Abstract
The assembly of infectious human immunodeficiency virus (HIV) requires that Gag transport and oligomerization be coordinated with its association with other viral proteins, viral RNAs, and cellular membranes. We have developed a replication-competent HIV type 1 molecular clone that carries a Gag-internal or interdomain green fluorescent protein (iGFP) fusion to reveal a physiologically accurate temporal sequence of Gag localization and oligomerization during the formation of infectious HIV. This recombinant HIV is as infectious as native HIV in single-round infectivity assays, validating its use for trafficking studies. It replicates robustly in permissive MT4 cells and is infectious, yet it spreads poorly in other T-cell lines. Immunofluorescence of Gag-iGFP showed a pattern very similar to that of native Gag. However, the intense plasma membrane Gag-iGFP fluorescence contrasts markedly with its immunofluorescence at this site, indicating that many Gag epitopes can be masked by oligomerization. Consistent with this, fluorescence resonance energy transfer studies visualized intense Gag oligomerization at the plasma membrane and weaker oligomerization at cytoplasmic sites. Four-dimensional, time-lapse confocal imaging reveals a temporal progression of Gag distribution over hours in which Gag is initially diffusely localized within the cytoplasm. Plasma membrane signals then accumulate as Gag levels increase and vesicular association appears late, only after plasma membrane site signals have reached high intensity. Lastly, the cell rounds up and HIV protease activation induces diffuse fluorescence throughout the cell. These distinct phases reveal a natural progression of Gag trafficking during the viral gene expression program. HIV Gag-iGFP is a useful tool for dissecting mechanisms of viral assembly and transmission.
The assembly of infectious human immunodeficiency virus type 1 (HIV-1) involves complex interactions between Gag and numerous viral and host factors (26). While HIV Gag can direct the formation of virus-like particles in the absence of other viral proteins, the interactions of Gag with viral components such as GagPol, Env, and the viral RNA are necessary to produce infectious virus. HIV assembly also is spatially regulated, occurring at specific cellular membrane compartments. In T cells and epithelial cells, assembly occurs at the plasma membrane. In macrophages, late endosomes were proposed to be major sites of virus production (36, 41), yet recent studies suggest that the plasma membrane also is the major site of productive assembly in these cells (17, 54). Gag and Gag-green fluorescent protein (GFP) fusions often are localized to late endosomal sites and associate with late endosomal machinery, suggesting that late endosomes play some role in HIV assembly (8, 30, 45). Other recent work suggests that Gag directly engages the plasma membrane without an endosomal intermediate (17, 29, 43). Many of our conclusions about Gag localization and oligomerization are based upon studies of Gag proteins expressed outside of the full proviral context.
Gag-GFP fusions are studied largely with the assumption that they function within the cell like native Gag (7, 12, 37, 44, 45). Using a biarsenical-tetracysteine tag approach, two different groups have found that Gag may either directly bind the plasma membrane or initially bind to an intracellular compartment prior to transport to the plasma membrane (38, 43). The varied designs of these tagged proteins indicate that the protein tags may alter the behavior of the fusion proteins. A limitation of most current fluorescent Gag fusions is that they are not capable of producing infectious virus without untagged helper Gag.
To express HIV Gag-GFP fusions in the absence of other viral proteins, the Gag-coding region must be codon optimized or otherwise made Rev independent to avoid oversplicing of the Gag mRNA. These expression-optimized Gag-GFP fusions can be packaged into virus-like particles and do not interfere with viral infectivity (46). However, particularly in the absence of coexpressed native Gag, they exhibit functional differences in protein localization or sensitivity to inhibition by dominant-negative proteins (30, 47). Virus-like particles made of Gag-GFP exhibit abnormal densities in electron microscopy examinations (19, 40).
The presence of other viral components can have a large effect on studies of the localization of Gag. Env can act in a dominant manner to recruit assembly in a polarized location in epithelial cells or neurons (23, 34, 35, 53). The native viral RNA can influence the trafficking of Gag proteins within the cell (2, 47). GagPol can induce Gag to partition into detergent-resistant membrane compartments (10). Viral accessory genes such as vpu and nef also alter protein-sorting pathways that may affect viral assembly (5, 14, 15, 29, 48, 49). In T cells, HIV assembly is directed towards the site of cell-cell contact in a manner that facilitates uptake of virus into neighboring T cells (16, 39). Therefore, to understand how viral assembly and transmission is regulated, it is optimal to study Gag in a native proviral context.
Here, we examine a strategy to insert a fluorescent tag into HIV Gag with minimal perturbation of viral functions. Insertion of GFP at the C terminus of Gag is incompatible with GagPol frameshifting, while insertion at the N terminus would disrupt myristoyl switch membrane-binding functions of MA. A feasible alternative is to insert GFP internally into Gag (27). The Krausslich group reported an HIV-1 clone carrying GFP in the C terminus of MA. This approach was compatible with assembly and processing of virus particles, yet the virus was poorly infectious in the absence of helper Gag.
In this study, we present a novel interdomain strategy for tagging HIV Gag with GFP in a proviral context. Rather than inserting GFP into one of the Gag subdomains, we placed it between the MA and CA domains flanked by two functional protease cleavage sites to allow processing of MA and CA into native-sized polypeptides. The approach retains high infectivity without helper virus and serves as a functional marker of Gag localization. We use spectral fluorescence resonance energy transfer (FRET) to measure oligomerization and localization of Gag during the formation of infectious virus and present long-duration time-lapse imaging to illustrate the continuous sequence of Gag localization in a single infectious HIV-producing cell.
MATERIALS AND METHODS
Plasmids.
HIV-1 molecular clone NL4-3 (1) was modified with standard overlap extension PCR methods to insert a polylinker between the MA and CA domains of Gag.
The fluorescent proteins enhanced GFP (EGFP) (Clontech), Venus yellow fluorescent protein (YFP) (28), and monomeric Cerulean cyan fluorescent protein (CFP) (42) were PCR amplified and inserted into the polylinker. The resulting clone expresses an 8-amino-acid repeat, SQNYPIVQ, flanking either side of the EGFP or GFP variant. The following sequence encodes the junction between MA and EGFP: 5′-CAGGAAACAACAGCCAGGTCtcgcagaactatccaattgtacaaacgcgtATGGTGAGCAAGGGCGAGGAGCTG-3′. The MA coding sequence (uppercase) is followed by a synthetic sequence encoding SQNYPIVQ (lowercase), followed by the 5′ EGFP sequence (uppercase and underlined). The following sequence encodes the junction between EGFP and CA: 5′-ATGGACGAGCTGTACAAGTCTAGAAGCCAAAATTACCCTATAGTGCAG-3′. The 3′ EGFP gene sequence (underlined) is followed by the HIV CA sequence. All PCR-amplified regions were sequenced. A monomeric Venus variant was employed (A206K) (56); a protease catalytic mutant carries a double alanine substitution (D25A/T26A) (33); I-domain mutants in SP1, L364A, and M368A (9) were based on cited references. For virus production, standard calcium phosphate transfection of 293T cells was used. Transfection of cells for microscopy was performed with Lipofectamine 2000 (Invitrogen).
ELISA and Western blot analyses.
A quantitative luminescent enzyme-linked immunosorbent assay (ELISA) with antibody reagents from Aalto Bio Reagents was adapted from previously described methods (24, 25). For particle analyses, virus was pelleted through a 20% sucrose cushion, and immunoblots were probed with anti-HIV AIDS patient serum (AIDS Reference and Reagent Program [ARRP]) or rabbit anti-GFP (Amersham).
Infectivity analyses.
Cells were exposed to supernatants with the indicated amounts of p24 antigen. At 48 h postinfection, flow cytometry was performed on trypsinized, paraformaldehyde-fixed HOST4 cells. HeLa CD4 MAGI (from Michael Emerman, through the ARRP) was used to determine the titers of the virus without fluorescence (18).
Microscopy.
Confocal fluorescence microscopy was conducted on a Zeiss 510 LSM META Axiovert 200 microscope. Anti-p24 clone 24-3 (from Michael Malim, through the ARRP; catalog no. 6458) and anti-p17 clone 32/1.24.89 (Zeptometrix) monoclonal antibodies were developed with anti-mouse Alexa Fluor 488 or Alexa Fluor 546 (Molecular Probes). The membrane dye CM-DiI (Molecular Probes) was used to stain cells at 37°C for 15 min prior to fixation with 4% paraformaldehyde. FRET analysis was performed on fixed cells, using the spectral imaging mode of the Zeiss LSM 510 META detector. Pixelwise linear unmixing using the 12-bit emission profiles of Cerulean and Venus was performed with the Zeiss LSM software, v3.2. Image analysis and concentration-normalized FRET (NFRET) calculations (55) were conducted with the ImageJ (http://rsb.info.nih.gov/ij) and Volocity (Improvision) programs. For live imaging, 293T cells were plated in a poly-l-lysine-coated, sealed, gas-permeable microchamber (μ-Slide I; Ibidi Integrated BioDiagnostics), and the temperature was maintained at 37°C.
RESULTS
Gag-iGFP insertion strategy is compatible with viral assembly.
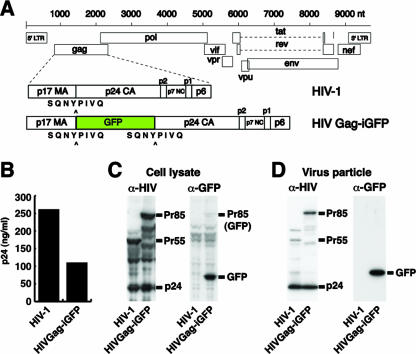
Our recent studies have found Gag to be remarkably tolerant to protein insertions between MA and CA (13). We therefore created an HIV clone with GFP placed between MA and CA and flanked by protease cleavage sites, as if it were an additional domain of Gag. An insertion into a flexible, protease-accessible linker region may be less disruptive of Gag folding or function. The viral construct is called HIV Gag-internal or interdomain green fluorescent protein (iGFP) (Fig. 1A). After assembly, the iGFP is designed to be cleaved from the Gag-iGFP precursor to permit normal processing, particle maturation, and early functions of MA and CA.
FIG. 1.
Construction and expression of proviral clone HIV Gag-iGFP. (A) GFP is inserted into HIV-1(NL4-3) between the MA and CA domains of Gag with protease cleavage sites (SQNYPIVQ) created at both ends of GFP. (B) Viral production from HIV Gag-iGFP or native HIV-1 was determined by p24 ELISA of supernatants from provirus-transfected 293T cells. (C) Cell lysates from HIV- or HIV Gag-iGFP-transfected 293T cells were examined by Western blotting with anti-HIV patient serum (left) or anti-GFP antibody (right). (D) Virus particles purified through a 20% sucrose cushion were analyzed by Western blotting with anti-HIV serum (α-HIV) (left) and anti-GFP (α-GFP) (right). Viral supernatants and cell lysates were collected at 48 hpt. LTR, long terminal repeat.
HIV Gag-iGFP produced high levels of virus particles when transfected into 293T cells, only modestly lower than the level produced by native HIV-1 (Fig. 1B). A Western blotting of transfected 293T cell lysates with anti-HIV patient serum detected the Gag-GFP fusion as the full-length precursor of the expected size of 85 kDa (Fig. 1C, left). A processed form of p24 CA that comigrated with native p24 also was observed in the HIV Gag-iGFP lane. This indicated that the viral protease recognized and cleaved the Gag-iGFP precursor. The ratio of the precursor Gag-iGFP to p24 was similar to that of native p55 Gag to p24, suggesting that the efficiency of Gag processing was not greatly perturbed by the GFP insertion. When probing with an anti-GFP antibody, the processed form of GFP was preferentially detected with a weak recognition of the Gag-iGFP Pr85 fusion (Fig. 1C, right).
To confirm that the p24 antigen released from the cells had formed virus particles, we ultracentrifuged viral supernatants through 20% sucrose. In virus particles, Gag-iGFP was efficiently processed by the viral protease, producing cleaved p24 CA at a level similar to that of native HIV (Fig. 1D, left). The GFP in virus particles again was detected primarily in the processed form (Fig. 1D, right). The internal insertion strategy therefore was compatible with viral assembly and resulted in virus particles carrying native-sized MA and CA and stoichiometric quantities of GFP.
HIV Gag-iGFP exhibits high levels of infectivity.
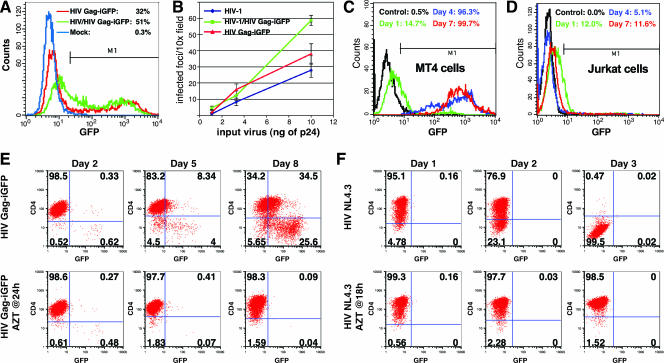
To examine the infectivity of the HIV Gag-iGFP, virus was produced in 293T cells in the presence or absence of helper provirus, transfecting a constant level of plasmid DNA. HeLa T4 cells were exposed to equal supernatant volumes without normalizing for viral production. HIV Gag-iGFP infected 32% of HeLa T4 cells at 48 h postinfection (Fig. 2A). The addition of helper virus at a 1:1 ratio modestly improved the titer to 51%. These results indicated that HIV Gag-iGFP is infectious without a requirement for helper virus.
FIG. 2.
High level of infectivity of HIV Gag-iGFP. (A) Infection of HeLa T4 cells with HIV Gag-iGFP or HIV Gag-iGFP made in the presence of helper virus. Infected cells were analyzed by flow cytometry at 48 hpi. (B) Infection of a HeLa T4 MAGI indicator cell line with viral supernatants carrying the indicated amounts of p24. Infected cells were measured by counting infected cell foci per 10× magnification field. (C) Infection of CD4+ MT4 T-cell lines with 10 ng of p24 was monitored over time by flow cytometry. (D) Infection of Jurkat CD4+ T cells was monitored over time as described for panel C. (E) Infection of MT4 cells with 30 ng of HIV Gag-iGFP virus was monitored by GFP and CD4 fluorescence over time. Cells were washed at 24 h and were treated with AZT (2 μM) or were left untreated. (F) Infection of MT4 cells with HIV(NL4-3) (10 ng of p24) was monitored indirectly by monitoring CD4 downmodulation in the infected cells.
To compare the infectivity of HIV Gag-iGFP to that of native HIV-1, we diluted viral supernatants to equivalent levels of p24 antigen and infected HeLa MAGI indicator cells (18). Titers of HIV Gag-iGFP and HIV Gag-iGFP in the presence of helper virus were similar to those of the native HIV(NL4-3) under nonsaturating conditions (Fig. 2B). Thus, in single-round infectivity assays, HIV Gag-iGFP produced virus particles with a titer comparable to that of its parental clone.
To determine the infectivity of HIV Gag-iGFP in human T-cell lines, we infected the highly permissive CD4+ lymphoblastoid T-cell line MT4 and monitored viral spread by flow cytometry at 3-day intervals. MT4 supported rapid viral spread, completely infecting the culture in 7 days (Fig. 2C), with prominent cytopathic effects. In contrast, the infection of Jurkat T cells (Fig. 2D) generated a similar small fraction of GFP-positive cells at day 1 but without viral spread on days 4 and 7. Thus, HIV Gag-iGFP was replication competent in a cell-type-dependent manner. The infection of primary CD4+ cells was similar to that of the Jurkat cells (data not shown).
Because the spread of HIV Gag-iGFP was rapid, we performed an azidothymidine (AZT) interruption experiment to see whether the spread of HIV Gag-iGFP fluorescence in culture was dependent upon continued reverse transcriptase activity after an initial infection period without the inhibitor. Viral spread was monitored by flow cytometry measuring Gag-iGFP green fluorescence and CD4-phycoerythrin immunostaining to observe the downmodulation of CD4 during infection (Fig. 2E). Initial signs of infection by day 2 showed a small fraction of infected cells of less than 2% in both the untreated and AZT-treated cells. Viral spread of HIV Gag-iGFP could be blocked by treatment of the cells with AZT at 24 h following infection, suggesting that the rapid spread in the cells can be interrupted by blocking later rounds of ongoing viral reverse transcription. The ability of native HIV to spread through the culture of MT4 cells was monitored indirectly by monitoring CD4 downmodulation (Fig. 2F). Native HIV spread even more rapidly than HIV Gag-iGFP, downregulating CD4 in nearly all the cells within 3 days. This experiment suggested that full viral spread in HIV Gag-iGFP in MT4 cells required continued rounds of reverse transcription and infection and showed that the parental virus replicates significantly faster in MT4 cells than does HIV Gag-iGFP. We conclude that although attenuated relative to the parental virus, HIV Gag-iGFP can replicate in multiple rounds in MT4 cells but not other cell lines, such as Jurkat.
Distribution of Gag-iGFP resembles that of native Gag.
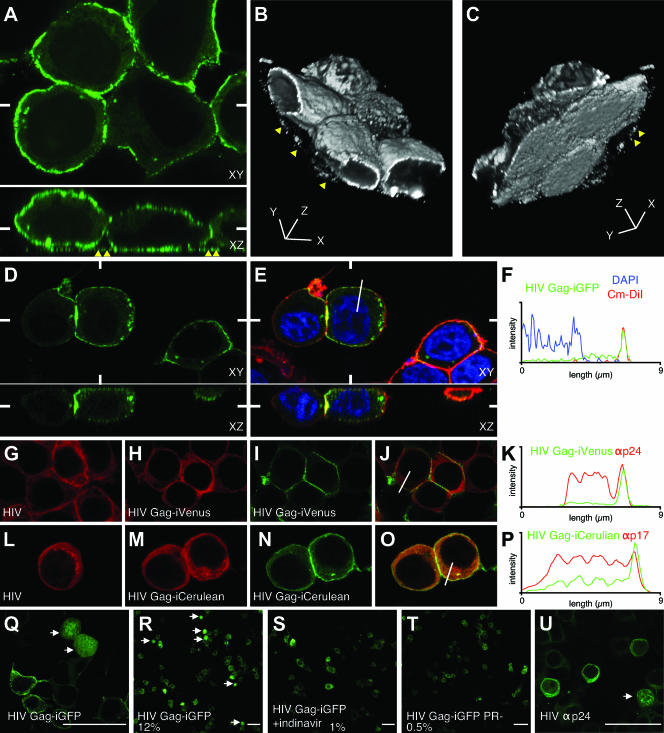
We performed laser-scanning confocal microscopy of HIV Gag-iGFP-expressing cells to visualize the cellular distribution of Gag-iGFP. At 24 h posttransfection (hpt), Gag-iGFP was predominantly associated with the plasma membrane (Fig. 3A). Intense dots of fluorescence also were evident in the focal plane of the coverslip and are highlighted in the x-z view (Fig. 3A, bottom) and in three-dimensional reconstructions of the images (Fig. 3B and C; also see movie S1 in the supplemental material). The intense membrane signal was revealed to be membrane associated by colocalization in three dimensions with the membrane dye Cm-DiI (Fig. 3E). A profile of the Gag-iGFP fluorescence intensity showed an intense plasma membrane signal that coincided nearly perfectly with the plasma membrane Cm-DiI signal (Fig. 3F). In addition, the profile showed a weak GFP signal in cytoplasmic sites and exclusion of Gag-iGFP fluorescence from the 4′,6′-diamidino-2-phenylindole (DAPI)-positive nuclear regions. The prominent membrane-associated signal and nuclear exclusion of Gag-iGFP suggest that the cleaved GFP that is found in cell lysates (Fig. 1C) is not freely diffusible and may be trapped within the confines of intact virus particles.
FIG. 3.
Confocal fluorescence microscopy of HIV Gag-iGFP validates the use of Gag-iGFP as a marker to study native Gag localization. (A) Single-slice, confocal images of HIV Gag-iGFP-transfected 293T cells that were fixed at 24 hpt. Bottom panels represent a slice through the reconstructed x-z axes. The white marks indicate the position of the x-z slice with respect to the x-y plane and vice versa. Yellow triangles point out dots of intense fluorescence bound to the coverslip that are not associated with cells. (B) The top view of a three-dimensional reconstruction of the image stack in panel A. (C) Rotated three-dimensional view showing the bottom view of cells shown in panel B. (D) Green fluorescence of an HIV Gag-iGFP-transfected 293T cell that was costained with membrane and nuclear dyes (see Movie S1 in the supplemental material). (E) Three-color representation of the image in panel D showing colocalization of Gag-iGFP (green) with the membrane dye CM-DiI (red) and exclusion from the nucleus, demarcated by DAPI (blue). (F) Three-color intensity profile indicating the fluorescence intensity of HIV Gag-iGFP (green), CM-DiI (red), and DAPI (blue) along the 9-μm line drawn on the cell in panel E. (G) HIV(NL4-3)-transfected cells stained with anti-p24 monoclonal antibody (red). (H, I, and J) HIV Gag-iVenus-transfected cells stained with anti-p24 (red) (H) and Venus fluorescence (green) (I), with a overlay shown in panel J. (K) Two-color intensity profile showing the relationship between anti-p24 antibody staining and Gag-iVenus signals along the line from panel J. (L) HIV(NL4-3)-transfected cells stained with anti-p17 monoclonal antibody (red). (M) HIV Gag-iCerulean-transfected cells stained with anti-p17 (red) and Cerulean fluorescence (green) (N), with an overlay shown in panel O. (P) Two-color intensity profile of the relationship between anti-p17 antibody staining and Gag-iCerulean signals along the line from panel O. (Q) High-magnification view of HIV Gag-iGFP, with arrows pointing to cells that do not maintain nuclear exclusion of green fluorescence. (R) Low-magnification view shows that the cells that lack nuclear exclusion are a small fraction of the total cell population (12%). (S) Treatment with indinavir inhibits the production of cells with nuclear fluorescence. (T) HIV Gag-iGFP with a point mutation in the viral protease (PR−) also fails to produce cells with strong nuclear fluorescence. (U) A small fraction of HIV(NL4-3) expressing 293T cells stained with an anti-p24 antibody also show Gag localization in the nucleus. (Q to U) The white bar represents 50 μm.
To test the utility of HIV Gag-iGFP as a marker for Gag localization, we compared the immunofluorescence of Gag-iGFP to that of native Gag. Anti-p24 immunostaining of native Gag revealed a strong cytoplasmic signal without a prominent plasma membrane signal (Fig. 3G). The same antibody gave a similar pattern for cells expressing HIV Gag-iVenus, a clone carrying Venus YFP in place of GFP. These cells also lacked an intense plasma membrane signal (Fig. 3H). We conclude that the Gag-iGFP insertion did not perturb the distribution of p24 epitopes in cells expressing Gag-iGFP. In contrast, the Venus fluorescence on the same cells showed a prominent plasma membrane signal (Fig. 3I). In comparisons of anti-p24 fluorescence to Venus fluorescence (Fig. 3J), the Venus signal prominently highlighted the membrane, whereas the anti-p24 immunofluorescence showed little contrast between the membrane and intracellular sites (Fig. 3K). We next tested an anti-p17 antibody that recognizes both p55 precursor and processed p17 (31). Anti-p17 immunofluorescence revealed similar patterns for HIV Gag-iCerulean, a CFP variant, and native HIV-1 (Fig. 3L and M). Again, the profile of the fluorescence yielded a low level of contrast between the membrane and cytoplasmic signals. In contrast, the direct Cerulean fluorescence featured a prominent membrane signal (Fig. 3N, O, and P). Based on these results and on a recent report (32), it is likely that immunofluorescence routinely underestimates the levels of Gag at the plasma membrane due to epitope-masking effects. Another possible explanation for the differences in fluorescence patterns is attributable to the maturation (protein folding) of GFP in the Gag-iGFP constructs. In either case, the GFP fluorescence from Gag-iGFP and antibody fluorescence clearly reveal different fractions of Gag in the cell.
Because Western blots on HIV Gag-iGFP-transfected cell lysates showed a high level of cleaved GFP (Fig. 1C), we performed experiments to determine what fraction of the total fluorescent signal is attributable to HIV protease-liberated, free GFP. A small fraction (about 12%) of HIV Gag-iGFP-transfected cells exhibited a bright, diffuse green fluorescence that was not excluded from the nucleus (Fig. 3Q and R). These uniformly fluorescent cells were not observed in cells treated with an HIV protease inhibitor or in cells expressing a catalytically inactive protease mutation (Fig. 3S and T). Anti-p24 immunostaining suggested that a similar protease-dependent change in localization occurs with capsid epitopes in native HIV-1 (Fig. 3U). These results suggest that in most virus-expressing cells, the viral protease activation does not alter the distribution of Gag, further indicating that Gag-iGFP is a faithful mimic of Gag precursor localization. We conclude that the cleaved GFP in the cell lysates likely is attributable to mature virus particles that may have been reinternalized (29) or to the 12% of cells that exhibit protease-dependent cytoplasmic processing of Gag, pools that do not affect the overall pattern of Gag localization in most cells.
Visualizing Gag oligomerization in virus-producing cells by FRET.
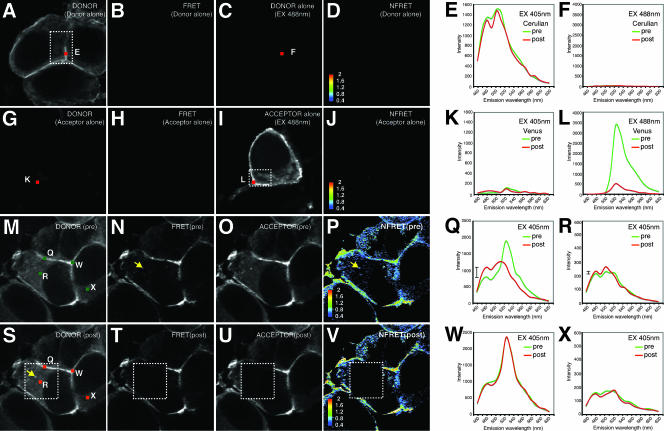
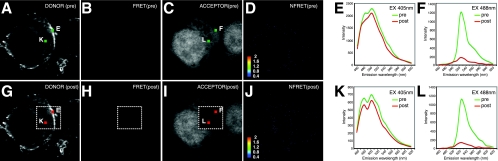
To study Gag oligomerization during infectious particle formation, we utilized HIV Gag-iGFP constructs carrying spectral variants of GFP, monomeric Cerulean (42) CFP and monomeric Venus (28) YFP. These proteins are an ideal donor/acceptor pair for FRET, a fluorescence-based method to monitor protein interactions in cells (42). Spectral confocal imaging (57) of singly transfected HIV Gag-iCerulean (Fig. 4A) or HIV Gag-iVenus (Fig. 4I) cells gives a fluorescence emission spectrum characteristic of Cerulean (Fig. 4E) or Venus proteins, respectively (Fig. 4L). When excited with light at 405 nm, the Cerulean fluorophore (Fig. 4A), but not the Venus fluorophore (Fig. 4G and K), is activated. Cerulean excitation will produce a spectral shift towards the Venus emission profile only if a Venus acceptor fluorophore is within the 50-Å Forster radius of a Cerulean donor fluorophore (52). A Venus-shifted emission upon stimulation at 405 nm indicates close apposition of the two fluorophores, which can be used as a surrogate measure of oligomerization (7, 20).
FIG. 4.
Spectral FRET analysis of Gag-iCerulean donor and Gag-iVenus acceptor reveals intense membrane FRET and weaker cytoplasmic FRET. (A through F) Images and emission profiles for HIV Gag-iCerulean alone. HIV Gag-iCerulean excited at 405 nm (EX 405nm) yields a strong linear unmixed Cerulean donor signal (Cerulean) (A) but no FRET signal (Venus profile at 405 nm excitation) (B). HIV Gag-iCerulean excited at 488 nm produces no background Venus yellow fluorescence image (C) and no NFRET signal (D). (E) Emission spectrum of HIV Gag-iCerulean stimulated at 405 nm gives no donor signal at region E before (pre) or after (post) photobleaching. (F) Emission spectra of HIV Gag-iCerulean stimulated at 488 nm before and after photobleaching show no increase in Cerulean signal (no pseudo-dequenching). (G through L) Images and emission profiles for HIV Gag-iVenus alone. HIV Gag-iVenus excited at 405 nm does not yield a linear unmixed donor signal (Cerulean) (G) or FRET signal (Venus profile at 405 nm excitation) (H). HIV Gag-iVenus excited at 488 nm produces a characteristic Venus yellow fluorescence image (I) but no NFRET signal (J). (K) Emission spectrum of HIV Gag-iVenus stimulated at 405 nm gives no donor signal at region K before or after photobleaching. (L) Emission spectrum of HIV Gag-iVenus stimulated at 488 nm before and after photobleaching reveals efficient loss of Venus signal. (M through X) Data from cells cotransfected with HIV Gag-iCerulean and HIV Gag-iVenus. (M) Linear unmixed donor image of cells excited at 405 nm before acceptor photobleach. (N) Excitation at 405 nm results in a FRET signal. (O) Linear unmixed image of the acceptor signal at 488 nm before photobleaching. (P) NFRET index of cells calculated from linear unmixed images. A yellow arrow indicates an intracellular vesicle. (Q) Emission profile upon excitation at 405 nm of membrane region Q before and after acceptor photobleaching. The hash mark to the left of the profiles indicates the size of the dequenching of the 480-nm peak. (R) Emission profile upon excitation at 405 nm of cytoplasmic region R before and after acceptor photobleaching. The small hash mark indicates the size of the dequenching at 480 nm. (S) Linear unmixed donor image after acceptor photobleaching in the dotted square. (T) The FRET signal is lost in the bleach area. (U) Depletion of the acceptor signal after acceptor photobleaching. (V) NFRET index of cells after acceptor photobleaching. (W) The spectral profile of membrane region W outside the bleach zone shows no change. (X) The spectral profile of the cytoplasmic region X outside the bleach zone shows no change.
When the iCerulean and iVenus viral constructs were cotransfected, excitation at 405 nm of the Cerulean donor revealed a strong Venus-shifted emission indicative of FRET at the plasma membrane (Fig. 4N and Q). The NFRET index was calculated (55) to normalize the FRET signal as a function of the concentration of the donor and acceptor fluorophores (Fig. 4P). The NFRET image showed a strong membrane signal and a smaller positive signal in the cytoplasm. The emission profiles of two spots on the membrane, labeled Q and W, show a prominent yellow fluorescent peak at 527 nm, indicative of strong FRET (Fig. 4Q and W). A small intracellular vesicle also was found to be FRET positive (Fig. 4P, arrowhead). With three-dimensional confocal images, we observe that these FRET-positive compartments are not contiguous with the plasma membrane (data not shown). Spectral analyses of the cytoplasmic regions R and X each showed a small 527-nm peak, indicative of FRET, when the donor wavelength was excited.
To examine whether low-level excitation of the acceptor fluorophore with light at 405 nm may contribute to an apparent FRET signal, control experiments cotransfecting HIV Gag-iCerulean with free Venus fluorescent protein were performed. These did not show evidence of FRET at the cytoplasm or membrane (Fig. 5A to D). We conclude that there is strong FRET on the membrane and weaker but true FRET in the cytoplasm, indicative of Gag oligomerization at both sites.
FIG. 5.
FRET analysis of HIV Gag-iCerulean and unconjugated Venus YFP. (A through L) Images and emission profiles of 293T cells cotransfected with HIV Gag-iCerulean and free Venus fluorescent protein. Control cells excited at 405 nm (EX 405nm) yield a strong linear unmixed Cerulean donor signal (A) but no FRET signal (Venus profile for excitation at 405 nm) (E). The free Venus excited at 488 nm produces a diffuse Venus image (C) but no NFRET signal (D). (E) Emission spectrum at membrane region E of cotransfected cells stimulated at 405 nm yields only Cerulean fluorescence with no evidence of Venus-shifted 527-nm peak before (pre) photobleaching and no dequenching at 480 and 500 nm after (post) photobleaching at 514 nm. (F) Emission spectrum of cotransfected cells at membrane region F stimulated at 488 nm shows the efficiency of acceptor photobleaching at 514 nm. (G) The donor signal is not changed after acceptor photobleaching. (H) The FRET signal is not changed after acceptor photobleaching. (I) Depletion of acceptor signal in the boxed region after photobleaching at 514 nm. (J) Lack of NFRET signal after photobleaching. (K) Emission spectrum at cytoplasmic region K of cotransfected cells stimulated at 405 nm yields only Cerulean fluorescence with no evidence of a Venus-shifted 527-nm peak before photobleaching and no dequenching at 480 and 500 nm after photobleaching at 514 nm. (L) Emission spectrum of cotransfected cells at cytoplasmic region L stimulated at 488 nm shows the efficiency of acceptor photobleaching at 514 nm.
Acceptor photobleaching analyses were used to measure FRET efficiency in the samples. After photobleaching the acceptor, the fluorescence of the donor fluorophore involved in FRET is recovered (donor dequenching). The Gag-iVenus acceptor was fully depleted in a selected region of interest, resulting in loss of Venus fluorescence when stimulated at 488 nm (Fig. 4U). Following this acceptor depletion, we observed a strong increase in Cerulean fluorescence, particularly at the plasma membrane (Fig. 4S and Q). To estimate the FRET transfer efficiency (E), we employed the equation E = 1 − Fda/Fd, where F is the fluorescence of the donor in the presence (Fda) or absence (Fd) of acceptor (3). The FRET transfer efficiency at the plasma membrane was 25 to 30% of donor energy transferring to acceptor, a figure that is comparable in intensity to those for published intramolecular CFP-YFP FRET systems (51).
Acceptor photobleaching at cytoplasmic regions also resulted in a loss of the small 527-nm peak and an increase in Cerulean fluorescence at 480 and 500 nm (Fig. 4R). In contrast, the spectral profile of the neighboring unbleached areas did not show donor dequenching (Fig. 4W and X). While donor dequenching at cytoplasmic sites was weak (Fig. 4R), no donor dequenching was observed when HIV Gag-iCerulean was cotransfected with free Venus protein (Fig. 5G to J).
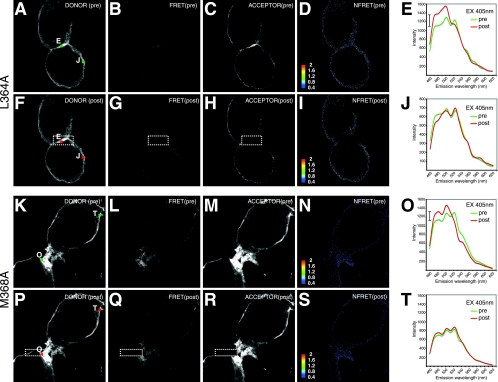
To further probe whether the spectral shifts we observed were mediated by Gag sequences, we tested two point mutants, with L364A and M368A mutations in the interaction (I) domain of Gag (9), for their ability to engage in FRET. The intensity of the membrane-associated FRET was severely reduced, but not totally eliminated, when either of the I-domain mutants was tested (Fig. 6). Evidence for the weak spectral FRET is best appreciated in the spectral profiles at the membrane regions E and J (for L364A) or O and T (for M368A) (Fig. 6E, J, O, and T). The third 527-nm peak is evident for both constructs at membrane-associated sites. Acceptor photobleaching in the regions of interest (Fig. 6H and R) revealed an increase in the donor fluorescence indicative of FRET in regions E and O (Fig. 6E and O). Thus, we found that the strong FRET at the membrane was I domain dependent, but some lower level of oligomerization was I domain independent. These results extend and clarify findings of earlier Gag-GFP FRET studies (7, 20) and provide evidence for unique oligomeric states of Gag at different cellular sites. These studies also are the first to study Gag FRET that is measured with a protein expressed within a full viral context.
FIG. 6.
Gag I-domain L364A and M368A mutants severely reduce but do not abolish FRET. (A through J) Images and emission profiles of 293T cells cotransfected with HIV Gag-iCerulean/Venus L364A mutants. L364A-transfected cells excited at 405 nm (EX 405nm) yield a strong linear unmixed Cerulean donor signal (A) but very little FRET signal (Venus profile for excitation at 405 nm) (B). The L364A acceptor Venus excited at 488 nm produces an image similar to the donor image (C), and a very weak NFRET signal is apparent (D). (E) Emission spectrum at membrane region E of L364A-transfected cells stimulated at 405 nm yields dominant Cerulean fluorescence with evidence of a weak Venus-shifted 527-nm peak before (pre) photobleaching, with modest dequenching at 480 and 500 nm after (post) photobleaching at 514 nm. The hash mark indicates the extent of dequenching at 480 nm. (F) Donor signal after acceptor photobleaching. (G) FRET signal after acceptor photobleaching. (H) Depletion of acceptor signal in the boxed region after photobleaching at 514 nm. (I) The low-level NFRET signal is lost after photobleaching. (J) The emission spectrum at unbleached membrane region J of L364A-transfected cells stimulated at 405 nm yields evidence of a weak Venus-shifted 527-nm peak before photobleaching that is unaffected after bleaching, because it lies outside of the bleached region. FRET images in this figure were leveled uniformly to better convey the localization of the weak FRET signals observed. (K through T) Images and emission profiles of the M368A mutant. M368A-transfected cells excited at 405 nm yield a strong linear unmixed Cerulean donor signal (K) but a weak FRET signal (Venus profile for excitation at 405 nm) (L). The M368A acceptor Venus excited at 488 nm produces an image similar to the donor image (M), and very weak NFRET signal is apparent (N). (O) Emission spectrum at membrane region O of M368A-transfected cells stimulated at 405 nm yields dominant Cerulean fluorescence with evidence of a weak Venus-shifted 527-nm peak before photobleaching, with modest dequenching at 480 and 500 nm after photobleaching at 514 nm. The hash mark indicates the extent of dequenching at 480 nm. (P) Donor signal after acceptor photobleaching. (Q) FRET signal after acceptor photobleaching. (R) Depletion of acceptor signal in the boxed region after photobleaching at 514 nm. (S) Low-level NFRET signal is lost after photobleaching. (T) Emission spectrum at unbleached membrane region T of M368A-transfected cells stimulated at 405 nm yields evidence of a weak Venus-shifted 527-nm peak before photobleaching that is unaffected after bleaching, because it lies outside of the bleached region.
Distinct stages of Gag-iGFP localization revealed by live four-dimensional fluorescent imaging in virus-producing cells.
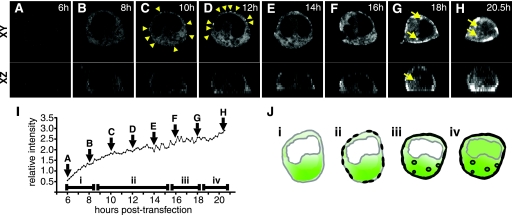
To monitor the progression of Gag localization during the lifetime of a virus-expressing cell, live confocal microscopy was performed on HIV Gag-iGFP-expressing cells. Image stacks were recorded every 10 min from 6 to 21 hpt to generate a four-dimensional movie (see movie S2 in the supplemental material). Low-intensity Gag-iGFP fluorescence was detected at 6 h as a diffusely cytoplasmic signal that was excluded from the nucleus (Fig. 7A). Small areas of exclusion of Gag also are apparent in a perinuclear zone and in small vesicle-sized structures, giving the cytoplasm a Swiss cheese-like appearance. The total fluorescence intensity of the HIV Gag-iGFP-expressing cell increased steadily over time (Fig. 7I). At 10 h, as the total cell Gag concentration increases (Fig. 7C), points of higher-intensity green fluorescence appear at the plasma membrane. During a window of 10 to 16 h (Fig. 7C to F), plasma membrane fluorescence increased steadily, while cytoplasmic fluorescence remained diffuse without evidence of accumulation at intracellular vesicles. By 18 h, high-intensity Gag fluorescence is observed at the plasma membrane and in intracellular vesicular pools (Fig. 7G and H). In the latest stages of viral gene expression, we observed a loss of nuclear exclusion of fluorescence (Fig. 7H), which resembled the pattern of a minority of cells transfected in the absence of protease inhibitors (Fig. 3Q and R). The images show a progression through four distinct patterns of Gag localization as the levels of total Gag fluorescence increase (Fig. 7J). The patterns observed were representative of three independent long-duration imaging sessions in which one or more cells were captured from early to late stages of this progression.
FIG. 7.
Time-lapse four-dimensional confocal fluorescence imaging of a living HIV Gag-iGFP-expressing cell reveals a temporal sequence of Gag localization. Confocal z stacks were acquired at 10-min intervals for 14.5 h. (A through H) Montage of fluorescence images illustrating changes in the fluorescence pattern of Gag-iGFP at 2-h intervals. x-y cross-sections through the middle of the cell (top) and x-z profiles (bottom) are indicated. Triangles in panels C and D highlight early membrane accumulations of Gag. Arrows in panels G and H highlight intracellular vesicular accumulations of Gag. (I) The total integrated fluorescence intensity of the cell is graphed over time. Letters A through H indicate the time point at which each representative image was acquired. (J) Gag localization in 293T cells progresses through four morphological stages: (i) diffuse cytoplasmic; (ii) diffuse cytoplasmic and punctate membrane staining; (iii) diffuse cytoplasmic, strong membrane, and vesicular staining; and (iv) loss of nuclear exclusion. Time-lapse imaging of the 293T cell is representative of more than three independent long-duration imaging experiments in which one or more cells were successfully captured over time (see Movie S2 in the supplemental material).
This approach documented the changes in Gag localization with continuous video microscopy of individual cells producing infectious virus particles. Gag-iGFP accumulation at plasma membrane sites preceded strong intracellular vesicular sites. These time-lapse images suggest that the plasma membrane is the predominant initial site of Gag accumulation under conditions in which there was production of infectious HIV-1. We conclude that trafficking patterns of Gag change as a function of the total viral gene expression program.
DISCUSSION
Fluorescent protein fusions provide a powerful methodology to reveal dynamic intracellular protein-trafficking pathways. However, a prerequisite to interpreting the significance of GFP fusion experiments is to determine the protein's ability to function in place of the native protein. To follow the dynamics of HIV Gag during assembly and transmission, we present the first autonomously infectious, provirally expressed Gag-GFP fusion. An iGFP insertion strategy minimizes disturbances to viral fitness. This tool allowed us to track the distribution and oligomerization of Gag during the formation of infectious virus particles without helper virus. The remarkable infectivity of the virus validates its use for studying Gag dynamics.
Immunofluorescence with anti-p17 and anti-p24 antibodies revealed that the localization of HIV Gag-iGFP was nearly identical to that of native HIV-1 Gag. Interestingly, however, the fluorescence at the membrane from GFP was much more pronounced than that seen by immunofluorescence. A recent study has demonstrated that many antibodies do not readily react with oligomerized forms of Gag that may be concentrated at the plasma membrane (32). It therefore is possible that standard immunostaining approaches routinely underestimate the levels of Gag at the plasma membrane. HIV Gag-iGFP will be useful in developing and testing quantitative models for Gag transport and viral assembly. Our FRET studies, discussed further below, confirmed that comparatively low levels of antibody reactivity at the plasma membrane correlated with intense Gag oligomerization at that site. As an additional consideration, the GFP tagging also may preferentially reveal more mature, or well folded, pools of Gag that are concentrated at the plasma membrane. The results further establish fluorescence tagging as an important adjunct to immunofluorescence for determining the cellular distribution of Gag.
The high level of infectivity is achieved in part by introducing GFP into the Gag precursor protein flanked by HIV protease cleavage sites, which effectively are processed in mature virus particles. The resulting processed virus particle thus carries unperturbed MA and CA, which limits the negative effects on viral infectivity. Single-round infectivity experiments showed no defect in infectivity of the HIV Gag-iGFP virus. We found that in a highly permissive T-cell line, MT4, the virus actually could replicate rapidly with full cytopathic effects. The spread of the virus in culture was further demonstrated by the requirement for ongoing reverse transcriptase activity to fully infect the culture. In other T cells, such as Jurkat cells and peripheral blood mononuclear cells, infection was observed at similar initial titers, but we did not observe rapid spread. These results suggest that the spacing between MA and CA is important for propagation in culture for certain T-cell lines.
This fusion strategy also allowed us to assess the extent to which the localization of the Gag precursor in producer cells is altered by the activity of HIV protease. Although cleaved Gag-iGFP was found in all cell lysates, intracellular protease activity did not alter the localization of Gag-iGFP in most cells. From this, we infer that protease is active only within virus particles, where cleaved GFP cannot diffuse. Our live imaging further suggested that intracellular cleavage of diffusible Gag occurs in the latest stage of viral gene expression in a small percentage of cells. We suggest that in these cells a diffuse GFP fluorescence and rounded morphology are indicative of the terminal effects of intracellular HIV protease activation (50). We conclude that HIV Gag-iGFP permits tracking of full-length unprocessed Gag within the majority of virus-producing cells, without complications of premature intracellular processing of Gag-iGFP.
Our FRET assays show a strong homooligomerization of Gag at the plasma membrane. FRET signal also was detected at intracellular vesicular compartments. Two previous FRET studies that examined Gag-GFP oligomerization in the absence of other viral proteins (7, 20) also found that Gag was strongly oligomeric at membrane sites. These studies differed in their view of whether any lower-order oligomerization occurs in the cytoplasm. With an NFRET index and acceptor photobleaching analyses, we show that nonvesicular Gag-iGFP within the cytoplasm is engaged in FRET. These studies also reveal that intense membrane FRET is dependent upon key residues within the I domain. Previous biochemical studies have speculated that prior to membrane engagement, Gag forms low-order oligomers (21, 22). Weak cytoplasmic FRET may be indicative of these assembly intermediates. A previous study of Gag-GFP did not observe nonvesicular, cytoplasmic FRET (7). Our contrasting result may be explained by the presence of other viral genes or may be due to differences in the construct design. Inserting GFP internally into Gag may create an organized lattice of GFP constrained between MA and CA that may enhance its ability to induce FRET. The high sensitivity of the Gag-iGFP FRET assay makes it attractive for use in high-throughput assays to identify inhibitors of HIV assembly.
GFP has revolutionized cell biological studies by enabling researchers to dynamically track the movements of proteins in living cells. This study validates a novel GFP fusion approach to track HIV Gag during the course of viral infection. Our single-cell, four-dimensional time-lapse fluorescence microscopy studies showed the progression of Gag distribution in cells through four discreet stages (Fig. 6F). The progression went from a diffuse cytoplasmic localization, to a pattern with additional plasma membrane accumulation, to later stages in which intense plasma membrane staining coincides with accumulations at intracellular vesicular compartments. Finally, in the most intensely expressing cells, we see nuclear invasion, loss of discreet compartments, and rounded cell morphology. Because we observe different patterns at different stages, we speculate that the intracellular dynamics are altered in a switch-like manner depending on the concentration of Gag (37) or other viral proteins.
There has been considerable debate regarding the pathway of Gag trafficking to the plasma membrane. Many suggest that the late endosome is a requisite trafficking step prior to Gag engagement of the plasma membrane (8, 30, 38). Recently, others have presented compelling data to suggest that Gag directly engages the plasma membrane (17, 29, 43). We show with continuous single-cell live imaging that as levels of Gag increase in a cell expressing all viral genes from a natural proviral context, accumulations of Gag at the plasma membrane precede accumulations of Gag at intracellular vesicular sites. As suggested recently, intracellular vesicle-associated Gag may represent endocytosed protein from the cell surface (11, 29). Our live-imaging results with HIV Gag-iGFP are supportive of the view that localization of Gag to the plasma membrane does not require a late endosomal intermediate (17). The acquisition of full three-dimensional data sets at each time point also allowed us to resolve that these intracellular accumulations of Gag were not contiguous with the plasma membrane, as has recently been described for macrophages (6, 54). Further analysis of HIV Gag-iGFP in other cell types may help to determine the extent to which Gag targeting and assembly are controlled differently according to cell type.
Lastly, we find that not only are the infected cells fluorescently labeled but the progeny viruses are also stoichiometrically endowed with green fluorescence. This virus will be particularly useful for visualizing intercellular structures that facilitate cell-to-cell transmission, called virological synapses (16). An accompanying paper in this issue illustrates the utility of HIV Gag-iGFP in quantifying the efficiency of cell-to-cell transfer between HIV-infected and uninfected T cells (4). Although the virus is not replication competent in all cell types, HIV Gag-iGFP still may provide a sensitive method to track the movements of infected cells and viral particles in animal models. Ultimately, a clearer understanding of the events that link assembly with viral transmission will be obtained by directly observing the events with live imaging of infected immune cells.
Supplementary Material
Acknowledgments
We thank Theresa Chang for helpful comments, David Piston for the gift of monomeric Cerulean, Atsushi Miyawaki for the gift of the Venus fluorescent protein, and Rumana Huq and Robert Hennigan for advice and support with confocal imaging.
This work was supported in part by NIH, NIAID grant AI-058856 and an American Foundation for AIDS Research (amfAR) Research Award (106660) to B.K.C. A.D.P. was supported by NIAID training grant T32AI007647. Confocal microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported by NIH-NCI shared resources grant R24 CA095823 and NSF Major Research Instrumentation grant DBI-9724504.
Footnotes
Published ahead of print on 29 August 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell. 5:161-174. [DOI] [PubMed] [Google Scholar]
- 3.Berney, C., and G. Danuser. 2003. FRET or no FRET: a quantitative comparison. Biophys. J. 84:3992-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P., W. Hübner, M. Spinelli, and B. K. Chen. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 81:12582-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, S. H., N. Van Damme, J. R. Day, C. M. Noviello, D. Hitchin, R. Madrid, S. Benichou, and J. C. Guatelli. 2005. Leucine-specific, functional interactions between human immunodeficiency virus type 1 Nef and adaptor protein complexes. J. Virol. 79:2066-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deneka, M., A. Pelchen-Matthews, R. Byland, E. Ruiz-Mateos, and M. Marsh. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177:329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, X., H. Li, A. Derdowski, L. Ding, A. Burnett, X. Chen, T. R. Peters, T. S. Dermody, E. Woodruff, J. J. Wang, and P. Spearman. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120:663-674. [DOI] [PubMed] [Google Scholar]
- 9.Guo, X., A. Roldan, J. Hu, M. A. Wainberg, and C. Liang. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halwani, R., A. Khorchid, S. Cen, and L. Kleiman. 2003. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J. Virol. 77:3973-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harila, K., I. Prior, M. Sjoberg, A. Salminen, J. Hinkula, and M. Suomalainen. 2006. Vpu and Tsg101 regulate intracellular targeting of the human immunodeficiency virus type 1 core protein precursor Pr55gag. J. Virol. 80:3765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hübner, W., and B. K. Chen. 2006. Inhibition of viral assembly in murine cells by HIV-1 matrix. Virology 352:27-38. [DOI] [PubMed] [Google Scholar]
- 14.Janvier, K., Y. Kato, M. Boehm, J. R. Rose, J. A. Martina, B. Y. Kim, S. Venkatesan, and J. S. Bonifacino. 2003. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes. J. Cell Biol. 163:1281-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin, Y. J., C. Y. Cai, X. Zhang, H. T. Zhang, J. A. Hirst, and S. J. Burakoff. 2005. HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J. Immunol. 175:3157-3164. [DOI] [PubMed] [Google Scholar]
- 16.Jolly, C., K. Kashefi, M. Hollinshead, and Q. J. Sattentau. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jouvenet, N., S. J. Neil, C. Bess, M. C. Johnson, C. A. Virgen, S. M. Simon, and P. D. Bieniasz. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson, D. R., M. C. Johnson, W. W. Webb, and V. M. Vogt. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. USA 102:15453-15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson, D. R., Y. M. Ma, V. M. Vogt, and W. W. Webb. 2003. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 162:1233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 23.Lodge, R., H. Gottlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKeating, J. A., A. McKnight, and J. P. Moore. 1991. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J. Virol. 65:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 26.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 27.Müller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai, T., K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, and A. Miyawaki. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87-90. [DOI] [PubMed] [Google Scholar]
- 29.Neil, S. J., S. W. Eastman, N. Jouvenet, and P. D. Bieniasz. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4:902-910. [DOI] [PubMed] [Google Scholar]
- 31.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono, A., A. A. Waheed, A. Joshi, and E. O. Freed. 2005. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131-14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6(Gag) protein produces a mutant that fails to package its envelope protein. J. Virol. 73:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owens, R. J., and R. W. Compans. 1989. Expression of the human immunodeficiency virus envelope glycoprotein is restricted to basolateral surfaces of polarized epithelial cells. J. Virol. 63:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owens, R. J., J. W. Dubay, E. Hunter, and R. W. Compans. 1991. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc. Natl. Acad. Sci. USA 88:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 78:9560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perlman, M., and M. D. Resh. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic 7:731-745. [DOI] [PubMed] [Google Scholar]
- 39.Piguet, V., and Q. Sattentau. 2004. Dangerous liaisons at the virological synapse. J. Clin. Investig. 114:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G. P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 42.Rizzo, M. A., G. H. Springer, B. Granada, and D. W. Piston. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22:445-449. [DOI] [PubMed] [Google Scholar]
- 43.Rudner, L., S. Nydegger, L. V. Coren, K. Nagashima, M. Thali, and D. E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 79:4055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 46.Stuchell, M. D., J. E. Garrus, B. Muller, K. M. Stray, S. Ghaffarian, R. McKinnon, H. G. Krausslich, S. G. Morham, and W. I. Sundquist. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059-36071. [DOI] [PubMed] [Google Scholar]
- 47.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varthakavi, V., R. M. Smith, S. P. Bour, K. Strebel, and P. Spearman. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100:15154-15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varthakavi, V., R. M. Smith, K. L. Martin, A. Derdowski, L. A. Lapierre, J. R. Goldenring, and P. Spearman. 2006. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic 7:298-307. [DOI] [PubMed] [Google Scholar]
- 50.Ventoso, I., J. Navarro, M. A. Munoz, and L. Carrasco. 2005. Involvement of HIV-1 protease in virus-induced cell killing. Antivir. Res. 66:47-55. [DOI] [PubMed] [Google Scholar]
- 51.Verrier, S. E., and H. D. Soling. 2006. Photobleaching of YFP does not produce a CFP-like species that affects FRET measurements. Nat. Methods 3:491-493. [DOI] [PubMed] [Google Scholar]
- 52.Vogel, S. S., C. Thaler, and S. V. Koushik. 2006. Fanciful FRET. Sci. STKE 2006:re2. [DOI] [PubMed] [Google Scholar]
- 53.Weclewicz, K., M. Ekstrom, K. Kristensson, and H. Garoff. 1998. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J. Virol. 72:2832-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welsch, S., O. T. Keppler, A. Habermann, I. Allespach, J. Krijnse-Locker, and H. G. Krausslich. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia, Z., and Y. Liu. 2001. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81:2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]
- 57.Zimmermann, T., J. Rietdorf, and R. Pepperkok. 2003. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 546:87-92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.