Abstract
Recovery of plants from virus-induced symptoms is often described as a consequence of RNA silencing, an antiviral defense mechanism. For example, recovery of Nicotiana clevelandii from a nepovirus (tomato black ring virus) is associated with a decreased viral RNA concentration and sequence-specific resistance to further virus infection. In this study, we have characterized the interaction of another nepovirus, tomato ringspot virus (ToRSV), with host defense responses during symptom induction and subsequent recovery. Early in infection, ToRSV induced a necrotic phenotype in Nicotiana benthamiana that showed characteristics typical of a hypersensitive response. RNA silencing was also activated during ToRSV infection, as evidenced by the presence of ToRSV-derived small interfering RNAs (siRNAs) that could direct degradation of ToRSV sequences introduced into sensor constructs. Surprisingly, disappearance of symptoms was not accompanied by a commensurate reduction in viral RNA levels. The stability of ToRSV RNA after recovery was also observed in N. clevelandii and Cucumis sativus and in N. benthamiana plants carrying a functional RNA-dependent RNA polymerase 1 ortholog from Medicago truncatula. In experiments with a reporter transgene (green fluorescent protein), ToRSV did not suppress the initiation or maintenance of transgene silencing, although the movement of the silencing signal was partially hindered. Our results demonstrate that although RNA silencing is active during recovery, reduction of virus titer is not required for the initiation of this phenotype. This scenario adds an unforeseen layer of complexity to the interaction of nepoviruses with the host RNA silencing machinery. The possibility that viral proteins, viral RNAs, and/or virus-derived siRNAs inactivate host defense responses is discussed.
RNA silencing is an innate and ubiquitous defense mechanism that is activated by double-stranded RNAs (dsRNAs) and has been implicated in antiviral defense (6, 7, 60, 61). The vast majority of plant viruses possess a positive-sense single-stranded RNA genome that is replicated through dsRNA intermediates (31). In addition, folding of self-complementary stretches within the single-stranded RNA genome allows the formation of stem-loop structures, which provide local regions of dsRNA (66). Once detected by the surveillance machinery of the plant cell, viral dsRNAs are cleaved by dicer-like (DCL) RNase III enzymes into 21- to 24-nucleotide (nt) small interfering RNAs (siRNAs) (20, 23, 59). The siRNAs guide RNA-induced silencing complexes (RISCs) to the target RNA in a sequence-specific manner (61). The target RNA is then cleaved by argonaute (AGO) proteins, which are RNase H-like enzymes and are associated with the RISC (9, 74). In addition, host RNA-dependent RNA polymerases (RDRs) have been proposed to play a crucial role in amplifying the RNA silencing signal (5, 8, 60, 65). They bind the cleaved fragment released by RISC and produce a new generation of dsRNAs. This is probably followed by a new round of dicer cleavage to produce secondary siRNAs. siRNAs are also likely to be the mobile signal that spreads the defense response with or ahead of the invading virus, thus preventing viral accumulation in systemic leaves (26).
In plants, several pathways associated with RNA silencing of viruses, transgenes, or endogenous genes have been identified (11, 58). These pathways are distinct but interconnected and include common as well as specific components. Genes for 4 DCL enzymes, 6 RDRs, and 10 AGO proteins have been identified in the Arabidopsis genome (11). Although specific DCL enzymes have been implicated in the response to different viruses, they also exhibit redundant effects in virus-infected plants (10, 20). Similarly, the biochemical activity of RDRs appears to be both virus specific and redundant (6, 7).
The characterization of recovery phenotypes observed in some natural virus infections or in transgenic plants engineered for viral resistance provided the first experimental evidence for a link between RNA silencing and an antiviral defense mechanism (17, 25, 36, 44, 45). Recovery was first described nearly 80 years ago and is characterized by an initial symptomatic infection that is followed by attenuation or elimination of the symptoms in newly emerging leaves (68). In plants infected with tobacco ringspot virus (a nepovirus), recovered leaves were resistant to secondary infection with the same virus, although the sap extracted from these leaves readily induced typical symptoms on young healthy plants (68). It was later shown that host recovery from symptoms induced by another nepovirus (tomato black ring virus [TBRV]), a caulimovirus, and a tobravirus is accompanied by a reduction in viral RNA concentration (17, 44, 45). Moreover, sequence-specific resistance to further virus infection was also observed, suggesting that RNA silencing is activated prior to recovery and may be responsible for the reduced virus accumulation and surveillance phenotype (17, 44, 45).
Many viral proteins are able to suppress the plant RNA silencing response (60). Inactivation of suppressors of silencing in mutant viruses can induce a recovery phenotype in plants that would otherwise respond with systemic symptoms to infection by the wild-type viruses (21, 54). Furthermore, incubation of plants at an elevated temperature correlates with increased RNA silencing activity, lowered virus accumulation, and attenuation of symptom development or induction of recovery phenotypes (15, 55). Thus, recovery of plants from natural virus infection is often described as a consequence of RNA silencing (7, 38). However, recovery is not necessarily accompanied by virus clearance, as recently demonstrated for plants infected with an ilarvirus (tobacco streak virus) (70). Therefore, other factors are likely to be involved in the induction of recovery.
Tomato ringspot virus (ToRSV; a nepovirus) is an important pathogen of small fruit and fruit trees in North America (48, 53). On herbaceous hosts, it induces necrotic lesions, followed by the emergence of symptom-free new leaves. In this study, we have characterized host responses induced in Nicotiana benthamiana plants during the course of infection with ToRSV. We determined that initial development of symptoms was accompanied by the induction of a necrotic response with characteristics of the hypersensitive response (HR). Later during infection, the disappearance of symptoms was accompanied by ToRSV-specific RNA silencing but not by a significant reduction in viral RNA levels. Our results demonstrate that induction of recovery does not require a reduction in viral titer and suggest that viral proteins, RNAs, or virus-derived siRNAs function to counteract the host defense responses.
MATERIALS AND METHODS
Plants and viruses.
Transgenic N. benthamiana lines 16c (63) and RDR6i (51) were kindly provided by David Baulcombe (Sainsbury Laboratory, Norwich, United Kingdom). Transgenic homozygous N. benthamiana plants containing the RDR1 gene from Medicago truncatula (R0 15-1 4-41) or the T-DNA without the RDR1 transgene (V12-1 5-3) (72) were generously provided by Richard Nelson (Samuel Robert Noble Foundation, Ardmore, OK). Plants were cultivated under greenhouse conditions except for experiments with RDR6i plants, which were conducted in Conviron climatic chambers set at 21 and 27°C with a 16-h day length. For virus inoculations, infected tissues were macerated in 0.5 M NaPO4 buffer (pH 7.2), and the extracts were rub inoculated onto Carborundum-dusted plants.
Agroinfiltration experiments and detection of proteins.
Plasmids containing green fluorescent protein (GFP) and tomato bushy stunt virus p19 have been described previously (73). Sensor constructs were created by replacing the KpnI-SstI fragment of plasmid pGFP-cNV3 (73) with an 81-nt fragment of ToRSV RNA 1 (nt 3713 to 3794) or Prunus necrotic ringspot virus (PNRSV) RNA 3 (nt 258 to 339), generated by PCR and containing the corresponding restriction sites. Agroinfiltration experiments were conducted as described previously (64). Five days after infiltration, ∼100 mg of leaf tissue was crushed in liquid nitrogen, supplemented with 500 μl of protein extraction buffer (10 mM KCl, 5 mM MgCl2, 400 mM sucrose, 100 mM Tris-HCl, pH 8.1, 10% [vol/vol] glycerin, 0.007% [vol/vol] β-mercaptoethanol), and centrifuged at 4°C for 5 min at 20,000 × g. Separation of proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting were conducted as described previously (27), using a mouse monoclonal anti-GFP antibody (BD Bioscience) or an anti-ToRSV coat protein (CP) rabbit polyclonal antibody (47). The secondary antibody was goat anti-mouse or goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Bio/Can). Coomassie staining of rubisco protein is shown as a loading control.
Isolation and detection of RNA.
Total RNA was extracted with TRIzol reagent (Invitrogen) as recommended by the manufacturer.
(i) Northern blot analysis.
Five micrograms of total RNA was separated in a 1.2% formaldehyde-agarose gel as described previously (2). Unless otherwise indicated, the 25S rRNA stained with ethidium bromide (EtBr) was used as a loading control. RNAs were blotted onto a nylon membrane (Hybond-N+; Amersham) by capillary action in 10× SSC buffer (1.5 M NaCl, 0.15 M trisodium citrate, pH 7.0). The membrane was incubated on a gel drier (Bio-Rad) at 80°C for 2 h. For probe preparation, 25 ng of DNA was labeled with [α-32P]dCTP (3,000 Ci/nmol; Perkin-Elmer), using a random-priming DNA labeling system (Invitrogen). Membranes were soaked in 1× PerfectHyb Plus buffer (Sigma) and hybridized at 65°C for 16 h. Two posthybridization washes were performed at 65°C for 30 min, using 2× SSC containing 0.5% sodium dodecyl sulfate. Hybridization signals were collected using a Cyclone Plus storage phosphor system (Perkin-Elmer).
(ii) siRNA detection.
Low-molecular-weight plant RNAs were enriched from total RNA by differential precipitation as described previously (56). The purified RNAs were separated by denaturing polyacrylamide gel electrophoresis and transferred to Hybond-N+ membranes by electroblotting as described previously (2). The RNAs were cross-linked to the membrane under UV light (310 nm) for 3 min, and hybridizations with radiolabeled probes were performed as described above, with the exception that the hybridization and washes were conducted at 40°C.
(iii) Mapping of siRNAs.
Fragments of approximately 1,000 nt were amplified from the ToRSV genome by reverse transcription-PCR using suitable primers (sequences are available upon request). The amplified cDNA fragments were separated in 1% agarose gels, eluted, and precipitated with ethanol (EtOH). Equal amounts of each fragment (140 ng) were separated in a new 1% agarose gel. The gel was equilibrated in transfer solution (120 mM NaCl, 80 mM NaOH) for 30 min, and the cDNA fragments were transferred to a Hybond-N+ membrane by capillary action for 12 h. The membrane was then incubated in neutralizing solution (1.5 M NaCl, 0.5 M Tris-HCl, pH 7.5) for 15 min. The probe was prepared using low-molecular-weight RNAs purified from ToRSV-infected N. benthamiana plants as described previously (15). Approximately 8 μg of low-molecular-weight RNAs was dephosphorylated with calf intestinal alkaline phosphatase (Invitrogen) and end labeled with [γ-32P]dATP (3,000 Ci/nmol; Perkin-Elmer), using polynucleotide kinase (Invitrogen). Hybridizations were conducted as described for siRNA detection.
Diagnosis of HR, cell death, and transport blockage.
Detection of hydrogen peroxide by 3,3′-diaminobenzidine (DAB) staining (57) and staining of starch (46) were done as described previously. Cell death diagnosis by trypan blue staining was modified from a previously described method (33) as follows. Leaves were boiled for 1 minute in trypan blue solution (12.5% phenol [wt/vol], 12.5% glycerol [vol/vol], 12.5% [vol/vol] lactic acid, 48% [vol/vol] EtOH, and 0.025% [wt/vol] trypan blue) and incubated overnight at room temperature. Samples were repeatedly destained in 70% chloral hydrate. For microscopic imaging, samples were analyzed with an Axiophot/Photomicroscope (Zeiss, Germany) under bright-field optics.
Electron microscopy.
Leaf sections were fixed in glutaraldehyde, postfixed in osmium tetroxide (OsO4), dehydrated in EtOH, and embedded in Spur's resin (Polyscience) following standard protocols (available upon request). Ultrathin sections were sliced with a Reichert-Jung Ultracut microtome (AO Instrument Company, NY), mounted on Formvar-coated nickel grids, stained with uranyl acetate, and examined using a JEM-1000CX II transmission electron microscope (JEOL Ltd., Japan).
Photography.
DAB-, trypan blue-, and iodine potassium iodide (JJK)-stained samples were photographed under trans-illumination. In the last case, plants were placed to float on water. For GFP imaging, plants were illuminated with a long-wavelength UV light (model B 100 A; Black Ray, Upland, CA) and photographed with a medium-wavelength yellow filter (Rodenstock, Munich, Germany). For microscopic imaging of GFP, we used an epifluorescence microscope (Axiophot) with the following settings: excitation filter, 450 to 490 nm; dichroic mirror, 510 nm; band-pass barrier filter, 515 to 565 nm; and beam splitter, 510 nm. Images were captured with a Nikon D-200 digital camera and processed using Nikon Capture NX software.
Nucleotide sequence accession number.
The sequence of a 700-nt fragment from the RDR1 gene of Nicotiana clevelandii was deposited in the NCBI database under accession number EF594060.
RESULTS
ToRSV induces an HR-like necrotic response in N. benthamiana.
N. benthamiana plants infected with ToRSV exhibit the following two characteristic symptoms: (i) inoculated leaves show concentric necrotic ring spots (Fig. 1a) and (ii) depending on the intensity of the infection, the leaves immediately above develop chlorotic and/or necrotic lesions surrounding the veins (Fig. 1b). Shortly after the appearance of the second symptom, plants recover and newly emerged leaves resemble healthy ones (Fig. 1c). The severity of infection is variable and is influenced by temperature, light, and relative humidity. If infection is severe (under conditions of low temperature, short illumination periods, and high humidity), plants may die or develop strong necrosis that usually follows the above-mentioned spatial pattern surrounding the veins. Moreover, ring spots may fail to develop if infection proceeds quickly.
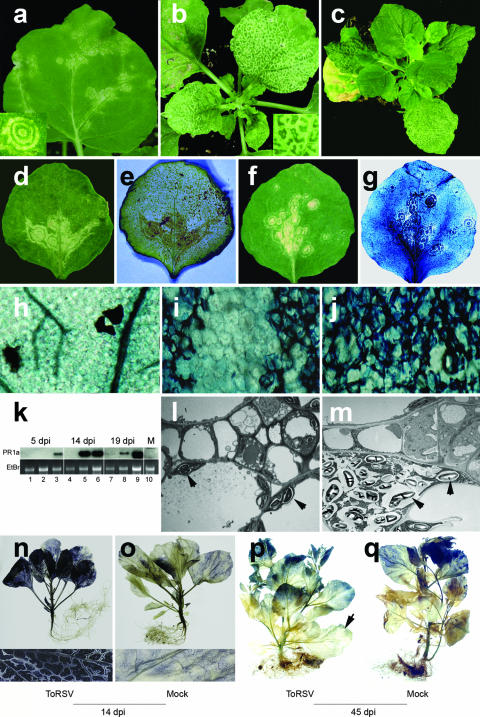
FIG. 1.
Characterization of the recovery phenotype in N. benthamiana plants infected with ToRSV. (a to c) Symptom development in ToRSV-infected plants. ToRSV-induced symptoms in inoculated (a) and first systemic (b) leaves are followed by the emergence of recovered leaves (c). (d and e) Colocalization of hydrogen peroxide production and necrotic symptoms. Symptomatic leaves were stained with DAB and photographed before (d) and after (e) staining. (f to j) Colocalization of cell wall collapse and necrotic symptoms. Symptomatic leaves were stained with trypan blue and photographed before (f) and after (g) staining. Close-up examinations of trypan blue-stained healthy tissues (h) and ToRSV-infected tissues [two adjacent necrotic rings separated by a nonnecrotic region (i) and a necrotic vein (j)] are shown. (k) Transcriptional activation of PR1a in ToRSV-infected tissues. For each sampling date, the inoculated leaf (lanes 1, 4, and 7), the first systemically infected leaf (lanes 2, 5, and 8), and the apical leaf (lanes 3, 6, and 9) were included. PR1a transcripts were detected by Northern blotting using a full-length N. benthamiana PR1a cDNA probe. Lane M, mock-inoculated plant. (l and m) Accumulation of starch grains in ToRSV-infected cells. Cells from mock-inoculated (l) or ToRSV-infected (m) plants were stained with uranyl acetate and analyzed by electron microcopy. Arrowheads indicate the locations of starch grains. (n to q) Impairment of starch translocation in ToRSV-infected plants. Accumulation of starch was revealed by JJK staining of ToRSV-infected or mock-inoculated plants at 14 or 45 dpi, as indicated. Lower insets are close-up magnifications of leaf sections from plants shown in panels n and o. The arrow in panel p points to a leaf that was initially symptomatic.
DAB staining revealed colocalization of hydrogen peroxide with ring spots in the inoculated leaves (Fig. 1d and e; specific DAB staining in Fig. 1e colocalizes with the symptoms shown in Fig. 1d) and necrotic veins in the first systemically infected leaves (data not shown). In addition, uptake of trypan blue was observed in cells with concentric ring spots (Fig. 1f, g, and i) and necrotic veins (Fig. 1j and data not shown). This is a sign of plasma membrane disruption, indicating that cell death had occurred. Microscopic analysis of two adjacent ring spots separated by an area of living cells (Fig. 1i) confirmed the specificity of this assay. Trypan blue uptake was not observed in leaves from mock-inoculated plants (Fig. 1h). Symptoms also correlated with the induction of mRNA of pathogenicity- related protein 1a (PR1a) (Fig. 1k). Plants exhibiting a mild symptomatic (5 days postinoculation [dpi]), strong symptomatic (14 dpi), or recovered (19 dpi) phenotype were sampled at the indicated time points. PR1a transcripts were first detected in the apical leaves of infected plants at 5 dpi (Fig. 1k, lane 3). By 14 dpi, PR1a mRNA levels increased in both the first systemically infected leaves (showing necrotic veins) and the uppermost leaves (Fig. 1k, lanes 5 and 6). By 19 dpi, PR1a mRNA was abundant only in the uppermost recovered leaf and had almost vanished from lower symptomatic leaves (Fig. 1k, lanes 7 to 9). In summary, PR1a induction moved with or ahead of the virus infection front and was present after the plants recovered. The induction of hydrogen peroxide, the transcriptional activation of PR1a, and the cell death associated with the initial stages of ToRSV infection are common features of the HR that some plants exhibit upon pathogen attack (28).
ToRSV infection causes starch transport blockage.
ToRSV-infected N. benthamiana plants are frequently shorter than healthy ones, and their leaves exhibit a darker, more coriaceous appearance. Staining of leaves from ToRSV-infected (Fig. 1n) or mock-inoculated (Fig. 1o) plants with JJK revealed a higher level of starch accumulation in ToRSV-infected leaves. Closer examination showed that in ToRSV-infected leaves, the area surrounding the veins was not loaded with starch (Fig. 1n, close-up). However, starch traces were visible inside the veins, suggesting that uploading of starch into the vasculature was impaired in ToRSV-infected leaves. Interestingly, the spatial pattern of starch exclusion bears a resemblance to that of the necrotic veins shown in Fig. 1b (inset). One possible interpretation of this result is that the boundary between bundle sheath and vascular parenchyma and/or companion cells is a checkpoint that hinders transport in symptomatic leaves infected with ToRSV. This suggestion is supported by the observation that starch grains accumulate in the vicinity of cells of the vasculature in ToRSV-infected plants (compare Fig. 1l and m). The blockage of starch transport was consistently observed at 14 dpi, when symptoms were strongest (Fig. 1n), but was largely alleviated by 45 dpi, when plants had recovered from symptoms (compare infected and healthy plants in Fig. 1p and q).
Chlorophyll degradation is a mechanism to stop the production of photosynthates, thereby preventing toxicity resulting from accumulation of carbohydrates in plants that suffer from transport blockage (30). The concentration of chlorophyll was found to be lower in ToRSV-infected leaves than in healthy ones (1.25 ± 0.16 mg/g versus 5.69 ± 0.51 mg/g), which correlates with the extreme chlorosis observed upon severe infection with ToRSV (data not shown). This result is consistent with the temporary transport blockage observed in ToRSV-infected N. benthamiana plants.
Recovery is not accompanied by a commensurate reduction in ToRSV RNA concentration.
We sought to investigate the role of RNA silencing during recovery of N. benthamiana from ToRSV infection. A time course analysis was performed to measure the steady-state concentration of ToRSV RNA during infection. Although samples were analyzed at 5, 14, 19, 24, and 30 dpi, only three representative time points are shown in Fig. 2a. To estimate the variation between plants, three plants were chosen for each sampling time. To assess the variation within plants, the inoculated leaves as well as the third and first leaves from the top were analyzed. The size of the first leaf was approximately one-eighth the size of fully expanded leaves. At 24 and 30 dpi, inoculated leaves were omitted from the analysis due to their evident senescence. The results indicated that ToRSV RNA accumulation did not significantly change between 5 and 30 dpi (Fig. 2a and data not shown). In this particular experiment, recovery was observed after 15 dpi. Only minor variations in viral RNA levels were detected between and within plants, which correlated with the amount of RNA loaded in each lane (Fig. 2a). This result, although surprising, is consistent with the fact that sap from recovered leaves is highly infectious. In fact, the percentage of infected plants, the time for symptom emergence, and the severity of the symptoms observed were indistinguishable for plants inoculated with sap extracted from recovered leaves at 120 dpi or from young symptomatic leaves at 7 dpi (data not shown).
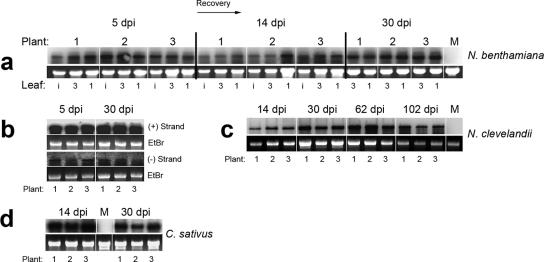
FIG. 2.
High steady-state levels of ToRSV RNA are found in time course experiments conducted on several plant species. (a) Time course analysis of ToRSV RNA concentration in infected N. benthamiana plants. Leaves from three individual plants were analyzed for each time point. Lane M, mock-inoculated plants; lanes i, inoculated leaf; lanes 3, third leaf from the top; lanes 1, apical leaf. (b) Detection of negative- and positive-strand ToRSV RNA in symptomatic (5 dpi) or recovered (30 dpi) leaves. (c and d) Time course analyses of viral RNA levels in N. clevelandii (c) and cucumber (d) plants. Fragments of ToRSV RNA corresponding to the CP and MP open reading frames (amplified with primers p47F [5′-GTCCCATGGGGTCTTCTCTAGGAACTCCTGGT] and p51R [5′-GTCCTCCAGGCCACGCCCGAAAGGAT]) were used for hybridization of blots in panels a, c, and d. The strand-specific probes used for panel b consisted of a 139-nt fragment from the 3′-nontranslatable region of ToRSV (67), which is identical in RNA 1 and RNA 2. Sense and antisense radiolabeled transcripts were generated in vitro using the T7 and SP6 polymerases and the cDNA fragment inserted between the corresponding promoters. Because the probes were produced using different polymerases, the ratio of positive- to negative-strand RNA could not be inferred directly.
The accumulation of viral RNA in systemic recovered leaves could either reflect active viral replication or be the result of transport of viral RNA from lower leaves. To determine whether viral replication occurred in systemic recovered leaves, we tested for the presence of ToRSV negative-strand RNA, using a strand-specific RNA probe. As shown in Fig. 2a, positive-strand RNA was readily detected at 5 and 30 dpi (Fig. 2b). Negative-strand viral RNA was also detected at both time points (Fig. 2b). Detection of negative-strand RNA at 30 dpi in systemic recovered leaves confirmed that viral replication is active in these leaves.
We next wanted to investigate whether ToRSV RNA stability was a peculiarity of the N. benthamiana-ToRSV pathosystem or was a more general phenomenon. We chose N. clevelandii because it was previously demonstrated to suppress RNA accumulation of another nepovirus through a sequence-specific RNA degradation mechanism (44). Symptoms induced by ToRSV in N. clevelandii were severe but disappeared shortly after their emergence, as described above for N. benthamiana. A time course analysis of ToRSV RNA revealed that there were no significant decreases in ToRSV RNA accumulation between 14 and 102 dpi (Fig. 2c). Cucumber (Cucumis sativus) is routinely used as an experimental host for ToRSV. ToRSV induced well-defined ring spots in cucumber cotyledons, and a vein-limited mosaic was visible in the first systemic leaf. Leaves that emerged subsequently were symptom-free. Northern blots revealed only a moderate reduction in virus RNA accumulation in recovered cucumber leaves compared to mosaic ones (Fig. 2d). Taken together, these results demonstrate that ToRSV is able to maintain relatively high steady-state levels of viral RNA in different families of plants.
N. benthamiana is known to be susceptible to a number of plant viruses. It has been shown that the RDR1 gene (referred to as NbRdRP1m or, here, as RDR1m) harbors an insertion that includes tandem premature in-frame stop codons that likely inactivate the encoded protein (72). The effect of RDR1m or any active RDR1 ortholog on ToRSV accumulation has not been determined. It was therefore possible that the accumulation of the ToRSV viral RNA in recovered leaves was due to a defect in the RDR1m gene which might reduce the efficiency of the RNA silencing machinery. A previous study has shown that an RDR1 ortholog from M. truncatula (MtRDR1) can functionally complement the N. benthamiana RDR1 mutation (72). Homozygous transgenic N. benthamiana plants in which the MtRDR1 gene was integrated were challenged with ToRSV. Plants transformed with the empty vector were used as a control. ToRSV-induced symptoms were not attenuated in MtRDR1 plants (Fig. 3a) and were similar to those observed in wild-type and empty vector control plants. The MtRDR1 plants recovered from ToRSV infection at a rate and time similar to those observed for the control plants (data not shown). No differences in viral RNA accumulation were observed between MtRDR1 and vector-transformed plants (Fig. 3b, top panel). As a complementary approach, we sequenced a 700-nt fragment from the RDR1 gene of N. clevelandii. In N. benthamiana, the homologous region contains the insertion that inactivates the RDR1 gene. Sequence alignments with tobacco (Nicotiana tabacum) and N. benthamiana RDR1 genes showed a high degree of identity among the three genes and revealed that the insertion present in N. benthamiana is not found in N. clevelandii (data not shown). Thus, it is likely that RDR1 is a functional gene in N. clevelandii, but it apparently does not affect ToRSV RNA accumulation.
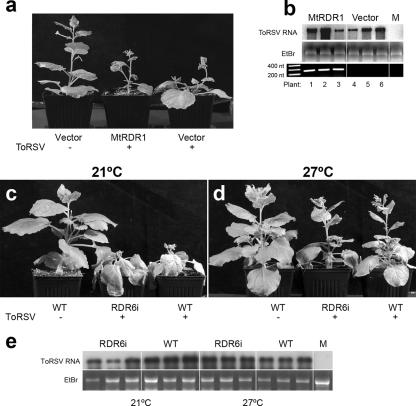
FIG. 3.
ToRSV accumulation is not affected by Medicago truncatula RDR1 or N. benthamiana RDR6 activity. (a) Symptoms induced by ToRSV on N. benthamiana plants expressing MtRDR1 or on plants transformed with an empty vector. (b) Concentration of ToRSV RNA in MtRDR1 or control (vector) plants, as determined by Northern blotting (top). Transcription of MtRDR1 was confirmed by reverse transcription-PCR, using primers mtPm5 and mtPm3 (72) (bottom). (c and d) ToRSV-induced symptoms in RdR6i or wild-type (WT) N. benthamiana plants grown at 21 or 27°C. (e) Northern blot of ToRSV RNA in RDR6i and wild-type plants cultivated at 21 or 27°C. ToRSV-specific probes for hybridization were prepared as described in the legend to Fig. 2. In every case, the topmost leaf was sampled at 14 dpi.
Other host RDRs have been implicated in the silencing of RNA viruses. The susceptibility of N. benthamiana to ToRSV was tested in transgenic lines in which the RDR6 gene was silenced (RDR6i lines) (51). The experiment was conducted at 21°C and 27°C, as it has been demonstrated that RDR6 activity is increased at 27°C (43). ToRSV virulence was increased in both wild-type and RDR6i lines at 21°C compared to that at 27°C (Fig. 3c and d). However, no obvious phenotypic differences were observed between RDR6i and wild-type plants infected with ToRSV at each temperature. Under both conditions, RNA hybridization experiments did not indicate any significant difference in virus accumulation between wild-type and RDR6i plants (Fig. 3e). Although the blot presented in Fig. 3e shows a lower concentration of ToRSV RNA in some RDR6i plants maintained at 21°C, this difference was not observed consistently in repeated hybridization experiments (data not shown).
Taken together, these results suggest that ToRSV RNA accumulation in recovered leaves is a general phenomenon and is independent of the activity of RDR1 and RDR6. There are at least two possible interpretations of these results, as follows: (i) the RNA silencing machinery fails to efficiently recognize ToRSV or (ii) ToRSV suppresses RNA silencing. We conducted a series of experiments (described below) to investigate these possibilities.
ToRSV triggers and is targeted by the RNA silencing machinery.
To determine if ToRSV RNA triggers the RNA silencing machinery, low-molecular-weight RNAs were purified from ToRSV-infected N. clevelandii and N. benthamiana plants. ToRSV-derived siRNAs were detected in both plant species (Fig. 4a). The stronger signal on the N. benthamiana blot was possibly due to the fact that a larger amount of RNA was loaded into the gel (see the legend to Fig. 4). We next compared the accumulation of viral siRNAs in plants infected with ToRSV or with potato virus X (PVX), a virus known to encode a weak suppressor of silencing (62). Under similar conditions, PVX-specific siRNAs were readily detected while the concentration of ToRSV-derived siRNAs was below the detection limit (Fig. 4b). This experiment confirmed that although ToRSV-derived siRNAs are present in infected plants (Fig. 4a), they are there at a low concentration (Fig. 4b).
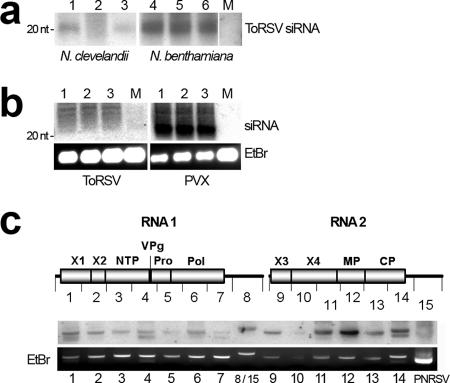
FIG. 4.
Characterization of ToRSV-derived siRNAs. (a) Detection of ToRSV-derived siRNAs in N. benthamiana and N. clevelandii plants. Thirty- and 45-μg samples of low-molecular-weight RNA from ToRSV-infected N. clevelandii (lanes 1 to 3) and N. benthamiana (lanes 4 to 6) plants, respectively, were analyzed. Lane M, equivalent amounts of low-molecular-weight RNA extracted from mock-inoculated plants. ToRSV-specific probes for hybridization were prepared as described in the legend to Fig. 2. The position of a 20-nt oligonucleotide used as a size marker is indicated to the left of the gel. (b) Comparison of viral siRNA concentrations in plants infected with ToRSV and with PVX. Eight micrograms of low-molecular-weight RNA was loaded into each well. As a loading control, 1/10 of each sample was run in a 1% agarose gel and stained with EtBr. For hybridization, labeled 1,000-nt cDNA fragments corresponding to nt 1001-2000 of the PVX or ToRSV genome were used as probes. The following primers were used for amplification: for PVX, p117F (5′- CGATTCTTAAGAAAACTATG) and p118R (5′-TCCCTCTGAATCTCCAGCGC); and for ToRSV, p122F (5′-TGTTGTCGCCCCCCTTGCC) and p123R (5′-CACTAGCCCATCGCCAATAG). An equal amount of labeled probe (0.15 × 109 cpm/μg) was added to each membrane. Membranes were processed in parallel, and hybridization signals were collected simultaneously using a single phosphorimage screen. (c) Mapping of siRNAs along the ToRSV genome. Amplified fragments of approximately 1,000 nt covering the entire RNA 1 and RNA 2 are depicted in the diagram. Each fragment (140 ng) was run in a 1% agarose gel. As a negative control, 1 μg of a fragment containing PNRSV MP was loaded in the rightmost lane. Fragments 8 and 15 correspond to the 3′-untranslated region, which is identical in ToRSV RNA 1 and RNA 2 (lane 8). Hybridization was conducted as described in Materials and Methods. EtBr staining of PCR-generated fragments is shown as a loading control.
To further characterize ToRSV-derived siRNAs, we amplified ∼1,000-nt cDNA fragments covering the entire ToRSV genome (Fig. 4c). Small RNAs from ToRSV-infected N. benthamiana plants were gel purified and end labeled with [γ-32P]dATP (54). In this assay, if ToRSV-derived siRNAs are present in the labeled pool, they will hybridize to the fragment of the virus genome from which they were derived. As a negative control, an excess of DNA corresponding to the movement protein (MP) of PNRSV was loaded in the rightmost lane. Reproducible results were obtained for three replicates of this experiment, and a typical result is shown in Fig. 4c. The labeled siRNAs hybridized more strongly to RNA 2-derived fragments, in particular to fragments 11, 12, 13, and 14, which correspond to the open reading frames for X4 (a protein of unknown function), MP, and CP and to a small stretch of the 3′-untranslated region. No specific signal was detected for PNRSV MP. Such an asymmetrical spatial distribution of siRNAs along the ToRSV genome might be congruent with the presence of local regions of secondary structure on the ToRSV genome. To explore this possibility, the secondary structure of the ToRSV RNA was predicted using the Mfold program (76). Fragments of approximately 800 nt covering the whole ToRSV genome were folded, and their secondary structures were compared visually. We did not find any obvious feature that would promote preferential generation of siRNAs from the MP and CP regions. Though nearly perfect hairpin structures were predicted in these regions, similar structures were also present in other regions that produced only weak signals in our siRNA mapping experiment.
Collectively, the results of these experiments provide the first evidence that ToRSV does not completely evade silencing of its genome and is susceptible to at least DCL activity, which is believed to be the initial step of programmed RNA silencing.
ToRSV does not prevent RISC assembly.
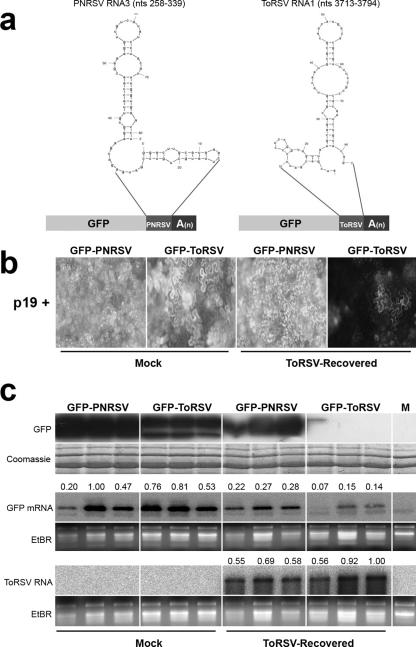
We next investigated whether active RISCs are specifically assembled in response to ToRSV infection. To do this, we used a sensor transgene approach. Eighty-one-nucleotide fragments derived from ToRSV RNA 1 or PNRSV RNA 3 (control) were fused to the 3′ end of the GFP open reading frame (Fig. 5a). Neither fragment possesses a predicted secondary structure that is likely to be a DCL substrate, since internal loops truncate the extension of double-stranded regions to a maximal length of 7 bp (Fig. 5a). These constructs were coinfiltrated with the tomato bushy stunt virus p19 protein into mock-inoculated N. benthamiana plants or N. benthamiana plants that had recovered from ToRSV infection (ToRSV-recovered N. benthamiana plants). The p19 protein binds double-stranded siRNAs in a nonspecific fashion, thereby preventing RISC assembly (49). However, it does not affect the activity of preassembled RISCs. Thus, preassembled RISCs should be solely responsible for degradation of the sensor constructs. The PNRSV fragment used as a control shares only 42.4% homology with the ToRSV fragment and does not include any 21-nt stretch in common with the ToRSV genome. Therefore, it is not a potential target of RISCs assembled in response to ToRSV infection.
FIG. 5.
ToRSV does not prevent assembly and/or affect the activity of RISCs. (a) Design of sensor constructs. Fragments (81 nt long) from PNRSV RNA 3 or ToRSV RNA 1 were fused in frame to the 3′ end of the GFP coding sequence and expressed under the control of the 35S promoter. The putative secondary structures of the fragments (as predicted by the Mfold program) are shown. (b) Relative fluorescence obtained after expression of sensor constructs in mock-inoculated or ToRSV-recovered leaves 5 days after agroinfiltration. (c) Comparative analysis of the concentrations of GFP (top), GFP mRNA (middle), and ToRSV RNA (bottom) in mock-inoculated or ToRSV-recovered plants 5 days after agroinfiltration. For detection of GFP mRNA and ToRSV RNA, a full-length GFP cDNA (amplified with primers p18F [5′-ATGAGTAAAGGAGAAGAACT] and p19R [5′-CAAACTCAAGAAGGACCATG]) and the ToRSV CP open reading frame (amplified with primers p52F [5′-GTCTCTAGATGGGGCGGGTCCTGGCAAGAAGG] and p51R [described in the legend to Fig. 2]) were used as probes, respectively. For relative quantification of band intensities, digital light units were measured from each lane from both EtBr-stained gel pictures and phosphorimages from Northern blots, using OptiQuant 5.0 software (Perkin-Elmer). The relative RNA concentration was corrected using the following formula: values from the blots (intensities of RNA signals)/values from the EtBr-stained gel pictures (amounts of RNA loaded). The results were divided by the maximum value so that the relative concentrations of RNA are presented on a scale of 0 to 1 (shown above each lane of the GFP mRNA and ToRSV RNA Northern blots).
The activity of p19 was confirmed by evaluating its effect on the transient expression of unfused GFP. As expected, p19 increased GFP accumulation (data not shown). In mock-inoculated plants, both fusion proteins were expressed at high levels, as indicated by the bright fluorescence in the agroinfiltrated leaves observed under UV light (Fig. 5b). In ToRSV-recovered plants, fluorescence resulting from expression of the GFP-PNRSV control was decreased only slightly compared to that observed in mock-inoculated plants (Fig. 5b). In contrast, a severe reduction in the fluorescence resulting from expression of the GFP-ToRSV construct was observed in ToRSV-recovered leaves (Fig. 5b). Northern and Western blots revealed that reductions of GFP-PNRSV mRNA and protein in ToRSV-recovered plants were moderate in comparison to those in mock-inoculated plants (Fig. 5c) and may be attributed to the depressed physiological condition of infected plants. However, the accumulation of GFP-ToRSV protein and mRNA was significantly reduced in ToRSV-recovered plants. These results support the idea that ToRSV RNA is recognized by dicer and that ToRSV is unable to prevent the assembly of RISCs formed in response to virus infection. The results also indicate that while mRNAs containing ToRSV sequences are targeted by preassembled RISCs, ToRSV viral RNA accumulates to high levels throughout infected plants (Fig. 5c, bottom panel).
Systemic movement of the silencing signal is partially hindered in ToRSV-infected plants.
Many viral suppressors of silencing have been identified based on their ability to suppress silencing of the GFP reporter gene (4, 12, 18, 22, 24, 39, 41, 63, 71, 75). We decided to test if ToRSV could suppress the establishment, systemic spread, and/or maintenance of GFP silencing.
Several viral suppressors of silencing (including the potyvirus HC-Pro protein) have the ability to revert preestablished silencing (63). We took advantage of a previously described silencing reversion assay using N. benthamiana plants harboring a GFP transgene (line 16c) (63). Silencing of the GFP transgene can be induced by agroinfiltration of the same GFP cassette into the 16c plants (Fig. 6a1). After GFP silencing is established systemically (plants appear completely red under UV light), plants are inoculated with viruses (Fig. 6a2) and the ability of these viruses to revert silencing and reinstate GFP expression is tested. Fully silenced GFP transgenic lines were inoculated with ToRSV or potato virus Y (PVY; a potyvirus used as a positive control) or were mock inoculated. As described by others (63), mock-inoculated plants remained red under UV light until the end of the observation period (16 weeks after inoculation) (data not shown), while PVY-infected plants gradually recovered their fluorescence (Fig. 6a3). ToRSV-infected plants phenocopied mock-inoculated plants (Fig. 6a4). In three independent experiments, none of the 54 ToRSV- or mock-inoculated plants recovered GFP fluorescence, while all plants infected with PVY did (48/48 plants). Thus, ToRSV was not capable of reverting fully established silencing against the GFP transgene. This is consistent with a previous report on TBRV, another nepovirus that is also unable to revert established GFP silencing (63).
FIG. 6.
ToRSV does not prevent the establishment or maintenance of silencing of a GFP transgene but partially hinders systemic movement of the silencing signal. (a) Inability of ToRSV to revert fully established silencing of a GFP transgene. Silencing of the GFP transgene was induced in N. benthamiana 16c plants by agroinfiltration of a sense GFP transgene (a1). Systemically silenced GFP plants were challenged by inoculation with ToRSV or PVY (a2). Three to four weeks after inoculation, PVY-infected plants displayed green fluorescence (a3), while ToRSV-inoculated plants remained red (a4). PTGS, posttranscriptional gene silencing. (b) Delay of systemic silencing of the GFP transgene in ToRSV-infected plants. Silencing of GFP in 16c plants was induced as described for panel a1, and plants were inoculated with ToRSV or PVY when systemic leaves exhibited red veins under UV light, indicating that silencing was progressing into mesophyll cells (b1). By 35 dpi, all PVY-inoculated plants had completely green fluorescent leaves in the apical zone (b2), while all mock-inoculated plants had turned completely red (not shown). At 35 dpi, a subpopulation of ToRSV-infected plants showed various degrees of green fluorescence (b3), but it vanished a few days later (b4). (c) Partial inhibition of systemic silencing of GFP triggered in ToRSV-infected symptomatic leaves. Silencing of GFP was induced in leaves showing the initial symptoms associated with ToRSV infection (c1). Systemic silencing was assumed to have occurred when red veins were observed in systemic leaves under UV light (c2). Plants that did not show any red fluorescent veins in systemic leaves (c3) were counted as nonsilenced plants. (c4) According to this criterion, the systemic occurrence of silencing was evaluated in 36 ToRSV- or mock-inoculated plants at the intervals indicated.
Next, we investigated whether ToRSV could interfere with the systemic movement of the silencing signal. Plants were inoculated with virus at an earlier time point, before complete silencing was established (as shown by the partial silencing of GFP in Fig. 6b1). In this experiment, all mock-inoculated plants appeared red under UV light by 35 dpi (data not shown). PVY-infected plants never became completely red under UV light and, instead, only allowed silencing of areas in and around the veins of systemic leaves. A few weeks later, newly emerged PVY-infected leaves were completely green under UV light, indicating that the virus had suppressed silencing of GFP in these leaves (Fig. 6b2). Two different phenotypes were observed in ToRSV-infected plants. A subpopulation of plants that exhibited milder symptoms appeared red under UV light, indicating extensive silencing of GFP. A second subpopulation that showed more severe symptoms deployed a phenotype similar to that observed for PVY-infected plants at 35 dpi (Fig. 6b3). However, by 45 dpi, all ToRSV-infected plants had turned completely red under UV light (Fig. 6b4).
We also tested the ability of ToRSV to interfere with the establishment of local and systemic GFP silencing. In this new set of experiments, 16c plants were first inoculated with viruses. The GFP cassette was agroinfiltrated into symptomatic leaves shortly after the onset of symptoms (Fig. 6c1). All agroinfiltrated leaves in mock-inoculated plants showed the characteristic red spots associated with establishment of RNA silencing against the GFP transgene (data not shown). ToRSV- and PVY-infected agroinfiltrated leaves displayed similar phenotypes, indicating that they did not interfere with the establishment of silencing (data not shown). By 10 days after agroinfiltration, all mock-inoculated plants (n = 36) exhibited some degree of systemic GFP silencing, as indicated by the red veins on the green fluorescent background of systemic leaves (Fig. 6c2). As expected, none of the plants infected with PVY (positive control) showed any visual trace of systemic silencing (no red veins were observed) (data not shown). Only 40% of the ToRSV-infected plants became systemically silenced. The remaining plants did not show any signs of systemic silencing, as evidenced by the bright green fluorescence observed on these leaves (Fig. 6c3). The number of ToRSV-infected plants that became systemically silenced increased slightly over time but was still lower than 60% by 39 days after infiltration (Fig. 6c4).
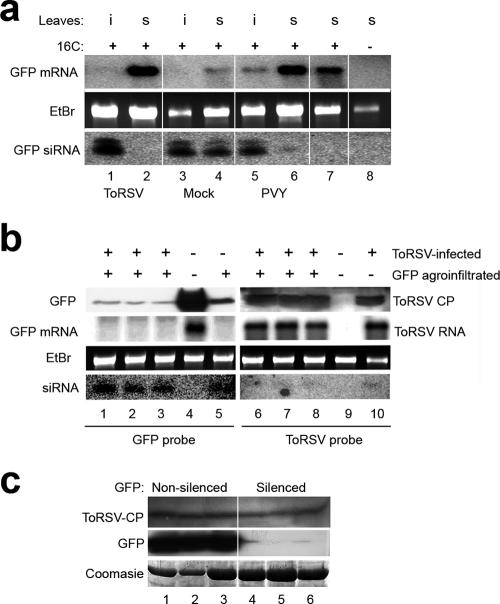
We next examined the levels of GFP mRNA and GFP-specific siRNAs in the infiltrated leaf (using the silenced region of the leaf, i.e., the spot that appeared red under UV light) and in systemic leaves at 18 days postagroinfiltration. In mock-inoculated plants, both the infiltrated (i) and systemic (s) leaves showed a high concentration of short-class siRNAs and a moderate abundance of long-class siRNAs (Fig. 7a, bottom panel, lanes 3 and 4). A concomitant reduction of GFP mRNA levels was observed (Fig. 7a, top panel, lanes 3 and 4). PVY did not prevent the establishment of silencing in infiltrated leaves but suppressed silencing in systemic leaves of all plants tested, as indicated by the high levels of GFP transcripts and low levels of the corresponding siRNAs in the systemic leaves (Fig. 7a, lanes 5 and 6). ToRSV did not prevent production of GFP-derived siRNAs or degradation of the GFP mRNA in agroinfiltrated leaves (Fig. 7a, lane 1), confirming the suggestion that ToRSV does not interfere with the establishment of silencing. As mentioned above, systemic GFP silencing was apparently inhibited in 40% of the plants infected with ToRSV. In these plants, red veins were not observed under UV light in systemic leaves, GFP-specific siRNAs were not detected, and GFP mRNA was abundant (Fig. 6c3 and 7a, lane 2). We concluded that in this subpopulation of plants, systemic movement of the GFP silencing signal was prevented. However, this effect was partial, as 60% of the ToRSV-infected plants did show signs of systemic silencing at 39 days postinfiltration (Fig. 6c4).
FIG. 7.
Evaluation of GFP and ToRSV concentrations in ToRSV-infected plants at various times after induction of GFP silencing. (a) Analysis of GFP mRNA and siRNAs in a subpopulation of ToRSV-infected plants in which silencing of GFP was hindered in systemic leaves. Plants (16c line) were first mock inoculated or inoculated with ToRSV or PVY. Silencing of the GFP transgene was induced after initial development of virus-induced symptoms as described in the legend to Fig. 6c. Plants were tested for the presence of GFP mRNA and siRNA in the agroinfiltrated (i) and systemic (s) leaves at 18 days postagroinfiltration. RNAs from GFP transgenic plants that had not been silenced (lane 7) and from wild-type plants (lane 8) were used as controls. (b) Accumulation of ToRSV in a subpopulation of ToRSV-infected plants in which systemic silencing of GFP was active. Concentrations of GFP protein, GPF mRNA, and GFP-derived siRNAs (lanes 1 to 3) and of ToRSV CP, ToRSV RNA, and ToRSV-derived siRNAs (lanes 6 to 8) were analyzed in three individual plants at 120 dpi. Mock-inoculated silenced (lane 5) or nonsilenced (lane 4) plants were used as controls for the analysis of GFP protein and RNAs. Mock-inoculated (lane 9) or ToRSV-infected (7 dpi) (lane 10) plants were included as controls for the analysis of ToRSV CP and RNAs. Detection of GFP mRNA and ToRSV RNA was done as described in the legend to Fig. 5. Five- and 50-μg samples of total RNA were used for Northern blotting and siRNA detection, respectively. (c) Accumulation of ToRSV in GFP-silenced or -nonsilenced branches late in infection. At 120 dpi, levels of GFP protein and ToRSV CP in red fluorescent (silenced) or green fluorescent (nonsilenced) branches of ToRSV-infected 16c plants were tested by Western blotting.
To investigate a possible correlation between virus concentration and the extent of transgene silencing, we examined the accumulation of ToRSV and GFP at a later time during infection. In this experiment, we used the subpopulation of ToRSV-infected 16c plants in which the GFP transgene had been silenced systemically. At 120 dpi, systemic leaves from these plants appeared completely red under UV light. In these leaves, the GFP protein was present at a very low concentration and the GFP mRNA was not detected, while GFP siRNAs were present at a high concentration (Fig. 7b, lanes 1 to 3). Similar results were obtained when mock-inoculated plants were examined (Fig. 7b, lane 5). In contrast, both the GFP protein and mRNA were abundant in nonsilenced plants of the same age, and GFP-derived siRNAs were not detected (Fig. 7b, lane 4). Virus accumulation was also examined in these plants. At 120 dpi, the viral concentration was high (Fig. 7b, lanes 6 to 8), as indicated by the detection of ToRSV CP and genomic viral RNA. Indeed, the levels of ToRSV RNA were similar at 120 dpi and in young plants at 7 dpi (Fig. 7b, compare lanes 6 to 8 with lane 10). ToRSV-specific siRNAs were detected at 7 and 120 dpi, but at a very low concentration. This result confirmed that ToRSV accumulates at high levels in leaves in which the silencing machinery is active, as demonstrated by silencing of the GFP transgene.
In some plants, both GFP-silenced and non-GFP-silenced branches were observed. Interestingly, ToRSV CP accumulation was comparable in silenced (red fluorescent) and nonsilenced (green fluorescent) branches of 16c plants (Fig. 7c, top panel), suggesting that movement of the systemic transgene silencing signal can be separated mechanistically from virus movement and/or accumulation.
Collectively, our results suggest that ToRSV does not carry a strong suppressor of transgene silencing. The partial inhibition of the systemic progress of silencing observed in ToRSV-infected plants could be attributed to the presence of a weak suppressor of silencing encoded by ToRSV. Alternatively, it could be an indirect result of the transport blockage observed in symptomatic leaves.
DISCUSSION
Induction of defense responses in ToRSV-infected plants.
In this study, we have characterized the host responses that are activated in N. benthamiana during infection with ToRSV. Symptom development was accompanied by induction of hydrogen peroxide, transcriptional activation of PR1a, cell death, and inhibited carbohydrate transport (Fig. 1). The fact that ToRSV induces a necrotic phenotype that recapitulates the HR suggests that ToRSV RNAs or proteins are perceived by the plant cell as elicitors of the HR. Increased transcription of the PR1a gene, which belongs to a cluster of salicylic acid (SA)-induced defense genes (19), was found in inoculated and distal leaves and seemed to accumulate with or ahead of the virus infection front.
Recently, an interplay between RNA silencing and more general defense mechanisms, such as the HR mediated by SA, has been suggested (60). Several findings support this notion. First, the activity of an RDR in N. tabacum (NtRDR1) is enhanced by treatment with SA or by virus infection (69). Second, several viral suppressors of silencing induce an HR-like phenotype (35, 50). Third, at least two suppressors of silencing have been shown to influence SA-mediated responses (32, 40). SA-mediated resistance and RNA silencing have been shown to cooperate in restricting the movement of plum pox virus in tobacco plants (3). It was suggested that the SA-mediated defense response prevents cell-to-cell movement of the virus from the site of inoculation (resulting in ring spots), while RNA silencing combined with the SA-mediated defense response prevents unloading of the virus from the vasculature into mesophyll cells (producing vein clearing). Analogously, both HR and RNA silencing may be implicated in symptom development in ToRSV-infected plants. Interestingly, although ToRSV elicited both an HR-like response and RNA silencing, these combined defense mechanisms were unable to restrict virus movement and the virus accumulated to high levels throughout the plant.
Interaction of ToRSV with the RNA silencing pathway.
The stable accumulation of ToRSV RNA in infected plants suggests that it evades or suppresses one or several steps of the RNA silencing machinery. For example, ToRSV RNA might be a poor substrate for dicer, AGO, and/or RDR enzymes. In addition, ToRSV RNA may evade recognition by dicer enzymes by hiding in sheltered subcellular compartments. In fact, this could explain the low abundance of ToRSV-derived siRNAs in infected cells (Fig. 4). Replication of ToRSV RNA probably occurs in membranous vesicles derived from the endoplasmic reticulum (27, 48), which may protect viral RNA against degradation (1). The RNA genome is thought to move from cell to cell as a virus-like particle through tubular structures that are induced by the virus and traverse the cell wall (67). Thus, exposure of the viral RNA to the silencing machinery could be rather limited during the course of infection. However, many other viruses use similar subcellular compartments for their replication and movement, including TBRV, a nepovirus that is apparently susceptible to RNA silencing, as indicated by the reduced concentration of viral RNA observed in recovered leaves (44). Thus, it is perhaps more likely that sufficient accumulation of a viral product (RNA or protein) is necessary to suppress or saturate the silencing machinery.
The viral suppressors of silencing characterized to date deploy diverse modes of action, including siRNA inactivation and direct inhibition of DCL and AGO proteins (42, 52, 60). The results from our experiments using the GFP reporter transgene indicate that the ToRSV genome does not encode a strong suppressor of silencing. ToRSV was unable to prevent the initiation of local silencing or to suppress fully established RNA silencing in the GFP reporter transgene assay (Fig. 6). Although systemic movement of the GFP silencing signal was delayed or hampered in ToRSV-infected N. benthamiana 16c plants (Fig. 6), this effect was partial and may have been the result of the transport blockage induced by ToRSV during the symptomatic phase (as shown in Fig. 1). However, we cannot disregard the possibility that a weak viral suppressor would affect systemic silencing. Although many viral suppressors of silencing have been identified based on their ability to suppress silencing of GFP, it has also been noted that such assays are not always able to identify suppressors that affect the systemic progress of silencing or that only weakly inhibit transgene silencing (34). Using an experimental setup similar to the one described in this study, three suppressors of silencing have been reported to affect systemic but not local silencing of a GFP transgene. These include citrus tristeza virus (CTV) CP, the rice yellow mottle virus P1 protein, and apple chlorotic leaf spot virus (ACLSV) MP (29, 37, 71). Ectopic expression of the rice yellow mottle virus P1 protein reduced accumulation of the 25-nt species of GFP-derived siRNAs (29), which has been implicated in systemic movement of the silencing signal (26). However, CTV CP and ACLSV MP did not have apparent effects on the accumulation of siRNAs (37, 71). Further experimentation will be required to determine if the ToRSV genome encodes a suppressor of systemic silencing that acts in a manner similar to that of CTV CP and ACLSV MP.
ToRSV did not have a strong effect on the silencing of the GFP transgene. However, in infected plants, the concentration of ToRSV-derived siRNAs was low (Fig. 4 and 7), and degradation of a sensor construct containing a ToRSV fragment was incomplete (Fig. 5). These observations raise the possibility that ToRSV suppresses a virus-specific silencing pathway. It has been demonstrated that different classes of siRNAs are produced in response to RNA viruses or transgenes, owing to the action of distinct DCL enzymes (20, 23). Thus, if ToRSV partially suppresses the activity of a virus-specific DCL, this effect will be undetectable in the GFP experimental system. Moreover, given the multiplicity of AGO proteins found in plants, it is also possible that ToRSV interferes with the function of one or several virus-specific AGO proteins, thereby preventing silencing of its genome. Interestingly, we found that ToRSV accumulation was not susceptible to N. benthamiana RDR6 or to M. truncatula RDR1 activity. Thus, the apparent resistance of ToRSV to the RNA silencing machinery may result from the inability of RDRs to mediate iterative degradation of ToRSV genomic RNA. It would be of interest to test if ToRSV has the ability to synergistically enhance the replication of other viruses and to determine if a ToRSV suppressor of silencing is directed to a virus-specific silencing enzyme.
In a previous study, it was shown that recovery of N. clevelandii from TBRV infection is accompanied by a reduced titer of the virus, suggesting that the virus is degraded by the silencing machinery (44). ToRSV (a subgroup C nepovirus) differs from TBRV (a subgroup B nepovirus) in that it has larger 3′-noncoding regions in the two RNAs and an additional protein domain in the N-terminal region of the RNA 2-encoded polyprotein (13). Further experiments will be necessary to determine if these or other differences can account for the divergent susceptibilities of the two viruses to RNA silencing.
Reassessing the role of RNA silencing in the recovery phenotype.
In our experiments, the concentration of ToRSV RNA did not correlate with the presence or absence of symptoms on infected leaves (Fig. 2), providing further support for earlier suggestions that virus clearance is not a strict requirement for the induction of recovery (14, 70). In fact, as early as 1928, it was suggested that although recovered leaves are asymptomatic and resistant to reinfection by the same virus, the sap from these leaves is highly infectious (68). More recently, a natural tobacco streak virus variant that does not induce recovery was characterized (70). Interestingly, the authors of that study noticed that the variant, which carried a single nucleotide substitution, accumulated to much lower levels than its wild-type counterpart, and they proposed that high levels of viral RNA and/or viral proteins may be required to suppress a symptomatic defense response. Interestingly, although a reduction of virus titer is not necessarily required for the induction of the recovery phenotype, in all cases studied to date, activation of virus-specific RNA silencing has been observed (15, 16, 44, 45, 70; this study). What, then, is the role, if any, of RNA silencing during recovery? An intriguing hypothesis is that virus-derived siRNAs down-regulate host genes implicated in the defense response. We are currently exploring this possibility.
Acknowledgments
We are indebted to David Baulcombe and Richard Nelson for their gift of materials (as indicated in Materials and Methods). We also thank Joan Chisholm and Guanzhi Zhang for helpful discussions and for providing some of the constructs used in our experiments. We thank Michael Weis for his expert assistance with microscopy and image processing. We are grateful to Richard Nelson and Shou-Wei Ding for critically reading the manuscript.
This work was supported by PPV-targeted funding from AAFC.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbergenov, R., A. Si-Ammour, T. Blevins, I. Amin, C. Kutter, H. Vanderschuren, P. Zhang, W. Gruissem, F. Meins, Jr., T. Hohn, and M. M. Pooggin. 2006. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 34:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alamillo, J. M., P. Saenz, and J. A. Garcia. 2006. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 48:217-227. [DOI] [PubMed] [Google Scholar]
- 4.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axtell, M. J., C. Jan, R. Rajagopalan, and D. P. Bartel. 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127:565-577. [DOI] [PubMed] [Google Scholar]
- 6.Baulcombe, D. 2005. RNA silencing. Trends Biochem. Sci. 30:290-293. [DOI] [PubMed] [Google Scholar]
- 7.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 8.Baulcombe, D. C. 2007. Molecular biology. Amplified silencing. Science 315:199-200. [DOI] [PubMed] [Google Scholar]
- 9.Baumberger, N., and D. C. Baulcombe. 2005. Arabidopsis Argonaute1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102:11928-11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blevins, T., R. Rajeswaran, P. V. Shivaprasad, D. Beknazariants, A. Si-Ammour, H. S. Park, F. Vazquez, D. Robertson, F. Meins, Jr., T. Hohn, and M. M. Pooggin. 2006. Four plant dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34:6233-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodersen, P., and O. Voinnet. 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22:268-280. [DOI] [PubMed] [Google Scholar]
- 12.Canizares, M. C., K. M. Taylor, and G. P. Lomonossoff. 2004. Surface-exposed C-terminal amino acids of the small coat protein of cowpea mosaic virus are required for suppression of silencing. J. Gen. Virol. 85:3431-3435. [DOI] [PubMed] [Google Scholar]
- 13.Carrier, K., Y. Xiang, and H. Sanfacon. 2001. Genomic organization of RNA2 of tomato ringspot virus: processing at a third cleavage site in the N-terminal region of the polyprotein in vitro. J. Gen. Virol. 82:1785-1790. [DOI] [PubMed] [Google Scholar]
- 14.Carrillo-Tripp, J., E. Lozoya-Gloria, and R. F. Rivera-Bustamante. 2007. Symptom remission and specific resistance of pepper plants after infection by pepper golden mosaic virus. Phytopathology 97:51-59. [DOI] [PubMed] [Google Scholar]
- 15.Chellappan, P., R. Vanitharani, F. Ogbe, and C. M. Fauquet. 2005. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 138:1828-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chellappan, P., R. Vanitharani, J. Pita, and C. M. Fauquet. 2004. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 78:7465-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner. 1997. Plants combat infection by gene silencing. Nature 385:781-782. [Google Scholar]
- 18.Cui, X., X. Tao, Y. Xie, C. M. Fauquet, and X. Zhou. 2004. A DNAbeta associated with tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78:13966-13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cutt, J. R., and D. Klessig. 1992. Pathogenesis-related proteins, p. 209-243. In T. Boller and F. Meins (ed.), Genes involved in plant defense (plant gene research). Springer-Verlag, Vienna, Austria.
- 20.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 21.Ding, S. W., W. X. Li, and R. H. Symons. 1995. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14:5762-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong, X., R. van Wezel, J. Stanley, and Y. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunoyer, P., C. Himber, and O. Voinnet. 2005. Dicer-like 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37:1356-1360. [DOI] [PubMed] [Google Scholar]
- 24.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, H. S., and J. A. Garcia. 1997. Delayed resistance to plum pox potyvirus mediated by a mutated RNA replicase gene: involvement of a gene silencing mechanism. Mol. Plant-Microbe Interact. 10:160-170. [Google Scholar]
- 26.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han, S., and H. Sanfacon. 2003. Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J. Virol. 77:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heath, M. C. 2000. Hypersensitive response-related death. Plant Mol. Biol. 44:321-334. [DOI] [PubMed] [Google Scholar]
- 29.Himber, C., P. Dunoyer, G. Moissiard, C. Ritzenthaler, and O. Voinnet. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hortensteiner, S. 2006. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57:55-77. [DOI] [PubMed] [Google Scholar]
- 31.Hull, R. 2002. Matthew's plant virology, 4th ed. Academic Press, San Diego, CA.
- 32.Ji, L. H., and S. W. Ding. 2001. The suppressor of transgene RNA silencing encoded by cucumber mosaic virus interferes with salicylic acid-mediated virus resistance. Mol. Plant-Microbe Interact. 14:715-724. [DOI] [PubMed] [Google Scholar]
- 33.Koch, E., and A. Slusarenko. 1990. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, H. W., A. P. Lucy, H. S. Guo, W. X. Li, L. H. Ji, S. M. Wong, and S. W. Ding. 1999. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindbo, J. A., L. Silva-Rosales, W. M. Proebsting, and W. G. Dougherty. 1993. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, R., A. Folimonov, M. Shintaku, W. X. Li, B. W. Falk, W. O. Dawson, and S. W. Ding. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. USA 101:15742-15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macdiarmid, R. 2005. RNA silencing in productive virus infections. Annu. Rev. Phytopathol 43:523-544. [DOI] [PubMed] [Google Scholar]
- 39.Merai, Z., Z. Kerenyi, A. Molnar, E. Barta, A. Valoczi, G. Bisztray, Z. Havelda, J. Burgyan, and D. Silhavy. 2005. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 79:7217-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruss, G. J., C. B. Lawrence, T. Bass, Q. Q. Li, L. H. Bowman, and V. Vance. 2004. The potyviral suppressor of RNA silencing confers enhanced resistance to multiple pathogens. Virology 320:107-120. [DOI] [PubMed] [Google Scholar]
- 41.Qi, Y., X. Zhong, A. Itaya, and B. Ding. 2004. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu, F., and T. J. Morris. 2005. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 579:5958-5964. [DOI] [PubMed] [Google Scholar]
- 43.Qu, F., X. Ye, G. Hou, S. Sato, T. E. Clemente, and T. J. Morris. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79:15209-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe. 1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 45.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinne, P. L., R. van den Boogaard, M. G. Mensink, C. Kopperud, R. Kormelink, R. Goldbach, and C. van der Schoot. 2005. Tobacco plants respond to the constitutive expression of the tospovirus movement protein NS with a heat-reversible sealing of plasmodesmata that impairs development. Plant J. 43:688-707. [DOI] [PubMed] [Google Scholar]
- 47.Sanfacon, H., A. Wieczorek, and F. Hans. 1995. Expression of the tomato ringspot nepovirus movement and coat proteins in protoplasts. J. Gen. Virol. 76:2299-2303. [DOI] [PubMed] [Google Scholar]
- 48.Sanfacon, H., G. Zhang, J. Chisholm, B. Jafarpour, and J. Jovel. 2006. Molecular biology of tomato ringspot nepovirus, a pathogen of ornamentals, small fruits and fruit trees, p. 540-546. In J. Teixeira da Silva (ed.), Floriculture, ornamental and plant biotechnology: advances and topical issues, 1st ed., vol. III. Global Science Books, London, United Kingdom. [Google Scholar]
- 49.Scholthof, H. B. 2006. The tombusvirus-encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 4:405-411. [DOI] [PubMed] [Google Scholar]
- 50.Scholthof, H. B., K. B. Scholthof, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silhavy, D., and J. Burgyan. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-83. [DOI] [PubMed] [Google Scholar]
- 53.Stace-Smith, R. 1984. Tomato ringspot virus. CMI/AAB descriptions of plant viruses 290. Commonwealth Mycological Institute, Kew, Surrey, United Kingdom.
- 54.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szittya, G., D. Silhavy, A. Molnar, Z. Havelda, A. Lovas, L. Lakatos, Z. Banfalvi, and J. Burgyan. 2003. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomson, J. M., J. Parker, C. M. Perou, and S. M. Hammond. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 1:47-53. [DOI] [PubMed] [Google Scholar]
- 57.Thordal-Christensen, H., Z. Zhang, Y. Wei, and D. Collinge. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11:1187-1194. [Google Scholar]
- 58.Vaucheret, H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20:759-771. [DOI] [PubMed] [Google Scholar]
- 59.Vazquez, F. 2006. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 11:460-468. [DOI] [PubMed] [Google Scholar]
- 60.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 61.Voinnet, O. 2005. Non-cell autonomous RNA silencing. FEBS Lett. 579:5858-5871. [DOI] [PubMed] [Google Scholar]
- 62.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 63.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 65.Wassenegger, M., and G. Krczal. 2006. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11:142-151. [DOI] [PubMed] [Google Scholar]
- 66.Waterhouse, P. M., M. W. Graham, and M. B. Wang. 1998. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95:13959-13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wieczorek, A., and H. Sanfacon. 1993. Characterization and subcellular localization of tomato ringspot nepovirus putative movement protein. Virology 194:734-742. [DOI] [PubMed] [Google Scholar]
- 68.Wingard, S. A. 1928. Hosts and symptoms of ring spot, a virus disease of plants. J. Agric. Res. 37:127-153. [Google Scholar]
- 69.Xie, Z., B. Fan, C. Chen, and Z. Chen. 2001. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA 98:6516-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xin, H. W., and S. W. Ding. 2003. Identification and molecular characterization of a naturally occurring RNA virus mutant defective in the initiation of host recovery. Virology 317:253-262. [DOI] [PubMed] [Google Scholar]
- 71.Yaegashi, H., T. Takahashi, M. Isogai, T. Kobori, S. Ohki, and N. Yoshikawa. 2007. Apple chlorotic leaf spot virus 50 kDa movement protein acts as a suppressor of systemic silencing without interfering with local silencing in Nicotiana benthamiana. J. Gen. Virol. 88:316-324. [DOI] [PubMed] [Google Scholar]
- 72.Yang, S. J., S. A. Carter, A. B. Cole, N. H. Cheng, and R. S. Nelson. 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 101:6297-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, S. C., G. Zhang, L. Yang, J. Chisholm, and H. Sanfacon. 2005. Evidence that insertion of tomato ringspot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79:11752-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, X., Y. R. Yuan, Y. Pei, S. S. Lin, T. Tuschl, D. J. Patel, and N. H. Chua. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zrachya, A., E. Glick, Y. Levy, T. Arazi, V. Citovsky, and Y. Gafni. 2007. Suppressor of RNA silencing encoded by tomato yellow leaf curl virus-Israel. Virology 358:159-165. [DOI] [PubMed] [Google Scholar]
- 76.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]