Abstract
The expression of herpes simplex virus (HSV) genomes in the absence of viral regulatory proteins in sensory neurons is poorly understood. Previously, our group reported an HSV immediate early (IE) mutant (d109) unable to express any of the five IE genes and encoding a model human cytomegalovirus immediate early promoter-green fluorescent protein (GFP) transgene. In cultured cells, GFP expressed from this mutant was observed in only a subset of infected cells. The subset exhibited cell type dependence, as the fractions of GFP-expressing cells varied widely among the cell types examined. Herein, we characterize this mutant in murine embryonic trigeminal ganglion (TG) cultures. We found that d109 was nontoxic to neural cultures and persisted in the cultures throughout their life spans. Unlike with some of the cultured cell lines and strains, expression of the GFP transgene was observed in a surprisingly large subset of neurons. However, very few nonneuronal cells expressed GFP. The abilities of ICP0 and an inhibitor of histone deacetylase, trichostatin A (TSA), to activate GFP expression from nonexpressing cells were also compared. The provision of ICP0 by infection with d105 reactivated quiescent genomes in nearly every cell, whereas reactivation by TSA was much more limited and restricted to the previously nonexpressing neurons. Moreover, we found that d109, which does not express ICP0, consistently reactivated HSV type 1 (KOS) in latently infected adult TG cultures. These results suggest that the state of persisting HSV genomes in some TG neurons may be more dynamic and more easily activated than has been observed with nonneuronal cells.
Infection with herpes simplex virus (HSV) in vivo occurs in three stages: primary (productive) infection, latency, and reactivation. During primary or lytic infection, the virus replicates and produces progeny in susceptible epithelial cells, an event which may or may not be accompanied by the appearance of mucocutaneous lesions, the hallmark of HSV infection (46). The primary infection almost invariably culminates in the infection of innervating sensory neurons, which then become sites of HSV latency. Throughout the lifetime of the host, HSV reactivates, both spontaneously and in response to specific reactivating stimuli, causing recurrent infections. These episodic reactivations often produce lesions at or near the primary site of infection, but are frequently asymptomatic (49, 89-91).
During productive infection, viral gene expression proceeds in a temporally controlled cascade of the immediate early (IE), early (E), and late (L) genes (38, 39). Viral gene expression during latency and reactivation, both of which occur in neurons, differs from that observed during lytic infection (19, 50, 51). Latency is characterized by the absence of productive viral transcripts, save for the latency-associated transcripts, whose exact function in maintaining or reactivating from the latent state is still without consensus (4, 5, 20, 35, 42, 45, 52, 58, 72-74, 81, 86). Viral gene expression during reactivation in neuronal cells may differ from that seen in either productive or latent infection, where the cascade is not so tightly controlled and E gene expression occurs simultaneously with or may even precede IE gene expression (30, 49, 70, 84).
HSV encodes five IE genes: ICP0, ICP4, ICP22, ICP27, and ICP47. Of these, only ICP4, the major viral transactivator, and ICP27 are essential (16, 17, 53, 64, 76, 79). Though nonessential (77, 83), ICP0 is a multifunctional zinc-binding RING finger protein that enhances HSV infectivity through a variety of actions (6, 22, 23). Although ICP0 does not bind DNA directly, it is an efficient, promiscuous transactivator of both viral and cellular messages in transient-transfection assays. ICP0 transactivates all three classes of HSV genes, and its presence is critical for effective reactivation from latency (8-10, 31, 32, 43). The effects of ICP0 expression on cells are varied, as ICP0 has been shown to disrupt nuclear domain 10 (ND10) structures (27, 63), block cells at the G1/S and G2/M checkpoints (36, 56), stabilize some cellular proteins (cyclins D1 [88] and D3 [48], EF-1d [48], and BMAL1 [47]), and, acting as a ubiquitin E3 ligase (87), direct the proteosome-dependent degradation of several others (the ND10 constituent proteins PML and Sp100 [11, 25, 26, 29, 62] and the centromeric proteins CENP-A [57] and CENP-C [24], as well as the DNA-dependent protein kinase [71]). ICP0 expression is also vital to circumvention of the antiviral effects of interferon, and it regulates the physical configuration of the genome during productive infection (21, 34, 41, 68, 69).
Despite exhaustive research concerning the effects of both its expression and absence, both in the presence and absence of other viral proteins, the mechanisms by which ICP0 may tip the balance from latency to reactivation and subsequent productive infection are still unclear. Putative mechanisms of ICP0 function have come from comparison of ICP0's effects with substances of known function, such as trichostatin A (TSA). TSA is a well-characterized inhibitor of histone deacetylases and exhibits many of the effects on cells that ICP0 exhibits. For instance, both upregulate transcription, halt the cell cycle at the G1/M and G2/M checkpoints (36, 37, 92, 95, 96), and induce p53-responsive genes, such as p21 and gadd45 (3, 36, 44). To date, though TSA has been shown to induce reactivation in latently infected explanted ganglia (2) and has induced transgene expression in neurons (1), its ability to do so in the complete absence or with the limited expression of IE genes has not been addressed. Critical to understanding mechanisms of latency and reactivation is a determination of how and to what extent neurons regulate latent viral genomes in both the absence and presence of key IE proteins, specifically ICP0.
In a previous study, we detailed the production and characterization of several HSV IE mutants varying in their abilities to express subsets of IE genes (78). Two of these mutants, d106 and d109, were particularly relevant to studies described here. Both mutants are isogenic variants expressing a human cytomegalovirus (HCMV) IE promoter-green fluorescent protein (GFP) transgene construct from the ICP27 locus but differ in the numbers of other IE genes that they express: d106 expresses only ICP0, while d109 expresses no IE genes. In the present study, we extend our characterization of the d109 mutant to include a variety of nonneuronal cell types and describe the characterization of both mutants in primary cultures of dissociated trigeminal ganglia (TG). Interested in how the mutant d109 might serve as both an effector and a target of reactivation in a neuronal environment, we also investigated its response to reactivation stimuli such as superinfection (with and without ICP0) and treatment with TSA, as well as the ability of both mutants to reactivate latent wild-type virus.
MATERIALS AND METHODS
Viruses and cells.
Wild-type KOS virus was propagated on Vero cells. The HSV type 1 (HSV-1) IE mutants d105, d106, d99, and d109 were propagated on E11 or FO6 cells as previously described (78). Vero, U2OS, 293, HeLa, COS-7, CaCO-2, MCF-7, and Raji cells, normal embryonic lung cells, and normal skin fibroblasts were propagated as recommended by the American Type Culture Collection (ATCC). The FO6, E11, and HT-116 cell lines were maintained and propagated as previously described (78).
Primary TG cultures.
Primary cultures of dissociated TG were prepared from adult or embryonic day 18 CD-1 mice using methods previously described (12, 30). Briefly, TG were removed from embryonic day 18 or adult mice, dissociated for approximately 20 minutes in dissociation medium (Hanks balanced salt solution [Invitrogen] supplemented with collagenases IV and XI [Sigma]; 50 mg of each/ml). Cells were spun briefly (5 min) to remove dissociation medium and then washed three times in L15 medium (Invitrogen). Following the final wash, the cells were then resuspended in neurobasal medium supplemented with 1× B27, 4 mM GlutaMAX (Invitrogen), and 50 ng/ml each of 2.5s and 7s neuronal growth factor (Becton Dickinson). Mechanical dissociation was then performed by repeatedly passing the cells through 18- and then 22-gauge needles. To remove large debris, cells were then filtered through a 70-μm nylon cell strainer (Falcon) twice. Cells (1 × 105/well) were then plated on 12-mm coverslips precoated with poly-d-lysine (Sigma)-laminin (Becton Dickinson) in 24-well plates. Cultures were incubated at 37°C. Cells were fed with neurobasal medium, supplemented as described above, every 2 to 3 days. For the first week of culture, the neurobasal medium was supplemented with 20 μM uridine (Sigma)-20 μM fluorodeoxyuridine (Sigma).
Production of latently infected TG cultures.
Where adult, latently infected mice were used, 6- to 8-week-old CD-1 mice (weighing 20 to 25 g) were anesthetized initially with the inhalative anesthetic isoflurane (Isoflo; Schering-Plough) until unconscious. The mice were then more fully anesthetized via injection intraperitoneally with 10 μl/g of 2.5% tribromoethanol (Avertin). Mice corneas were scarified with a 26-gauge needle and infected with KOS at a viral load of 2 × 106 PFU in each eye (total viral load per mouse, 4 × 106 PFU) in a viral inoculum of 10 μl per eye. Thirty days postinfection, mice were sacrificed and their ganglia removed and dissociated as detailed above, with the following exceptions: neurobasal A medium was substituted for neurobasal medium and acyclovir was substituted for 20 μM fluorodeoxyuridine-20 μM during the first week of plating.
Monitoring KOS reactivation.
To detect infectious reactivated KOS from the latently infected TG, on days 4 and 7 to 21 postplating, 100-μl samples of culture supernatant from the dissociated latently infected TG established above were used to infect matched Vero cell cultures. The cultures were then monitored for viral cytopathic effects.
Infection of neuronal cultures.
Neuronal cultures were prepared as described above and infected 7 to 14 days postplating. Viral inputs were calculated per well. Standing medium was removed and replaced with the appropriate virus diluted in neurobasal medium in a volume not to exceed 200 μl/well. Plates were incubated at 37°C for 1 hour with gentle rocking every 5 to 7 min.
Rescue of d109-infected cultures.
For immediate rescue, cultures of Vero or U2OS cells were grown in flasks and coinfected with d109 and KOS or d109 and d99 at the multiplicity of infection (MOI) indicated below. Monolayers were harvested approximately 20 hours postinfection, and the titers of the progeny were determined on FO6 cells. Results are expressed as numbers of fluorescent plaques versus the total number of plaques. For delayed rescue, cultures were infected with d109 and then coinfected with either KOS or d99 day 3 postinfection at the multiplicities indicated below. The titers of progeny were determined on FO6 cells, and results are expressed as numbers of fluorescent plaques divided by the total number of plaques.
Green/white-cell sorting of d109-infected monolayers.
Monolayers of the indicated cells were infected with d109 (MOI = 10 PFU/cell). Sixteen to 18 hours postinfection, the cells were harvested, fixed with 3% paraformaldehyde in phosphate-buffered saline, for 15 minutes at room temperature, and analyzed for GFP expression on a FACStar flow cytometer. For rescue of d109-infected cultures, Vero cells were infected with d109 and trypsinized, sorted, plated, and superinfected with either d99 or KOS on either day 1 or day 3 postinfection. Monolayers were then harvested approximately 20 to 24 h after the superinfection. The titers of progeny were determined on FO6 cells and plaques counted as described above.
RESULTS
Constitutive GFP expression from d109 genomes varies widely and is cell type dependent.
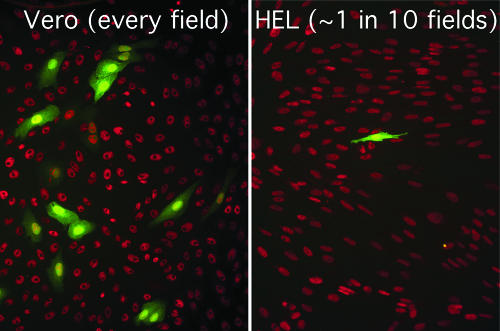
An initial characterization of d109 revealed that it was nontoxic to both Vero and HEL cells and persisted in the cells for an extended period of time, and despite low-level transgene expression overall, GFP was abundantly expressed in a subpopulation of cells, with the expressing subpopulation being higher in Vero than in HEL cells (Fig. 1) (78). To determine whether the expression pattern of d109 observed in Vero and HEL cells extended to other nonneuronal cells as well, a variety of commonly cultured cells were infected with d109 or d106, and the results are listed in Table 1. As is shown, expression from d106 genomes was consistently high among the majority of cell types tested, with the majority of cell types exhibiting at least 80% transgene expression. The percentages of d109-infected cells expressing GFP varied greatly from cell type to cell type. GFP expression from d109 genomes ranged from a low of less than 1% in HEL cells to a high of 99% in U2OS cells.
FIG. 1.
d109 transgene in Vero and HEL cells. Vero (left) and HEL (right) cells were infected with d109 at an MOI of 10 PFU/cell. Representative fields at 24 h postinfection are shown. The red nuclear fluorescence is from sites of rhodamine staining of Sir2 antibody reactivity.
TABLE 1.
Expression of the GFP transgene by d109 in different cell typesa
| Cell type | % of cells that were fluorescent inb:
|
|
|---|---|---|
| d109 | d106 | |
| U2OS | 99 | 96 |
| 293 | 52 | 80 |
| HeLa | 21 | 89 |
| COS-7 | 16 | 90 |
| HCT-116 | 7 | 79 |
| Vero | 6 | 87 |
| WS1 (normal skin fibroblasts) | 2 | 85 |
| MCF7 | 2 | 28 |
| HEL (normal embryonic lung fibroblasts) | <1 | 90 |
Cells were infected with the indicated virus at an MOI of 10. At 16 to 18 h postinfection, they were fixed with 3% paraformaldehyde and analyzed for GFP expression on a FACStar flow cytometer.
Percentage of cells that were fluorescent in a sample of 104 cells.
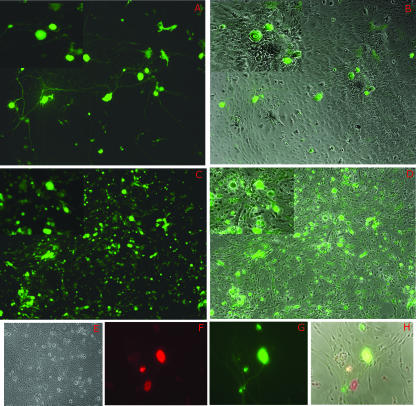
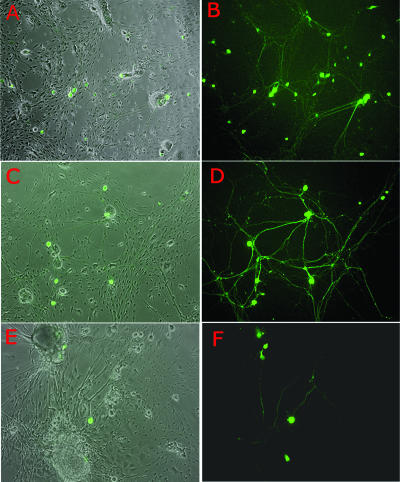
We then sought to extend these observations to cells in which HSV establishes latency in vivo. To model this, primary cultures of murine TG were established and the expression patterns of d109 and d106 were observed in these cultures. In primary cultures of TG cells, d106 and d109 again had considerably different expression patterns. Cultures infected with d106 displayed widespread transgene expression, as nearly every cell in the monolayer expressed some level of GFP (Fig. 2C and D). In addition, many cells rounded up and detached from the stratum, characteristic of ICP0-induced toxicity. This took between 2 and 5 days depending on the MOI and the cell type. In contrast, and as was reported previously with Vero cells, GFP expression from d109-infected cultures was restricted to a subpopulation. Interestingly, the majority of this subpopulation of GFP-expressing cells displayed a morphology consistent with that of neurons, in that they possessed large and relatively round cell bodies compared to those of the surrounding cells (Fig. 2A and B, insets). Staining of d109-infected cultures with a neuron-specific antibody (anti-HuC/D) confirmed their neuronal identity (Fig. 2F and G). In addition, many of the cells staining red for the neuron-specific marker also expressed GFP, while few or none of the cells that did not stain red expressed GFP. Toxicity was not observed in d109-infected cultures (Fig. 2E).
FIG. 2.
d109 transgene expression in fetal TG cultures. Primary cultures of embryonic TG were infected with either d109 (A, B, and F to H) or d106 (C and D) and photographed 24 h postinfection. Duplicate exposures of the same field showing either phase-contrast results together with GFP expression or GFP expression alone are shown. In order to provide greater detail, magnified insets of all fields are shown in the upper left corner of each exposure. (F, G, and H) Triplicate exposures of the same field showing either anti-HuC/D (neuron-specific antibody) expression alone (red in panel F), GFP expression alone (G), or phase-contrast exposure together with anti-HuC/D and GFP expression (H). (E) Mock-infected embryonic TG.
To determine whether the GFP-negative cells in d109-infected monolayers were unable to express the transgene because they lacked viral genomes, TG cultures infected with d109 were superinfected with d105. d105 is identical to d106 in that it expresses only ICP0 and not GFP. Thus, when used to superinfect cultures, d105 transactivates GFP expression most likely because it supplies ICP0, allowing easy identification of cells harboring quiescent d109 genomes. As can be observed below in Fig. 4, 5, and 6, upon superinfection with d105, nearly every cell in the monolayer expressed GFP. In fact, the monolayers looked very similar to those infected with d106 (Fig. 2), clearly demonstrating that the lack of transgene expression in d109-infected monolayers was not because d109 genomes were absent but because they were most likely repressed.
FIG. 4.
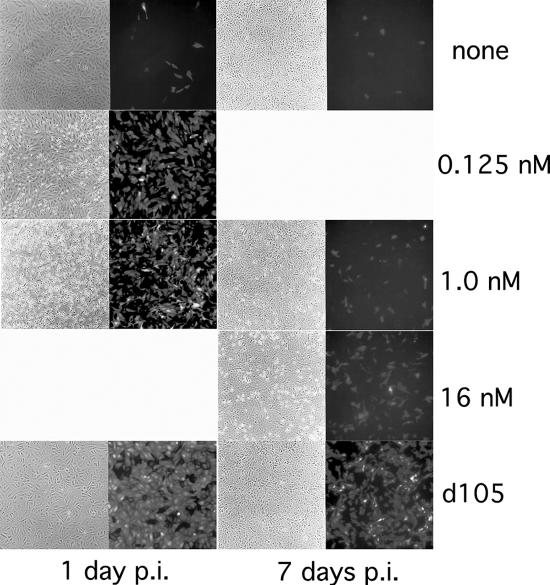
Derepression of d109 by TSA and d105 in Vero cells. Monolayers of Vero cells were infected with d109 at an MOI of 10 PFU/cell. The indicated amounts of TSA were added at either day 1 or day 7 postinfection (p.i.). d105 was used to superinfect d109-infected cells 1 and 7 days after d109 infection. Fields were photographed 24 hours posttreatment.
FIG. 5.
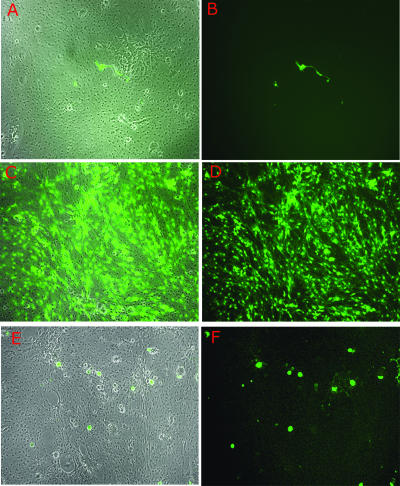
Derepression of d109 by TSA and d105 in TG cultures 24 hours postinfection. Embryonic cultures were infected with d109 (A through F). Twenty-four hours postinfection, the cultures were either left untreated (A and B), superinfected with d105 (2 × 106 PFU/well) (C and D), or treated with TSA (330 nM) (E and F). Shown are duplicate exposures depicting either phase-contrast results and GFP expression or GFP expression alone. Fields were photographed 18 to 24 h after superinfection or treatment.
FIG. 6.
Derepression of d109 by TSA and d105 in TG cultures at 22 days postinfection. Embryonic TG cultures were infected with d109 (A through F). Twenty-two days postinfection, the cultures were either left untreated (A and B), superinfected with d105 (2 × 106 PFU/well) (C and D), or treated with TSA (330 nM) (E and F). Shown are duplicate exposures depicting either phase-contrast results and GFP expression or GFP expression alone. Fields were photographed 18 to 24 h after superinfection or treatment.
As in nonneuronal cells and cell lines (Fig. 1; Table 1), expression of GFP in neuronal cells from d109 occurs in only a fraction of the cells, even though all or most of the cells possess genomes. The proportion of cells expressing GFP from d109 varies from cell type to cell type, with TG neurons exhibiting a relatively high proportion of GFP-expressing cells.
d109 transgene expression persists in neurons over time.
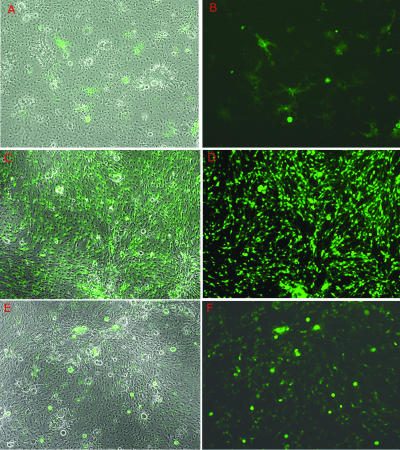
To observe the characteristics of d109 in neural cells, primary cultures of embryonic TG were infected with d109 and monitored over time for viral toxicity, GFP expression, and retention of the resident viral genomes (Fig. 3). As described above, GFP expression was initially observed in a subset of neurons, their processes, and a limited number of support cells. Over time, however, transgene expression in these support cells quickly declined, while GFP expression continued almost exclusively in neurons and their processes. By approximately 7 days postinfection (Fig. 3C and D), the majority of cells expressing GFP were neurons. This level of constitutive GFP expression in neurons was maintained for approximately 10 to 14 days, after which time transgene expression gradually diminished by neuron (Fig. 3E and F), until it was no longer seen at 53 days postinfection (data not shown). In keeping with the nontoxic nature of the virus, the monolayers at this and all time points postinfection showed no signs of cytopathic effect and were indistinguishable from mock-infected cultures, despite the high input of virus (1 × 10 7 PFU/well).
FIG. 3.
Persistence of d109 in neuron cultures. Cultures of embryonic TG were infected with d109 and photographed over time. Shown are duplicate exposures of d109 infection depicting either phase-contrast results and GFP expression or GFP expression alone. d109 is shown at day 1 postinfection (A and B), at day 7 postinfection (C and D), and at day 38 postinfection (E and F).
To gain a more quantitative understanding of the proportion of cells exhibiting transgene expression from d109, many microscopic fields from the experiment discussed above were examined and the numbers of neurons and GPF-positive neurons were quantified. Neurons were identified as those having the same morphology as those that stained with the neuron-specific antibody (anti-HuC/D) used in the experiment represented in Fig. 2. Table 2 gives the fractions and percentages of neurons that were GFP positive relative to the total number of neurons counted for 1, 7, and 38 days postinfection. Table 2 also gives an indication of the variability possible from field to field. However, fields such as those shown in Fig. 2 are most often seen and are fairly representative of the average number of GFP-positive neurons. The results in Table 2 show that the proportion of neurons expressing the GFP transgene from the d109 genome is greater than that seen in a number of cultured cell lines (Table 1). It also far exceeds that seen in diploid human fibroblasts (Table 1). Therefore, while cell lines with the more transformed nature (U2OS, 293, HeLa) seem to be more permissive for expression from d109 and cell strains such as WS1 and HEL are less permissive, the primary diploid TG neuron is surprisingly more permissive.
TABLE 2.
Expression of the GFP transgene from d109 in neurons over timea
| Day postinfection | No. of GFP+ neurons/total no. of neurons (%) | Range (%)b |
|---|---|---|
| 1 | 100/679 (14.7) | 9.0-23 |
| 7 | 92/513 (18) | 4.2-40 |
| 38 | 10/234 (4.3) | 3.8-5.0 |
Cultures of embryonic TG were infected with d109 and photographed over time.
The ranges for numbers of GFP+ neurons over numbers of total neurons (in percentages) for each counted field are given.
Derepression of d109 genomes: ICP0 versus TSA.
The previous studies provide insight into the quiescent state of d109 genomes in a number of cell types, including primary TG neurons using GFP expression from the HCMV promoter as a surrogate indicator of nonrepressed genomes. The following studies compare the abilities of two agents known to reverse cell-mediated repression of gene expression, ICP0 and TSA, to alter the quiescent state as a function of cell type.
Considering that ICP0 is sufficient to reactivate quiescent genomes and that its expression in the absence of other HSV IE genes reportedly mimics the effects of the histone deacetylase inhibitor TSA, we sought to determine if TSA, like ICP0, could also reactivate quiescent d109 in Vero cells. Vero cells were infected with d109 and then treated with various amounts of TSA at 1 or 7 days postinfection (Fig. 4). At early times postinfection, TSA-induced derepression was potent, with only small amounts of TSA being required to produce significant derepression. Once repression was established, however, the derepressive capabilities of TSA diminished, and drastically increasing the amount of TSA resulted in only marginal GFP expression. The amounts of TSA required to derepress the majority of quiescent d109 genomes at 1 week postinfection were in excess of 10-fold that required to achieve similar levels of derepression observed at 24 h postinfection. In contrast, time played no role in the derepressive capabilities of ICP0. Addition of ICP0 via d105 superinfection efficiently derepressed d109 at both early and late times postinfection (Fig. 4). Derepression mediated by ICP0 at 7 days postinfection was marginally reduced from that observed at 24 hours postinfection but was still significantly more robust than derepression induced by TSA at the same time postinfection.
To extend these studies to neuronal cells, embryonic TG cultures were infected with d109 and then were either superinfected with d105 or treated with TSA at the times postinfection indicated below. The reactivation patterns of cultures to either superinfection with d105 or treatment with TSA varied both in magnitude and with respect to the affected cell types within the culture (Fig. 5). When the derepressive stimuli were added 24 hours postinfection, the overall effect was quite dramatic (Fig. 5C through F). Superinfection with d105 (ICP0) resulted in extensive GFP expression (Fig. 5C and D) such that the cultures compare quite closely to those directly infected with d106, while treatment of cultures with TSA (Fig. 5E and F) resulted in a more limited increase in transgene expression. Upon closer inspection, however, there appears to be a subtle difference in cell populations derepressed by ICP0 or TSA. Overall, derepression induced by d105 (ICP0) was widespread, affecting nearly every cell in the monolayer. In comparison, the cell subpopulation derepressed following TSA treatment appeared to be skewed, with more neurons than support cells being induced to express the transgene.
Though the cell type preferences of ICP0 and TSA appear somewhat subtle when reactivation stimuli are added 24 hours postinfection, these differences are more pronounced and easily discernible when quiescence is more firmly established. Figure 6 illustrates the effects of superinfection with d105 or treatment with TSA at 22 days postinfection with d109. Much like with the results observed at 24 hours postinfection, d105 (ICP0) induced extensive activation of the transgene in the vast majority of cells. In contrast to that observed 24 hours postinfection, TSA-induced derepression of d109 genomes occurred almost exclusively in neurons at 22 days postinfection. It should be noted that in Vero cells, larger amounts of TSA were required to activate GFP expression at longer times postinfection (Fig. 4). It may be that GFP could be activated in more nonneuronal cells in these TG cultures with larger amounts of TSA.
When ICP0 was provided by infection with d105, activation of the GFP transgene from d109 was widespread, regardless of cell type (Vero cell, neuron, support cell). TSA efficiently activated GFP expression in Vero cells early after infection and in neurons. TSA also very poorly derepresses gene expression in HEL cells, even when added very early after infection (unpublished observations). Therefore, while the ability of ICP0 to activate the transgene appears to be ubiquitous, the ability of TSA to activate the quiescent GFP gene is cell type dependent and may reflect the trend seen in the previous section. The cell types in which more cells express the transgene from the d109 genome are more easily derepressed by TSA. Perhaps this suggests that in cell types such as HEL cells, there exists additional, or different, barriers to gene expression than histone deacetylation.
Rescue of quiescent and latent virus as a function of ICP0.
The previous two sections examine expression from quiescent virus as a function of ICP0, TSA, and time using GFP driven off the HCMV promoter as a marker for derepressed genomes. The HCMV promoter possesses a strong enhancer, so that as long as the genomes are not repressed, it will be highly active in the absence of viral activators, such as ICP4 and VP16. However, it is of interest to know how the HSV genes are expressed in such experiments. While small levels of HSV gene expression can be observed in the absence of ICP4 when ICP0 is present or when any other condition operationally substitutes for ICP0 (40, 80), we sought to examine the question of overall genome activity by assaying the rescue of viral genomes in both nonneuronal (Vero) and neuronal (latently infected TG) cells.
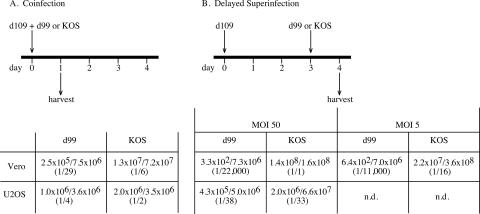
We first examined the efficiency of rescue of d109 genomes in Vero and U2OS cells by superinfection as a function of ICP0 and if this efficiency was affected by time. U2OS cells can operationally substitute for ICP0 (94). Vero or U2OS cells were infected with d109 and either (i) coinfected with either wild-type KOS or d99, an HSV-1 mutant which expresses the full complement of HSV IE genes except ICP0, or (ii) superinfected at a later time (3 days later) via superinfection with either KOS or d99. In either case, virus suspensions were prepared from cultures harvested 1 day after coinfection or superinfection and plated on F06 cells for the formation of PFU. The ability to rescue d109 genomes by complementation or recombination was determined by comparison of the ratios of green to white plaques. When KOS or d99 was added coincident to d109, rescue of d109 genomes was significantly reduced in the absence of ICP0 (Fig. 7A). Coinfection of Vero cells with d109 and KOS resulted in approximately 16% of plaques (1 of every 6) expressing GFP, whereas coinfection of Vero cells with d109 and d99 resulted in approximately 3% (1 of every 29) expressing GFP. As was expected, coinfection of U2OS cells with d109 and either d99 or KOS resulted in similar ratios of green to white plaques. When d109-infected cells were superinfected with KOS or d99 3 days after d109 infection, rescue of d109 was drastically impaired in the absence of ICP0 (Fig. 7B). When d109-infected cells were superinfected with KOS 3 days postinfection, the ratio of green to white cells was marginally less (1/16) than that observed during the d109/KOS coinfection (1/6). However, superinfection with d99 3 days postinfection resulted in a precipitous drop in green plaques (1/11,000) compared to that observed during the d109/d99 coinfection (1/29). This dramatic decrease in rescued virus could not be overcome by increasing the superinfecting input of d99. When the inputs of d99 or KOS 3 days postinfection were increased to an MOI of 50, there was an increase in the proportion of rescued (green) plaques when KOS was the superinfecting virus (1/1) but not when d99 was the superinfecting virus (1/22,000) (Fig. 7B).
FIG. 7.
Rescue of d109 genomes as a function of time, the presence of ICP0, and cell type. (A). Vero and U20S cells were coinfected with d109 and d99 or d109 and KOS. The MOI for d109 was 5 PFU/cell. Twenty-four hours after the coinfection, monolayers were harvested and the titers of the resulting progeny were determined on FO6 cells. Reported ratios represent the numbers of fluorescent plaques over the total number of plaques on FO6 cells. (B) The experiment was carried out as described for panel A except that the d109-infected cells were superinfected with KOS or d99 3 days after the d109 infection instead of being coinfected.
The rescue of d109 genomes by superinfection with d99 most likely represents the ability of the other viral regulators of gene expression to function in trans on the d109 genome. The decrease in the ability of d99 to rescue d109 from day 1 to day 3 likely represents a change in availability of d109 to the trans-acting activators supplied by d99. Interestingly, this change is also reflected in the decrease in the efficiency of the TSA-mediated activation of GFP expression from d109 (Fig. 4).
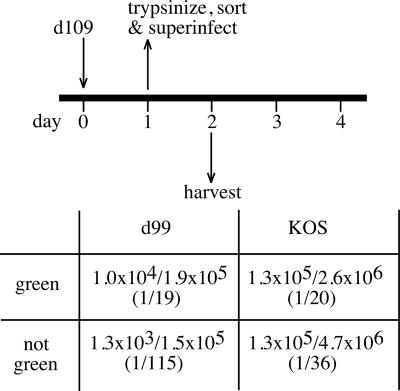
For the results depicted in Fig. 7, it should be noted that in both the 1-day coinfection and 3-day superinfection with d99, a small amount of green plaques resulted, indicative of a small amount of rescue in the absence of ICP0. d109 genomes are usually repressed in cells; however, the subpopulation of green d109-infected cells could possibly have been due to these cells existing in a derepressed state. The fraction of green d109-infected Vero cells decreases over time (78). Therefore, the subpopulation of d109-infected cells constitutively expressing GFP might be more easily “reactivated” than d109-infected cells not expressing the transgene. In other words, for d109-infected cultures, where virtually all cells harbor d109 genomes, we hypothesized that when exposed to reactivating stimuli in the absence of ICP0, the green cells would be more likely to reactivate than the white cells. To this end, Vero cells were infected with d109 and then trypsinized, sorted by color, and superinfected with either KOS or d99. The cells were then harvested again and plated on F06 cells, where the rescue of d109 was assessed by counting resulting green and white plaques. Results presented in Fig. 8 indicate that when d99 was used as the superinfecting virus, green cells produced more green plaques (1/19) than when white cells were used as the starting point (1/115). As predicted, KOS was able to “reactivate,” or rescue, d109 from both green and white cells with equal efficiencies. Therefore, we hypothesize that the green cells represent d109 genomes that are not as repressed as the white cells and therefore can be “reactivated” by superinfection in the absence of ICP0.
FIG. 8.
Rescue of d109 from green and white infected cells. Vero cells were infected with d109, trypsinized, and sorted with respect to GFP expression. The green and white populations were each then superinfected with either d99 or KOS. Monolayers were harvested 24 hours after the superinfection, and the titers of the resulting progeny were determined on FO6 cells. Reported ratios represent the numbers of fluorescent plaques over the total number of plaques on FO6 cells.
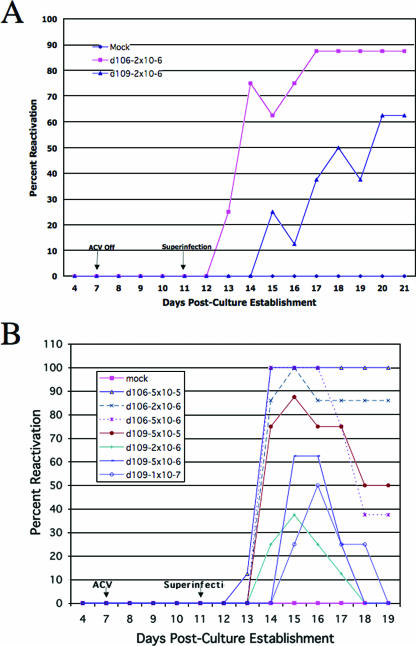
The previous studies demonstrate that there appears to be a relatively large proportion of d109-infected TG neurons that express GFP and that some of these persist for more than a month (Fig. 3). In addition, d109-infected Vero cells that express GFP were more easily reactivated by superinfection with d99 than were white neurons. If these GFP-expressing d109-infected cells truly represent derepressed genomes that are more accessible to transacting factors, then a significant proportion of wild-type virus present in latently infected TG neurons may be susceptible to reactivation via superinfection in the absence of ICP0. To test this hypothesis, d109 and d106 were used to superinfect cultures of TG established from adult mice latently infected with wild-type KOS. Thirty days postinfection, TG were isolated, dissociated, and used to establish cultures. Analysis of culture media for infectious virus following superinfection revealed that superinfection of latent cultures with d106 induced reactivation of wild-type KOS in 87% of the TG cultures (Fig. 9A). Superinfection with d109 reactivated latent KOS genomes in more than 60% of the cultures (Fig. 9A). To determine whether the ability of d109 to reactivate latent KOS was dose dependent, a dose-response experiment was conducted. It was performed as described above but this time using a range of reactivating doses of both d106 and d109. As observed in Fig. 9B, no clear dose response could be established with respect to reactivation induced by d109. In fact, the amount of reactivation observed appeared to increase with decreasing amounts of superinfecting d109. For instance, d109 induced greater reactivation at an inoculum of 5 × 105 PFU/well than when the inoculum was increased 10-fold. Therefore, consistent with the observation that there may be a considerable number of viral genomes in TG neurons that are not as repressed as standard tissue culture lines, as indicated by the proportion of GFF-expressing, d109-infected cells, latent wild-type virus resident in TG neurons is readily reactivated by superinfection in the absence of ICP0.
FIG. 9.
Reactivation of latent KOS by d109 and d106. TG cultures were made from mice latently infected (30 days postinfection) with HSV-1 (KOS). Upon establishment, cultures were superinfected with one (A) or several (B) doses of either d106 or d109. On days 4 and 7 to 21, culture supernatants were assayed for the presence of infectious wild-type virus. ACV, acyclovir.
DISCUSSION
We have examined the expression characteristics of a previously described, HSV IE early gene mutant incapable of expressing any of the five IE genes both in nonneuronal, cultured cell lines and in primary cultures of dissociated TG. The major findings of the work presented here specifically illustrate how cell type and possibly the state of the cell affect the expressiblity of viral genomes in the absence of IE proteins. These results confirm the significant contribution of ICP0 in counteracting the latent or quiescent state but also reveal ICP0-independent pathways of reactivation or cellular derepression. We also provide support for previous findings that repression is a gradual process, that a component of the virion, possibly VP16, is sufficient to reactivate latent KOS in dissociated ganglion cultures, and that TSA reactivates viral genomes in subpopulations of cultured epithelial cells and preferentially in neurons in TG cultures.
d109 expression in neural versus nonneural cells.
The impaired virus used in these studies, d109, served as a sentinel of sorts for the identification of cells capable of expressing genes from its severely impaired genome, from which no IE proteins are expressed. Consistently with its initial characterization in Vero cells, we found that d109 genomes were repressed in most of the nonneuronal cell lines tested here, save for a derepressed subpopulation which exhibited constitutive GFP expression. The size of this derepressed subpopulation was variable, cell type dependent, and most abundant in U2OS cells, which express an activity analogous to ICP0 (94). 293 cells were the second-most-permissive cell line tested for d109-driven GFP expression, which is particularly interesting because 293 cells, an adenovirus-transformed cell line derived from human embryonic kidney cells, were originally isolated and characterized as having an epithelial-cell-type morphology but have actually been shown to be of neuronal lineage (82). The ability of d109 to produce significant, constitutive GFP expression in this cell population in the absence of IE gene expression complemented our findings in neuronal cultures.
In the dissociated cultures of primary neurons, GFP expression from d109 genomes varied in intensity especially in neurons. Given the many different types of cells and subtypes of neurons within primary cultures of TG, it was somewhat surprising that upon infection with d109, neurons primarily served as the derepressed population. The constitutive expression of GFP preferentially in neurons presents a twofold paradox: (i) latency via genome repression is typically the invariant consequence of abortive infections where IE genes are silenced (75), and (ii) neurons are thought to be the sites of latency because of their naturally repressive environment. While neurons almost exclusively comprised the derepressed population, not all neurons were derepressed. The idea of neurons differing in their abilities to repress viral genomes is consistent with previous reports concerning the ability of neuronal subtypes to differentially activate certain IE promoters (54, 55, 59, 67), differentially splice latency-associated transcripts and ICP0 transcripts (59), and preferentially support either lytic or latent infection (28, 60, 61, 93). As such, functional differences between neuronal subtypes may make some neuron populations more or less relevant to HSV latency or productive infection. The present studies do not address this possibility; therefore, this is an area for further study.
Significance of GFP expression.
Our working hypothesis to explain constitutive ICP0-independent GFP expression, and thus the reason d109 infection is not totally silent, is that the said cells (green cells) exist in a relaxed or derepressed state in which the need for viral proteins, specifically ICP0, to maintain a transcriptionally active genome is reduced. To determine this, we tested whether reactivation efficiency in the presence or absence of ICP0 segregated with the green or white phenotype in d109-infected cells. Consistently with our hypothesis, in the absence of ICP0, not only did green (derepressed) cells reactivate at a significantly higher frequency than did their white (repressed) counterparts, they did so at a rate nearly identical to that observed with wild-type KOS. Moreover, while the addition of ICP0 significantly increased the reactivation efficiency of white, repressed cells, no such improvement was observed in green, derepressed cells. The nonadditive effect of ICP0 in derepressed cells demonstrates that the state of the cell supporting reactivation can compensate for or at least greatly influence the repertoire of viral genes required for reactivation. Alternatively, this state of derepression may be mechanistically identical to or may utilize pathways or portions of pathways redundant to ICP0 function. Whether this derepressed or relaxed state is induced by the presence of an ICP0-like protein or the absence of a requisite repressor function is unknown at present.
The mechanism(s) of action for ICP0 is also unclear, as ICP0 could serve to induce transcription from the viral genome directly, or alternatively, it could inactivate or target for degradation a cellular repressor. Work by Hancock et al. (33) showed, at least with respect to HEL cells, that lack of permissiveness is a function of active repressors. The presence or absence of repressors in derepressed neuron populations has yet to be determined. What is clear from results presented here and by others is that ICP0 can induce a derepressed or relaxed state sufficient to support viral gene expression and ultimately reactivation (31, 32). What is clear from results presented here is that significant numbers of cells exist in this state in its absence. We have also shown that in cultures of dissociated ganglia, neurons predominate the derepressed population and that in these derepressed cells, the viral regulatory genes requisite for genomic expression are obviously altered and do not include ICP0. Consistent with this interpretation are reports that ICP0 may not be required for initiation of reactivation in vivo (13, 53, 85).
TSA activation.
Histone deacetylase inhibitors such as TSA have previously been shown to reactivate HSV genes (2, 15). Overall, TSA reactivated GFP expression from quiescent d109 genomes in substantial numbers of cells, whether cultured cells or cells from primary TG cultures. This portion of reactivated cells varied in size, dependent on the time postinfection when TSA was added. When added shortly after infection, its effects were far more substantial. This decreased efficiency of TSA-mediated reactivation as a function of time was likely caused by global repression of quiescent d109 genomes. In keeping with observations of others that ICP0 is required for efficient reactivation (32, 66), the ability of ICP0 to reactivate quiescent d109 genomes was widespread and not dependent on the timing of the reactivation stimulus postinfection.
In primary cultures of dissociated TG, however, TSA reactivation of GFP expression was substantial at early times postinfection, with equal numbers of support cells and neurons expressing GFP. At late times postinfection, it was primarily neurons that responded to TSA. This last observation is similar to that previously reported (2). Thus, again at early times after d109 infection, the viral genomes are not as repressed in all cell types as they are in most cells as time advances. In addition, the enhanced ability to activate d109 genomes exclusively in neurons at late times may reflect the same mechanisms that allow d109 to be exclusively expressed in neurons in the absence of ICP0 or TSA. The differences between TSA- and ICP0-mediated reactivation indicate that there are different pathways to reactivation or, at the very least, suggest that histone acetylation is but one step in the pathway to reactivation.
KOS reactivation.
Input d109 genomes were able to reactivate latent KOS from adult trigeminal neurons. The efficiency was not as great as that for d106 but was significant. Since d109 dos not express any viral proteins, the reactivation must be a function of a virion component(s). Thus, this could be a result of VP16 in the superinfecting d109 particles. While other possibilities formally exist, we note that the VP16 activation domain has been shown to allow activators to gain access to the DNA in a chromatinized genome (7) and that the viral genome is packaged in nucleosomes during latency (18). It has been shown that VP16 expressed from adenovirus can reactivate latent HSV; however, an ICP0-deficient HSV very poorly reactivated latent HSV (32). It has also been reported that VP16 and ICP0 overcome an innate nuclear barrier to viral gene expression (33). It is consistent with our studies described above that VP16 may activate “latent HSV” if it is present in an infected cell where the viral genome is not repressed, for example, as with the population of “green cells” described above. ICP0 simply puts all the cells in this state such that productive-state activators of gene expression may more efficiently function.
With d109, the lack of a dose response in KOS reactivation could be a function of a struggle between reactivation and an antiviral interferon response induced by d109 (21). Such a response could offset the induction of productive cycle gene expression mediated by VP16 and explain why increases in the input of d109 were not directly proportional to reactivation rates, which were most efficient at lower inputs. ICP0 mediates repression of the interferon response (21, 69). Therefore, at higher d109 inocula, interferon induction and the resultant antiviral state of the cell could attenuate the reactivation process. Consistently with this possibility, wells reactivated with d109 showed recovery, but those mediated by d106 did not. Decreases in reactivation efficiency mediated by d106 were due to total destruction of the reactivated monolayer, not recovery.
In conclusion, the existence of derepressed neurons could theoretically provide an environment wherein reactivation is much more likely, possibly creating a reservoir for virus production and shedding. As such, these derepressed cells are most likely sites of spontaneous reactivation or are first responders to reactivation stimuli. With different types of nervous tissue being more or less ideal for HSV gene expression, neuronal permissiveness is much more dynamic than initially thought, thus making the presence or role of derepressed neurons even more critical in discussions of latency and reactivation. Results presented here not only are consistent with those concerned with further defining the role of ICP0 in reactivation and latency (65, 85) but also provide support for the growing hypothesis that reactivation is restricted to a select group of neurons (14, 59).
Acknowledgments
This work was supported by NIH grant AI44812.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Arthur, J. L., R. Everett, I. Brierley, and S. Efstathiou. 1998. Disruption of the 5′ and 3′ splice sites flanking the major latency-associated transcripts of herpes simplex virus type 1: evidence for alternate splicing in lytic and latent infections. J. Gen. Virol. 79:107-116. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, J. L., C. G. Scarpini, V. Connor, R. H. Lachmann, A. M. Tolkovsky, and S. Efstathiou. 2001. Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro. J. Virol. 75:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blagosklonny, M. V., R. Robey, D. L. Sackett, L. Du, F. Traganos, Z. Darzynkiewicz, T. Fojo, and S. E. Bates. 2002. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 1:937-941. [PubMed] [Google Scholar]
- 4.Block, T. M., S. Deshmane, J. Masonis, J. Maggioncalda, T. Valyi-Nagi, and N. W. Fraser. 1993. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology 192:618-630. [DOI] [PubMed] [Google Scholar]
- 5.Block, T. M., J. G. Spivack, I. Steiner, S. Deshmane, M. T. McIntosh, R. P. Lirette, and N. W. Fraser. 1990. A herpes simplex virus type 1 latency-associated transcript mutant reactivates with normal kinetics from latent infection. J. Virol. 64:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunker, C. A., and R. E. Kingston. 1996. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc. Natl. Acad. Sci. USA 93:10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X., J. Li, M. Mata, J. Goss, D. Wolfe, J. C. Glorioso, and D. J. Fink. 2000. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J. Virol. 74:10132-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements, G. B., and N. D. Stow. 1989. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latency-competent in mice. J. Gen. Virol. 70:2501-2506. [DOI] [PubMed] [Google Scholar]
- 14.Danaher, R. J., R. J. Jacob, and C. S. Miller. 2006. Reactivation from quiescence does not coincide with a global induction of herpes simplex virus type 1 transactivators. Virus Genes 33:163-167. [DOI] [PubMed] [Google Scholar]
- 15.Danaher, R. J., R. J. Jacob, M. R. Steiner, W. R. Allen, J. M. Hill, and C. S. Miller. 2005. Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J. Neurovirol. 11:306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ecob-Prince, M. S., C. M. Preston, F. J. Rixon, K. Hassan, and P. G. Kennedy. 1993. Neurons containing latency-associated transcripts are numerous and widespread in dorsal root ganglia following footpad inoculation of mice with herpes simplex virus type 1 mutant in1814. J. Gen. Virol. 74:985-994. [DOI] [PubMed] [Google Scholar]
- 21.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everett, R., P. O'Hare, D. O'Rourke, P. Barlow, and A. Orr. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 69:7339-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 24.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feldman, L. T., A. R. Ellison, C. C. Voytek, L. Yang, P. Krause, and T. P. Margolis. 2002. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc. Natl. Acad. Sci. USA 99:978-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, H., and B. Roizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halford, W. P., B. M. Gebhardt, and D. J. Carr. 1996. Mechanisms of herpes simplex virus type 1 reactivation. J. Virol. 70:5051-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halford, W. P., C. D. Kemp, J. A. Isler, D. J. Davido, and P. A. Schaffer. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 75:6143-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock, M. H., J. A. Corcoran, and J. R. Smiley. 2006. Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology 352:237-252. [DOI] [PubMed] [Google Scholar]
- 34.Harle, P., B. Sainz, Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293:295-304. [DOI] [PubMed] [Google Scholar]
- 35.Ho, D. Y., and E. S. Mocarski. 1989. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc. Natl. Acad. Sci. USA 86:7596-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hobbs, W. E., II, and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imbalzano, A. N., D. M. Coen, and N. A. DeLuca. 1991. Herpes simplex virus transactivator ICP4 operationally substitutes for the cellular transcription factor Sp1 for efficient expression of the viral thymidine kinase gene. J. Virol. 65:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javier, R. T., J. G. Stevens, V. B. Dissette, and E. K. Wagner. 1988. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology 166:254-257. [DOI] [PubMed] [Google Scholar]
- 43.Jordan, R., and P. A. Schaffer. 1997. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J. Virol. 71:6850-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju, R., and M. T. Muller. 2003. Histone deacetylase inhibitors activate p21(WAF1) expression via ATM. Cancer Res. 63:2891-2897. [PubMed] [Google Scholar]
- 45.Kang, W., R. Mukerjee, and N. W. Fraser. 2003. Establishment and maintenance of HSV latent infection is mediated through correct splicing of the LAT primary transcript. Virology 312:233-244. [DOI] [PubMed] [Google Scholar]
- 46.Kaufman, R. H. 1986. Clinical features of herpes genitalis. J. Reprod. Med. 31:379-383. [PubMed] [Google Scholar]
- 47.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 50.Kramer, M. F., S. H. Chen, D. M. Knipe, and D. M. Coen. 1998. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J. Virol. 72:1177-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kramer, M. F., and D. M. Coen. 1995. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 69:1389-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krause, P. R., L. R. Stanberry, N. Bourne, B. Connelly, J. F. Kurawadwala, A. Patel, and S. E. Straus. 1995. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J. Exp. Med. 181:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loiacono, C. M., R. Myers, and W. J. Mitchell. 2004. The herpes simplex virus type 1 early gene (thymidine kinase) promoter is activated in neurons of brain, but not trigeminal ganglia, of transgenic mice in the absence of viral proteins. J. Neurovirol. 10:116-122. [DOI] [PubMed] [Google Scholar]
- 55.Loiacono, C. M., R. Myers, and W. J. Mitchell. 2002. Neurons differentially activate the herpes simplex virus type 1 immediate-early gene ICP0 and ICP27 promoters in transgenic mice. J. Virol. 76:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 58.Maggioncalda, J., A. Mehta, Y. H. Su, N. W. Fraser, and T. M. Block. 1996. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology 225:72-81. [DOI] [PubMed] [Google Scholar]
- 59.Maillet, S., T. Naas, S. Crepin, A. M. Roque-Afonso, F. Lafay, S. Efstathiou, and M. Labetoulle. 2006. Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts. J. Virol. 80:9310-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Margolis, T. P., C. R. Dawson, and J. H. LaVail. 1992. Herpes simplex viral infection of the mouse trigeminal ganglion. Immunohistochemical analysis of cell populations. Investig. Ophthalmol. Vis. Sci. 33:259-267. [PubMed] [Google Scholar]
- 61.Margolis, T. P., F. Sedarati, A. T. Dobson, L. T. Feldman, and J. G. Stevens. 1992. Pathways of viral gene expression during acute neuronal infection with HSV-1. Virology 189:150-160. [DOI] [PubMed] [Google Scholar]
- 62.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 63.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 64.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller, C. S., R. J. Danaher, and R. J. Jacob. 2006. ICP0 is not required for efficient stress-induced reactivation of herpes simplex virus type 1 from cultured quiescently infected neuronal cells. J. Virol. 80:3360-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minaker, R. L., K. L. Mossman, and J. R. Smiley. 2005. Functional inaccessibility of quiescent herpes simplex virus genomes. Virol. J. 2:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitchell, W. J. 1995. Neurons differentially control expression of a herpes simplex virus type 1 immediate-early promoter in transgenic mice. J. Virol. 69:7942-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nichol, P. F., J. Y. Chang, E. M. Johnson, Jr., and P. D. Olivo. 1996. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J. Virol. 70:5476-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perng, G. C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perng, G. C., C. Jones, J. Ciacci-Zanella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 74.Perng, G. C., S. M. Slanina, A. Yukht, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J. Virol. 74:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 76.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sedarati, F., K. M. Izumi, E. K. Wagner, and J. G. Stevens. 1989. Herpes simplex virus type 1 latency-associated transcription plays no role in establishment or maintenance of a latent infection in murine sensory neurons. J. Virol. 63:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869-871. [DOI] [PubMed] [Google Scholar]
- 83.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 84.Tal-Singer, R., T. M. Lasner, W. Podrzucki, A. Skokotas, J. J. Leary, S. L. Berger, and N. W. Fraser. 1997. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 71:5268-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thompson, R. L., and N. M. Sawtell. 2006. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J. Virol. 80:10919-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICPO. J. Virol. 75:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wald, A., J. Zeh, S. Selke, R. L. Ashley, and L. Corey. 1995. Virologic characteristics of subclinical and symptomatic genital herpes infections. N. Engl. J. Med. 333:770-775. [DOI] [PubMed] [Google Scholar]
- 90.Wald, A., J. Zeh, S. Selke, T. Warren, A. J. Ryncarz, R. Ashley, J. N. Krieger, and L. Corey. 2000. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N. Engl. J. Med. 342:844-850. [DOI] [PubMed] [Google Scholar]
- 91.Wolff, M. H., J. Schmitt, M. Rahaus, H. Dudda, and W. Hatzmann. 2002. Clinical and subclinical reactivation of genital herpes virus. Intervirology 45:20-23. [DOI] [PubMed] [Google Scholar]
- 92.Yamashita, Y., M. Shimada, N. Harimoto, T. Rikimaru, K. Shirabe, S. Tanaka, and K. Sugimachi. 2003. Histone deacetylase inhibitor trichostatin A induces cell-cycle arrest/apoptosis and hepatocyte differentiation in human hepatoma cells. Int. J. Cancer 103:572-576. [DOI] [PubMed] [Google Scholar]
- 93.Yang, L., C. C. Voytek, and T. P. Margolis. 2000. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 74:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoshida, M., and T. Beppu. 1988. Reversible arrest of proliferation of rat 3Y1 fibroblasts in both the G1 and G2 phases by trichostatin A. Exp. Cell Res. 177:122-131. [DOI] [PubMed] [Google Scholar]
- 96.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]