Abstract
Hepatitis C virus (HCV) has a linear positive-stranded RNA genome of ∼9,600 nucleotides in length and displays a high level of sequence diversity caused by high mutation rates and recombination. However, when we performed long distance reverse transcription-PCRs on HCV RNA isolated from serum of chronic HCV patients, not only full-length HCV genomes but also HCV RNAs which varied in size from 7,600 to 8,346 nucleotides and contained large in-frame deletions between E1 and NS2 were amplified. Carefully designed control experiments indicated that these deletion mutants are a bona fide natural RNA species, most likely packaged in virions. Moreover, deletion mutants were detected in sera of patients infected with different HCV genotypes. We observed that 7/37 (18.9%) of genotype 1, 5/43 (11.6%) of genotype 3, and 4/13 (30.7%) of genotype 6 samples contained HCV deletion mutant genomes. These observations further exemplify HCV's huge genetic diversity and warrant studies to explore their biological relevance.
Hepatitis C virus (HCV) is an enveloped virus that belongs to the family Flaviviridae. Its linear positive-stranded RNA genome of approximately 9.6 kb in length encodes both structural (core, E1, and E2) and nonstructural (p7, N2, NS3, NS4a/b, and NS5a/b) proteins in a single open reading frame, flanked by short conserved untranslated regions (UTRs) located at the 5′ and 3′ ends of the genome that are required for viral replication and protein translation (5, 7, 8). One of the most striking characteristics of HCV is its capacity to cause a chronic liver disease in a high percentage of individuals (24). To do so, HCV encodes proteins which promote persistence, and sequence variation, especially in the envelope genes (E1 and E2), results in escape from adaptive immune responses (9, 27).
Lack of proofreading ability of the viral RNA-dependent RNA polymerase is the driving force behind HCV's genetic diversity. As a result of the large amount (1012) of virions produced each day in chronic hepatitis C patients and the rate of incorrect nucleotide insertions, which reaches the order of 10−3 to 10−4 base substitutions per site per year, HCV quasispecies are generated (22, 26). Recombination may be another mechanism by which genetic diversity is driven, given the recent identification of naturally occurring intergenotypic recombinant viruses (13, 21). Because of the huge genetic diversity, HCVs are currently categorized into six major genotypes and more than 80 subtypes (25).
HCV genetic variation has been studied in relation to epidemiology, response to antiviral therapy, and clinical parameters, using different techniques that have focused on short genomic regions. However, analysis of full-length viral genomes may be necessary to better understand the characteristics of HCV. Previously, we analyzed sera from HCV RNA-positive blood donors from Ho Chi Minh City, Vietnam, in order to analyze the molecular heterogeneity of HCV in Southeast Asia (21). Based on sequence analysis of core and NS5b regions in a set of sera, two samples were identified which contained viruses of different genotypes. Whole-genome analysis and bootscan analysis of one particular sample revealed a recombinant virus with genotype 2i and genotype 6p sequences at the 5′ and 3′ ends, respectively. Partial characterization of the other sample revealed the presence of a full-length genotype 2i virus and another genotype 6 virus which could be characterized only partially. In the present study, we further characterized this virus by a large-fragment amplification method. Surprisingly, we identified a naturally occurring HCV deletion mutant genotype 6h virus that contained a large in-frame deletion of E1 and E2 genes. Further studies confirmed the existence of circulating HCV E1-E2 deletion mutants in a substantial percentage of chronic hepatitis C patients.
MATERIALS AND METHODS
Samples.
HCV-positive sera of blood donors from Ho Chi Minh City, Vietnam, and Bangkok, Thailand, were obtained during the period from 2000 to 2002. Samples were genotyped based on sequence analysis as described previously (21). Thirteen samples of HCV genotype 6 were selected and characterized in this study. Other sera from chronic hepatitis C patients were obtained from the Erasmus MC, Rotterdam, The Netherlands; 37 plasma samples of HCV genotype 1 and 43 samples of genotype 3 were randomly selected for this study. All samples were kept at −80°C until further analysis.
RNA extraction and large-fragment PCR amplification.
Total RNA was isolated from 100 μl serum by proteinase K digestion and phenol-chloroform extraction as previously described (23) and dissolved in 20 μl sterile water. To generate a long fragment of cDNA, reverse transcription (RT) was performed on 10 μl RNA with Expand reverse transcriptase (Roche Diagnostics GmbH) and primer 5460 or 8625 or the 3′UTR, according to the manufacturer's instructions, in a total volume of 30 μl at 42°C for 2 h as described previously (21). Ten-μl volumes of cDNA were amplified with 2.5 units of Expand high-fidelity enzyme mixture (Roche Diagnostics GmbH), 1× Expand PCR buffer, 0.4 mM of each dideoxynucleotide, and 0.4 μM outer primer pairs with a thermal profile that was described previously (21). First-round amplification from each cDNA reaction resulted in three overlapping PCR fragments (primers 16 and 5460, 3227F and 8625, and 4039 and 3′UTR). Next, 2-μl volumes of amplified product were subjected to nested PCR with inner primers to generate overlapping genome fragments (primer pairs s17 and 3277R, 66 [5′-TCCCGCGAGAGCCATAGT-3′] and 3636, 1992 and 4662, 4039 and 7100, 5930 and Pr2, and Pr3 and 9325) under similar conditions as employed for the first PCR. Amplified products were gel purified with the QIAquick gel extraction kit (QIAGEN) and ligated directly into plasmid pCR2.1 (TA cloning kit; Invitrogen). For each amplicon, five positive clones were selected and sequenced.
To determine the sensitivity of HCV deletion mutants to RNase, we performed an assay as previously described (14). HCV plasma samples were treated with or without 0.1% Triton X-100 in phosphate-buffered saline at 37°C for 1 h; subsequently, 5 U of RNase A (QIAGEN) was added and incubation continued for 1 h. In order to degrade RNase A before RNA extraction, proteinase K was added to each reaction mixture and incubated at 37°C for 15 min prior to adding lysis buffer. RNA was extracted as previously described (23) and was analyzed by RT-PCR.
Large-fragment PCR and deletion mutant amplification using specific primers.
In order to avoid mismatch of degeneracy primers, specific primers were designed for large-fragment PCR of specific samples. Moreover, primers overlapping the deletion region of specific samples were designed and used for the deletion mutant amplification described in this study (Table 1). Conditions for RT-PCR were identical to the protocol described above.
TABLE 1.
Primers used for amplification HCV of specific samples
| Primera | Sequence (5′-3′) | Positionb | Polarity | Sample |
|---|---|---|---|---|
| 3277R | CTRATRABYTTVDTCTCCAT | 3277-3296 | Reverse | All |
| 3277RP21 | GTGATGATCTTGGTCTCCAT | 3277-3296 | Reverse | P21 |
| 3277RR30 | GTGATGACCTTAATTTCCAT | 3277-3296 | Reverse | R30 |
| 3277RD33 | GTCATGAGCTTGGTCTCCAT | 3277-3296 | Reverse | D33 |
| 3277RD88 | GTGATAACCTTCTTCTCCAT | 3277-3296 | Reverse | D88 |
| DelFP21 | TGCGGATCTGTTTTCTGTG | 1182//2538 | Forward | P21 |
| DelFR30 | GCTGATGACGTTATGATGG | 987//2253 | Forward | R30 |
| DelFD33 | CTATGAGGCGGATATACC | 980//2389 | Forward | D33 |
| DelFD88 | CCCTGCACTATCCCCAAC | 913//2757 | Forward | D88 |
| DelFD54 | GCAGCATCGGTTATGAGG | 970//3003 | Forward | D54 |
The specific primer used for amplification of samples.
The nucleotide position of the specific primer designed over the cross-junction site of the deletion region. The nucleotide position is according to the numbering system for the prototype strain HCV H (accession number M67463). “//” indicates discontinuous sequences of the deletion regions.
cDNA synthesis using high-thermostability reverse transcriptase.
To prevent HCV RNA secondary structure formation during RT, we performed cDNA synthesis using high-thermostability reverse transcriptase enzyme. Transcriptor reverse transcriptase (Roche) and Superscript III reverse transcriptase (Invitrogen) were used for cDNA syntheses, according to the manufacturers’ instructions. Protector RNase inhibitor (Roche) was used in this experiment because of its heat-resistant property. Conditions of the RT-PCR were identical to the protocol described above, except for the incubation times: Transcriptor reverse transcriptase was incubated at 50°C for 5 min and 55°C for 50 min, while Superscript III was used at 55°C for 50 min.
Southern blot hybridization.
Ten-μl aliquots of PCR amplification products were loaded on a 0.8% agarose-Tris-borate-EDTA buffer gel. DNA samples were denatured in 0.5 N NaOH and 1.5 M NaCl for 45 min and neutralized in 1 M Tris-HCl (pH 7.4) and 1.5 M NaCl for 45 min before transfer to Hybond N+ membranes (Amersham) by electroblotting (Amersham) in 1× Tris-borate-EDTA buffer according to the manufacturer's instructions. The blots were air dried for 30 min and UV irradiated for 10 min. The hybridization procedure was conducted as described elsewhere (6) with biotin-labeled probe specific for the 5′UTR (5′-ATTCCGGTGTACTCACCGGTTCCG-3′, nucleotides 149 to 174) at 50°C for 3 h. Membranes were washed and incubated with streptavidin-beta peroxidase conjugate (Roche) at 42°C for 45 min. After several washes the blots were visualized with enhanced chemiluminescence detection reagents (Roche) according to the protocol of the manufacturer and by exposure to hyperfilm (Amersham) for 1 to 5 min.
Preparation of synthetic complete and deletion mutant HCV RNA.
HCV large-fragment amplicons were cloned into plasmid pCR2.1 (TA cloning kit; Invitrogen) under control of the T7 promoter. Plasmids were linearized with BamHI and purified by QIAquick PCR purification (QIAGEN), and in vitro transcription was performed as described elsewhere (2). Reaction mixtures contained ∼ 1 μg of linearized DNA template, 5.0 mM ribonucleoside triphosphate mix, 40 U RNaseOUT (Invitrogen), 1× reaction buffer, and 50 U T7 RNA polymerase (Invitrogen) in a total volume of 20 μl. After 2 h at 37°C, 5 U DNase I (QIAGEN) was added and incubation was prolonged for 1 h. DNase-treated synthetic RNA was purified by phenol-chloroform extraction and used as a template for RT-PCR.
DNA sequencing and sequence analysis.
Sequence reactions on PCR products and plasmids were performed using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems) and an ABI Prism 3100 autosequencer (Applied Biosystems). Multiple sequence alignments were generated with the BioEdit program (version 7.0.1).
Nucleotide sequence accession numbers.
New sequences reported in this study have been submitted to GenBank and have been assigned the following accession numbers: RP21.complete (EF420126), RP21.deletion (EF420127), RP30.complete (EF420128), RP30.deletion (EF420129), D88.complete (EF420130), D88.deletion (EF420131), D33.complete (EF420132), D33.deletion (EF420133), and D54.deletion (EF420134).
RESULTS
Identification of an HCV genotype 6 deletion mutant.
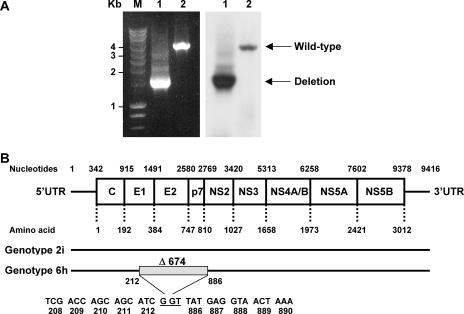
In order to study the genetic diversity of HCV in Southeast Asia, we analyzed serum samples obtained from Vietnamese blood donors. Previously, we reported that cloning of the 5′UTR-to-core region of one specific sample (D54) revealed the presence of genotype 2i and 6h strains, and the whole genome of the genotype 2i virus was published previously (21). However, short-fragment amplification of nucleotides 17 to 2085 (according to the numbering system of the prototype strain, M67463, genotype 1a) and nucleotides 1992 to 3277 only amplified genotype 2i sequences; genotype 6h sequences were never obtained (21). Remarkably, large-fragment amplification of nucleotides 17 to 3277 always amplified a smaller product of approximately 1,200 bp. A subsequent effort to amplify a fragment from nucleotides 66 to 3636 again revealed the amplification of a small PCR product (approximately 1,500 bp). In contrast, some other samples, such as D42, showed the expected amplicon of 3,600 bp (Fig. 1A). Southern blot hybridization using an HCV-specific probe revealed the specificity of the PCR products. Subsequent nucleotide sequence analysis of the obtained amplicons revealed a 2,022-nucleotide (674- amino-acid) in-frame deletion in E1/E2/p7 and part of the NS2 region (Fig. 1B) in the genotype 6h strain. Because of the nature of the PCR method, the smaller genotype 6h fragment is amplified preferentially over the larger strain 2i fragment. Thus, both a full-length 2i strain and a deletion mutant of the 6h strain seem to be present in sample D54.
FIG. 1.
Identification of an HCV genotype 6h virus with a deletion in the E1-NS2 region in a patient (D54) doubly infected with genotypes 2i and 6h. (A) RT-PCR analysis (nucleotides 66 to 3636) of HCV RNA from sample D54 (lane 1) and sample D42 (lane 2) and the subsequent Southern blot analysis are shown in the left and right panels, respectively. Amplicons were separated in 0.8% agarose gels along with a marker (M) for which the molecular masses are given on the left side (in kilobase pairs). The deletion mutant and wild-type HCV genome are indicated on the right side. (B) The upper panel depicts the genome organization of HCV, with boxes indicating the coding regions for the core protein (C), envelope 1 and 2 (E1 and E2) proteins, p7, and nonstructural proteins 1 to 5 (NS1 to NS5). Also indicated are the 5′ and 3′ UTRs and the nucleotide and amino acid numbers, according to the numbering system for the prototype strain HCV-H (accession number M67463). In the lower panel, black lines indicate the D54 full-length (genotype 2i, DQ155561) and deletion (genotype 6h) sequences. The shaded box displays the position of the in-frame deletion region (Δ674).
Detection of HCV deletion mutants in chronically infected hepatitis C patients.
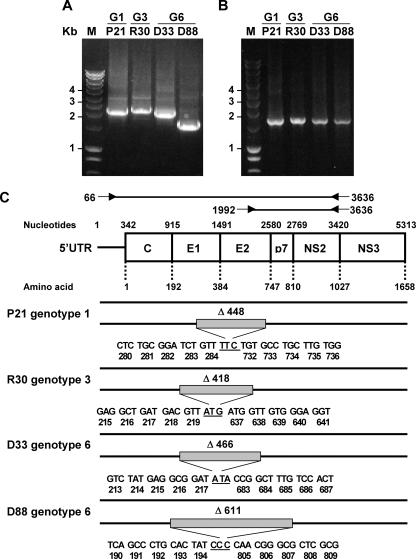
To determine whether HCV deletion genomes can be detected in other hepatitis C patients, we randomly selected plasma samples obtained from blood donors or chronic hepatitis C patients infected with HCV genotypes 1, 3, or 6 and performed a long-fragment amplification of the HCV genome (nucleotides 66 to 3636). We observed that 7/37 (18.9%) of genotype 1, 5/43 (11.6%) of genotype 3, and 4/13 (30.7%) of genotype 6 samples contained HCV deletion mutant genomes. RT-PCR analysis performed on at least two independent RNA isolations from the same patient sample resulted in the same deletion products. Four samples (genotypes 1, 3, and 6) were selected and found to contain deletion mutants of varying size (Fig. 2A). In contrast, amplicons generated by primer 1992 (located in the deleted region) and 3636 showed bands of the expected size (1,644 bp) for all samples, indicating that nondeleted genomes were present in plasma as well (Fig. 2B). HCV specificity of the amplified bands was confirmed by Southern blot analysis (data not shown). Sequencing of the fragments confirmed the presence of in-frame deletions in the E1-p7 region (Fig. 2C). Sequence analysis of three to five clones per sample revealed that the sequence of the deletion junction was identical in all clones for a given sample. Moreover, there were minor differences in the sequences of regions flanking the deletion junctions, implying independent quasispecies diversification of the deletion variants relative to their wild-type parent (Fig. 3).
FIG. 2.
HCV deletion mutants occur in the serum of patients chronically infected with HCV genotypes 1, 3, or 6. (A and B) RT-PCR products obtained from HCV genotype 1 (G1; P21), genotype 3 (G3; R30), and genotype 6 (G6; D33 and D88) serum samples, using degenerate primers for amplification of HCV nucleotides 66 to 3636 (A) and 1992 to 3636 (B), were separated on 0.8% agarose gels. The molecular mass marker (M) is indicated on the left side in kilobase pairs. (C) Schematic representation of complete and deletion genomes determined for the different HCV genotypes. The upper panel depicts the genome organization of HCV as in Fig. 1. Forward and reverse primers to detect complete and deletion HCV genomes are presented. In the lower panel, black lines indicate the full-length and deletion sequences. The shaded boxes display the positions of the in-frame deletion region.
FIG. 3.
Consensus sequence alignments of different HCV full-length complete genomes (com) and deletion mutants (del). Nucleotide differences between complete and deletion mutant sequences are shown in bold. Residues identical to the major sequence are indicated by a dash, the deletion region is indicated by dots, and slashes indicate discontinuous sequences of the deletion regions. The nucleotide positions are presented above the sequences (numbering system for the prototype strain HCV-H), and sample names are indicated in the left-hand column. Complement nucleotide codes: R = T or C; Y = G or A.
Experimental confirmation of HCV E1-E2 deletion mutants.
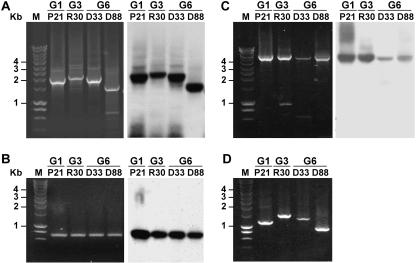
To confirm that the deletion genomes were not RT-PCR artifacts, we performed a wide array of experiments to prove that the deletion mutant genomes are bona fide RNA species present in the serum of chronically infected patients. First of all, the initial large-fragment PCR analysis utilized degenerate primers for amplification (21). To avoid the possibility of primer mismatch during amplification, reverse primers 3277R (nucleotides 3277 to 3296) specific for sample P21 (genotype 1), R30 (genotype 3), and D33 and D88 (genotype 6) were used (Table 1). Subsequent analysis using these specific primers revealed similar results to those observed in the previous experiment (Fig. 4A). In some but not all experiments, the full-length genome could be detected although at a lower level, given the preferential amplification of the deletion mutant genomes. Moreover, amplification of the 5′UTR-to-core (nucleotides 17 to 750) and NS3-to-NS5 (nucleotides 4039 to 8625) regions showed no evidence for the presence of deletions in these parts of the HCV genome (Fig. 4B and C). Next, we designed isolate-specific primers overlapping the deleted region (Table 1). As presented in Fig. 4D, these primers amplified HCV-specific products with appropriate lengths in the samples they were designed for, but not in others (not shown).
FIG. 4.
Experimental confirmation of HCV E1-E2 deletion mutants. (A to C) RT-PCR (left panel) and Southern blot analysis (right panel) of HCV genotype 1 (P21), genotype 3 (R30), and genotype 6 (D33 and D88), using sequence-specific primers to amplify the regions 5′UTR to NS3 (nucleotides 17 to 3277) (A), 5′UTR to core (nucleotides 17 to 750) (B), and NS3 to NS5 (nucleotides 4039 to 8625) (C). Amplicons were separated in 0.8% agarose gels along with a marker (M) for which the molecular masses are given on the left side in kilobase pairs. (D) Gel electrophoresis of RT-PCR products amplified from RNA of samples P21, R30, D33, and D88 with primer 3636 and a junction site primer overlapping the deleted region that was specifically designed for each sample (Table 1). The molecular mass marker (M) is indicated on the left side (in kilobase pairs).
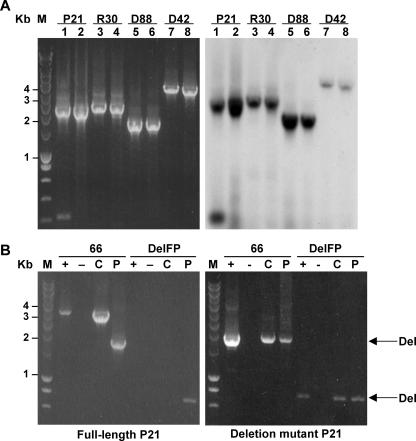
To minimize the frequency of template switching during cDNA synthesis (18), we used (RNase H-deficient) high-thermostability reverse transcriptase enzymes to perform the cDNA synthesis. Using these enzymes, small-sized nonspecific bands disappeared but deletion genome fragments were detected using both Transcriptor reverse transcriptase (Fig. 5) and Superscript III reverse transcriptase (not shown). Southern blot analysis confirmed the specificity of the amplified products (Fig. 5A). In addition, we designed a simulation experiment in which we performed an RT-PCR assay with in vitro-transcribed RNA obtained from cloned DNA templates derived from the full-length or deletion genome of sample P21. Amplification of the full-length RNA template revealed only bands for full-length template (no deletion form) (Fig. 5B), whereas the template for the deletion genome only showed bands for the deletion form (Fig. 5C). Based on all of the control experiments above, we conclude that the observed deletion mutants of HCV are bona fide natural RNA species and that the deletions are found only in the region encoding the E1, E2, p7, and NS2 proteins.
FIG. 5.
HCV deletion mutants are bona fide RNA species. (A) Comparison of RT-PCR amplification of HCV nucleotides 66 to 3636 (left panel) and Southern blot analysis (right panel) of HCV genotype 1 (P21), genotype 3 (R30), and genotype 6 (D33 and D88) serum samples, using Expand reverse transcriptase (lanes 1, 3, 5, and 7) and Transcriptor reverse transcriptase (lanes 2, 4, 6, and 8). (B) RT-PCR analysis of in vitro-transcribed full-length HCV RNA (left panel) and deletion mutant RNA (right panel) of sample P21 (genotype 1), using primer 3636 and either primer 66 or a junction site primer overlapping the deleted region (DelFP). Reactions were performed in the presence (+) or absence (−) of T7 RNA polymerase. As controls, a cDNA clone containing either full-length or deletion mutant HCV (C) or RNA isolated from the P21 serum sample (P) was used. The molecular mass marker (M) is shown in kilobase pairs on the left side. The amplicons representing deletion forms (Del) are indicated with an arrow.
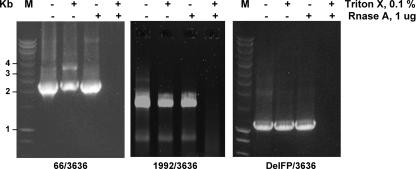
Circulating HCV deletion mutants are protected from RNase.
In case HCV deletion mutants in plasma originate from replicating viruses in the liver, viral RNAs should be packaged in viral particles and resist degradation by RNase A. Plasma was treated with Triton X-100 to disintegrate membranes, and the sample was treated with RNase A. As shown in Fig. 6, similar to HCV full-length genomes, HCV deletion mutants were detected in samples treated with RNase A but not when Triton X-100 was added concomitantly. These results were confirmed using deletion-specific primers (Fig. 6). Thus, circulating HCV deletion mutant genomes are most likely located within viral particles.
FIG. 6.
The HCV RNA genotype 1 (P21) deletion mutant circulating in plasma is protected from RNase A digestion. The P21 serum sample was treated with or without 0.1% Triton X-100 and 1 μg RNase A before RNA extraction. RT-PCR amplifications of nucleotides 66 to 3636 (deletion mutant; left panel) or 1992 to 3636 (full-length; middle panel) or of the deletion mutant with a junction site primer overlapping the deleted region (DelFP) and primer 3636 (deletion mutant; right panel) are shown. The molecular mass marker (M) is indicated on the left side (in kilobase pairs).
DISCUSSION
In this study, we identified and characterized circulating HCV deletion mutants in plasma of chronically infected HCV patients. These HCV RNAs always coexisted with full-length genomes and contained a large in-frame deletion that varied in size from 1,254 to 2,022 nucleotides located in the E1-NS2 region of the genome. We concluded that these deletion mutants are naturally occurring RNA species on the basis of carefully designed control experiments using deletion-specific primers for amplification, elevated temperatures for cDNA synthesis, and in vitro-transcribed RNA as template. Moreover, samples have their own unique deletion mutant that contains nucleotide substitutions compared to the full-length genome, suggesting independent evolution of the full-length genome and deletion mutant. Efforts to detect the full- length and deletion mutants by Northern blot techniques were unsuccessful. Therefore, accurate estimations of the ratios of both forms present in the plasma are difficult to make. The fact that only one dominant deletion form is detected per sample suggests that the “fittest” defective virus is selected. Last but not least, deletion mutants likely occur in all HCV genotypes, as we identified deletion mutants in ∼20% of chronic HCV patients infected with genotypes 1, 3, and 6. It is of note that our discovery seems to validate the claims of subgenomic HCV in liver biopsy specimens of chronic HCV patients and liver transplant recipients (12, 29). The region where the deletion occurs is similar in all studies, but in contrast to our findings, those authors reported also the presence of out-of-frame deletion genomes. Moreover, our findings generalize the detection of HCV deletion mutants to a significant portion of the chronic hepatitis C patients infected with different genotypes.
The deletion genomes contain essential parts for autonomous HCV replication (5′UTR, core, and NS2-NS5-3′UTR), regions similar to what has been described for the artificially constructed subgenomic replicons (17, 28), and may therefore represent naturally occurring replicons. Interestingly, both full-length genome and mutant genome were resistant to degradation by RNase A, suggesting that both RNA forms are packaged in virions. Assuming that HCV particle assembly requires functional E1/E2 glycoproteins, the deletion mutants can only be packaged and secreted from cells when cells are coinfected with a wild-type virus with a full-length genome.
The HCV deletion mutants are highly reminiscent of defective interfering (DI) particles described for many viruses (10), including members of the Flaviviridae family, such as tick-borne encephalitis virus, Murray Valley encephalitis virus, Dengue virus, West Nile virus, and Japanese encephalitis virus (1, 3, 15, 19, 30). DI viruses are mutants that arise spontaneously when the standard wild-type virus is passaged in tissue culture at high multiplicities of infection. The DI genomes contain cis-acting signals required for replication but lack part or most of the region encoding viral proteins (4). These smaller viruses can be complemented by coinfection with a helper (wild-type) virus and might interfere with the replication of helper virus through competition for limiting factors in the host cell (16). To our knowledge, most, if not all, DI particles have been discovered in laboratory settings, and HCV is the first positive-stranded RNA virus for which defective genomes have been described in a natural human infection in vivo.
Methods to quantify and determine the heterogeneity of HCV genomes are currently used routinely to guide antiviral therapy and to study the evolution and epidemiology of this virus. The existence of deletion mutants in chronic hepatitis C patients further exemplifies the huge diversity of HCV and has some major implications. First, viral load quantification methods based on amplification of the 5′UTR (20) detect both defective and complete genomes. Considering a high amount of circulating HCV deletion mutant, it may be of interest to differentiate between these genomic HCV forms in order to further unravel the pathogenesis of HCV and response to antiviral therapy. However, given the fact that only a fraction of the patients harbor deletion mutants, a large panel of samples from well-characterized patients needs to be screened. Last, but not least, it has been proposed that defective viruses may play a significant role in the establishment and maintenance of chronic infection in vivo (11), despite the fact that most if not all data come from in vitro experiments (15, 30). Whether HCV deletion genomes play a role in the persistence of HCV infection remains to be delineated.
Acknowledgments
This work was supported by the Commission of the European Community (HECSA Project, ICA4-CT-1999-10009).
We thank Vu Thuy Yen, Jan Brouwer, Paula van Luijt, Kavita Bedi, and Apiradee Theamboonlers for technical assistance and Ron Fouchier for valuable discussions.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Aaskov, J., K. Buzacott, H. M. Thu, K. Lowry, and E. C. Holmes. 2006. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311:236-238. [DOI] [PubMed] [Google Scholar]
- 2.Bredenbeek, P. J., A. F. Noten, M. C. Horzinek, and W. J. Spaan. 1990. Identification and stability of a 30-kDa nonstructural protein encoded by mRNA 2 of mouse hepatitis virus in infected cells. Virology 175:303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinton, M. A. 1983. Analysis of extracellular West Nile virus particles produced by cell cultures from genetically resistant and susceptible mice indicates enhanced amplification of defective interfering particles by resistant cultures. J. Virol. 46:860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann, A. J. (ed.). 2000. RNA viruses: a practical approach. Oxford University Press, Oxford, England.
- 5.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier, R. A., T. M. Bestebroer, S. Herfst, L. Van Der Kemp, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland, J. J. 1990. Defective viral genomes, p. 151-165. In B. N. Fields, D. M. Knipe, et al. (ed.), Fields virology, 2nd ed. Raven Press Ltd., New York, NY.
- 11.Huang, A. S., and D. Baltimore. 1970. Defective viral particles and viral disease processes. Nature 226:325-327. [DOI] [PubMed] [Google Scholar]
- 12.Iwai, A., H. Marusawa, Y. Takada, H. Egawa, K. Ikeda, M. Nabeshima, S. Uemoto, and T. Chiba. 2006. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J. Viral Hepat. 13:523-531. [DOI] [PubMed] [Google Scholar]
- 13.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 15.Lancaster, M. U., S. I. Hodgetts, J. S. Mackenzie, and N. Urosevic. 1998. Characterization of defective viral RNA produced during persistent infection of Vero cells with Murray Valley encephalitis virus. J. Virol. 72:2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazzarini, R., J. D. Keene, and M. Schubert. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145-154. [DOI] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 18.Luo, G., and J. Taylor. 1990. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 64:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolte, F. S. 2001. Hepatitis C virus genotyping: clinical implications and methods. Mol. Diagn. 6:265-277. [DOI] [PubMed] [Google Scholar]
- 21.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80:7569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto, H., M. Kojima, S. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, D. A. Peterson, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 23.Rispeter, K., M. Lu, S. Lechner, A. Zibert, and M. Roggendorf. 1997. Cloning and characterization of a complete open reading frame of the hepatitis C virus genome in only two cDNA fragments. J. Gen. Virol. 78:2751-2759. [DOI] [PubMed] [Google Scholar]
- 24.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 26.Smith, D. B., S. Pathirana, F. Davidson, E. Lawlor, J. Power, P. L. Yap, and P. Simmonds. 1997. The origin of hepatitis C virus genotypes. J. Gen. Virol. 78:321-328. [DOI] [PubMed] [Google Scholar]
- 27.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagi, S., K. Mori, E. Tanaka, A. Matsumoto, F. Sunaga, K. Kiyosawa, and K. Yamaguchi. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 77:399-413. [DOI] [PubMed] [Google Scholar]
- 30.Yoon, S. W., S. Y. Lee, S. Y. Won, S. H. Park, S. Y. Park, and Y. S. Jeong. 2006. Characterization of homologous defective interfering RNA during persistent infection of Vero cells with Japanese encephalitis virus. Mol. Cell 21:112-120. [PubMed] [Google Scholar]