Abstract
Influenza virus mRNAs bear a short capped oligonucleotide sequence at their 5′ ends derived from the host cell pre-mRNAs by a “cap-snatching” mechanism, followed immediately by a common viral sequence. At their 3′ ends, they contain a poly(A) tail. Although cellular and viral mRNAs are structurally similar, influenza virus promotes the selective translation of its mRNAs despite the inhibition of host cell protein synthesis. The viral polymerase performs the cap snatching and binds selectively to the 5′ common viral sequence. As viral mRNAs are recognized by their own cap-binding complex, we tested whether viral mRNA translation occurs without the contribution of the eIF4E protein, the cellular factor required for cap-dependent translation. Here, we show that influenza virus infection proceeds normally in different situations of functional impairment of the eIF4E factor. In addition, influenza virus polymerase binds to translation preinitiation complexes, and furthermore, under conditions of decreased eIF4GI association to cap structures, an increase in eIF4GI binding to these structures was found upon influenza virus infection. This is the first report providing evidence that influenza virus mRNA translation proceeds independently of a fully active translation initiation factor (eIF4E). The data reported are in agreement with a role of viral polymerase as a substitute for the eIF4E factor for viral mRNA translation.
Viruses do not possess the required components to initiate mRNA translation; thus, they are obligated to utilize host cell factors and therefore to compete for and manipulate the translation apparatus to its own benefit. Cellular mRNAs possess a 7-methyl guanine structure (cap) at their 5′ ends, which plays a critical role in the recruitment of the ribosome to the mRNA. The cap structure is recognized by eIF4E, the cytosolic cap-binding protein that, together with eIF4G and the eIF4A helicase, forms the eIF4F complex. The cap structure is bound in the nucleus by the nuclear cap-binding complex, and in order to translate a given mRNA, the cap-binding complex must be replaced by eIF4E at the 5′ terminus. The eIF4F complex, through eIF4G, recruits the 40S ribosomal subunit bound to eIF3 (for a review, see reference 23). The binding of eIF4G to eIF4E triggers a conformational change in both polypeptides that enhances the association of eIF4E with the cap and stabilizes the complex (25).
The influenza virus RNA polymerase is a complex composed of three subunits: PA, PB1, and PB2. The synthesis of capped and polyadenylated viral mRNAs is primed by short capped oligonucleotides of around 10 to 12 nucleotides, which are scavenged from host cell nuclear mRNAs by viral polymerase endonuclease activity (46). The PB2 subunit is responsible for the binding of the cap structures (4), while it is thought that the endonucleolytic activity required for the “cap-snatching” process lies in the PB1 subunit (36). Influenza virus mRNAs therefore contain host cell-derived sequences at their 5′ ends followed by a highly conserved sequence that is common to all viral genes. It has been reported that the influenza virus polymerase complex binds to this common sequence in vitro (51). This association increases the cap-binding activity of the polymerase complex and enhances its binding to the capped viral mRNAs, protecting them from the cap-snatching process (51). The 3′ end of viral messengers is polyadenylated by a reiterative copy of a U5-7 track present near the 5′ end of the viral RNA (38). Consequently, although different pathways synthesize cellular and viral mRNAs, both types of mRNAs are structurally similar.
Influenza virus efficiently shuts off host cell protein synthesis (21). Upon infection of susceptible cells, initiation and elongation steps of translation of cellular mRNAs are inhibited (31). This translational control is accompanied by a selective translation of viral mRNAs, with the sequences within the 5′ untranslated regions (UTRs) playing a critical role (21). The NS1 viral protein is important for the selective translation of viral messengers, especially for the late ones, by increasing their rate of initiation (10, 14, 31, 43). This process is mediated by its functional interaction with the 5′-terminal conserved sequences of viral mRNAs (10, 43). We have identified two cellular targets of NS1 that support its role in protein translation: the eIF4GI subunit of eIF4F (1) and poly(A) binding protein 1 (PABP1) (5). However, there are NS1 mutant viruses (point mutations or partial or total deletions of the NS1 protein) that present alterations in protein synthesis but that are still capable of inducing cellular shutoff and performing selective translation of their own mRNAs (17, 19, 26, 48). These data suggest that other viral factors should be involved in the translation of viral mRNAs. Influenza virus infection alters the translation initiation eIF4F complex: the cap-binding protein eIF4E becomes underphosphorylated, and the factor eIF4GI becomes hyperphosphorylated (18). On the other hand, influenza virus infection cannot proceed in poliovirus-infected cells, where the eIF4G factor is cleaved (20). This suggests that viral mRNA translation requires full-length eIF4G and therefore does not proceed by an internal cap-independent translation initiation pathway. To elucidate the mechanisms involved in the selective translation of viral messengers during infection, we examined whether viral polymerase, as a cap-binding protein complex, functions as the cap-binding factor for viral mRNA translation, allowing functional independence from the cellular cap-binding protein eIF4E.
MATERIALS AND METHODS
Biological materials.
Influenza virus strains A/Victoria/3/75 (VIC), A/Puerto Rico/8/34 (PR8), and PR8 lacking NS1 (delNS1) (a gift of A. García-Sastre) and coronavirus strain HCoV-229E (a gift of L. Enjuanes) were used. To reconstitute viral RNP, plasmids pCMVPA, pCMVPB1, pCMVPB2, pCMVNP, and pHHNS, which were previously described (17), were used. To analyze the translation initiation complex-viral polymerase association, plasmids pCMVPAΔUTR, pCMVPB1ΔUTR, and pCMVPB2ΔUTR that express PA, PB1, and PB2 polymerase subunits but that do not contain the 5′ and 3′ influenza virus UTR sequences were used and kindly provided by P. Resa. HEK293T, HeLa, and A549 cell lines were used throughout. Vero and ST-hAPN cells were used to amplify recombinant influenza virus lacking NS1 (48) and human coronavirus HCoV-229E (34), respectively. Monoclonal antibody against green fluorescent protein (GFP) and complete protease and RNase (human placenta RNAse inhibitor) inhibitors were obtained from Roche. Rapamycin was obtained from Calbiochem.
Construction of plasmids.
For the construction of 4E-BP1-expressing plasmids, human 4E-BP1 cDNA was used as a template for PCR mutagenesis to mutate T37, T46, S65, and T70 to alanine. Wild-type or mutated sequences were inserted in frame into vector pcDNA3-3HA (which contains an N-terminal fusion with a tag comprising three hemagglutinin [HA] epitopes).
The eIF4E-silencing plasmid pSUPERretroNeoGFP-4E (pSUPER-GFP-4E) expressing the short hairpin RNA corresponding to positions 447 to 465 of eIF4E mRNA was generated according to the manufacturer's instructions (Oligoengine). This short hairpin showed no homology to other gene sequences when using BLAST. Control plasmid pSUPERretroNeo-GFP-TM (a gift of A. Rodriguez) expresses a nonsilencing short hairpin RNA derived from a transcript of the bacterium Thermotoga maritima and does not target any known mammalian gene (the targeted sequence is AATTCTCCGAACGTGTCACGT).
Transfection and virus infection.
All infections were carried out at a multiplicity of infection of 5 to 10 PFU/cell. Where needed, HEK293T or HeLa cells were previously transfected by the calcium-phosphate method (52) or using FUGENE HD reagent (Roche Applied Science), respectively. At different times postinfection, the cells were used for studies using sucrose gradient separation, immunofluorescence, metabolic labeling, or binding to m7GTP resins (cap resins). In eIF4E gene silencing experiments, when transfection efficiency was lower than 70%, the cells were separated using GFP fluorescence by a MoFlo cell sorter (DAKO Cytomation). Viral RNPs were reconstituted as previously described (44).
Western blotting.
Western blotting was done as described previously (1). The following primary antibodies were used: a mixture of four rabbit polyclonal antibodies (1:8,000 each) was used for translation initiation factor eIF4GI (1), a monoclonal antibody from Transduction Laboratories (1:2,000) was used for eIF4E, monoclonal antibodies 2 and 14 (1:20 each) (28) were used for PA, a rat polyclonal antibody (1:1,000) (a gift of J. Ortín) was used for PB1, monoclonal antibodies 8 and 28 (1:100 each) (28) were used for PB2, a rat polyclonal antibody (1:1,000) (a gift of J. Ortín) was used for the NP protein, and a rabbit polyclonal antibody (1:10,000) (1) was used for the NS1 protein. For β-actin, a mouse monoclonal antibody (1:50,000) from Sigma was used; for 4E-BP1, a rabbit polyclonal antibody (1:1,000) from Cell Signaling Technology was used; for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), a rabbit polyclonal antibody (1:5,000) was used; for RNA polymerase II, a mouse monoclonal antibody (1:500) from Covance was used; for HA, a mouse monoclonal antibody (1:1,000) from Abcam was used; and for total eIF2α, a rabbit polyclonal antibody from Santa Cruz (1:2,000) was used, and for its phosphorylated form (eIF2α [pS52]), a rabbit polyclonal antibody from Biosource (1:200) was used.
Immunofluorescence.
HEK293T or HeLa cells were fixed, permeabilized, and incubated with the following primary antibodies: anti-NP (1:2,000), anti-coronavirus S protein (1:20) (a gift of L. Enjuanes), anti-GFP (1:1,000), and anti-HA (1:1,000 or 1:500 when using monoclonal or polyclonal antibodies, respectively). Microscopy was performed with a Leica DMRX epifluorescence microscope or with a Bio-Rad Radiance 2100 confocal laser scanning system on a Zeiss Axiovert 200 microscope.
Analysis of viral proteins associated with translation initiation complexes.
HEK293T cells were mock or influenza virus infected, and at 7 h postinfection (hpi), the cells were collected and lysed in buffer A (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 8.5], 0.2% Igepal) with protease (complete) and phosphatase (5 mM Na3VO4, 5 mM β-glycerophosphate, 5 mM sodium molibdate) inhibitors plus the RNase inhibitor human placenta RNAse inhibitor (1:1,000). The lysate was centrifuged at 10,000 × g, and the supernatant was loaded into a 7 to 47% sucrose gradient and centrifuged for 14 h at 24,000 rpm at 4°C in a SW41Ti (Beckman) rotor. Fractions were collected from the top of the gradient and resuspended in Laemmli sample buffer or used for immunoprecipitation studies. For coimmunoprecipitation, the corresponding fractions were extensively dialyzed in buffer A without detergent and incubated with specific anti-eIF4GI antibody or preimmune serum as reported previously (1). The immunocomplexes were washed five times with buffer A and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting.
To analyze the association of influenza virus polymerase subunits with translation initiation complexes and to study the contribution of viral mRNA to this association, p100 plates of HEK293T cells were transfected with 3 μg of plasmids pCMVPB1ΔUTR and pCMVPB2ΔUTR and 0.6 μg of plasmid pCMVPAΔUTR without the 5′ and 3′ influenza virus UTR sequences. Sixteen hours later, cytosolic extracts were prepared in buffer A containing proteases, phosphatases, and RNase inhibitors; after centrifugation at 10,000 × g, the supernatants were collected and used for coimmunoprecipitation studies as described above.
Cap-binding assays.
HEK293T cells were left untransfected or were transfected with empty plasmid or plasmids expressing wild-type or mutated 4E-BP1 proteins. Next, they were mock or influenza virus infected. At different times, the cells were collected and lysed in buffer A with proteases, phosphatases, and RNase inhibitors as described above. The lysates were centrifuged at 10,000 × g, and the supernatants were incubated with Sepharose-m7GTP (Amersham) or Sepharose-4B (Sigma), as a negative control, overnight at 4°C. The resins were washed five times with buffer A, resuspended in Laemmli sample buffer, and analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting.
Metabolic labeling.
For continuous labeling, HEK293T cells that were mock infected or infected with the VIC strain of influenza virus were incubated with medium containing 30 μCi/ml of [35S]Met-Cys during the last 4 to 5 h of infection. For pulse experiments, 100 μCi/ml of [35S]Met-Cys was added during 30 min or 1 h at the indicated times.
RESULTS
Influenza virus polymerase associates with translation initiation complexes.
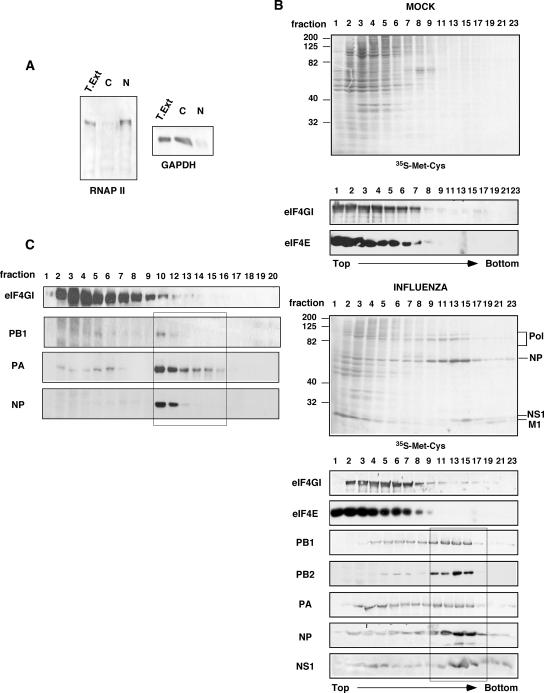
To study whether viral polymerase is involved in viral mRNA translation, we tested if the polymerase complex associates with translation initiation complexes. In the infected cell, influenza virus polymerase can be found as a trimeric complex or as ribonucleoproteins together with the nucleoprotein and the viral RNA. Previous reports have shown that a cytoplasmic extract from influenza virus-infected cells depleted of nucleocapsids and subjected to centrifugation on a sucrose gradient allows the characterization of polymerase subunits not associated with viral nucleocapsids (11). Furthermore, using specific antibodies against different polymerase subunits, it has been shown that the three polymerase subunits not present in RNPs were found together in complexes ranging from about 11S to 22S (11). Thus, to separate viral polymerase from viral RNPs, we performed a sucrose gradient separation of cytoplasmic extracts of either mock- or influenza virus-infected HEK293T cells with the VIC strain. First, to control the effectiveness of the subcellular fractionation, Western blot assays using specific antibodies against nuclear (RNA polymerase II) and cytosolic (GAPDH) proteins were performed. The results (Fig. 1A) show that the cytosolic fraction used in this study is free of nuclear proteins. Analysis of viral proteins in the sucrose gradients (Fig. 1B) using either in vivo metabolic labeling with [35S]Met-Cys or Western blot assays showed that PA, PB1, PB2, NP, and NS1 were mainly present in two regions of the gradient. These were fractions 2 to 8, which represent free proteins and viral polymerase free of RNPs, and fractions 9 to 17, which represent viral RNPs. The eIF4E and eIF4GI proteins of the translation initiation factor eIF4F were mainly present between fractions 1 and 9. Additional information about RNP distribution in sucrose gradients was obtained when an individual viral RNP was reconstituted as previously reported (44), using a model viral RNA encoding NS1 (Fig. 1C). A clear separation was found between the migration of this small viral RNP that begins in fraction 10 and the polymerase subunits not associated with RNPs, as reflected by the near absence of NP protein in fractions previous to fraction 10 (Fig. 1C). Combining these data with the distribution pattern of the rRNA measured by the absorbance at 260 nm (data not shown), we decided to join fractions 7 to 9 of the sucrose gradients. These fractions contain part of the translation preinitiation complexes and viral polymerase, and in addition, they would be almost free of viral RNPs. The fractions were dialyzed and used for coimmunoprecipitation assays with antibodies against eIF4GI, analyzing the presence of polymerase subunits in the immunocomplexes. The results (Fig. 2A) show that the three subunits were specifically coimmunoprecipitated with the eIF4GI protein but not with a preimmune control antibody. The presence of viral NP and NS1 proteins was also examined: whereas NS1 was coimmunoprecipitated, NP was absent, indicating that the presence of polymerase in the immunocomplexes is not due to RNP contamination.
FIG. 1.
Separation of influenza virus polymerase from viral RNPs. HEK293T cells were mock or influenza virus infected for 7 h with the VIC strain at 5 to 10 PFU/cell. (A) Subcellular fractionation. Nuclear (N) and cytosolic (C) fractions from total extracts (T. Ext.) were separated and analyzed by Western blotting with specific antibodies against RNA polymerase II (RNAP II) or GAPDH. (B) Separation of viral polymerase subunits from viral RNPs. HEK293T cells were labeled in vivo during the last 4 h of infection (Promix; Amersham). Next, the cells were collected and processed as described in Materials and Methods, and the labeled proteins were analyzed by autoradiography ([35S]Met-Cys). Samples of the same fractions were analyzed by Western blotting with specific antibodies against the indicated proteins. (C) Viral RNP expressing NS1 was reconstituted in vivo and processed as described above (B). The corresponding proteins were analyzed by Western blotting.
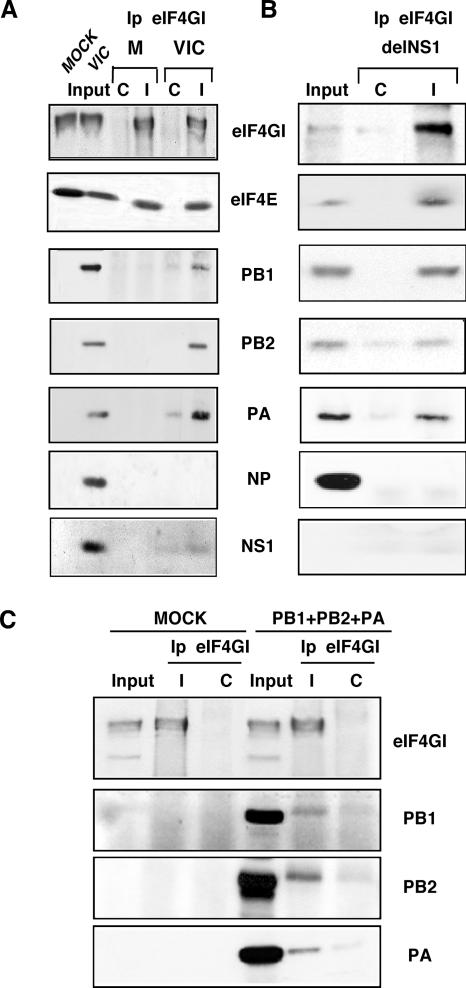
FIG. 2.
Influenza virus polymerase subunits associate with translation initiation complexes. (A) Cytosolic extracts from mock-infected or VIC-infected HEK293T cells were applied to sucrose gradients and processed as described in Materials and Methods. Samples were used for immunoprecipitation studies (Ip) using specific antibodies against the eIF4GI protein (I) or the preimmune serum (C). (B) HEK293T cells were infected with the delNS1 strain, and cytosolic extracts were immunoprecipitated as described above (A) to analyze the associated proteins by Western blotting. (C) HEK293T cells were untransfected (MOCK) or cotransfected with plasmids expressing PB1, PB2, and PA (PB1+PB2+PA), and cytosolic extracts were prepared and immunoprecipitated with eIF4GI antiserum as described above. The polymerase proteins associated with eIF4GI were analyzed by Western blotting.
To exclude that NS1 could work as a link between the polymerase and the translation initiation complexes, similar experiments were carried out in HEK293T cells infected with an influenza virus-rescued virus lacking NS1 (delNS1). Infection with PR8, the parental strain of delNS1, was used as a control. Similar to infection with the VIC strain, the polymerase subunits of both delNS1 (Fig. 2B) and PR8 (data not shown) were capable of associating with eIF4GI. Again, the NP protein was absent in the eIF4GI immunocomplexes. These results indicate that viral polymerase associates with translation preinitiation complexes independently of the presence of the NS1 protein and the viral strain used.
To further characterize the association of viral polymerase with translation initiation complexes outside the context of viral infection, we coexpressed PB1, PB2, and PA polymerase subunits in HEK293T cells and performed coimmunoprecipitation assays. Moreover, to study the contribution of viral mRNAs to the association, we used plasmids that express the polymerase subunits but that do not contain the 5′ and 3′ UTR sequences of influenza virus mRNAs, and therefore, viral mRNAs were not present in the assay. The results are presented in Fig. 2C. The three polymerase subunits were found in the immunocomplexes together with the eIF4GI protein, although the efficiency of coimmunoprecipitation was lower than that obtained when influenza virus-infected cells were used. This could be due to the fact that not all the coexpressed polymerase subunits are forming polymerase complexes, since an important fraction remains as free subunits. Nevertheless, these results indicate that PB1, PB2, and PA coimmunoprecipitate with eIF4GI in the absence of viral mRNAs, corroborating the finding that polymerase subunits interact with translation initiation complexes and indicating that viral mRNA is not required for this association.
Translation of influenza virus mRNAs occurs under conditions of functional impairment of cellular cap-binding protein eIF4E.
The above-described results raised the possibility that viral polymerase could replace the cellular cap-binding protein eIF4E. This replacement would allow the translation of viral mRNAs without the contribution of this cellular factor. This possibility was examined using three different approaches.
(i) Translation of influenza virus mRNAs is rapamycin insensitive.
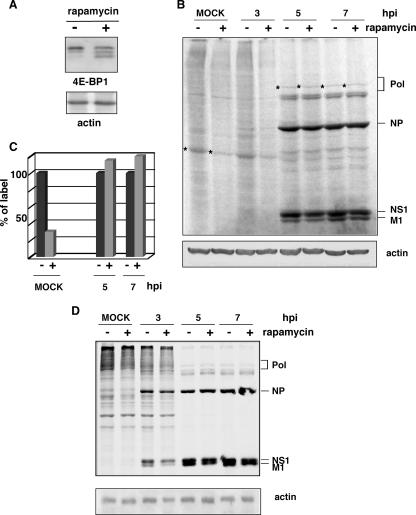
The mTOR protein kinase regulates protein synthesis through the phosphorylation and inactivation of the repressor of cap-dependent translation, the 4E-binding protein (4E-BP), and through the phosphorylation and activation of S6 kinase. Phosphorylation of 4E-BP is inhibited in vivo by the drug rapamycin, leading to a reduction in cap-dependent translation (reviewed in reference 24). HeLa cells were preincubated with or without rapamycin at a concentration of 20 ng/ml in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum for 12 h. To control the effectiveness of the drug, we analyzed the degree of phosphorylation of 4E-BP1 and observed bands with higher electrophoretic mobility corresponding to more dephosphorylated isoforms upon rapamycin treatment (Fig. 3A). For loading controls in Western blots, we analyzed the β-actin protein, whose half-life is greater than 100 h (7). No variations were found in its accumulation levels after the drug treatment. Treated and untreated cells were subsequently infected with influenza virus in the presence or absence of the drug. At different hpi, the cells were metabolically labeled with [35S]Met-Cys for 30 min, and the synthesized proteins were analyzed by SDS-polyacrylamide denaturing gels (Fig. 3B). In agreement with previous results (3, 39), rapamycin led to a reduction in global protein synthesis in mock-infected cells of approximately 50% (measured by label quantitation). In contrast, efficient viral protein translation was observed under these conditions. To estimate the effect of rapamycin on the protein synthesis of defined proteins, we selected an abundant cellular protein and the viral polymerase subunits to perform quantitative analysis. The results (Fig. 3C) show that the synthesis of the cellular protein in cells treated with rapamycin is around 30% of the synthesis in control cells. In contrast, the synthesis of viral polymerase is unaffected under these conditions, indicating that viral translation is insensitive to drug treatment.
FIG. 3.
Influenza virus infection progresses efficiently in rapamycin-treated cells. (A) HeLa cells treated with (+) or without (−) rapamycin for 12 h were subjected to Western blotting against 4E-BP1 or actin. (B) HeLa cells treated with (+) or without (−) rapamycin for 12 h were mock infected (MOCK) or infected with the VIC strain, maintaining the rapamycin condition. At the indicated hpi, the cells were metabolically labeled, and the proteins were analyzed by SDS-polyacrylamide gels and autoradiography. (C) Quantitation of the incorporated label in specific cellular and viral proteins (marked with asterisks) from B. (D) A549 cells treated with (+) or without (−) rapamycin were processed as indicated above (B).
To investigate the action of rapamycin upon influenza virus infection in a more-biological system, we performed similar experiments in A549 lung epithelial cells. The results (Fig. 3D) showed that at early times, such as 3 hpi, the translation of viral proteins is partially affected, but as the infection proceeds, viral translation takes place efficiently in the rapamycin-treated cells. These results indicate that viral infection can progress properly in different types of cells under conditions of impaired cellular translation.
(ii) Translation of influenza virus mRNAs is not affected in eIF4E-silenced cells.
The next approach was the use of gene-silencing experiments using RNA interference. With this aim, HEK293T cells were transfected with a control silencing plasmid (pSUPER-GFP-TM) or with a plasmid specific for eIF4E silencing (pSUPER-GFP-4E). To avoid the contribution of eIF4E from untransfected cells, they were sorted 12 h after transfection by using GFP fluorescence. Selected cells were plated again and infected with influenza virus 36 h posttransfection. At the indicated hpi, aliquots were used for Western blotting, metabolic labeling with [35S]Met-Cys, and immunofluorescence studies. As can be seen in Fig. 4A, the eIF4E-silencing plasmid efficiently decreased the accumulation levels of the eIF4E protein compared with the control plasmid. The recognition of the interfering double-stranded RNA by oligoadenylate synthetase (2′,5′-oligoadenylate synthetase) and PKR pathways of the innate cellular defense system could result in nonspecific translation inhibition (12) due to the general RNA degradation or phosphorylation of the eIF2α factor, respectively. The integrity of the cellular RNA was evaluated by ethidium bromide staining of total RNA isolated from transfected HEK293T cells, and no degradation was observed under any experimental condition (data not shown). On the other hand, the down-regulation of eIF4E did not increase the degree of phosphorylation of the eIF2α protein (Fig. 4A). Similar accumulation levels of the eIF2α protein were obtained in control and silenced cells using an antibody that recognizes total levels of the eIF2α protein (Fig. 4A). These results indicate that the activation of neither PKR nor oligoadenylate synthetase pathways takes place in our gene-silencing conditions. Total protein synthesis upon eIF4E silencing is shown in Fig. 4A. As a loading control, we used the accumulation of total eIF2α. Quantitation analysis showed that transfection with plasmid pSUPER-GFP-4E significantly inhibited the translation of cellular mRNAs, presenting a 30 to 35% reduction compared with the translational rate of control cells. This inhibition is in agreement with data from previous reports analyzing either total cellular protein synthesis (13) or the translation of defined cellular mRNAs (42). In contrast, the synthesis of viral proteins (Pol, NP, NS1, and M1) was unaffected. To estimate the effect of eIF4E silencing on the protein synthesis of defined proteins, we selected cellular and viral (HA and neuraminidase) proteins to perform a quantitative analysis. The results in Fig. 4B show that the synthesis level of the cellular protein in eIF4E-silenced cells is around 50% of the synthesis in control cells, whereas the synthesis of viral proteins is unaffected, indicating that viral translation occurs normally upon eIF4E silencing. Immunofluorescence studies were also carried out, and we observed that influenza virus infection takes place in eIF4E-silenced cells with normal kinetics of viral RNP production and export (data not shown).
FIG. 4.
Gene silencing of the eIF4E factor does not affect influenza virus protein synthesis but inhibits cellular protein translation. HEK293T cells were transfected with control pSUPER-GFP-TM (TM) or pSUPER-GFP-4E (4E) plasmids. Twelve hours after transfection, the cells were selected by cell sorting using the GFP fluorescence and plated again. Thirty six hours posttransfection, cells were infected with influenza virus. (A) At the indicated hpi, aliquots were taken and used for Western blotting against the indicated proteins and metabolic labeling with [35S]Met-Cys. (B) Quantitation of the incorporated label in specific cellular and viral proteins (marked with asterisks) from the [35S]Met-Cys panel.
(iii) Overexpression of constitutively unphosphorylated 4E-BP1 does not prevent the translation of influenza virus mRNAs.
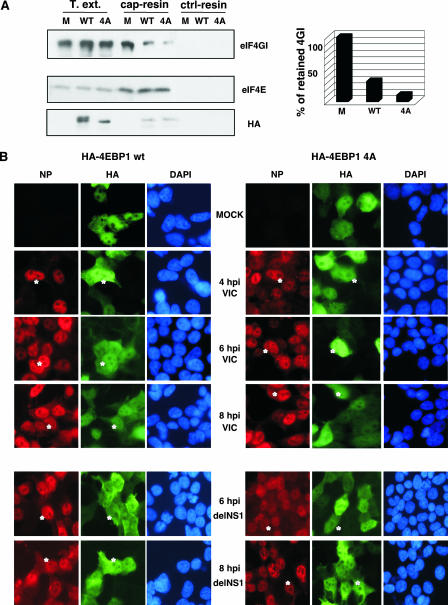
The eIF4E-binding proteins (4E-BPs) are a family of three small polypeptides that inhibit cap-dependent translation by binding to eIF4E and obstructing its interaction with eIF4G (22). The binding of the 4E-BPs to eIF4E is reversible: whereas hypophosphorylated 4E-BPs bind avidly to eIF4E, 4E-BP hyperphosphorylation abrogates this interaction (for a review, see reference 23). Five mTOR-dependent phosphorylation sites have been identified in 4E-BP1: T37, T46, S65, T70, and S83 (16). Phosphorylation of the first four sites is required for the release of 4E-BP1 from eIF4E (40). To analyze the effect of eIF4E sequestration on influenza virus mRNA translation, we infected HEK293T cells previously transfected with plasmids expressing HA-tagged wild-type 4E-BP1 or a nonphosphorylatable 4E-BP1 mutant with alanine substitutions at T37, T46, S65, and T70 (HA-4E-BP1 4A). The 4E-BP1 mutant associates with eIF4E very strongly, prevents the formation of the eIF4E/eIF4G complex, and impairs protein translation (37, 47). Previous studies reported that the binding of eIF4E to cap structures occurs normally upon its binding to the 4E-BP protein (47, 54). To analyze the dissociation of the eIF4E-eIF4G complex caused by 4E-BP1 overexpression, we performed eIF4E pull-down assays using m7GTP resins. As can be observed in Fig. 5A, similar amounts of eIF4E bound to the resin were found under the different experimental conditions. However, the amount of eIF4GI bound to the cap resin clearly diminished upon the overexpression of both forms of HA-4E-BP1. Quantitative data for eIF4GI retention are shown in Fig. 5A (right). Next, the course of infection of HEK293T cells previously transfected with the corresponding HA-4E-BP1 constructs was analyzed by immunofluorescence. The results (Fig. 5B) show that the overexpression of wild-type or mutated 4E-BP1 proteins had no effect on viral infection. To exclude the possible contribution of NS1 to this process, HEK293T cells were transfected as described above and infected with the delNS1 virus. Immunofluorescence studies (Fig. 5B) show that influenza virus infection with delNS1 virus proceeds equally under all conditions used.
FIG. 5.
Overexpression of HA-4E-BP1 proteins does not affect influenza virus infection in HEK293T cells. (A) HEK293T cells were untransfected (M) or transfected with plasmids expressing HA-tagged wild-type 4E-BP1 (WT) or nonphosphorylatable 4E-BP1 (4A) protein. At 36 h posttransfection, total cell extracts (T.ext.) were used to study eIF4GI, eIF4E, and HA-4E-BP1 (HA) retention either to cap resins or to Sepharose-4B control resins (ctrl-resin) by Western blotting. Quantitation of eIF4GI protein retained on the cap resins is shown on the right. (B) HEK293T cells were transfected with plasmid pcDNA3-HA-4E-BP1 wt or pcDNA3-HA-4E-BP1 4A, and 36 h posttransfection, the cells were mock or influenza virus infected with either the VIC (top) or delNS1 (bottom) strain. At the indicated hpi, cells were fixed and used for immunofluorescence using antibodies against HA to monitor plasmid transfection and NP protein to monitor influenza virus infection. Asterisks indicate transfected and infected cells. DAPI, 4′,6′-diamidino-2-phenylindole.
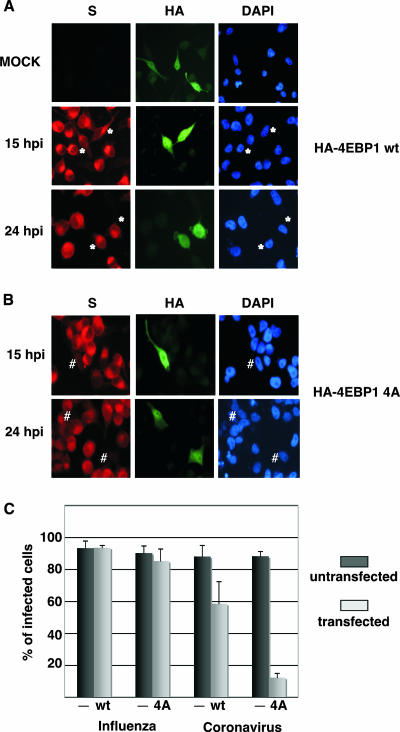
To obtain a functional control for the impairment of regular viral 4E-dependent translation, we performed similar experiments using a human coronavirus (HCoV-229E) to infect cells transfected with the different HA-4E-BP1 constructs. Coronavirus genome replication takes place exclusively in the cytoplasm. Its genome is a positive-stranded RNA that contains an m7GpppN cap structure at the 5′ end of the mRNA and initiates translation in a cap-dependent manner (15, 33). As the human coronavirus does not grow efficiently in HEK293T cells, we used HeLa cells for this experimental approach. HeLa cells were then transfected with wild-type HA-4E-BP1 or HA-4E-BP1 4A plasmids, and at 36 h posttransfection, the cells were infected with either influenza virus (data not shown) or human coronavirus (Fig. 6). As previously observed for HEK293T cells, influenza virus infection in transfected HeLa cells was unaffected. Infection with human coronavirus in cells that overexpressed wild-type HA-4E-BP1 was partially affected. Moreover, in coronavirus-infected HeLa cells expressing the mutant HA-4E-BP1 protein, the infection was severely impaired (Fig. 6). A quantitation of influenza virus and coronavirus infection in HeLa cells transfected with the different HA-4E-BP1-expressing plasmids is presented in Fig. 6C. More than 120 transfected and untransfected cells of the same plate were counted for every experimental condition, and the experiment was performed three times. As can be observed, influenza virus infection progressed normally under all experimental conditions. In contrast, coronavirus infection efficiency was reduced from 88% in nontransfected cells to 58% and to 12% in cells transfected with the wild-type or mutated forms of the HA-4E-BP1 protein, respectively. Collectively, these results indicate that the translation of influenza virus mRNAs can take place under conditions where the availability of the cap-binding protein eIF4E is compromised and suggest that their translation could be eIF4E independent.
FIG. 6.
Overexpression of underphosphorylated 4E-BP1 protein impairs human coronavirus infection. HeLa cells were transfected with plasmid pcDNA3-HA-4E-BP1 wt (A) or pcDNA3-HA-4E-BP1 4A (B), and 36 h posttransfection, the cells were mock or coronavirus infected. At the indicated hpi, cells were used for immunofluorescence using antibodies against HA to monitor plasmid transfection and S protein to monitor coronavirus infection. Asterisks indicate transfected and infected cells. # indicates cells that were transfected but uninfected. DAPI, 4′,6′-diamidino-2-phenylindole. (C) Quantitation of the efficiency of infection in untransfected cells (−) and in pcDNA3-HA-4E-BP1 wt (WT)- or pcDNA3-HA-4E-BP1 4A (4A)-transfected cells subsequently infected with influenza virus (data not shown) or human coronavirus HCoV-229E.
Translation initiation factor eIF4GI is recruited to cap resins in influenza virus-infected cells with a reduced availability of the eIF4E factor. (i) Viral cytosolic polymerase is retained by cap resins.
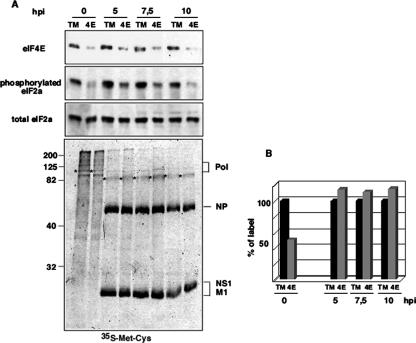
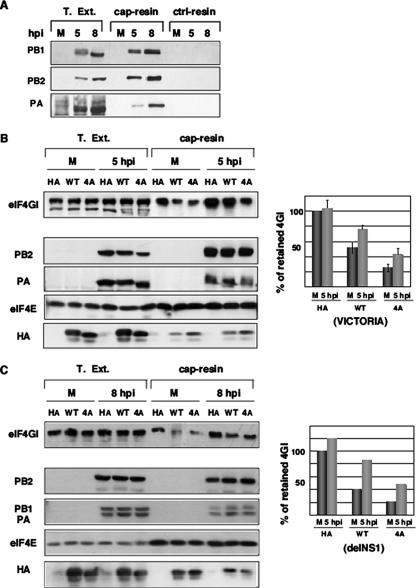
Influenza virus polymerase is associated with translation initiation complexes (Fig. 2), and viral infection occurs normally in cells with reduced eIF4E activity and impaired cap-dependent translation (Fig. 3 to 6). Consequently, we asked whether eIF4GI associates with cap structures under conditions of low eIF4E accessibility in influenza virus-infected cells. Since the nuclear PB2 subunit has the ability to recognize and bind type 1 cap structures (4), we previously assayed if viral polymerase present in the cytosol maintained the cap-binding capacity. Thus, we assayed the retention of the viral polymerase by m7GTP affinity columns (Fig. 7A). The three polymerase subunits were retained on these columns and were absent in the Sepharose-4B control columns. None of the resins retained viral NP protein, whereas m7GTP affinity columns specifically retained both eIF4E and eIF4GI translation factors (data not shown). These results indicate that viral polymerase present in the cytosol can associate with cap analogs.
FIG. 7.
Influenza virus infection enhances the recruitment of eIF4GI to cap resins. (A) Cytosolic extracts (T. Ext.) of mock-infected (M) or influenza virus-infected (5 and 8 hpi) HEK293T cells were applied to m7GTP-Sepharose resins (cap resins) or to Sepharose-4B control resins (ctrl-resins), and the indicated proteins were analyzed by Western blotting. (B) HEK293T cells were transfected with plasmid pcDNA3-HA (HA), pcDNA3-HA-4E-BP1 wt (WT), or pcDNA3-HA-4E-BP1 4A (4A), and 36 h posttransfection, the cells were mock (M) or influenza virus infected (5 hpi). Cytosolic extracts (T. Ext.) were prepared and applied to m7GTP-Sepharose resins (cap-resin). Retention of the indicated proteins was evaluated by Western blot analysis, and quantitation is shown on the right (means and standard deviations). (C) Experiment similar to that performed in B but using recombinant delNS1 influenza virus. On the right side, the quantitation of the eIF4GI protein retained in the cap resins as described in the text is shown.
(ii) Influenza virus infection enhances the recruitment of eIF4GI to cap resins under conditions of low eIF4E availability.
To study whether eIF4GI is recruited to cap structures during infection under conditions of limiting amounts of free eIF4E, HEK293T cells were transfected with the control empty plasmid pcDNA3-HA or with the different HA-4E-BP1 constructs and infected with influenza virus. At different times postinfection, cytosolic extracts were prepared and used to analyze eIF4GI binding to cap analogs. The results are shown in Fig. 7B. Detection of transfected HA-4E-BP1 proteins in total extracts using anti-HA antibodies showed the presence of HA-4E-BP1 forms with lower electrophoretic mobility under conditions of wild-type HA-4E-BP1 overexpression compared with the overexpression of the HA-4E-BP1 4A construct (Fig. 7B). In agreement with the mutations at T and S residues inserted into the HA-4E-BP1 4A mutant, these forms would correspond to 4E-BP1 isoforms with a lower degree of phosphorylation. On the other hand, PA, PB2 (Fig. 7B), and PB1 (data not shown) influenza virus proteins were bound to cap resins in infected cells. In uninfected cells, the overexpression of HA-4E-BP1 induced the dissociation between eIF4E and eIF4G as indicated by the decreased levels of eIF4G retained in the cap resin in this situation. This decreased retention was attenuated by infection (5 hpi). Quantitation of the retained eIF4GI protein coming from three different experiments (Fig. 7, right) indicated that influenza virus infection increased 54% and 75% the presence of eIF4GI in the cap resins in cells transfected with the wild-type or mutated forms of the HA-4EBP1 protein, respectively. To eliminate the possible contribution of NS1 in eIF4GI binding to the cap resin, similar experiments were carried out using the recombinant influenza delNS1 virus. Analogous results were found in this situation, indicating that NS1 is not involved in the observed cap-eIF4GI binding (Fig. 7C). The experiment was repeated twice, and quantitation of the retained eIF4GI from a representative experiment is shown in Fig. 7C (right). These results indicate that under conditions of decreased eIF4GI-eIF4E association, eIF4GI of influenza virus-infected cells is retained efficiently on cap resins in an NS1-independent way, suggesting that viral polymerase can act as a link between eIF4GI and cap structures.
DISCUSSION
Many viruses have developed varied and sophisticated mechanisms to specifically repress cellular mRNA translation and concomitantly allow the selective translation of viral mRNAs. Translation initiation of most cellular mRNAs requires the cap-binding complex eIF4F. This circumstance has provided viruses with exceptional opportunities to develop specific translation mechanisms that circumvent or decrease the requirement for this complex. Thus, viruses frequently inactivate the cap initiation complex by altering some of its key components.
Different viral mechanisms are used to efficiently translate viral mRNAs.
A wide range of picornaviruses have mRNAs in which initiation occurs downstream of the 5′ end, on internal ribosome entry sites, allowing cap-independent initiation. These picornaviruses encode a protease that cleaves the eIF4G protein into two polypeptides and separates the eIF4E-binding domain from the eIF3-binding domain. This causes the uncoupling of cellular mRNA recognition (via the cap structure and the eIF4E protein) and small ribosomal subunit recruitment (via ribosome-bound eIF3), thereby impairing cap-dependent initiation (for reviews, see references 6 and 50).
Among the viruses whose mRNAs possess a cap structure, a well-understood example is the case of rotavirus (a double-stranded RNA virus), which encodes mRNAs that are capped but not polyadenylated. The viral nonstructural protein NSP3 binds to eIF4G and the 3′ end of rotavirus mRNAs, disrupting the eIF4G-PABP1 interaction that stimulates initiation and thereby inhibits cellular mRNA translation (45). Surprisingly, the list of viruses whose mRNAs hold a 5′ cap structure and show an unexpected low dependence of the functionally active eIF4E factor is growing quickly. Infection with adenovirus provokes robust eIF4E dephosphorylation, which is consistent with its inhibition of host protein synthesis. This virus encodes a 100-kDa protein that binds to eIF4G and displaces the eIF4G-Mnk1 interaction, thus eliminating the ability to phosphorylate eIF4E (9). Adenovirus promotes selective translation through a cap-dependent translation mechanism known as ribosome shunting. In this case, there is a loading of 40S ribosome subunits onto the mRNA and then a translocation of 40S ribosome subunits to the initiation codon (50, 53). Infection with vesicular stomatitis virus produces the dephosphorylation of the eIF4E and 4E-BP1 proteins, which results in a reduced eIF4E-eIF4G association (8). An alternative translation mechanism has not yet been described for this virus. Recently, it has been reported that dengue virus translation takes place under conditions of limited amounts of eIF4E where cap-dependent translation is compromised (13). In this case, it has been proposed that a decrease in the concentration of eIF4E prompts a reorganization of the viral RNP complexes bridging the 5′ and 3′ UTRs, allowing the recruitment of factors such as eIF4G and bypassing the requirement for the eIF4E factor (13).
Translation of influenza virus mRNAs.
Translation of cellular mRNAs is strongly inhibited in influenza virus-infected cells. Some of the viral activities preclude the translation of de novo-synthesized cellular mRNAs. Thus, infection decreases the synthesis of cellular mRNAs, probably as a consequence of virally induced cap-snatching activity, and inhibits the nucleocytoplasmic transport of cellular mRNAs (32). Later in the infection, there is cytoplasmic degradation of cellular mRNAs (2, 29). Additionally, the virus has developed mechanisms to discriminate and selectively translate its 5′-capped and 3′-polyadenylated mRNAs among previously accumulated cellular mRNAs. As mentioned previously, the NS1 protein has an important role in the efficient translation of late viral mRNAs. However, in view of the phenotypes exhibited by several NS1 mutant viruses, other viral mechanisms need to operate to discriminate and efficiently translate viral mRNAs.
The fact that influenza virus infection cannot proceed in poliovirus-infected cells where the eIF4G initiation factor is cleaved (20) has traditionally led to the concept that influenza virus translation initiation occurs using the full eIF4F complex (eIF4E, eIF4G, and eIF4A) bound to capped viral messengers. Since viral polymerase binds to cap structures, we have considered that this association could block eIF4E accessibility to the cap. Here, we provide data indicating that viral translation occurs under various conditions of impairment of functional eIF4E such as rapamycin treatment, eIF4E gene silencing, and overexpression of constitutively underphosphorylated 4E-BP1. Additional data support the independence of viral mRNA translation of a fully active eIF4E factor. First, coinfection of influenza virus and adenovirus can simultaneously occur despite the strong dephosphorylation of eIF4E that causes adenovirus infection. Moreover, influenza virus infection also produces eIF4E dephosphorylation (18, 30). In line with the role of eIF4E dephosphorylation, adenovirus infection promotes the inhibition of host protein synthesis. Phosphorylation of eIF4E strongly correlates with the rate of translation in many systems, although its reduced phosphorylation does not affect the rate of translation in certain situations (49). Second, the expression of the antiviral molecule ISG15 and protein modification by ISG15 (ISGylation) are strongly activated by interferon and viral infection. It has been reported that as early as 3 days after infection, influenza virus-infected mice express large amounts of both free ISG15 and ISG15 conjugates in the lung (35). Very recently, it has been shown that 4EHP is modified by ISG15 and ISGylated (41). 4EHP is an mRNA 5′ cap-binding protein expressed ubiquitously and acts as a translation suppressor of cap-dependent translation by competing with eIF4E for binding to the cap structure. The ISGylation of 4EHP drastically increases its cap-binding activity, suggesting that 4EHP may play an important role in the regulation of cap-dependent translation (41). These data indicate that influenza virus mRNAs would be translated even under conditions of high competition of eIF4E binding to cap structures due to ISGylation of the 4EHP protein.
We have shown that viral polymerase binds to translation initiation complexes (Fig. 2) and that influenza virus infection triggers the eIF4G association with cap structures (Fig. 7). Collectively, these results support a model in which influenza virus polymerase, bound to the viral 5′ UTR common sequence, would remain associated with the capped 5′ end of the viral mRNAs, avoiding the replacement by eIF4E and recruiting translation initiation complexes. In this context, it should be mentioned that m7GTP is a 200-fold-less-potent cap-binding inhibitor with influenza virus polymerase than with eIF4E factor (27), suggesting that viral polymerase binds to cap structures with greater affinity than eIF4E does. Although viral polymerase associates with eIF4GI-containing complexes, the direct partner of the viral polymerase among the translation initiation factors still remains unknown, and we are conducting experiments to elucidate the possible role of eIF4GI in this association. In addition, the interaction of NS1 with the translation initiation factors eIF4GI and PABP1 could help the formation of a “closed loop” between the 5′ and 3′ ends of the viral mRNA. Thus, both activities would cooperate actively to recruit translation initiation complexes and would enhance the efficiency of viral mRNA translation. At the initial step of infection, virion RNPs are transported into the nucleus, where they start viral transcription and replication. Viral mRNAs from primary transcription are then translated to generate new polymerase subunits and NP protein to perform influenza virus genome amplification. Previous studies have shown that the viral polymerase complex not associated in viral RNPs can be detected in the cytoplasm of infected cells at approximately 2.5 to 3.5 hpi (11). Thus, it is conceivable that translation at very early times postinfection takes place without the contribution of viral proteins, and as the infection proceeds and the eIF4E factor becomes dephosphorylated, the viral polymerase localized in the cytoplasm helps to recruit translation initiation complexes to viral mRNAs. This proposal is in agreement with the partial impairment of viral protein synthesis observed at early times postinfection in influenza virus-infected cells treated with rapamycin (Fig. 3). Further characterization of the role of polymerase in viral translation could facilitate an understanding of the underlying mechanisms involved in the preferential translation of viral mRNAs during influenza virus infection.
Concluding remarks.
Unlike other alternative viral translation mechanisms that elude cap utilization, here, we propose an alternative cap-dependent way to initiate influenza virus mRNA translation. The results presented showing normal progression of influenza virus infection during functional impairment of cap-binding eIF4E factor are in agreement with a growing list of reports pointing out the reduced dependence of the cellular cap-binding factor for viral cap mRNA translation exerted by different viruses with 5′ cap structures (adenovirus, vesicular stomatitis virus, and dengue virus). They also appear to indicate that this mechanism is more widespread than previously thought and could be a system developed by certain viruses to evade the requirement of a fully active cap initiation complex, contributing to the repression of host cell mRNA translation.
Acknowledgments
We are indebted to J. Ortin, U. Garaigorta, A. Rodriguez, and T. Lutz for critical reviews of the manuscript. The technical assistance of Y. Fernández and N. Zamarreño is gratefully acknowledged. I.B. thanks A. L. Corbí for allowing the time to perform experiments for this report.
This work was supported by Ministerio de Educación y Ciencia (grants BMC2002-01141 and BFU2005-02834).
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Aragón, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beloso, A., C. Martínez, J. Valcárcel, J. Fernández-Santarén, and J. Ortín. 1992. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J. Gen. Virol. 73:575-581. [DOI] [PubMed] [Google Scholar]
- 3.Beretta, L., A. C. Gingras, Y. V. Svitkin, M. N. Hall, and N. Sonenberg. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15:658-664. [PMC free article] [PubMed] [Google Scholar]
- 4.Blaas, D., E. Patzelt, and E. Keuchler. 1982. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10:4803-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgui, I., T. Aragón, J. Ortín, and A. Nieto. 2003. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 6.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, F. F., and D. Doyle. 1985. Turnover of plasma membrane proteins in rat hepatoma cells and primary cultures of rat hepatocytes. J. Biol. Chem. 260:3097-3107. [PubMed] [Google Scholar]
- 8.Connor, J. H., and D. S. Lyles. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76:10177-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Luna, S., P. Fortes, A. Beloso, and J. Ortin. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Detjen, B. M., C. St. Angelo, M. G. Katze, and R. M. Krug. 1987. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J. Virol. 61:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Veer, M. J., C. A. Sledz, and B. R. Williams. 2005. Detection of foreign RNA: implications for RNAi. Immunol. Cell Biol. 83:224-228. [DOI] [PubMed] [Google Scholar]
- 13.Edgil, D., C. Polacek, and E. Harris. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 80:2976-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami, K., T. A. Sato, S. Nakada, and M. Enami. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enjuanes, L., F. Almazan, I. Sola, and S. Zuniga. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60:211-230. [DOI] [PubMed] [Google Scholar]
- 16.Fadden, P., T. A. Haystead, and J. C. Lawrence, Jr. 1997. Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J. Biol. Chem. 272:10240-10247. [DOI] [PubMed] [Google Scholar]
- 17.Falcón, A., R. Marión, T. Zürcher, P. Gomez, A. Portela, A. Nieto, and J. Ortín. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feigenblum, D., and R. J. Schneider. 1993. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J. Virol. 67:3027-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garaigorta, U., A. M. Falcón, and J. Ortín. 2005. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J. Virol. 79:15246-15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garfinkel, M. S., and M. G. Katze. 1992. Translational control by influenza virus. Selective and cap-dependent translation of viral mRNAs in infected cells. J. Biol. Chem. 267:9383-9390. [PubMed] [Google Scholar]
- 21.Garfinkel, M. S., and M. G. Katze. 1993. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J. Biol. Chem. 268:22223-22226. [PubMed] [Google Scholar]
- 22.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 24.Gingras, A. C., B. Raught, and N. Sonenberg. 2004. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279:169-197. [DOI] [PubMed] [Google Scholar]
- 25.Gross, J. D., N. J. Moerke, T. von der Haar, A. A. Lugovskoy, A. B. Sachs, J. E. McCarthy, and G. Wagner. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115:739-750. [DOI] [PubMed] [Google Scholar]
- 26.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooker, L., R. Sully, B. Handa, N. Ono, H. Koyano, and K. Klumpp. 2003. Quantitative analysis of influenza virus RNP interaction with RNA cap structures and comparison to human cap binding protein eIF4E. Biochemistry 42:6234-6240. [DOI] [PubMed] [Google Scholar]
- 28.Huarte, M., A. Falcón, Y. Nakaya, J. Ortín, A. García-Sastre, and A. Nieto. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inglis, S. C. 1982. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza and herpes simplex virus. Mol. Cell. Biol. 2:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katze, M. G., Y. T. Chen, and R. M. Krug. 1984. Nuclear-cytoplasmic transport and VAI RNA-independent translation of influenza viral messenger RNAs in late adenovirus-infected cells. Cell 37:483-490. [DOI] [PubMed] [Google Scholar]
- 31.Katze, M. G., D. DeCorato, and R. M. Krug. 1986. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J. Virol. 60:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katze, M. G., and R. M. Krug. 1984. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol. Cell. Biol. 4:2198-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassnig, C., A. Kolb, B. Strobl, L. Enjuanes, and M. Muller. 2005. Studying human pathogens in animal models: fine tuning the humanized mouse. Transgen. Res. 14:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin IV. 2007. From the cover: IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, S., N. Sonenberg, A. C. Gingras, M. Peterson, S. Avdulov, V. A. Polunovsky, and P. B. Bitterman. 2002. Translational control of cell fate: availability of phosphorylation sites on translational repressor 4E-BP1 governs its proapoptotic potency. Mol. Cell. Biol. 22:2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, G. X., W. Luytjes, M. Enami, and P. Palese. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 65:2861-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeshima, Y., A. Sudhakar, J. C. Lively, K. Ueki, S. Kharbanda, C. R. Kahn, N. Sonenberg, R. O. Hynes, and R. Kalluri. 2002. Tumstatin, an endothelial cell-specific inhibitor of protein synthesis. Science 295:140-143. [DOI] [PubMed] [Google Scholar]
- 40.Mothe-Satney, I., D. Yang, P. Fadden, T. A. Haystead, and J. C. Lawrence, Jr. 2000. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol. Cell. Biol. 20:3558-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okumura, F., W. Zou, and D. E. Zhang. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21:255-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oridate, N., H. J. Kim, X. Xu, and R. Lotan. 2005. Growth inhibition of head and neck squamous carcinoma cells by small interfering RNAs targeting eIF4E or cyclin D1 alone or combined with cisplatin. Cancer Biol. Ther. 4:318-323. [DOI] [PubMed] [Google Scholar]
- 43.Park, Y. W., and M. G. Katze. 1995. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 270:28433-28439. [DOI] [PubMed] [Google Scholar]
- 44.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. Fernandez-Santarén, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847-858. [DOI] [PubMed] [Google Scholar]
- 47.Ptushkina, M., T. von der Haar, M. M. Karim, J. M. Hughes, and J. E. McCarthy. 1999. Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J. 18:4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvatore, M., C. F. Basler, J.-P. Parisien, C. M. Horvath, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheper, G. C., and C. G. Proud. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269:5350-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 51.Shih, S. R., and R. M. Krug. 1996. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology 226:430-435. [DOI] [PubMed] [Google Scholar]
- 52.Wigler, M., A. Pellicer, S. Silverstein, R. Axel, G. Urlaub, and L. Chasin. 1979. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc. Natl. Acad. Sci. USA 76:1373-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xi, Q., R. Cuesta, and R. J. Schneider. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18:1997-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youtani, T., K. Tomoo, T. Ishida, H. Miyoshi, and K. Miura. 2000. Regulation of human eIF4E by 4E-BP1: binding analysis using surface plasmon resonance. IUBMB Life 49:27-31. [DOI] [PubMed] [Google Scholar]