Abstract
The TRIM5 family of proteins contains a RING domain, one or two B boxes, and a coiled-coil domain. The TRIM5α isoform also encodes a C-terminal B30.2(SPRY) domain, differences within which define the breadth and potency of TRIM5α-mediated retroviral restriction. Because Macaca nemestrina animals are susceptible to some human immunodeficiency virus (HIV) isolates, we sought to determine if differences exist in the TRIM5 gene and transcripts of these animals. We identified a two-nucleotide deletion (Δ2) in the transcript at the 5′ terminus of exon 7 in all M. nemestrina TRIM5 cDNA clones examined. This frameshift results in a truncated protein of 300 amino acids lacking the B30.2(SPRY) domain, which we have named TRIM5θ. This deletion is likely due to a single nucleotide polymorphism that alters the 3′ splice site between intron 6 and exon 7. In some clones, a deletion of the entire 27-nucleotide exon 7 (Δexon7) resulted in the restoration of the TRIM5 open reading frame and the generation of another novel isoform, TRIM5η. There are 18 amino acid differences between M. nemestrina TRIM5η and Macaca mulatta TRIM5α, some of which are at or near locations previously shown to affect the breadth and potency of TRIM5α-mediated restriction. Infectivity assays performed on permissive CrFK cells stably transduced with TRIM5η or TRIM5θ show that these isoforms are incapable of restricting either HIV type 1 (HIV-1) or simian immunodeficiency virus infection. The expression of TRIM5 alleles incapable of restricting HIV-1 infection may contribute to the previously reported increased susceptibility of M. nemestrina to HIV-1 infection in vivo.
The human immunodeficiency virus (HIV) pandemic is the result of cross-species transmissions of simian immunodeficiency virus (SIV) from non-human primates to humans (28). The recent discovery of human T-cell leukemia virus types 3 and 4 suggests that cross-species transmission of retroviruses is not an infrequent occurrence (33). However, to establish a productive infection in a new species, it is necessary for retroviruses to evade host-specific restriction factors. A broad range of antiretroviral host restriction factors has been identified in mammalian species. The best-described primate host restriction factors are APOBEC3F/G and TRIM5α. APOBEC3F/G are cytidine deaminases which exert a late block to retroviral replication, inhibiting the virus in target cells rather than producer cells (29), whereas TRIM5α exerts a dominant block to infection immediately after viral entry into the cell through the inhibition of reverse transcription (30).
TRIM5α is a member of the tripartite motif family of proteins, also called RBCC proteins, because of the presence of a RING domain (C3HC4 type), one or two B boxes, and a coiled-coil region in an ordered arrangement from N terminus to C terminus (23). Six isoforms of TRIM5 have been identified in mammals (26). These proteins have been shown to form homo- and heteromultimers, although only homomultimers of TRIM5α have been shown to restrict lentiviruses (3, 8, 15, 30, 37).
The antiretroviral activity of TRIM5α is mediated through an interaction between the viral capsid and the B30.2(SPRY) domain (12, 20, 21, 31). The B30.2(SPRY) domain is the primary determinant of species-specific differences in the breadth and potency of TRIM5α antiretroviral activity. For instance, changing the arginine found in human TRIM5α at position 332 to the proline residue found in rhesus TRIM5α enables human TRIM5α to restrict HIV type 1 (HIV-1) nearly as efficiently as rhesus TRIM5α. Additionally, the ability of African green monkeys to restrict SIVmac has been mapped to a 37-amino-acid insertion in the B30.2(SPRY) domain (16, 36). Efficient binding of capsid-nucleocapsid complexes by TRIM5α requires coiled-coil-mediated trimerization, presumably to increase the avidity between the B30.2(SPRY) domain and capsid (10). Disruptions to the RING and B-box domains can also inhibit the antiviral activity of TRIM5α through currently undefined mechanisms (15, 22, 27).
Our laboratory has shown that Macaca nemestrina animals are susceptible to infection with a primary HIV-2 isolate, HIV-2/EHO (14, 25). M. nemestrina macaques have also been reported to be susceptible to some HIV-1 isolates (1, 4-7, 19) although this idea remains controversial. These observations led us to examine the TRIM5 gene of M. nemestrina for species-specific differences that may explain the apparent susceptibility of this macaque species to infection with HIV.
MATERIALS AND METHODS
Animals.
Fourteen outbred M. nemestrina animals bred at the Washington National Primate Research Center, University of Colorado, Tulane University, Yerkes Regional Primate Research Center, and Labs of Virginia were used in this study. Whole blood was collected from these animals in heparinized Vacutainer tubes in accordance with approved Institutional Animal Care and Use Committee protocols.
RNA and DNA samples.
The TRIM5 gene from 14 M. nemestrina animals (identified as A01150, A99030, J98259, K98171, A99024, J95306, A99029, A02105, J95315, J98316, J96082, A99216, J91216, and 97119) housed at the Washington National Primate Research Center was cloned and sequenced at genomic and cDNA levels. Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood by Lymphoprep density centrifugation. Total RNA was isolated from 1 × 107 PBMCs using an RNeasy Mini Kit (QIAGEN) following the manufacturer's suggested protocol. DNA was isolated from 5 × 106 PBMCs using a Puregene DNA purification kit (Gentra Systems) following the manufacturer's suggested protocol.
PCR and cloning. (i) cDNA amplification.
Primers designed for the amplification of the complete coding region of Macaca mulatta TRIM5α were modified to introduce unique restriction sites flanking the M. nemestrina TRIM5 gene as follows: forward, 5′-TCGACTAGATCTCTATGGCTTCTGGAATCCTGGTTAATGTAAAG-3′; reverse, 5′-ATATATGCGGCCGCTCAAGAGCTTGGTGAGCACAGAGTCATGGG-3′. Amplification of cDNA from total RNA was performed using SuperScript III One-Step reverse transcription-PCR (RT-PCR) with Platinum Taq kit (Invitrogen), a 0.2 μM concentration of each primer, and approximately 100 ng of total RNA from each macaque. The reaction mixture was incubated at 55°C for 30 min and then at 94°C for 2 min, followed by 40 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 90 s, with a hold at 68°C for 5 min and storage at 4°C. Amplification products were stored at −20°C prior to cloning.
(ii) Genomic DNA amplification.
Amplification of genomic DNA was performed from exon 5 to exon 8 with a forward primer complementing all of exon 5 (5′-GGTGTGGATGGCATCATTAAAAG) and the reverse primer described above. Each reaction mixture contained a final concentration of 1× PCR buffer (Roche), a 0.2 μM concentration of each deoxynucleoside triphosphate, a 0.2 μM concentration of each primer, 25 U of Taq DNA polymerase, and 100 ng of DNA from each macaque. The reaction mixture was incubated at 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min, with a hold at 72°C for 7 min and storage at 4°C. Amplification products were stored at −20°C prior to cloning. One microliter of undiluted PCR product from each amplification was cloned into the pSC-A vector using a Strataclone PCR cloning kit (Stratagene) following the manufacturer's suggested protocol. Plasmids were isolated from multiple individual colonies derived from each animal using a QIAprep Spin Miniprep Kit (QIAGEN) and stored at −20°C prior to sequencing.
(iii) Southern blotting.
Southern blot analysis was performed with a Detector AP Chemiluminescent Blotting Kit (KPL, Inc) following the manufacturer's protocol. Briefly, following agarose gel electrophoresis, DNA was depurinated in 0.25 N HCl, rinsed, denatured in 0.5 N NaOH-1.5 M NaCl, and then transferred to a Hybond-N+ nylon transfer membrane (Amersham Biosciences) using alkaline transfer buffer (0.75 M NaCl-75 mM sodium citrate-10 mM NaOH). After transfer, the membrane was blocked with sheared salmon sperm DNA and then probed with 50 ng/ml denatured, biotinylated rhesus TRIM5α probe generated with an NEBlot Phototope kit (New England Biolabs) following the manufacturer's protocol. After overnight incubation at 42°C, bound TRIM5α probe was detected by an alkaline phosphatase-streptavidin conjugate. TRIM5-specific sequences were visualized following reaction with CDP-Star chemiluminescent substrate.
Sequencing and data analysis. (i) cDNA sequencing.
Multiple TRIM5 cDNA clones derived from four animals were sequenced by the DNA Sequencing and Gene Analysis Center (Department of Pharmaceutics, University of Washington) using a BigDye 3.1 reaction kit (Applied Biosystems) on an Applied Biosystems 3100 genetic analyzer. Primers were designed to sequence the complete coding region, exons 2 through 8. The primer sequences are the following: M13R, 5′-CACACAGGAAACAGCTATG-3′; M13F, 5′-GTTGTAAAACGACGGCCAGTG-3′; T5F83.100, 5′-TGGACTGCGGCCACAGCT-3′; T5F356.375, 5′-GTGAGCGGCTCAGGAGCAC-3′; MneT5ex4F, 5′-CATCTCAGATCTGGAGCATC-3′; MneT5ex6F, 5′-TGAAAGGAATGCTAGACATG-3′; MneT5ex8F, 5′-CTGGCTCCAAACAACATTTC-3′; T5F1205.1224, 5′-GCTTCCAACCTGATGCAATG-3′; T5R217.198, 5′-GCCGATTAGGCCGTATGTTC-3′; MneT5ex4R, 5′-TCCTCCTTCTCCAGGTTCTG-3′; MneT5ex6R, 5′-TTGGCTTCTTCAAGGTCATG-3′; MneT5ex8R1, 5′-GAGCTCACTTGTCTCTTATC-3′; T5R1554.1535, 5′-TCAAGAGCTTGGTGAGCACA-3′. Sequencing products were assembled using Sequencher software, version 4.1 (Gene Codes). The resulting genetic and predicted amino acid sequences were analyzed using Vector NTI Advance, version 10.1.1 (Invitrogen).
(ii) Genomic sequencing.
Multiple genomic DNA clones from each animal were sequenced by the DNA Sequencing and Gene Analysis Center (Department of Pharmaceutics, University of Washington). Each clone was sequenced from exon 6 to exon 8 in both directions. The primers used were M13F, M13R, MneT5ex6R, MneT5ex6F, MneT5ex8R1, MneT5ex8F, 3TrmMCSex8Not1 (all as described above), MneT5Int5F2 (5′-CCTCTCTTGATATGTCTCAG-3′), and T5Aex5.1.23 (5′-GGTGTGGATGGCATCATTAAAAG-3′).
Sequencing was performed at the DNA Sequencing and Gene Analysis Center (Department of Pharmaceutics, University of Washington) and then analyzed as described above.
Stable transduction of CrFK Cells with TRIM5 genes.
CrFK and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-10% fetal bovine serum (FBS). Primers were used to introduce a unique 5′ XhoI site and a hemagglutinin (HA) epitope tag (XholHA5TRIM, 5′-CTAGATCTCGAGTACCCATATGACGTTCCAGACTACGCGGCTTCTGGAATCCTGGTTAATG TAAAG) and a unique 3′ NotI restriction site into the Homo sapiens and M. mulatta TRIM5α and M. nemestrina TRIM5η alleles (3TRIMNotI, 5′-ATCTAGGCGGCCGCTTAAGAGCTTGGTGAGCACAGAGTCATG) or the TRIM5θ allele (3pT5gNotI, 5′-ATCTAGGCGGCCGCTTAACCCAGCAGCGTCGGACATCTGTTA). The resulting amplification products were cloned into the pLPCX retroviral expression vector. 293T cells were used to produce retroviral vectors as previously described (2). Briefly, 293T cells were cotransfected with pLPCX containing the gene of interest, the murine leukemia virus Gag polymerase expression vector mGP, pL-VSV-G (where VSV-G is vesicular stomatitis virus G protein), and pCMV-tat (where CMV is cytomegalovirus). The resulting supernatants were used to infect CrFK cells, and 24 h after infection 4 μg/ml puromycin was added to the medium to select for cells stably transduced with the gene of interest.
Immunoblotting.
HA-tagged TRIM5 proteins expressed by stably transduced CrFK cells were detected by Western blotting of whole-cell lysates. Cells were lysed in Laemmli-sodium dodecyl sulfate (SDS) sample buffer (Bio-Rad). Samples were normalized by total protein concentration determined by optical density at 280 nm. SDS-polyacrylamide gel electrophoresis was performed with 4 to 12% NuPAGE denaturing gels (Invitrogen), and separated bands were transferred onto a nitrocellulose membrane (Bio-Rad). The primary antibody for detection of the HA tag was HA.11 (Covance, Berkeley, CA). Following the binding of primary antibody, the membrane was incubated with a goat anti-mouse alkaline phosphatase-conjugated secondary antibody (Sigma-Aldrich).
Flow cytometry and immunofluorescence assay (IFA).
Expression and localization of HA-tagged TRIM5 proteins inside the expressing cells were analyzed by staining with anti-HA AlexaFluor 488 conjugate (Invitrogen). Stably transduced CrFK cells were permeabilized using a BD Cytofix/Cytoperm Plus intracellular staining kit (Becton Dickinson). An aliquot of 106 cells was analyzed by flow cytometry with a FACScan instrument (Becton Dickinson) to enumerate cells expressing the HA-tagged TRIM5 protein. Another portion of cells was grown on glass slides, permeabilized, and stained as described above, and then cells were analyzed and photographed on an inverted microscope (Leica Microsystems, Switzerland) with a green fluorescence filter set.
Virus production.
Stocks of HIV-1 pNL4-3Δenv-eGFP (where eGFP is enhanced green fluorescent protein) and env-negative SIVmac239 eGFP were pseudotyped with VSV-G essentially as described previously (2). Briefly, 293T cells were transfected with a 2:1 ratio of backbone to VSV-G using polyethyleneimine as described previously (24). Three days posttransfection, the virus stock was concentrated 100-fold by ultracentrifugation in a Beckman SW41Ti rotor at 23,000 rpm for 1.5 h. Concentrated viral stocks were titrated by a reverse transcriptase assay (colorimetric) (Roche Applied Science), following the manufacturer's protocol.
Infectivity assays.
CrFK cells stably transduced with the TRIM5 genes of interest or empty vector were seeded in six-well culture plates in DMEM-10% FBS supplemented with 4 μg/ml puromycin at a density of 2 × 105 cells per well. After 24 h, the cells were washed in phosphate-buffered saline, and infected with threefold dilutions of VSV-G-pseudotyped HIV-1 pNL4-3 or SIVmac239 in 1 ml of DMEM-10% FBS supplemented with 5 μg/ml polybrene. Two hours postinfection, the virus-containing supernatant was replaced with 2 ml of DMEM-10% FBS. At 48 h postinfection, infected cells expressing GFP were enumerated by flow cytometric analysis using a FACScalibur (Becton Dickinson) and analyzed by FlowJo software (FlowJo).
Nucleotide sequence accession numbers.
Ten TRIM5 cDNA sequences representing both TRIM5η and TRIM5θ were deposited in the GenBank database from animal A01150 (accession numbers EU169825 and EU169826), animal A99030 (EU169827, EU169828, EU169829, and EU169830), animal J98259 (EU169831 and EU169832), and animal K98171 (EU169833 and EU169834). One genomic DNA sequence from each animal was also deposited (EU169835, EU169836, EU169837, and EU169838, respectively).
RESULTS
M. nemestrina expresses two novel TRIM5 isoforms, TRIM5η and TRIM5θ.
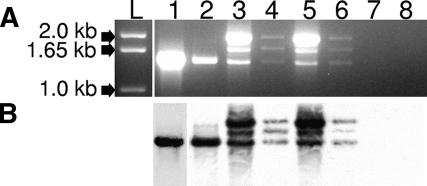
To evaluate species-specific differences in M. nemestrina TRIM5, we isolated total RNA from PBMCs and amplified the complete TRIM5 coding region from exon 2 to exon 8. A single 1.5-kb cDNA was amplified from H. sapiens (data not shown), M. mulatta, or Macaca fascicularis total RNA; however, three amplification products of approximately 1.5 kb, 1.7 kb, and 1.9 kb were consistently amplified from M. nemestrina (Fig. 1A). We confirmed that all three of these amplification products were TRIM5 derivatives by Southern blotting using a biotin-labeled probe derived from M. mulatta TRIM5α (Fig. 1B). Sequence analysis of the 1.7-kb amplification product showed that it was a splice intermediate retaining intron 7 (data not shown). The 1.9-kb amplification product was presumed to be a splicing intermediate and was not further characterized. Instead, we focused on the 1.5-kb amplification product, most likely representing the mature transcript.
FIG. 1.
(A) Amplification of M. nemestrina TRIM5 RNA. Total RNA from four M. nemestrina animals was isolated and amplified by RT-PCR using primers specific for exons 2 to 8 of the TRIM5 gene. Three amplification products of approximately 1.5 kb, 1.7 kb, and 1.9 kb were detected in all four M. nemestrina animals. Lane L, kilobase ladder; lane 1, M. mulatta TRIM5α; lane 2, M. fascicularis TRIM5α; lanes 3 to 6, respectively, amplification products from outbred M. nemestrina animals 97119, A99216, J91216, and J96082; lane 7, CrFK total RNA; lane 8, empty lane. (B) Southern blot analysis of RT-PCR amplification products described above. A biotin-labeled oligonucleotide specific for the M. mulatta TRIM5α sequence was used as the probe.
The 1.5-kb amplification product from four animals was gel purified, cloned, and sequenced. All 10 cDNA clones we sequenced had a 2-nucleotide deletion (Δ2) in their cDNAs between exon 6 and exon 7 (Fig. 2). This deletion in the cDNA may result from a deletion in the genomic DNA or from alternative splicing of the mRNA. These possibilities will be examined in the following section. The 2-nucleotide frameshift in the Δ2 transcript is predicted to truncate the protein at 300 amino acids, encoding only the TRIM domains, the linker region, and a 10-amino-acid “tail” that is the result of the frameshift in exon 7. We have named this isoform TRIM5θ (see Fig. 4A).
FIG. 2.
Alignment of the M. nemestrina and M. mulatta TRIM5 cDNA sequences. The top line shows the M. mulatta TRIM5α cDNA sequence with the junction between exon 6 and exon 7 indicated by the vertical line. The middle line shows the M. nemestrina Δ2 allele with the 2-nucleotide (AG) deletion between exons 6 and 7. The bottom line shows the Δexon7 allele of the M. nemestrina TRIM5 transcript. Residues present in M. mulatta but absent in M. nemestrina are indicated by dashes in the sequences.
FIG. 4.
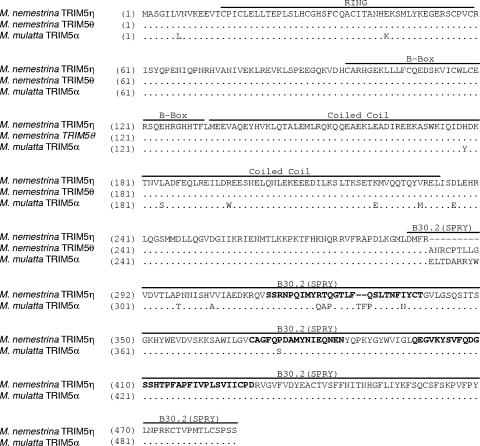
Alignment of the predicted amino acid sequences of M. nemestrina TRIM5η and TRIM5θ and M. mulatta TRIM5α. The RING domain, B box, coiled coil, and B30.2(SPRY) domains are indicated by labeled bars over the sequences. Variable regions in the B30.2(SPRY) domain are in bold.
In addition to the Δ2 transcript, we also identified a transcript missing all 27 nucleotides of exon 7 (Δexon7) in three of the four animals we examined (Fig. 2). The Δexon7 transcript restores the open reading frame that is disrupted by the Δ2 frameshift. This transcript was present in all four clones from A99030, one of two clones from J98259, and one of two clones from K98171. It is predicted to encode a 486-amino-acid protein that expresses the TRIM domains and the majority of the B30.2(SPRY) domain expressed by TRIM5α. We have named this novel isoform TRIM5η.
Transcription of TRIM5η and TRIM5θ is the result of a SNP at the intron 6 splice acceptor.
We did not identify any wild-type TRIM5α in the four M. nemestrina animals we analyzed. Therefore, we examined the M. nemestrina TRIM5 genomic sequence to determine whether the Δ2 transcript results from a frequent splice variant or whether the mutation is encoded at the genomic level. Genomic M. nemestrina TRIM5 sequences from exon 6 to exon 8 were cloned, analyzed, and aligned with the M. mulatta TRIM5 genomic sequence (http://www.hgsc.bcm.tmc.edu/projects/rmacaque/). This alignment revealed a change of G to T in all four M. nemestrina animals at the intron 6 splice acceptor site commonly used in M. mulatta (Fig. 3). Because the first two bases of exon 7 are also AG, the G-to-T single nucleotide polymorphism (SNP) in M. nemestrina is likely to cause the first two bases of exon 7 to be recognized as the preferred splice acceptor, resulting in the 2-nucleotide deletion found in TRIM5θ. Presumably, the use of a downstream splice acceptor results in the removal of exon 7 observed in TRIM5η. To confirm these results in a larger population, we performed the same analysis on an additional 10 M. nemestrina animals and identified the same G-to-T SNP in the canonical intron 6 splice acceptor in all animals (data not shown).
FIG. 3.
Alignment of the M. mulatta and M. nemestrina TRIM5 genomic sequences at the intron 6 and exon 7 junction. Lowercase letters designate intron sequences, and capital letters indicate exon sequences. The shaded box highlights the canonical M. mulatta intron 6 splice acceptor and the SNP (G to T) found in M. nemestrina. The open box highlights the downstream alternate splice acceptor used in M. nemestrina. The M. nemestrina sequence shown is present in all 14 animals examined. The M. mulatta sequence was obtained from online data from the Rhesus Macaque Genome Project at the Baylor College of Medicine at http://www.hgsc.bcm.tmc.edu/projects/rmacaque/.
M. nemestrina TRIM5 proteins have several amino acids that differ from the sequences in M. mulatta.
Alignment of the predicted protein sequences of M. nemestrina TRIM5η and TRIM5θ with M. mulatta TRIM5α identified 18 amino acid differences: 1 in the RING domain, 5 in the coiled coil, 2 in linker regions, and 10 in the B30.2(SPRY) domain (Fig. 4A). In addition, we identified 13 nonsynonymous amino acid polymorphisms among the individual M. nemestrina animals we examined.
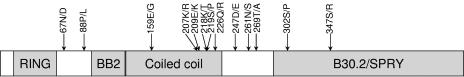
The lone amino acid difference in the RING domain, K44E, is directly downstream of a partially defective human allele, H43Y (27). In the coiled coil, amino acid differences between M. mulatta and M. nemestrina at Y178H, S184L, W196R, E222K, and M230V occur at potentially structurally important sites in this domain. Interestingly, only Y178H has not been reported as a site of intraspecies variation in M. mulatta. Of the remaining four sites, the major variant in M. nemestrina is the minor variant encoded by M. mulatta, and in the case of S184L and W196R it is also the minor variant encoded by Cercocebus atys. As reported in other primate species, M. nemestrina intraspecies polymorphisms tend to accumulate in the coiled coil, with 6 of the 13 variable positions occurring in this domain (Fig. 5). The intraspecies variations we have identified in M. nemestrina appear to occur at positions without significant variability in M. mulatta or C. atys (17).
FIG. 5.
Intraspecies polymorphisms in M. nemestrina TRIM5η. A diagram of the 486-amino-acid TRIM5η is shown. Functional domains are indicated by shaded boxes. Sites of nonsynonymous polymorphisms identified in four animals are indicated by arrows.
In the B30.2(SPRY) domain, most amino acid differences occur in the V1 region, including 323RTQ, corresponding to the important 332QAP motif in M. mulatta, and the two-amino-acid deletion (indicated by hyphens) 330- -Q, which is a minor species in M. mulatta (13). Amino acid changes in these regions have been shown to affect TRIM5α potency in other species (32, 36). Intriguingly, unlike other primate species, there are few intraspecies polymorphisms in the B30.2(SPRY) domain (Fig. 5). Only 298P and 330Q represent amino acid differences that correspond to identified minor variants in M. mulatta. Polymorphism has been observed at position 324 in M. mulatta; however, the threonine residue we found at this site in M. nemestrina has not been identified in M. mulatta. The potential effects of the additional B30.2(SPRY) domain differences T307P, A313V, N347I, and S385P on TRIM5-mediated restriction are not yet known. As previously noted, TRIM5η also lacks the 9 amino acids encoded by exon 7. In two other, unrelated proteins, GUSTAVUS and PSD, these amino acids are predicted to encode an N-terminal α-helix in the B30.2(SPRY) domain (34).
In vitro expression and intracellular localization of TRIM5 isoforms.
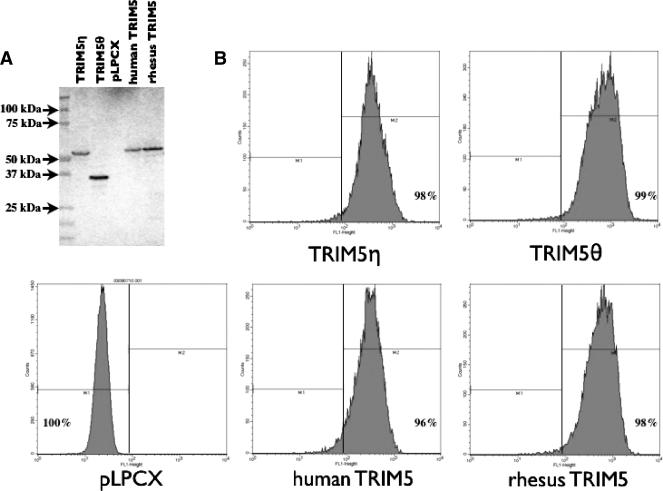
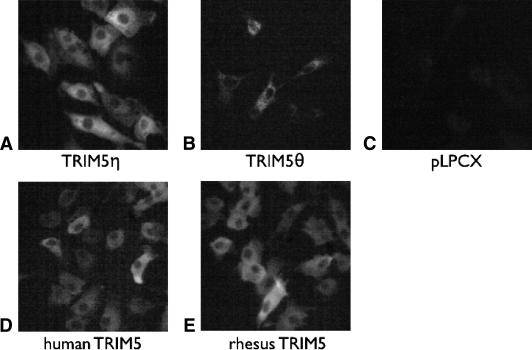
We generated CrFK cell lines stably transduced with either TRIM5η or TRIM5θ to investigate their biological properties. The expression level and size of each HA-tagged TRIM5 isoform in the newly established CrFK cell lines were confirmed by flow cytometric analysis, immunoblotting, and IFA using anti-HA monoclonal antibodies. To verify that the full-length TRIM5 proteins were expressed, cell lysates were analyzed by immunoblotting with an antibody that recognizes the HA tag. The results indicate that TRIM5η and TRIM5θ of the predicted sizes are expressed and that the expression levels are within two- to threefold of each other (Fig. 6A). As determined by flow cytometric analysis, almost 100% of the cells stably express TRIM5 molecules in comparison to empty vector-transduced cells (Fig. 6B). The intracellular localization of each TRIM5 isoform was visualized by IFA. As previously observed in other primate species, all of the TRIM5 isoforms were localized within the cytoplasm of stably expressing cells (Fig. 7), corresponding to the natural localization of TRIM5 proteins in vivo.
FIG. 6.
Expression of TRIM5 molecules by stably transformed CrFK cell lines. (A) Immunoblot analysis of HA-tagged TRIM5 proteins in whole-cell lysates. Lanes 2 to 6 correspond to TRIM5η, TRIM5θ, pLPCX empty vector, H. sapiens TRIM5α, and M. mulatta TRIM5α proteins, respectively. (B) Histograms of cells analyzed by flow cytometry with anti-HA monoclonal AlexaFluor conjugate 488. The percentage of cells expressing the HA-tagged TRIM5 protein is indicated in each panel.
FIG. 7.
The intracellular localization of HA-tagged TRIM5 proteins. HA-tagged TRIM5 proteins were visualized by staining fixed CrFK cells with anti-HA AlexaFluor conjugate 488. Expressed TRIM5 proteins are indicated below the panels. pLPCX, cells transduced with empty vector.
M. nemestrina TRIM5 isoforms do not restrict HIV-1 replication in vitro.
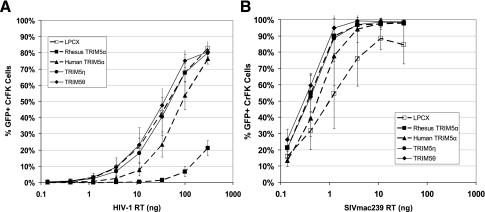
To assess the biological effect of TRIM5η and TRIM5θ on HIV-1 and SIVmac in vitro, we infected CrFK cells stably expressing TRIM5 isoforms with HIV-1 pNL4-3Δenv-eGFP or env-negative SIVmac239 eGFP and quantified GFP-positive CrFK cells 48 h postinfection. As expected, human TRIM5α-expressing CrFK cells restricted HIV-1 infectivity by approximately threefold, while rhesus TRIM5α-expressing CrFK cells restricted HIV-1 approximately 30-fold. As would be predicted by the absence of a B30.2(SPRY) domain, TRIM5θ does not restrict HIV-1 replication. We also determined that TRIM5η does not restrict HIV-1 infection when expressed in CrFK cells. This finding is somewhat surprising, as a previous study demonstrated that a chimeric human TRIM5α protein expressing M. nemestrina exon 8 was capable of restricting HIV-1 in vitro (18). As expected, none of the TRIM5 species we analyzed exerted an appreciable effect on SIVmac239 replication in vitro (Fig. 8).
FIG. 8.
Infectivity of HIV-1 and SIVmac in stably transduced CrFK cells. CrFK cells stably transduced with TRIM5 isoforms were infected with serial threefold dilutions of VSV-G-pseudotyped HIV-1 pNL4-3Δenv-eGFP (A) or env-negative SIVmac239 eGFP (B), normalized to reverse transcriptase (RT) input. The percentage of infected cells was determined by flow cytometric analysis of eGFP expression 48 h postinfection. Error bars represent the standard deviations of values obtained from three experiments.
DISCUSSION
In this paper we describe two novel isoforms, TRIM5η and TRIM5θ, and examine the basis of the Δ2 variant of TRIM5 in M. nemestrina. This is the first example of an Old World primate that does not express TRIM5α. Although TRIM5η expresses exon 8, which encodes the majority of the B30.2(SPRY) domain, this protein does not restrict HIV-1 replication in vitro. The discovery of these novel TRIM5 isoforms suggests the possibility of TRIM5 functional diversity even among closely related primate species.
The Δ2 variant of the TRIM5 transcript is predicted to encode a 300-amino-acid protein possessing the tripartite motif but lacking the B30.2(SPRY) domain necessary for lentiviral restriction. This Δ2 variant occurs rarely in M. mulatta (T. Kodama, unpublished data). The published M. mulatta genomic DNA sequence does not include the G-to-T mutation found in M. nemestrina (Fig. 3). Therefore, the M. mulatta Δ2 transcript is likely the result of a rare utilization of the AG motif at the 5′ end of exon 7 as an alternate splice site rather than the AG sequence at the 3′ end of intron 6. However, we detected only the Δ2 or Δexon7 variants in every M. nemestrina animal we examined. The presence of larger TRIM5 transcripts amplified from M. nemestrina total RNA suggests that incomplete or incorrectly processed transcripts accumulate to a greater degree in M. nemestrina than in M. mulatta or M. fascicularis. The SNP at the intron 6 splice acceptor may result in the preferential use of the alternative splicing site at the 5′ end of exon 7, leading to the transcription of either Δ2 or Δexon7. Because this SNP is encoded at the genomic level, it is unlikely that M. nemestrina is capable of expressing wild-type TRIM5α.
Multiple studies have demonstrated that the B30.2(SPRY) domain is necessary for association of TRIM5α with the retroviral capsid. Differences such as QAP332RTQ and TFP339- -Q in the variable regions of the TRIM5 B30.2(SPRY) domain have been shown to affect TRIM5α potency (13, 32, 36). However, an experiment by Ohkura and colleagues demonstrates that a chimera consisting of human TRIM5α exons 2 through 7 and M. nemestrina exon 8 is capable of restricting HIV-1, although less efficiently than M. mulatta TRIM5α (18). This result suggests that the inability of TRIM5η to restrict HIV-1 is not due to amino acid differences in the B30.2(SPRY) domain encoded by exon 8. Together, these observations indicate a potentially important role of exon 7 or amino acid polymorphism in the N-terminal portion of the protein.
The sole amino acid difference between the M. mulatta and M. nemestrina TRIM5 RING domain is K44E. The human allele H43Y eliminates the ability of human TRIM5α to restrict HIV-1, potentially by impacting E3 ubiquitin ligase activity (27). Because the H43Y allele impacts the restrictive capacity of TRIM5α without disrupting the Zn2+ coordinating motif, it is important to determine if K44E plays a similar role in M. nemestrina. Amino acid differences in the coiled coil, such as S184L, W196R, E222K, and M320V, may affect the structural stability of the coiled coil. However, the coiled coil is necessary but not sufficient for TRIM5 trimerization. Deletion of the region between the coiled coil and the B30.2(SPRY) domain, which includes the N-terminal portion of exon 7, also prevents trimerization of TRIM5α and binding to HIV-1 capsid (10). It is possible that these amino acid differences may contribute to the inability of TRIM5η to restrict HIV-1.
Crystallographic structures of the B30.2(SPRY) domain containing proteins GUSTAVUS and PSD reveal that this domain is composed of one or two N-terminal α-helices and 13 β-strands. The α-helices interact with and may stabilize the β-strands in a complex β-sandwich fold which forms a conformationally rigid pocket that is involved in protein-protein interactions. Sequence alignment of TRIM family proteins suggests that they adopt a structural conformation similar to PSD (34, 35). This TRIM5 N-terminal α-helix, predicted to be encoded primarily by exon 7, is absent in TRIM5η. It is possible that the loss of this potentially stabilizing α-helix alters the conformation of the β-sandwich, preventing TRIM5η from recognizing the HIV-1 capsid. Alternatively, amino acid differences in the N-terminal domains contribute to the inability of TRIM5η to restrict HIV-1 replication. It will be important for future studies to determine the contribution of amino acid differences in the RING domain, the coiled coil, and the B30.2(SPRY) domain to the defective restriction of HIV-1 by TRIM5η.
The lack of TRIM5α expression and the inability of TRIM5η or TRIM5θ to restrict HIV-1 may provide at least a partial explanation for the increased susceptibility of M. nemestrina to infection with HIV-1 isolates. Recently, HIV-1 constructs designed to evade M. mulatta TRIM5α- and APOBEC3G-mediated restriction by replacing the gag and vif genes with their SIVmac counterparts have been shown to productively infect M. mulatta lymphocytes (9, 11). It is possible that HIV-1 must only escape restriction by the APOBEC3 family of proteins in order to establish productive infection in M. nemestrina. However, it is also possible that these animals may express other, yet to be identified, TRIM transcripts, some of which may be capable of restricting HIV-1 to fill the role of TRIM5α.
Acknowledgments
We thank Joseph Sodroski for providing the rhesus TRIM5α-pLPCX plasmid, Welkin Johnson and Ruchi Newman for their help with the establishment of stably transduced CrFK cells, David Evans for providing the env-negative SIVmac239 eGFP plasmid, and Michael Emerman and Shari Kaiser for plasmids mGP, pL-VSV-G, pCMV-tat, and helpful discussions.
This work was supported by NIH grants P51 RR00016 to the Washington National Primate Research Center and K08 AI061738 to G.B.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Agy, M. B., L. R. Frumkin, L. Corey, R. W. Coombs, S. M. Wolinsky, J. Koehler, W. R. Morton, and M. G. Katze. 1992. Infection of Macaca nemestrina by human immunodeficiency virus type 1. Science 257:103-106. [DOI] [PubMed] [Google Scholar]
- 2.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 3.Berthoux, L., S. Sebastian, D. M. Sayah, and J. Luban. 2005. Disruption of human TRIM5α antiviral activity by nonhuman primate orthologues. J. Virol. 79:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, M. L., A. Schmidt, M. B. Agy, L. E. Kimball, and W. R. Morton. 1997. Infection of Macaca nemestrina neonates with HIV-1 via different routes of inoculation. AIDS 11:1555-1563. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, M. L., A. Schmidt, J. Chen, M. J. Florey, M. Agy, and W. R. Morton. 2000. Enhanced replication of HIV-1 in vivo in pigtailed macaques (Macaca nemestrina). J. Med. Primatol. 29:107-113. [DOI] [PubMed] [Google Scholar]
- 6.Frumkin, L. R., M. B. Agy, R. W. Coombs, L. Panther, W. R. Morton, J. Koehler, M. J. Florey, J. Dragavon, A. Schmidt, and M. G. Katze. 1993. Acute infection of Macaca nemestrina by human immunodeficiency virus type 1. Virology 195:422-431. [DOI] [PubMed] [Google Scholar]
- 7.Gartner, S., Y. Liu, M. G. Lewis, V. Polonis, W. R. Elkins, P. M. Zack, J. Miao, E. A. Hunter, J. Greenhouse, and G. A. Eddy. 1994. HIV-1 infection in pigtailed macaques. AIDS Res. Hum. Retrovir. 10(Suppl. 2):S129-S133. [PubMed] [Google Scholar]
- 8.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., M. Princiotta, M. Piatak, Jr., F. Yuan, F. Zhang, J. D. Lifson, and P. D. Bieniasz. 2006. Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314:95. [DOI] [PubMed] [Google Scholar]
- 10.Javanbakht, H., W. Yuan, D. F. Yeung, B. Song, F. az-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234-246. [DOI] [PubMed] [Google Scholar]
- 11.Kamada, K., T. Igarashi, M. A. Martin, B. Khamsri, K. Hatcho, T. Yamashita, M. Fujita, T. Uchiyama, and A. Adachi. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. USA 103:16959-16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, Y., X. Li, M. Stremlau, M. Lee, and J. Sodroski. 2006. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 80:6738-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, H. L., Y. Q. Wang, C. H. Liao, Y. Q. Kuang, Y. T. Zheng, and B. Su. 2005. Adaptive evolution of primate TRIM5α, a gene restricting HIV-1 infection. Gene 362:109-116. [DOI] [PubMed] [Google Scholar]
- 14.McClure, J., A. M. Schmidt, M. A. Rey-Cuille, J. Bannink, L. Misher, C. C. Tsai, D. M. Anderson, W. R. Morton, and S. L. Hu. 2000. Derivation and characterization of a highly pathogenic isolate of human immunodeficiency virus type 2 that causes rapid CD4+ cell depletion in Macaca nemestrina. J. Med. Primatol. 29:114-126. [DOI] [PubMed] [Google Scholar]
- 15.Mische, C. C., H. Javanbakht, B. Song, F. az-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama, E. E., H. Miyoshi, Y. Nagai, and T. Shioda. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79:8870-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman, R. M., L. Hall, M. Connole, G. L. Chen, S. Sato, E. Yuste, W. Diehl, E. Hunter, A. Kaur, G. M. Miller, and W. E. Johnson. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5α. Proc. Natl. Acad. Sci. USA 103:19134-19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohkura, S., M. W. Yap, T. Sheldon, and J. P. Stoye. 2006. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80:8554-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otten, R. A., B. G. Brown, M. Simon, L. D. Lupo, B. S. Parekh, M. D. Lairmore, C. A. Schable, G. Schochetman, and M. A. Rayfield. 1994. Differential replication and pathogenic effects of HIV-1 and HIV-2 in Macaca nemestrina. AIDS 8:297-306. [DOI] [PubMed] [Google Scholar]
- 20.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy, B. A., L. D. Etkin, and P. S. Freemont. 1992. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem. Sci. 17:344-345. [DOI] [PubMed] [Google Scholar]
- 24.Reed, S. E., E. M. Staley, J. P. Mayginnes, D. J. Pintel, and G. E. Tullis. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138:85-98. [DOI] [PubMed] [Google Scholar]
- 25.Rey-Cuille, M. A., and S. L. Hu. 2001. Conserved CXCR4 usage and enhanced replicative capacity of HIV-2/287, an isolate highly pathogenic in Macaca nemestrina. AIDS 15:2349-2357. [DOI] [PubMed] [Google Scholar]
- 26.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer, S. L., L. I. Wu, J. M. Akey, M. Emerman, and H. S. Malik. 2006. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5α in humans. Curr. Biol. 16:95-100. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. R. Soc. Lond. B 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 30.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 31.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. az-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe, N. D., W. Heneine, J. K. Carr, A. D. Garcia, V. Shanmugam, U. Tamoufe, J. N. Torimiro, A. T. Prosser, M. Lebreton, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. M. Switzer. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 102:7994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo, J. S., J. H. Imm, C. K. Min, K. J. Kim, S. S. Cha, and B. H. Oh. 2006. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 25:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo, J. S., H. Y. Suh, S. Y. Park, and B. H. Oh. 2006. Structural basis for protein recognition by B30.2/SPRY domains. Mol. Cell 24:967-976. [DOI] [PubMed] [Google Scholar]
- 36.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 37.Ylinen, L. M., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5α alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]