Abstract
Directed release of human immunodeficiency virus type 1 (HIV-1) into the cleft of the virological synapse that can form between infected and uninfected T cells, for example, in lymph nodes, is thought to contribute to the systemic spread of this virus. In contrast, influenza virus, which causes local infections, is shed into the airways of the respiratory tract from free surfaces of epithelial cells. We now demonstrate that such differential release of HIV-1 and influenza virus is paralleled, at the subcellular level, by viral assembly at different microsegments of the plasma membrane of HeLa cells. HIV-1, but not influenza virus, buds through microdomains containing the tetraspanins CD9 and CD63. Consequently, the anti-CD9 antibody K41, which redistributes its antigen and also other tetraspanins to cell-cell adhesion sites, interferes with HIV-1 but not with influenza virus release. Altogether, these data strongly suggest that the bimodal egress of these two pathogenic viruses, like their entry into target cells, is guided by specific sets of host cell proteins.
The surfaces of cells, even of those with no manifest polarization, can be transiently partitioned into discrete domains for the execution of specific functions. This principle was elegantly demonstrated some 20 years ago in a study that investigated the directionality of virus budding (46). It was shown that influenza virus, an enveloped virus that buds from the apical side of polarized epithelial cells, is released from free surfaces of such cells before they form polarized monolayers. In contrast, vesicular stomatitis virus, which buds at the basolateral side of a cell monolayer, was found to be released at cell-cell contact sites within aggregates composed of only a few, nonpolarized cells. Release from either surface likely secures efficient spread of these viruses. It was presumed that cytoskeletal changes, together with a redistribution of membrane proteins involved in organizing activities at either free cell surfaces or at the adhesion sites, were responsible for the functional separation of the two plasma membrane areas, but the molecular details were not investigated at the time.
Dissemination of human immunodeficiency virus type 1 (HIV-1) in infected individuals is thought to be driven both by virions released into the cleft of the so-called virological synapse (22) and by infection of cells with cell-free virus (for a review, see reference 43). However, in vitro replication studies clearly show that this virus, like vesicular stomatitis virus, is released preferentially at the basolateral side of epithelial cells and evolved to be transmitted most efficiently at cell-cell interfaces (9, 48), for instance, when replicating in lymphocytes (for reviews, see references 21 and 40). One group of membrane proteins that has been implicated in organizing cell surfaces, particularly at sites of cell-cell contact, includes the so-called tetraspanins (recently reviewed in references 18, 25, and 50). It is thought that the propensity of these small proteins to self-aggregate and to associate with other transmembrane proteins allows them to build temporary scaffolds for activities such as antigen presentation. Recently, we demonstrated that tetraspanins, including CD9 and CD63, coaccumulate at the plasma membrane, thus forming tetraspanin-enriched microdomains (TEMs) (35). Our report also described that newly produced components of HIV-1 colocalize with surface TEMs in epithelial cells and in T lymphocytes. Together with other recent reports which showed budding of HIV-1 through tetraspanin-rich membrane segments in T cells, macrophages, and dendritic cells (2, 7, 23, 53), our data suggested that tetraspanins are constituents of a universal HIV-1 exit gateway.
Here we provide further evidence in support of this hypothesis. Incubation of HIV-1-producing cells with the anti-CD9 antibody K41 leads to extensive aggregation of tetraspanins at cell-cell contact sites. This process, which is accompanied by clustering of viral components at the same sites, results in inhibition of HIV-1 release. Additionally, the infectivity of virus released from K41-treated cells is diminished. In contrast, egress of influenza virus is not inhibited by K41. Evidently, influenza virus, whose spread in infected individuals does not depend on transmission at the cell-cell interface, exits through microdomains that are clearly distinct from surface TEMs through which HIV-1 buds.
MATERIALS AND METHODS
Cell culture, plasmids, transfections, and antibodies.
HeLa, 293T, and CrFK cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (Invitrogen). E6-1 Jurkat T cells (NIH AIDS Research and Reference Reagent Program, Rockville, MD) were grown in RPMI supplemented with 10% fetal bovine serum. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Jurkat cells were electroporated using a Bio-Rad Gene Pulser. The following plasmids were used: pNL4-3, pGagopt (codon-optimized Gag), pSRalpha-Env, pGagopt-CFP, and TSG101-YFP (kind gift of W. Mothes, Yale University School of Medicine, New Haven, CT). In experiments analyzing influenza virus release, cells were infected with influenza virus stocks [strain A/Udorn/72 (Ud) (H3N2); gift from R. Lamb, Northwestern University].
The following antibodies were used: anti-CD9 (K41 [BMA Biomedicals AG, Augst, Switzerland] and H110 [Santa Cruz Biotechnology]), anti-CD63 (H5C6 [Hybridoma Bank, Iowa City, IA]), anti-CD81 (JS-81 [BD Pharmingen, San Diego, CA]), anti-CD82 (B-L2 [Diaclone, Besancon, France]), anti-Env (B12 [NIH AIDS Research and Reference Reagent Program, Rockville, MD]), anti-p6 (AIDS Vaccine Program, National Cancer Institute—Frederick, Frederick, MD), anti-Ud goat antibody (recognizing HA, NP, and M1 of influenza virus; kindly provided by R. Lamb, Northwestern University), anti-FIV PAK1-2B2 and anti-HIV CA monoclonal antibody (MAb) 183-H12-5C (NIH AIDS Research and Reference Reagent Program, Rockville, MD) and CA of HIV-1 (ABI, Columbia, MD). The secondary antibodies used were anti-mouse, anti-rabbit, anti-human, and anti-goat conjugated to Alexa Fluor 488, 594, and 647 (Molecular Probes, Eugene, OR).
Immunofluorescence microscopy.
HeLa cells were plated in 35-mm glass-bottom culture dishes (MatTek, Ashland, MA). Transfection and staining were performed directly in these dishes. Cells treated or not with K41 or immunoglobulin G (IgG) antibodies for 4 h, precooled on ice, washed with cold phosphate-buffered saline (PBS), and incubated for 1 h at 4°C with anti-CD9 in complete medium supplemented with 1% Ca2+. The cells were washed twice with cold PBS and fixed with 3.7% paraformaldehyde prior to secondary staining with Alexa Fluor 594-conjugated anti-mouse antibody. In the cases of CD63, CD81, or CD82 dual labeling with CD9, the former were coupled to Alexa Fluor 488 fluorophore using the Zenon technology (Molecular Probes) and incubated with the cells on ice after the CD9 staining was completed. After washes with PBS, cells were fixed with 3.7% paraformaldehyde.
HeLa cells expressing pNL4-3, pGag-opt, or pSRalphaEnv or influenza virus were treated or not with 1.5 μg/ml K41 or IgG antibodies overnight, precooled on ice, washed with cold PBS, and incubated for 1 h at 4°C with anti-CD9 and anti-Env/anti-Ud (where applicable) in complete medium supplemented with 1% Ca2+. After washes with PBS, cells were fixed with 3.7% paraformaldehyde and, following a blocking step with bovine serum albumin (BSA), were incubated with appropriate secondary antibodies. To stain for Gag, cells were then permeabilized using Triton X-100, blocked with BSA, and incubated with an anti-p6 antibody for 1 h followed by appropriate secondary antibodies. For TSG101 recruitment, cells cotransfected with yellow fluorescent protein (YFP)-TSG101 and Gagopt cyan fluorescent protein (CFP) were treated with K41 for 4 h followed by staining for surface CD9 as above. K41-pretreated cells were stained only with secondary antibody for visualization of its antigen.
The slides were examined on a Delta Vision workstation (DV base 3/3.5 Nikon Eclipse TE200 epifluorescence microscope fitted with an automated stage; Applied Precision Inc., Issaquah, WA), and images were captured in z-series with a charge-coupled-device digital camera (CH350E or CoolSnap HQ). Out-of-focus light was digitally reassigned using Softworx deconvolution software (Applied Precision Inc.). Images were processed using Adobe Photoshop.
Incorporation of tetraspanins into virions.
HeLa cells were transfected with equal amounts of pNLC4-3 and pNLC4-3 bearing a green fluorescent protein (GFP) fused to Gag MA (33) (kind gift of B. Muller and H.-G. Krausslich), thus producing GFP-labeled virions, or infected with influenza virus. After 24 h, supernatant was collected and either incubated directly on poly-l-lysine-coated coverslips or virions were pelleted through a 20% sucrose cushion, resuspended in PBS, and then adhered to poly-l-lysine-coated coverslips. Virions were then fixed with 4% paraformaldehyde, stained with appropriate antibodies, and visualized by fluorescence microscopy. To calculate the percentages of virions bearing CD63 and CD9, the images were analyzed with Volocity software (Improvision) as follows. The total number of objects for each channel was found using the classifier module set to 2 standard deviations above the mean channel intensity. The total number of objects colocalizing between the two channels was determined using the colocalization module. This number was divided by the total number of objects for the virion channel, resulting in the percent virions bearing the marker analyzed. Five images were analyzed for each condition, resulting in a total of approximately 1,000 virions analyzed per condition.
Release inhibition studies.
HeLa cells cultured in 35-mm dishes, 18 h posttransfection with pNL4-3 DNA, were labeled with 50 μCi/ml of Easy Tag Express [35S]methionine-cysteine protein labeling mix (NEN) in the presence of 1.5 μg/ml of K41, anti-CD63, anti-CD81, anti-CD82, or IgG isotype control antibody added in the culture medium. Harvesting was done at 16 h posttreatment. Supernatants were centrifuged at 4,000 rpm for 10 min followed by centrifugation at 16,000 × g for 90 min through a 20% sucrose cushion. The viral pellet was resuspended in radioimmunoprecipitation buffer. Cells were lysed in radioimmunoprecipitation buffer for 10 min, and lysates were centrifuged at 14,000 rpm for 10 min. Lysates were immunoprecipitated with an anti-p24 (capsid) antibody. Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed by PhosphorImager (Bio-Rad) using Quantity One software. For the dose-response experiments, cells were treated with increasing amounts of K41 and harvesting was done 8 h posttreatment. IgG was used at the highest equivalent concentration of K41, i.e., 6 μg/ml. For the time curve, 1.5 μg/ml K41 was used, and harvesting was done 4, 8, 12, and 24 h later. For CrFK cells, FIV-PPR or pNL4-3 DNA was used to transfect the cells. At 18 h posttransfection, cells were treated or not with 1.5 μg/ml of K41 for 16 h and harvested as above, and proteins were visualized by Western blot analysis.
Effect of cell density on release inhibition.
HeLa cells cultured in 100-mm dishes were transfected with pNL4-3 and then split into 35-mm dishes after 24 h, at low and high cell density (50,000 and 400,000 cells per dish, respectively). The cells were then treated or not with K41 (1.5 μg/ml) and harvested 8 h later, as described above. The samples were analyzed by Western blotting, using anti-CA MAb 183.
Release from Jurkat cells.
E6.1 Jurkat cells electroporated with pNL4-3 DNA were used on day 8, when the cells were 70 to 80% Gag positive. Two million cells were washed and resuspended in 1 ml of fresh medium with or without 6 μg/ml K41 and harvested 16 h later as described above. The samples were analyzed by Western blotting, using anti-CA MAb 183.
Release of influenza virus.
Influenza virus release assay was performed essentially as described elsewhere (5). Briefly, HeLa or 293T cells were infected at a multiplicity of infection of 2, treated or not with 1.5 μg/ml K41, and incubated for 16 h. For Western blot analysis, the supernatant was collected, pelleted through a sucrose cushion, resuspended, and further fractionated on a sucrose gradient, then pelleted again, resuspended in lysis/loading buffer, and run on an SDS gel. To determine the cellular fraction, the cells were also lysed and run on an SDS gel. The proteins were transferred to a nitrocellulose membrane, incubated with polyclonal anti-Ud goat serum (kind gift of R. Lamb), then anti-goat-horseradish peroxidase, and then detected on film by chemiluminescence. The film was scanned, and the relative amounts of proteins were determined using ImageJ software (NIH).
Electron microscopy analysis.
HeLa cells cotransfected with Gag and Env plasmids were incubated with 3 μg of K41 or IgG isotype control antibodies in the culture medium for 12 h at 37°C. Cells preincubated with K41 antibodies were then washed in cold PBS-1% BSA and directly labeled with secondary 10-nm gold-conjugated antibodies (British Biocell Intl., Cardiff, United Kingdom) for 90 min at 4°C. Cells incubated with IgG isotype control antibodies were first incubated 2 h at 4°C with K41 antibodies in cold PBS-1% BSA, washed, and then labeled with secondary 10-nm gold-conjugated antibodies for 90 min at 4°C. Immunogold-labeled cells were then fixed, dehydrated, and processed for electron microscopy as described previously (11). Localization of CD9-associated gold particles and Gag-containing virus-like particles (VLPs) was then quantitatively analyzed on cells considered well preserved by using a Technai 20 electron microscope (FEI Company, Eindhoven, The Netherlands).
RESULTS
Assembly and release of HIV-1 and influenza virus take place at separate microdomains.
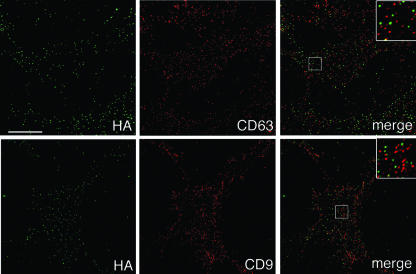
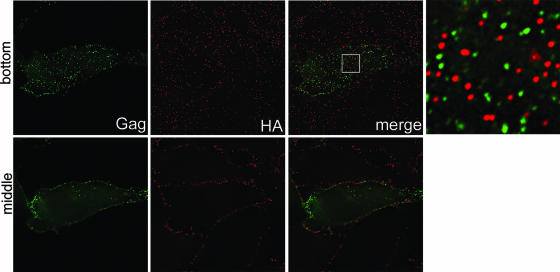
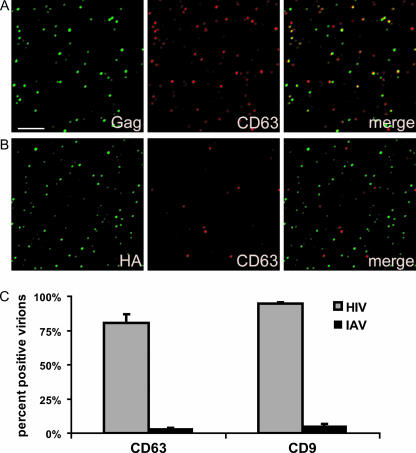
Previously, we demonstrated that the HIV-1 components Gag and envelope glycoprotein (Env) colocalize with CD9 and CD63 at the plasma membrane (35), while influenza virus hemagglutinin (HA), if expressed independently of the other viral components, did not show significant colocalization with these tetraspanins. Data shown in Fig. 1 now establish that HA does not accumulate at CD9- or CD63-containing surface TEMs, even if it is expressed in the context of the entire influenza virus, suggesting that this virus does not exit at these microdomains. To corroborate that HIV-1 and influenza virus assemble at different microdomains, even if generated simultaneously within the same cell, HeLa cells were infected with influenza virus and transfected with a HIV-1 Gag-GFP expression vector, thus producing both influenza virus particles and HIV-1 VLPs, and the putative viral egress sites were analyzed by fluorescence microscopy. Figure 2 documents that there was virtually no overlap between HIV-1 Gag and influenza virus HA, strongly suggesting that the two viruses exit through different plasma membrane microsegments. Confirmation for this assumption comes from the quantitative analysis of host cell protein acquisition by either of the two viruses (Fig. 3): while most HIV-1 particles incorporate the tetraspanins CD9 and CD63, these proteins are clearly absent from most influenza virus envelopes.
FIG. 1.
Influenza virus HA exhibits little colocalization with TEMs containing CD9 or CD63. HeLa cells infected with influenza virus were surface stained for HA and CD63 (top panels) or HA and CD9 (bottom panels). The bottom sections of cells are shown. Bar, 10 μm. The blow-ups show 3×-magnified views of the boxed regions in the merge panels. Quantitative analysis showed ∼ 4% and ∼ 8% colocalization of HA with CD63 and CD9, respectively.
FIG. 2.
HIV-1 and influenza virus exit through separate plasma membrane microdomains. HeLa cells infected with influenza virus and expressing HIV-1 Gag-GFP were surface stained for influenza virus HA. The blow-up shows a 7.3×-magnified view of the boxed region in the merge panel. The middle and bottom sections (upper and lower panels, respectively) are shown. Bar, 10 μm.
FIG. 3.
Tetraspanins are incorporated into HIV but not influenza virus. (A and B) Virions isolated from pNL4-3-transfected (A) or influenza virus-infected (B) HeLa cells were adhered to coverslips and stained with the indicated antibodies as described in Materials and Methods. Bar, 5 μm. (C) Quantification of incorporation of CD63 and CD9 into HIV-1 and influenza virus (IAV) particles. The percentages of Gag-positive or HA-positive particles bearing detectable levels of CD63 and CD9 were calculated from multiple images such as the ones shown in panels A and B, using Volocity software (see Materials and Methods). Error bars represent standard deviations.
Inhibition of HIV-1 egress but not influenza virus release by an antitetraspanin antibody.
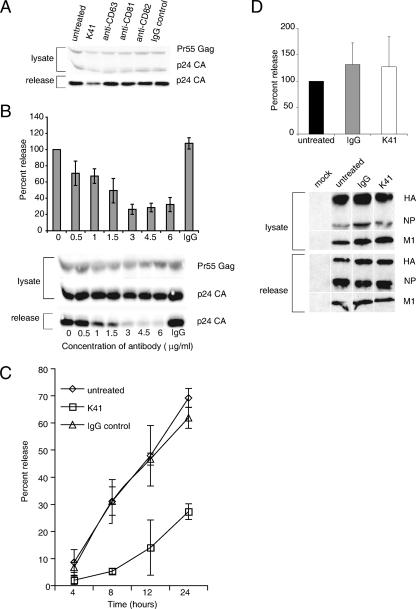
To test if TEMs do indeed function as gates through which HIV-1 exits from infected cells, we sought to analyze if interference with tetraspanin function affects virus egress. Cells expressing HIV-1 were treated with antibodies against the tetraspanins CD9, CD63, CD81, and CD82. As documented in Fig. 4A, incubation of HeLa cells with the anti-CD9 antibody K41 led to a significant reduction of particle release while not affecting Gag protein production. Neither the isotype control antibody nor any of the antibodies against the other tetraspanins, even when used at higher concentrations (data not shown), inhibited HIV-1 egress. Release inhibition for K41 was dose and time dependent, reaching its maximum effect around 8 h after the initiation of treatment (Fig. 4B and C). Further, utilizing a single-round infectivity assay (24), we found that HIV-1 virions shed from K41-treated HeLa cells were only half as infectious as virions released from untreated cells (data not shown). Overall, infectious virus output from cells incubated with K41 was thus reduced by approximately 1 log.
FIG. 4.
The anti-CD9 antibody K41 inhibits the release of HIV-1 but not of influenza virus. HeLa cells expressing HIV-1 were radiolabeled and treated or not with 1.5 μg/ml of anti-CD63, anti-CD81, anti-CD82, anti-CD9 K41, or isotype control antibody (A), with different concentrations of K41 or isotype control antibody used at 6 mg/ml (B), or were treated for different time periods using 1.5 μg/ml of K41 or isotype control antibody (C). Viral harvest and quantification were done as described in Materials and Methods. Error bars represent standard deviations (n = 3). A representative gel is shown in panels A and B. (D) HeLa cells infected with influenza virus were treated or not with 1.5 μg/ml K41 and harvested for analysis of viral proteins. Note that the different bands shown were regrouped from different parts of the gel. Error bars represent standard deviations (n = 4). A representative gel is shown.
Interestingly, anti-CD9 antibodies have previously been shown to negatively affect the propagation of other viruses. Vpg15, an anti-feline CD9 antibody, severely inhibited release of feline immunodeficiency virus (FIV) from chronically infected feline cells (8), and K41, which cross-reacts with CD9 from various species, inhibited release of and cell-cell fusion induced by canine distemper virus (26; reviewed in reference 27). Indeed, before embarking on the analysis of how K41 affects release of HIV-1 from human cells, we tested its potential (and also the potential of Vpg15) to inhibit FIV and HIV-1 release from CrFK cells transiently producing these viruses. Both antibodies were found to inhibit particle release of these two retroviruses (data not shown). We therefore asked if K41 indiscriminately prohibits the egress of enveloped viruses. Figure 4D demonstrates that this is not the case. Influenza virus release remained unaltered if infected HeLa cells (or infected 293T cells [data not shown]) were incubated with K41, in line with the finding presented in Fig. 1 that the tetraspanin CD9 is almost entirely absent from plasma membrane segments through which influenza virus exits.
Incubation of cells with the anti-CD9 antibody K41 leads to tetraspanin clustering.
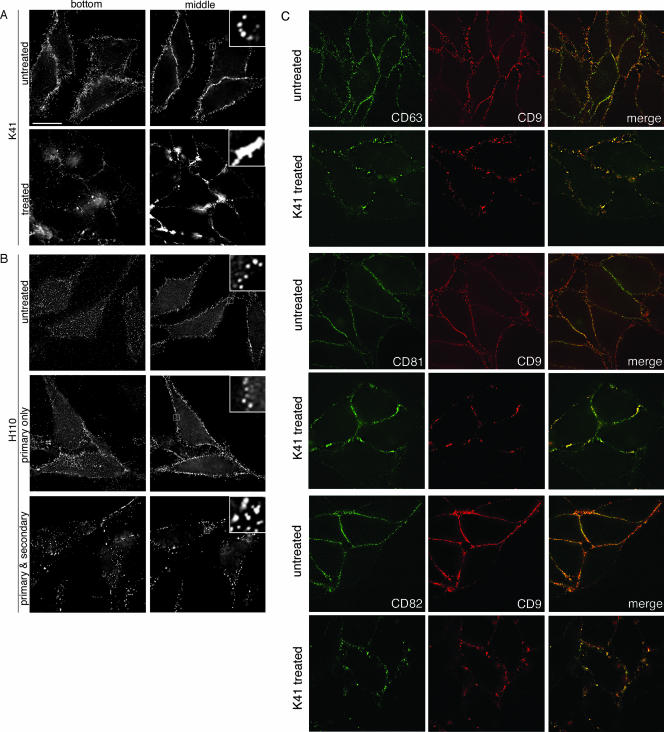
As a first step towards analyzing how K41 treatment inhibits HIV-1 release, we sought to visualize if this antibody alters the surface distribution of CD9. Figure 5A shows that this antigen was reorganized into large peripheral clusters in cells incubated with K41 (lower panels), while CD9-containing surface TEMs remained evenly distributed in untreated cells (upper panels) or in cells incubated with the isotype control antibody (data not shown). In contrast, when cells were treated with any of the anti-CD63, anti-CD81, or anti-CD82 antibodies that leave HIV-1 release unaffected (Fig. 4A), the surface distribution of their respective antigens remained unaltered (data not shown). Movies S1A and B in the supplemental material show three-dimensional renderings of cells left untreated (supplemental movie S1A) or incubated with K41 (supplemental movie S1B), visualizing that K41-induced CD9 clustering occurred primarily at cell-cell interfaces. Notably, incubation of cells with Vpg15, which also inhibits HIV-1 release (as mentioned above), also led to tetraspanin clustering, while with the treatment with an anti-CD9 antibody different from K41 (H110), which does not inhibit HIV-1 release, no redistribution of the antigen was apparent. If H110 was cross-linked with an appropriate secondary antibody, CD9 formed mid-sized clusters. Such CD9 cross-linking, however, unlike K41-induced clustering, was not limited to cell-cell adhesion sites but took place all over the cell surface (Fig. 5B) and was not associated with release inhibition (data not shown). The differential outcomes of incubations with the antibodies K41, Vpg15, and H110 are possibly due to differential interferences of these antibodies with the interaction of CD9 and cellular partner proteins, but testing of that hypothesis will have to await the outcome of the mapping of the epitopes for these different antibodies.
FIG. 5.
Differential clustering of CD9 by different anti-CD9 antibodies. (A) HeLa cells, left untreated or treated with K41, were surface stained for CD9. (B) HeLa cells were left untreated or were treated with anti-CD9 antibody H110 for 4 h or were treated for 1 h with H110 followed by 3 h with secondary antibody to induce microcapping. Following fixation, cells were stained with secondary antibodies. Bottom sections are shown in the left panels and the middle sections of the right panels. Bar, 10 μm. The blow-ups show 6×-magnified views of the boxed regions. (C) Untreated HeLa cells (top panel in a pair) or K41-treated cells (lower panel in a pair) were stained for surface CD9 and either CD63, CD81, or CD82. Middle sections are shown.
Since we previously documented that substantial fractions of CD63, CD81, and CD82 colocalize with CD9 in surface TEMs, we examined if the distribution of these tetraspanins was affected by K41 treatment. Figure 5C reveals that these other tetraspanins also underwent clustering at sites of cell-cell contact and, thus, that K41 affects the overall organization of surface TEMs.
Together, these data suggest, firstly, that K41-induced CD9 clustering is not due to antibody-mediated cross-linking of the tetraspanin and, secondly, that the retargeting of CD9 and other tetraspanins to the cell-cell interface is necessary for the antiviral activity of this antibody.
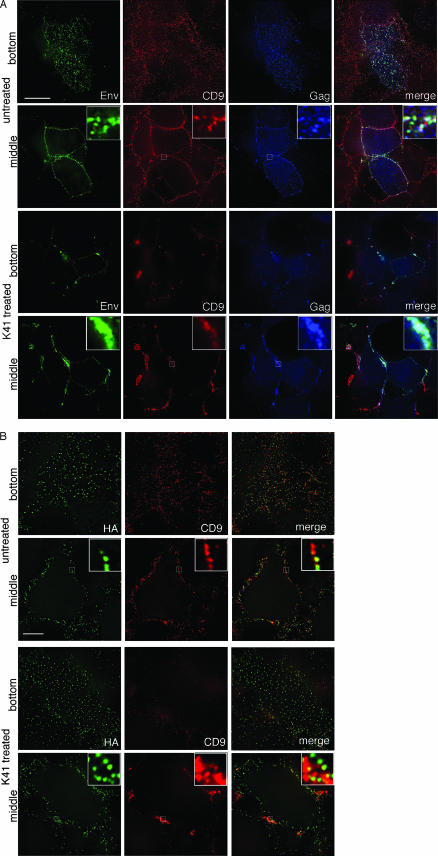
K41 treatment of cells results in the clustering of HIV-1 but not influenza virus components.
As suggested, for example, by the data shown in Fig. 3, CD9-containing TEMs serve as exit gateways for HIV-1. Not unexpectedly, therefore, K41-induced tetraspanin retargeting was found to also affect the distribution of HIV-1 Gag and Env. Indeed, these viral components formed massive clusters at peripheral sites of cell-cell contact in treated but not in untreated cells (Fig. 6A). K41-induced clustering of Gag was also observed if Gag was expressed alone (i.e., without Env) (unpublished data), and release of virus-like particles from such cells was inhibited by K41 (data not shown). In contrast, if Env was expressed alone, a situation in which it displays very little colocalization with surface TEMs (35), its distribution remained largely unaltered in K41-treated cells (data not shown). Likewise, the distribution of the influenza virus envelope glycoprotein HA, whether expressed alone (data not shown) or expressed together with the other viral components (Fig. 6B), remained unaltered in K41-treated cells. Note, for example, that the bottom sections of cells stained for HA show no clustering of that antigen (Fig. 6B, K41-treated cells in the upper left panel), whereas the bottom sections of HIV-1 Env-stained cells documented that virtually all Env puncta have disappeared and that Env was visible only in large clusters at cell-cell contact sites (Fig. 6A, K41-treated cells in the upper left panel). Also, the magnified images in the insets of Fig. 6B revealed that HA puncta were still clearly separate entities, even within CD9 clusters induced by the K41 treatment.
FIG. 6.
Differential effects of tetraspanin clustering on localization of HIV-1 Gag and Env and influenza virus HA. (A) HeLa cells expressing full-length HIV-1 (NL4-3) were left untreated (upper two panels) or were treated with K41 (lower two panels), followed by staining for surface CD9, Env, and (after permeabilization) Gag. The bottom and middle sections are shown in the upper and lower panels, respectively, for each pair. (B) HeLa cells infected with influenza virus were left untreated (upper two panels) or were treated with K41 (lower two panels) followed by staining for surface CD9 and HA. The bottom and middle sections are shown alternately. Bar, 10 μm. The blow-ups in the merge panels show 6×-magnified views of the boxed regions.
How does K41 interfere with HIV-1 release?
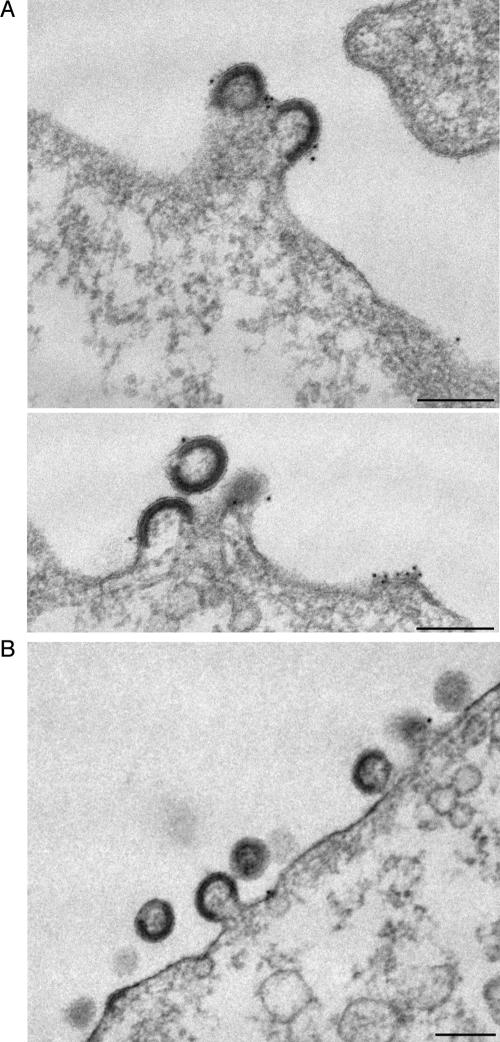
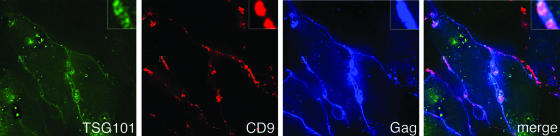
Together, the data presented in Fig. 4 through 6 strongly suggested that K41-induced clustering of the viral antigens is responsible for reduced HIV-1 release. K41 treatment of cells did not affect steady-state levels of Gag (Fig. 4A), and results of pulse-labeling experiments in which the rates of Gag production in treated cells were compared to those in untreated cells revealed that K41 did not affect the onset of Gag expression (data not shown). Given that it takes several hours until full-fledged K41-induced tetraspanin clustering is reached (data not shown), it is conceivable that either preexisting and/or newly synthesized viral components are (re)targeted to cell-cell contact sites. To investigate at what step K41 blocks HIV-1 release, we performed an ultrastructural analysis of cells coexpressing Gag and Env from separate plasmids (to reach high levels of expression) and thus producing HIV-1 VLPs. In both untreated (Fig. 7A) and K41-treated (Fig. 7B) cells, viral buds studded with anti-CD9 antibodies (visualized by using 10-nm gold particles) were found surrounded by plasma membrane with little CD9. However, quantification of VLPs budding at the plasma membrane also demonstrated that while budding of VLPs still took place in K41-treated cells, the number of buds per cell was considerably reduced (by about 70%) compared to untreated cells. Consistent with the finding that the budding process per se was not blocked, the cellular ESCRT1 component TSG101, which is required for HIV-1 budding (12, 28, 51; reviewed in references 1 and 32), was found to still be recruited to the sites where tetraspanins and viral components cluster upon K41 treatment (Fig. 8). Also, contrary to what is seen with so-called L-domain mutant Gag (for a recent review, see reference 1), which gives rise to the production of particles that remain tethered to the plasma membrane and whose processing is incomplete, we found that Gag was processed efficiently in K41-treated cells (Fig. 4 and data not shown). However, if HIV-1 particle formation per se is not inhibited in K41-treated cells, why are fewer virions released from these cells? Perhaps the K41 treatment, by inducing the redistribution of tetraspanins and virus components from free surfaces to cell-cell contact sites, physically prevents efficient particle morphogenesis in HeLa cells. Consistent with that hypothesis, we observed that HIV-1 release was not inhibited by K41 treatment of cells if they were grown subconfluently, i.e., if contacts between cells were rare and thus no significant clustering of tetraspanins and viral components could take place (Fig. 9A). Also, incubation of Jurkat T lymphocytes, which form only transient cell-cell contacts if grown under standard cell culture conditions, did not affect release of HIV-1 (Fig. 9B). This does not exclude the possibility that virus transmission is affected by K41 treatment.
FIG. 7.
Ultrastructural analysis of cells producing HIV-1 VLPs: K41 does not block the budding process per se. HeLa cells expressing HIV-1 Gag and Env were left untreated (A) or were treated with K41 (B) and were processed for electron microscopy analysis as described in Materials and Methods. Bars, 200 nm.
FIG. 8.
Effect of K41 treatment on recruitment of TSG101 to Gag-containing surface TEMs. HeLa cells expressing TSG101-YFP and Gagopt-CFP were treated with K41, followed by surface staining of CD9. The middle section is shown. The blow-ups in the merge panel show 5×-magnified views of the boxed regions.
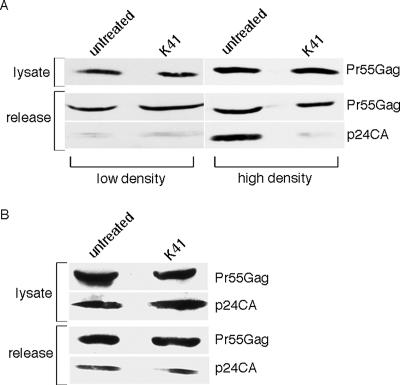
FIG. 9.
The effect of K41 is dependent on cell density and cell type. (A) HeLa cells expressing HIV-1 were split into dishes at low and high cell confluence prior to treating them with K41. Viral Gag in the lysate and supernatant were then harvested and analyzed by Western blotting as described in Materials and Methods. For the lower-cell-density part of the experiment, a longer exposure of the gel is shown to enable visualization of the signal. (B) E6-1 Jurkat T cells expressing HIV-1 were treated with 6 μg/ml K41 for 16 h and harvested for analysis of viral proteins in the supernatant and lysate by Western blotting as described in Materials and Methods.
DISCUSSION
The tetraspanin CD63, a resident of late endosomes/lysosomes with only minor representation at the plasma membrane, was shown more than 10 years ago to be acquired by HIV-1 particles (14, 31, 37; reviewed in reference 38), but the significance of such incorporation remained unclear. More recent work by various groups suggested that CD63 may be one of several members of the tetraspanin family that contributes to the formation of HIV-1 budding platforms in diverse cell types. Here we provide further evidence in support of this hypothesis by showing that an anti-CD9 antibody (K41) which clusters tetraspanins and HIV-1 Gag and Env inhibits particle release. We also demonstrate that influenza virus, the release of which remains unaffected by K41, assembles at domains distinct from those through which HIV-1 buds.
Both HIV-1 and influenza virus are known to exit through so-called lipid rafts (now called membrane rafts [42], local clusters of sphingolipids and cholesterol) (3, 36, 49). It was originally thought that proteins, merely by virtue of their affinity for raft lipids, would accumulate in specific membrane segments that can serve, for example, as platforms for signaling at immunological synapses (20) or during cell migration (15). Further, it was proposed (41) that lipid rafts serve as sites at which components of diverse viruses are concentrated for subsequent particle morphogenesis. However, the fact that HIV-1 and influenza virus exit through separate sites at the plasma membrane, as made evident in this report (Fig. 1 through 3), argues that the clustering of viral components at discrete microdomains is driven primarily by their affinities for specific cellular proteins located at those sites. Together with other recent reports, e.g., on protein sorting during T-cell signaling (10) or cell migration (13), these data provide further support for a refined raft concept (17, 44) which proposes that specific protein-protein interactions are critical for the functional compartmentalization of membranes by stabilizing inherently unstable lipid assemblies at discrete sites. Note that the finding that HIV-1 and influenza virus exit through different plasma membrane microdomains seemingly is in conflict with recent reports showing that HA can be incorporated into HIV-1- or SIV-based vectors (6, 47). However, in both of those studies, pseudotyping was achieved in an overexpression system (in 293T cells) where viral exit sites cover a higher percentage of the plasma membrane, thus increasing the likelihood that the HIV-1 and the influenza virus egress sites partially overlapped. Also, it should be pointed out that the titer of the HA-pseudotyped HIV-1 vector was shown to be considerably lower than the titers of HIV-1 vectors carrying retroviral envelope glycoproteins (6).
It already has been established that members of the tetraspanin protein family play a role(s) during the early phase of the viral replication cycle (16, 19, 52), though much remains to be learned about how tetraspanins contribute to the viral attachment/fusion and/or postfusion steps. In contrast, even though tetraspanins were shown to colocalize with retroviral components during egress (2, 7, 23, 29, 30, 35, 39, 45, 53), it remained unclear if they were functionally involved in the assembly/release process. CEM T lymphocytes, for example, do not express CD9 (4), yet they support robust HIV-1 replication. Further, overexpression or small interfering RNA-mediated knock-downs of CD9 or CD63 affected HIV-1 egress only marginally (data not shown), presumably because of redundancy among different tetraspanins (18). However, the findings that K41-induced accumulation of CD9 and other tetraspanins results in the clustering of HIV-1 components at cell-cell junctions, and that such clustering correlates with reduced virus release (Fig. 4 to 6), strongly suggest that tetraspanins are not just bystanders in HIV-1 egress. Rather, these proteins contribute to the buildup of budding platforms for HIV-1, perhaps by providing a scaffold for viral and cellular components necessary for particle assembly and shedding.
How does K41-induced tetraspanin clustering result in reduced HIV-1 release? Are TEMs rendered nonfunctional with respect to their role in particle assembly and release? Data presented in Fig. 7 and 8 of this report demonstrate that this is not the case. Particle assembly and the recruitment of cellular elements necessary for viral budding can still take place in the presence of K41. However, accumulation of the viral components followed by particle budding appears limited to cell-cell junctions in antibody-treated cells, and such spatially restricted exit may result in reduced yield of viral particles. In agreement with this, if cells are grown at low density, the K41 effect is diminished (Fig. 9A). Interestingly, we observed that the tetraspanin clusters induced by K41 treatment shown in this report for cells grown at high density (Fig. 5) are indeed tight microvilli zippers between adjacent cells when viewed by scanning electron microscopy (K. Singethan et al., submitted for publication). Further, HIV-1 release from T lymphocytes that express only moderate amounts of CD9 (compared to HeLa cells) and which do not form stable cell-cell junctions if grown under standard cell culture conditions, i.e., in suspension, was not inhibited by K41 (Fig. 9B). Given that, in vivo, HIV-1 is thought to be transmitted to a significant extent at cell-cell junctions, at virological synapses, should one thus expect that K41 would not affect virus release, or that it would even enhance virus spread in intact tissue, e.g., in lymph nodes, where synaptic transmission may be prevalent? The more likely scenario is that treatment with K41-like antitetraspanin antibodies would also be detrimental to HIV-1 spread under those circumstances, as tetraspanins have been demonstrated to prevent fusion of producer and target cell (16). Such fusion inhibition is critical for the sustained spread of viruses such as HIV-1, because the formation of syncytia that include producer and target cells most likely provides a dead end to virus replication. Finally, we surmise that influenza virus, which is released from free surfaces of infected cells (for a recent review, see reference 34), may not be affected by antitetraspanin antibodies, as it may have evolved to exit through microdomains built with a different set of cellular proteins. However, to the best of our knowledge, this has not yet been systematically analyzed.
In conclusion, we found that HIV-1 and influenza virus utilize distinct and separate plasma membrane microdomains for their egress. The composition of these sites appears tailored to the needs of these two enveloped viruses with HIV-1, but not influenza virus, being gated through areas enriched in tetraspanins including CD9 and CD63. Presumably, these sites ensure efficient transmission of HIV-1 particles to the target cell, thus allowing for rapid systemic spread of this virus.
Supplementary Material
Acknowledgments
We thank Benjamin Chen and Robert Lamb for reagents and technical advice, Geneviève Porcheron-Berthet for assistance in the electron microscopy analysis, and the PFMU at the Geneva Medical Faculty for access to electron microscope and ancillary equipment. Jürgen Schneider-Schaulies and Alan Howe are acknowledged for discussions and for critical reading of the manuscript, respectively.
This work was supported by grants RO1 AI 47727 and R03 AI 060679 to M.T. and SNSF 3100A0-104489/1 to M.F.
Footnotes
Published ahead of print on 12 September 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Booth, A. M., Y. Fang, J. K. Fallon, J. M. Yang, J. E. Hildreth, and S. J. Gould. 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172:923-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brugger, B., B. Glass, P. Haberkant, I. Leibrecht, F. T. Wieland, and H. G. Krausslich. 2006. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA 103:2641-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charrin, S., F. Le Naour, M. Oualid, M. Billard, G. Faure, S. M. Hanash, C. Boucheix, and E. Rubinstein. 2001. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276:14329-14337. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B. J., M. Takeda, and R. A. Lamb. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79:13673-13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deneka, M., A. Pelchen-Matthews, R. Byland, E. Ruiz-Mateos, and M. Marsh. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177:329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Parseval, A., D. L. Lerner, P. Borrow, B. J. Willett, and J. H. Elder. 1997. Blocking of feline immunodeficiency virus infection by a monoclonal antibody to CD9 is via inhibition of virus release rather than interference with receptor binding. J. Virol. 71:5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitrov, D. S., R. L. Willey, H. Sato, L. J. Chang, R. Blumenthal, and M. A. Martin. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67:2182-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglass, A. D., and R. D. Vale. 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 121:937-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foti, M., A. Mangasarian, V. Piguet, D. P. Lew, K. H. Krause, D. Trono, and J. L. Carpentier. 1997. Nef-mediated clathrin-coated pit formation. J. Cell Biol. 139:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Gaus, K., S. Le Lay, N. Balasubramanian, and M. A. Schwartz. 2006. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Mouton, C., J. L. Abad, E. Mira, R. A. Lacalle, E. Gallardo, S. Jimenez-Baranda, I. Illa, A. Bernad, S. Manes, and A. C. Martinez. 2001. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc. Natl. Acad. Sci. USA 98:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon-Alonso, M., M. Yanez-Mo, O. Barreiro, S. Alvarez, M. A. Munoz-Fernandez, A. Valenzuela-Fernandez, and F. Sanchez-Madrid. 2006. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 177:5129-5137. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, J. F. 2006. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 7:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemler, M. E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6:801-811. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. H., F. Martin, A. Higginbottom, L. J. Partridge, V. Parthasarathy, G. W. Moseley, P. Lopez, C. Cheng-Mayer, and P. N. Monk. 2006. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 80:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janes, P. W., S. C. Ley, and A. I. Magee. 1999. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147:447-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolly, C., K. Kashefi, M. Hollinshead, and Q. J. Sattentau. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolly, C., and Q. J. Sattentau. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J. Virol. 81:7873-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136-148. [DOI] [PubMed] [Google Scholar]
- 26.Loffler, S., F. Lottspeich, F. Lanza, D. O. Azorsa, V. ter Meulen, and J. Schneider-Schaulies. 1997. CD9, a tetraspan transmembrane protein, renders cells susceptible to canine distemper virus. J. Virol. 71:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, F., D. M. Roth, D. A. Jans, C. W. Pouton, L. J. Partridge, P. N. Monk, and G. W. Moseley. 2005. Tetraspanins in viral infections: a fundamental role in viral biology? J. Virol. 79:10839-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazurov, D., G. Heidecker, and D. Derse. 2006. HTLV-1 Gag protein associates with CD82 tetraspanin microdomains at the plasma membrane. Virology 346:194-204. [DOI] [PubMed] [Google Scholar]
- 30.Mazurov, D., G. Heidecker, and D. Derse. 2007. The inner loop of tetraspanins CD82 and CD81 mediates interactions with human T cell lymphotrophic virus type 1 Gag protein. J. Biol. Chem. 282:3896-3903. [DOI] [PubMed] [Google Scholar]
- 31.Meerloo, T., H. K. Parmentier, A. D. Osterhaus, J. Goudsmit, and H. J. Schuurman. 1992. Modulation of cell surface molecules during HIV-1 infection of H9 cells. An immunoelectron microscopic study. AIDS 6:1105-1116. [DOI] [PubMed] [Google Scholar]
- 32.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 33.Muller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nydegger, S., S. Khurana, D. N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173:795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orentas, R. J., and J. E. Hildreth. 1993. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res. Hum Retrovir. 9:1157-1165. [DOI] [PubMed] [Google Scholar]
- 38.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 39.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips, D. M. 1994. The role of cell-to-cell transmission in HIV infection. AIDS 8:719-731. [DOI] [PubMed] [Google Scholar]
- 41.Pickl, W. F., F. X. Pimentel-Muinos, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pike, L. J. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47:1597-1598. [DOI] [PubMed] [Google Scholar]
- 43.Pope, M., and A. T. Haase. 2003. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat. Med. 9:847-852. [DOI] [PubMed] [Google Scholar]
- 44.Rajendran, L., and K. Simons. 2005. Lipid rafts and membrane dynamics. J. Cell Sci. 118:1099-1102. [DOI] [PubMed] [Google Scholar]
- 45.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Boulan, E., K. T. Paskiet, and D. D. Sabatini. 1983. Assembly of enveloped viruses in Madin-Darby canine kidney cells: polarized budding from single attached cells and from clusters of cells in suspension. J. Cell Biol. 96:866-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandrin, V., and F. L. Cosset. 2006. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J. Biol. Chem. 281:528-542. [DOI] [PubMed] [Google Scholar]
- 48.Sourisseau, M., N. Sol-Foulon, F. Porrot, F. Blanchet, and O. Schwartz. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81:1000-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarrant, J. M., L. Robb, A. B. van Spriel, and M. D. Wright. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24:610-617. [DOI] [PubMed] [Google Scholar]
- 51.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Lindern, J. J., D. Rojo, K. Grovit-Ferbas, C. Yeramian, C. Deng, G. Herbein, M. R. Ferguson, T. C. Pappas, J. M. Decker, A. Singh, R. G. Collman, and W. A. O'Brien. 2003. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J. Virol. 77:3624-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welsch, S., O. T. Keppler, A. Habermann, I. Allespach, J. Krijnse-Locker, and H. G. Krausslich. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.