of the related 33-kDa protein did not complement the assembly function of the 22-kDa protein (24). Moreover, although the 22-kDa and 33-kDa proteins share an amino terminus, EMSA analysis with nuclear extracts from cells transiently expressing either protein with IVa2 confirmed that formation of the x complex requires only the 22-kDa protein, correlating binding activity of the 22-kDa protein to packaging (24). Lastly, viruses possessing mutations in the L1 52/55-kDa protein, which formed complexes on the packaging sequence independently from the IVa2 protein, assemble empty capsids but are partially or completely defective at packaging DNA (13, 16, 28). The existence of a protein complex containing the L1 52/55-kDa and IVa2 proteins therefore further indicates that the IVa2 protein is important for packaging (14). These studies, together with the EMSA analyses, suggest binding of the IVa2 and 22-kDa proteins to the A repeats plays an important role in packaging and thus validate further characterization of their interaction.
This report extends EMSA analysis of the interaction of the IVa2 and 22-kDa proteins with the A1 and A2 repeats in cell extracts by comparing binding of purified recombinant proteins to those expressed during Ad5 infection. This system also permitted experimentation with a probe encompassing the complete packaging sequence, which revealed that additional A repeats enhance the binding of the IVa2 and 22-kDa proteins. The data obtained from these analyses uphold the importance of a dynamic interaction between the IVa2 and 22-kDa proteins and the A repeats in adenovirus DNA packaging.
MATERIALS AND METHODS
Plasmid construction.
The open reading frame encoding the 22-kDa protein was amplified using primers 22kDafor (5′-ACCCATGGCACCCAAAAAGAAGC-3′) and 22kDarev (5′-TTCTCGAGTCCGGTCGCCTTTGCTTC-3′) from the previously created cAd212 cosmid (22). The resulting PCR product was inserted into the pCR II-Blunt-TOPO vector (Invitrogen) according to the manufacturer's protocol to create pCRBT-L422kDa. The plasmid was digested with NcoI and XhoI (New England Biolabs [NEB]), and the PCR product was inserted into the corresponding sites of pTYB4 (NEB) to make pTYB4-L422kDa. The integrity of the inserts was confirmed by DNA sequencing (University of Michigan Sequencing Core).
Cells, bacterial strains, and virus.
The Escherichia coli strains, One Shot Chemically Competent TOP10 cells (Invitrogen) and BL21-Codon-Plus RIL (Stratagene), were used for cloning purposes and recombinant protein expression, respectively. 293 cells, transformed human embryonic kidney cells expressing the E1A and E1B proteins (12), were grown and maintained as described previously (13). Wild-type Ad5 was manipulated and propagated as described previously (11).
Nuclear extracts.
Nuclear extracts were prepared from 293 cells 24 h postinfection with 5 PFU per cell of Ad5 as previously described (32).
Protein expression and purification.
Recombinant 22-kDa protein was expressed using the IMPACT-CN E. coli expression system (NEB). This system utilizes an inducible self-cleaving intein tag that is fused to the C terminus of the protein of interest. E. coli BL21 cells transformed with pTYB4-L422kDa were grown in 2 × 500 ml of LB broth to an optical density at 600 nm of 0.80 at 37°C in the presence of 100 μg/ml ampicillin and 20 μg/ml chloramphenicol. The cultures were then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside for approximately 16 h at 4°C. Cells were pelleted by centrifugation and stored at −80°C until lysis. For purification, pellets from the two cultures were combined and lysed in 25 ml of lysis buffer (20 mM HEPES, pH 7.9, 0.5 mM EDTA, 0.1% Triton X-100, 20 μM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin) containing 500 mM NaCl by using a French press. Cellular debris was removed by centrifugation at 20,000 × g for 10 min at 4°C, and the cleared lysate was then added to 6 ml of a 50% slurry of chitin beads (NEB) equilibrated in lysis buffer plus 500 mM NaCl. Protein lysate was allowed to bind to the beads for 1 h at 4°C with continuous rocking, after which the protein-bound beads were packed into a 3-ml column. Unbound proteins were allowed to flow through, and the beads were subsequently washed with nine column volumes of lysis buffer containing 1 M NaCl, followed by three column volumes lysis buffer containing 150 mM NaCl. The 22-kDa protein was eluted from the column by incubation for 24 h at 4°C in lysis buffer containing 150 mM NaCl and 50 mM dithiothreitol (DTT) (Invitrogen). The purified 22-kDa protein was concentrated using an Amicon Ultra Centrifugal 30K filter device (Millipore) and further purified by gel filtration using an Amersham HiPrep 16/60 Sephacryl S-200 high-resolution 120-ml column. The recombinant protein was filtered using a 0.22-μm filter prior to injection into the column and eluted in 2-ml fractions at a flow rate of 0.2 to 0.3 ml/min. An aliquot of each fraction was analyzed on a 9% sodium dodecyl sulfate (SDS) polyacrylamide gel, followed by detection using a ProtoBlue Coomassie stain (National Diagnostics). The fractions of interest were pooled and stored at −80°C in lysis buffer plus 150 mM NaCl and 10% glycerol. A Bio-Rad protein assay using bovine serum albumin (Sigma) as a standard was used to determine the protein concentration.
Purification of the IVa2 protein was performed with the IMPACT-CN system as previously described (31), except that rather than using cation exchange chromatography for the final step, gel filtration was performed as described above for the 22-kDa protein. The purity of the proteins was determined by SDS-polyacrylamide gel electrophoresis (PAGE), staining with ProtoBlue Coomassie stain, and densitometry.
Monoclonal antibody production.
Five 8-week-old female BALB/c mice (Jackson Laboratories) were immunized by intraperitoneal injection with 25 μg of purified 22-kDa protein in phosphate-buffered saline (PBS) and emulsified in complete Freund's adjuvant (Pierce). Mice were boosted at 4 and 6 weeks with 25 μg of the 22-kDa protein in incomplete Freund's adjuvant. Blood was collected from the retro-orbital sinus at 2 and 5 weeks, and serum was separated from blood cells before storage at −80°C. Four days after a final boost with antigen, the mouse with the highest titer of antibodies against the 22-kDa protein, as determined by indirect enzyme-linked immunosorbant assays, was given to the University of Michigan Hybridoma Facility for production of monoclonal antibodies using standard technology (8, 21). Enzyme-linked immunosorbant assays were also used to screen the hybridomas as follows. Immulon 96-well Maxisorp plates (Nalge Nunc) were coated with 0.1 μg IMPACT-purified 22-kDa protein/well in 100 μl PBS overnight at 4°C. Wells were washed twice with PBS containing 0.1% Tween 20 (PBST) and blocked for 30 min at room temperature using 5% dry milk in PBST (PBSTM), after which 50 μl of undiluted hybridoma supernatant was applied to the wells and incubated for 90 min at 37°C. Wells were washed three times with 400 μl of PBST, and 100 μl of horseradish peroxidase (HRP)-linked anti-mouse immunoglobulin G (IgG) (GE Bioscience), diluted 1:5,000 in PBSTM, was added to each well. After incubation for 1 h at room temperature, wells were washed three times with PBST, followed by one wash with PBS. One hundred microliters of HRP substrate (Bio-Rad Laboratories) was added to each well and agitated for 20 min at room temperature before the reaction was stopped by the addition of 100 μl of 2% oxalic acid. A SpectraMax 190 plate reader (Molecular Devices) was used to read absorbance at 415 nm. Positive supernatants were defined as those giving readings twofold above the background level, which was determined by adding hybridoma supernatant to a well coated with a nonspecific antigen. Positive lines were subcloned to generate clonal hybridomas. The hybridoma supernatants were concentrated using Amicon Ultra Centrifugal 30K filter devices for supershift analysis.
Western blotting.
Ten micrograms of nuclear extract or 25 ng of purified protein was boiled in 3× SDS sample buffer, separated by 9% SDS-PAGE, and transferred to a nitrocellulose membrane. To detect the 22-kDa protein, supernatant from an anti-22-kDa hybridoma, 7B4.3, was diluted 1:200 in PBSTM and used as the primary antibody, followed by anti-mouse IgG HRP-conjugated secondary antibody at a dilution of 1:10,000. ECL Plus (Amersham) was used as the developing agent. The IVa2 protein was detected as previously described (28).
Mass spectrometric analysis.
Seven micrograms of the 22-kDa protein were resolved by 10% SDS-PAGE, followed by detection with ProtoBlue Coomassie staining. The band of interest was excised and sent to the Michigan Proteome Consortium, where trypsin digestion and tandem mass spectrometry analysis were performed.
EMSA.
Double-stranded DNA probes of the wild-type and mutant A1-A2 repeats (nucleotides 238 to 279) and complete packaging sequence (nucleotides 200 to 397) were prepared and labeled as described previously (31, 32). DNA binding was assessed as previously described (31, 32) with the following modifications. All reactions were carried out in a final volume of 20 μl containing the following: 10 mM HEPES (pH 7.9), 20 mM KCl, 3 mM MgCl2, 10 mM EDTA, 12% glycerol, 50 μg/ml bovine serum albumin fraction V (Sigma), and 1 mM DTT. Purified IVa2 and 22-kDa proteins or 4 μg of nuclear extract was added to each reaction and incubated for 15 min at 23°C. Supershift analysis was performed by adding antibodies during the incubation. To form protein-DNA complexes, 100 pM radiolabeled probe mixed with 40-fold excess poly(dI/dC) (Amersham Biosciences) by mass was then added to the reaction and incubated for an additional 15 min before electrophoresis. For experiments comparing binding of purified proteins to nuclear extracts, 10 nM radiolabeled probe and 250 ng of poly(dI/dC) per reaction were used instead. Cold competition analysis was performed by mixing unlabeled oligonucleotides with the radiolabeled probe and poly(dI/dC) prior to incubation with proteins. Protein-DNA complexes were resolved by electrophoresis at 200 V at 4°C on a 6% (A1-A2 probe) or 4% (complete packaging sequence probe) native polyacrylamide (37.5:1, acrylamide/bisacrylamide) gel in 0.5× Tris-borate-EDTA buffer for 60 min or 90 min, respectively. For experiments comparing binding of purified proteins to nuclear extracts, 4.5% gels (40:1, acrylamide/bisacrylamide) were run for 90 min. Dried gels were analyzed by autoradiography.
RESULTS
Purification of 22-kDa protein.
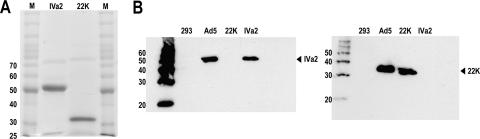
We used the IMPACT-CN system to obtain expression in E. coli of the recombinant 22-kDa protein as a fusion protein with a C-terminal chitin binding domain tag. Intein-mediated cleavage with DTT released the 22-kDa protein from a chitin column untagged with the exception of four residues: LKPG. This system yielded approximately 1.5 mg of the 22-kDa protein from 1 liter of LB broth. Following additional purification by gel filtration to remove contaminants, we estimated the purity of the 22-kDa and IVa2 proteins to be 87% and 95%, respectively, as determined by SDS-PAGE and staining (Fig. 1A). Compared to untagged protein standards, the 22-kDa protein migrated anomalously above the 30-kDa marker, similar to results described in a previous report (24). Excision of the major band from the gel followed by mass spectrometry analysis confirmed it contained the 22-kDa protein. We performed Western blotting analysis with supernatants from a newly derived anti-22-kDa hybridoma and purified polyclonal rabbit antibodies previously raised against an N-terminal peptide of the IVa2 protein to compare the migration of the purified 22-kDa and IVa2 proteins to that of proteins expressed during infection (Fig. 1B). The results of this analysis also verified the specificity of each antibody for use in subsequent supershift assays. The purified IVa2 and 22-kDa proteins each migrated as a single distinct band with mobility comparable to that of their respective bands in infected nuclear extracts. Given that the anti-22-kDa hybridoma was produced against the purified 22-kDa protein, displayed affinity to purified 22-kDa protein, and detected only a single protein in the infected nuclear extracts, we concluded that it does not recognize the viral 33-kDa protein, which shares its amino terminus with the 22-kDa protein. The 22-kDa protein in infected nuclear extracts had a slightly altered mobility relative to that of the purified protein, possibly due to posttranslation modification in the infected cell.
FIG. 1.
Characterization of 22-kDa and IVa2 proteins. Proteins were purified as described in Materials and Methods, separated by SDS-PAGE, and analyzed by ProtoBlue Coomassie staining (A) or Western blotting with anti-IVa2 (left panel) or anti-22 kDa (right panel) antibodies (B). 293, mock-infected extracts; Ad5, Ad5-infected extracts. The sizes of the molecular weight markers are indicated to the left of each gel.
Identification of a ternary complex between the IVa2 and 22-kDa proteins and the A1-A2 probe.
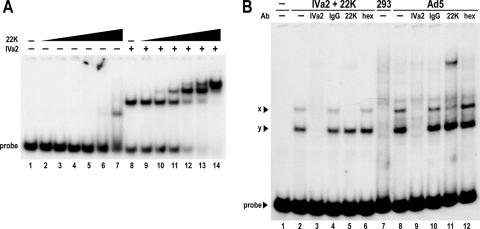
We wished to explore if the purified 22-kDa protein could interact with the A repeats directly or if it required the IVa2 protein to form complexes, as previously indicated in experiments with nuclear extracts (24). To test this hypothesis, we added the A1-A2 probe to EMSA binding reactions containing increasing concentrations of the 22-kDa protein in the presence or absence of the IVa2 protein (Fig. 2A). The 22-kDa protein alone yielded a weak, shifted complex detectable only at very high protein concentrations. This complex was also detected when probes A1-A2 m1 and m2, containing mutations in the TTTGN8CG consensus, were used (data not shown), and it does not correspond to any of the complexes formed when infected nuclear extracts are assayed. In contrast, when the same concentrations of the 22-kDa protein were added to reactions containing concentrations of the IVa2 protein near its half-maximal level of binding (10 nM), the remaining free probe shifted into complexes. Addition of the 22-kDa protein resulted in the formation of two new complexes that migrated slower than the complex with the IVa2 protein alone. A single new complex was first detected at 0.3 nM, 22 kDa, while increasing the protein concentration to 8.0 nM produced an additional new complex with even slower mobility. Higher concentrations of protein drove the majority of the A1-A2 probe into the slowest-migrating complex. The formation of three distinct complexes with purified proteins, one of which depended on the IVa2 protein alone and two of which required the IVa2 and 22-kDa proteins, paralleled the results previously obtained by Ostapchuk et al. using infected nuclear extracts (24).
FIG. 2.
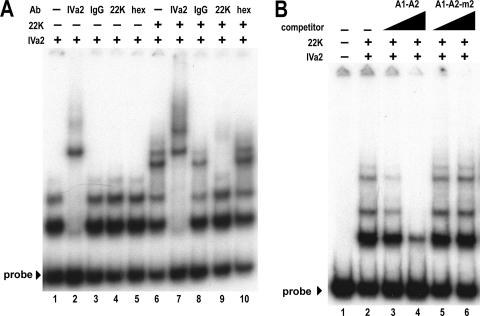
Interaction of the 22-kDa and IVa2 proteins with the A1-A2 probe. (A) Autoradiogram from EMSA comparing binding of the 22-kDa (22K) protein to the A1-A2 probe in the presence (+) or absence (−) of a 10 nM concentration of the IVa2 protein (IVa2). The dash above the first lane of each set indicates that the reaction contains no 22-kDa protein. Triangles above the subsequent lanes represent increasing concentrations of the 22-kDa protein (0.1, 0.3, 1.6, 8, 40, and 200 nM). (B) Supershift analysis. The labeled bold lines above the lanes indicate proteins or nuclear extracts added to reactions as follows: −, no protein; IVa2 + 22K, 50 nM IVa2 and 1.6 nM 22-kDa protein; 293, 4 μg mock-infected 293 nuclear extract; Ad5, 4 μg Ad5-infected 293 nuclear extract. The antibodies incubated in each reaction are indicated below the bold lines: −, no antibodies; IVa2, purified anti-IVa2 peptide rabbit IgG; IgG, purified preimmune rabbit IgG; 22K, anti-22-kDa hybridoma supernatant; hex, anti-hexon hybridoma supernatant. The x and y complexes are indicated with labeled arrowheads.
Given the apparent similarity between the complexes formed with purified proteins and those detected in nuclear extracts, we wished to compare the complexes directly. The complex formed by the IVa2 protein alone and the first complex formed by the IVa2 and 22-kDa proteins on the A1-A2 probe migrated exactly as did the y and x complexes in infected nuclear extracts, respectively (Fig. 2B). Both complexes were absent from control reactions containing probe alone or mock-infected nuclear extract. We also performed supershift analysis with the antibodies used in the Western blots shown in Fig. 1B to compare the protein compositions of the x and y complexes formed with the purified IVa2 and 22-kDa proteins to those formed by Ad5-infected nuclear extracts. Addition of the anti-IVa2 antibodies to the reactions disrupted both the x and y complexes, indicating that both complexes contain the IVa2 protein, as previously reported (32). The anti-22-kDa antibodies supershifted the x complex but had no effect on the y complex. Control antibodies did not affect formation of either complex. These results support the earlier conclusion that the 22-kDa protein becomes incorporated into slower-migrating complexes with the IVa2 protein. These data also demonstrated that the IVa2 and 22-kDa proteins formed complexes with the A repeats independently of other viral proteins and the cellular milieu.
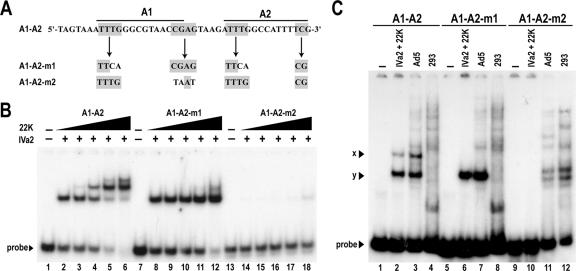
It has previously been demonstrated using EMSA analysis with infected nuclear extracts that formation of complexes on the A1-A2 probe depends on the TTTGN8CG consensus: mutation of the TTTG motif disrupted only the x complex, whereas mutation of the CG motif disrupted both the x and y complexes (32). We performed EMSA analysis using A1-A2 probes possessing mutations in these motifs to test the sequence specificity of the complexes formed by the purified 22-kDa and IVa2 proteins (Fig. 3). As before, three distinct complexes formed when the IVa2 protein and increasing concentrations of the 22-kDa protein were incubated with the wild-type A1-A2 probe (Fig. 3B). However, mutation of the TTTG motif prevented formation of the slower-migrating complexes, indicating that formation of complexes by the 22-kDa protein depends on this motif. As previously reported (32), mutation of the CG motif eliminated formation of all complexes. Even with an unaltered TTTG motif, the 22-kDa protein could not bind to the m2 probe in solution with the IVa2 protein or restore binding of the IVa2 protein to the probe, confirming that the formation of complexes by the 22-kDa protein depends on the IVa2-A repeat interaction. Additional EMSA analysis comparing nuclear extracts to the purified proteins, using the wild-type and mutant A1-A2 probes, reproduced these results (Fig. 3C). Loss of the x complex or both the x and y complexes upon mutation of the TTTG or CG motifs, respectively, correlated identically to the binding pattern observed with purified proteins. These data corroborated those from supershift analysis showing that the x complex contains the IVa2 and 22-kDa proteins while the y complex contains only the IVa2 protein.
FIG. 3.
Interaction of the IVa2 and 22-kDa proteins with mutant A1-A2 probes. (A) Sequences of the wild-type and mutant probes. The bold lines designate the A-repeat consensus (TTTGN8CG), and the gray boxes indicate the TTTG and CG motifs. (B) Autoradiogram of EMSA comparing binding of the 22-kDa protein (22K) in the presence (+) of 10 nM IVa2 protein (IVa2) to the A1-A2 wild-type and mutant probes. Dashes above lanes indicate reactions that did not contain the indicated protein. Triangles above the lanes represent increasing concentrations of the 22-kDa protein (0.3, 1.6, 8, and 40 nM). (C) Autoradiogram of EMSA comparing binding of the purified IVa2 and 22-kDa proteins or nuclear extracts to the wild-type and mutant probes.
Interaction of the IVa2 and 22-kDa proteins with the complete packaging sequence.
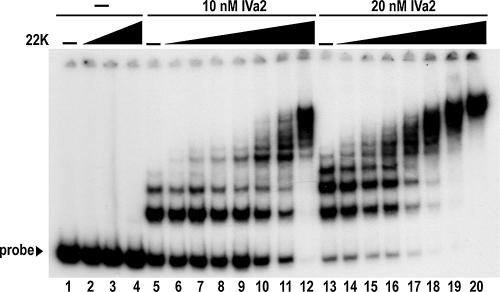
In a previous investigation of the interaction between the purified IVa2 protein and the A repeats, we extended EMSA analysis with the A1-A2 probe to experimentation with a probe containing all seven A repeats that spans the complete packaging sequence (31). In that study, the IVa2 protein assembled into multiple complexes on a packaging sequence probe with comparable half-maximal binding to the A1-A2 probe, 11.4 nM. Consequently, it was conceivable that the purified IVa2 and 22-kDa proteins together could form additional complexes on the complete packaging sequence probe. We performed EMSA analysis to test this hypothesis by replacing the A1-A2 probe with an equivalent concentration of the packaging sequence probe (Fig. 4). Addition of the 22-kDa protein to binding reactions alone did not shift the packaging sequence probe. However, in reactions with a 10 nM concentration of the IVa2 protein, addition of picomolar quantities of the 22-kDa protein yielded new complexes that migrated more slowly than those formed by IVa2 alone. As the concentration of the 22-kDa protein was increased from 0.1 nM to 8.0 nM, additional complexes formed that migrated at consecutively slower rates and ultimately incorporated the entire quantity of probe. This binding pattern resembled the interaction of the IVa2 and 22-kDa proteins with the A1-A2 probe but was detected in the presence of approximately 25-fold-lower concentrations of the 22-kDa protein in the reactions (compare Fig. 2A and 4). Additionally, the free probe and probe bound only to the IVa2 protein became completely incorporated into the slower-migrating complexes when concentrations of the 22-kDa protein increased fourfold, from 2.0 to 8.0 nM. The threshold quantity of the 22-kDa protein required to achieve this result decreased approximately twofold when the concentration of IVa2 was increased from 10 to 20 nM, indicating the binding activity depends on the concentration of both proteins.
FIG. 4.
Formation of multiple complexes by the IVa2 and 22-kDa proteins on the complete packaging sequence. Autoradiogram from an EMSA with the 22-kDa protein (22K), the IVa2 protein (IVa2), and a probe encompassing the entire packaging sequence from nucleotides 200 to 397. Dashes above lanes indicate reactions that do not contain the 22-kDa or IVa2 protein. The labeled bold lines above the lanes indicate the quantities of the IVa2 protein added to reactions. The leftmost triangle indicates increasing concentrations of the 22-kDa protein (2, 4, and 8 nM) in the absence of IVa2. The other two triangles indicate concentrations of the 22-kDa protein (0.1, 0.3, 0.5, 1, 2, 4, and 8 nM) in the presence of IVa2. Free probe is indicated with a labeled arrowhead.
To demonstrate that the slower-migrating complexes contained the 22-kDa protein, we performed supershift analysis (Fig. 5A). As before, the IVa2 protein formed multiple complexes on the packaging sequence probe alone, and addition of the 22-kDa protein in reactions with the IVa2 protein produced multiple slower-migrating complexes. Addition of anti-IVa2 antibodies to binding reactions retarded the mobility of every complex formed on the packaging sequence, indicating they all contain the IVa2 protein. The anti-22 kDa antibodies supershifted only complexes formed by the 22-kDa and IVa2 proteins, not those formed by the IVa2 protein alone, confirming that the 22-kDa protein becomes incorporated into these complexes with the IVa2 protein. Control antibodies had no effect on the formation or mobility of any complexes. Overall, these data correlated with the supershift analysis using the A1-A2 probe and similarly indicated that the formation of complexes on the A repeats by the 22-kDa protein depends on the IVa2 protein.
FIG. 5.
Supershift and cold competition analysis of IVa2 and 22-kDa protein binding to the complete packaging sequence. The plus and minus signs above lanes indicate the presence (+) or absence (−) of 10 nM IVa2 protein (IVa2) or 1.6 nM 22-kDa protein (22K). Free probe is indicated with a labeled arrowhead. (A) Supershift analysis. The antibodies described in the legend to Fig. 2 were added to the indicated reactions. (B) Competition analysis. The dashes represent reactions containing no competitor. The labeled triangles above lanes indicate 100-fold and 1,000-fold molar excess of unlabeled competitor corresponding to A1-A2 wild-type or m2 sequences shown in Fig. 3.
Considering that the binding of the IVa2 and 22-kDa proteins to the A1-A2 probe required the TTTGN8CG motif, we wished to assess if the complexes formed on packaging sequence probe were also sequence specific. To test this, we performed cold competition assays by adding unlabeled A1-A2 and A1-A2 m2 oligomers to binding reactions as sequence-specific and -nonspecific competitors (Fig. 5B). Unlabeled wild-type A1-A2 oligomer competed for interaction with the IVa2 and 22-kDa proteins when added to binding reactions at 100 and 1,000-fold molar excess over the labeled packaging sequence probe. However, the same quantities of mutant m2 oligomer, with which the IVa2 and 22-kDa proteins did not form complexes, had no effect on the interaction of the IVa2 and 22-kDa proteins with the packaging sequence probe. These results indicate that the IVa2 and 22-kDa proteins formed the additional slower-migrating complexes at specific sites on the packaging sequence probe.
DISCUSSION
We report that the adenovirus IVa2 and 22-kDa proteins recognize the viral packaging sequence through interdependent interactions. Purification of the 22-kDa protein permitted analysis using an EMSA system previously established with purified IVa2 (31). The 22-kDa protein alone could not interact in a sequence-specific manner with a probe encompassing the complete packaging sequence nor reproduce the virus-specific x and y complexes on the A1-A2 probe previously detected using infected nuclear extracts; however, the 22-kDa protein could interact with the DNA in the presence of IVa2. We observed multiple sequence-specific complexes on the packaging sequence or A1-A2 probe in the presence of picomolar concentrations of the 22-kDa protein and nanomolar concentrations of the IVa2 protein. Furthermore, although the 22-kDa protein required the IVa2 protein to form complexes, the presence of increasing concentrations of the 22-kDa protein with only a half-maximal binding concentration of IVa2 (10 nM) drove the entire quantity of packaging sequence or A1-A2 probe into complex, indicating that the 22-kDa protein also enhances binding of the IVa2 protein. The results also demonstrate that formation of these complexes correlates with previous genetic analysis of mutant viruses, reinforcing a collaborative role of the IVa2 and 22-kDa proteins in packaging.
In EMSA analysis with the A1-A2 probe, increasing concentrations of the 22-kDa protein, from picomolar to nanomolar, yielded a stepwise conversion of the y complex, which consists of the IVa2 protein binding to the A1-A2 probe alone, into the x complex and an additional slower-migrating complex. These complexes probably correspond to the three complexes described by Ostapchuk et al., 1 (same as y), 2 (same as x), and 3 (the largest of the three complexes) (24). The x and y complexes formed by the purified IVa2 and 22-kDa proteins migrated identically to the x and y complexes formed by infected nuclear extracts. The data from supershift analysis with anti-IVa2 and -22-kDa protein antibodies confirmed that the x complex contained the IVa2 and 22-kDa proteins, but the y complex contained only the IVa2 protein. Additionally, formation of the x and y complexes by IVa2 and the 22-kDa protein required the TTTGN8CG bipartite consensus crucial for packaging; although formation of the x complex required the TTTG motif, no interaction occurred unless the IVa2 protein formed the y complex through the CG motif. Therefore, we conclude the IVa2 and 22-kDa proteins must interact within the ternary x complex on the A repeats. Taken together, these results suggest that the purified IVa2 and 22-kDa proteins retained the same A-repeat binding properties as proteins expressed during Ad5 infection and that the x and y complexes seen in EMSA analyses with infected nuclear extracts resulted exclusively from interaction of the IVa2 and 22-kDa proteins with the A1-A2 probe.
During SDS-PAGE, both the purified and infected-cell-derived 22-kDa proteins migrated anomalously, slower than a 30-kDa marker. This result is analogous to that reported for the viral 33-kDa protein, which shares its N terminus with the 22-kDa protein (23). Previous studies using SDS-PAGE analysis with the 33-kDa protein attributed a similar anomalous migration of the 33-kDa protein at 39 kDa to the high glutamic acid and proline content of the common N terminus. The purified 22-kDa protein also migrated slightly faster than the 22-kDa protein expressed during infection. Studies of the 33-kDa protein identified a phosphorylated trypsin digest fragment corresponding to a portion of the amino terminus. A similar modification of the 22-kDa protein expressed during infection might explain its reduced migration relative to that of the 22-kDa protein purified from prokaryotic cells. Considering the purified 22-kDa protein exhibited the same activity in EMSAs as protein from infected nuclear extracts, the importance of posttranslational modifications of the 22-kDa protein in packaging and assembly remains unclear.
In contrast to a previous report (24) and the present data indicating that the purified 22-kDa protein is sufficient for forming the x complex with IVa2, a recent study suggested the 33-kDa protein also can form the x complex with the IVa2 protein on the A1-A2 probe (1). This study used nuclear extracts from cells stably expressing the IVa2 protein and transfected with an expression construct that potentially encodes both the 22-kDa and 33-kDa proteins. The results of reverse transcription-PCR performed with primers flanking this portion of the L4 transcript indicated that the transfected cells exported only the spliced transcripts encoding the 33-kDa open reading frame to the cytoplasm and, therefore, only the 33-kDa protein could be expressed. However, our data suggest that low concentrations of the 22-kDa protein, possibly expressed from undetectable levels of the unspliced transcript, could dramatically affect IVa2 binding. Furthermore, our data indicate that overexpression of the IVa2 protein during transfection would force the 22-kDa protein to form complexes on the A repeats at exceedingly low concentrations. The finding that the x complex can be detected in nuclear extracts from cells transfected with a vector that expresses only the 22-kDa protein strongly supports its role in packaging (24). Nonetheless, given that the two proteins share their amino-terminal 105 amino acids, it is possible that the 22-kDa and 33-kDa proteins have redundant functions.
EMSA analysis with the complete packaging sequence probe and the purified IVa2 and 22-kDa proteins provided additional evidence that the 22-kDa protein plays a role in packaging with IVa2. Such an analysis with nuclear extracts may otherwise become difficult to interpret due to interactions between cellular transcription factors and the overlapping E1A enhancer elements in the packaging sequence (18, 19). The IVa2 protein and increasing quantities of the 22-kDa protein formed multiple complexes in a stepwise manner on the packing sequence probe. The ability of unlabeled DNA containing the A1-A2 repeats, but not those possessing a mutation in the CG motif, to disrupt these complexes indicated the IVa2 and 22-kDa proteins interact with the available A-repeat binding sites on the full-length probe. We are unable to ascertain at this time whether the 22-kDa protein contacts the DNA directly or alters the conformation of the IVa2 protein such that it now interacts with the TTTG motifs. In addition, binding may certainly be influenced by other sequences in the packaging sequence. These data correlated with the requirement of multiple A repeats for efficient packaging observed in previous genetic studies with adenovirus (30). We posit that one function of the 22-kDa protein must be to enhance specific binding of the IVa2 protein to multiple A repeats and, thus, promote efficient packaging of the genome.
It was suggested in previous reports that efficient packaging requires threshold concentrations of the IVa2 and/or 22-kDa proteins. Complementation of an IVa2 null mutant virus in trans with an approximately 20-fold decrease in concentrations of the IVa2 protein relative to levels during infection with wild-type Ad5 resulted in particles with lower density and irregular shape (33). In another study, cotransfection of packaging sequence DNA with Ad5 genomes reduced production of infectious virus, probably by titrating the IVa2 and 22-kDa proteins to concentrations insufficient for packaging and assembly (10). The results of EMSA analysis with the IVa2 and 22-kDa proteins and the packaging sequence probe support the importance of threshold concentrations of the IVa2 and 22-kDa proteins in adenovirus packaging. A slight increase in the concentration of the 22-kDa protein produced several new complexes that ultimately shifted into a single slower-migrating complex. This threshold also varied inversely with the concentration of constant IVa2 in binding reactions. Thus, the expression of sufficient levels of the IVa2 and 22-kDa proteins together may be a timing mechanism that initiates adenovirus packaging late in infection. In addition, the relative amounts of the two proteins may be critical to achieve efficient encapsidation.
Although the exact structures of the complexes formed on the packaging sequence remain unknown, adenovirus may require a multiprotein complex for packaging, similar to the molecular motors of double-stranded DNA bacteriophages, which insert their genome into preformed capsids (3, 4). For example, the molecular motor of bacteriophage T4 depends on complexes containing crucial accessory proteins and an active ATPase (29), and PRD1, which has many structural similarities with adenovirus (2), has a unique portal that possesses ATPase activity (20). Although no ATPase activity has yet been ascribed to the IVa2 protein, mutations in a putative Walker A motif of this protein, which is characteristic of AAA+ ATPases (5), resulted in nonviable virus (27). It is conceivable then that expression of the 22-kDa protein past a threshold concentration might trigger the formation of an IVa2-containing portal through which the genome is inserted in an ATP hydrolysis-dependent manner. Additionally, the failure of an IVa2 null virus to produce empty capsids indicated that the IVa2 protein may have additional functions during assembly besides packaging (33); the present results indicate that the 22-kDa protein may also be involved prior to encapsidation. Further investigation of the interactions between the IVa2 and 22-kDa proteins and the viral packaging sequence will be required to elucidate the role that these proteins play in assembly and genome packaging.
Acknowledgments
We thank Silas Johnson for technical assistance, the University of Michigan Hybridoma Core for producing the hybridoma cell lines, the Michigan Proteomic Consortium for mass spectrometry analysis, David Friedman for use of his French press, and the members of the Imperiale laboratory for their insight and assistance.
This work was supported by grant AI052150 from the National Institutes of Health to M.J.I. and (in part) through the University of Michigan's Cancer Center Support Grant (P30 CA46592).
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Ali, H., G. LeRoy, G. Bridge, and S. J. Flint. 2007. The adenovirus L4 33-kilodalton protein binds to intragenic sequences of the major late promoter required for late phase-specific stimulation of transcription. J. Virol. 81:1327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 3.Black, L. W. 1988. DNA packaging in dsDNA bacteriophages, p. 321-374. In R. Calendar (ed.), The bacteriophages. Plenum Press, New York, NY.
- 4.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-92. In R. Calendar (ed.), The bacteriophages. Plenum Press, New York, NY.
- 5.Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93-114. [DOI] [PubMed] [Google Scholar]
- 6.Fessler, S. P., and C. S. Young. 1999. The role of the L4 33K gene in adenovirus infection. Virology 263:507-516. [DOI] [PubMed] [Google Scholar]
- 7.Finnen, R. L., J. F. Biddle, and J. Flint. 2001. Truncation of the human adenovirus type 5 L4 33-kDa protein: evidence for an essential role of the carboxy-terminus in the viral infectious cycle. Virology 289:388-399. [DOI] [PubMed] [Google Scholar]
- 8.Gefter, M. L., D. H. Margulies, and M. D. Scharff. 1977. A simple method for polyethylene glycol-promoted hybridization of mouse myeloma cells. Somat. Cell Genet. 3:231-236. [DOI] [PubMed] [Google Scholar]
- 9.Grable, M., and P. Hearing. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grable, M., and P. Hearing. 1992. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J. Virol. 66:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, F. L., and L. Prevec. 1991. Manipulation of adenovirus vectors. Methods Mol. Biol. 7:109-128. [DOI] [PubMed] [Google Scholar]
- 12.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 13.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustin, K. E., P. Lutz, and M. J. Imperiale. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammarskjold, M. L., and G. Winberg. 1980. Encapsidation of adenovirus 16 DNA is directed by a small DNA sequence at the left end of the genome. Cell 20:787-795. [DOI] [PubMed] [Google Scholar]
- 16.Hasson, T. B., P. D. Soloway, D. A. Ornelles, W. Doerfler, and T. Shenk. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hearing, P., and T. Shenk. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695-703. [DOI] [PubMed] [Google Scholar]
- 19.Imperiale, M. J., L. T. Feldman, and J. R. Nevins. 1983. Activation of gene expression by adenovirus and herpesvirus regulatory genes acting in trans and by a cis-acting adenovirus enhancer element. Cell 35:127-136. [DOI] [PubMed] [Google Scholar]
- 20.Karhu, N. J., G. Ziedaite, D. H. Bamford, and J. K. Bamford. 2007. Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6. J. Virol. 81:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearney, J. F., A. Radbruch, B. Liesegang, and K. Rajewsky. 1979. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J. Immunol. 123:1548-1550. [PubMed] [Google Scholar]
- 22.McConnell, M. J., X. Danthinne, and M. J. Imperiale. 2006. Characterization of a permissive epitope insertion site in adenovirus hexon. J. Virol. 80:5361-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oosterom-Dragon, E. A., and C. W. Anderson. 1983. Polypeptide structure and encoding location of the adenovirus serotype 2 late, nonstructural 33K protein. J. Virol. 45:251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostapchuk, P., M. E. Anderson, S. Chandrasekhar, and P. Hearing. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostapchuk, P., and P. Hearing. 2005. Control of adenovirus packaging. J. Cell Biochem. 96:25-35. [DOI] [PubMed] [Google Scholar]
- 26.Ostapchuk, P., J. Yang, E. Auffarth, and P. Hearing. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardo-Mateos, A., and C. S. Young. 2004. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 324:151-164. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Romero, P., R. E. Tyler, J. R. Abend, M. Dus, and M. J. Imperiale. 2005. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 79:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao, V. B., and M. S. Mitchell. 2001. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J. Mol. Biol. 314:401-411. [DOI] [PubMed] [Google Scholar]
- 30.Schmid, S. I., and P. Hearing. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyler, R. E., S. G. Ewing, and M. J. Imperiale. 2007. Formation of a multiple protein complex on the adenovirus packaging sequence by the IVa2 protein. J. Virol. 81:3447-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, W., and M. J. Imperiale. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]