Abstract
We used in vitro evolution to obtain RNA molecules that specifically recognize and bind with high affinity to the oxidative lesion 7,8-dihydro-8-hydroxy-2′-deoxyguanosine (8-oxodG) in DNA. A pool of ≈1015 RNA molecules containing a random insert of 45 nucleotides in length was subject to 10 successive rounds of chromatographic enrichment using an 8-oxodG affinity matrix, reverse transcription, PCR amplification, and RNA synthesis. Selected RNA molecules bind to 8-oxodG located at the 3′ terminus (Kd ≤ 270 nM) or in the center (Kd ≤ 2.8 μM) of a 19-nt strand of DNA, with no detectable affinity for the corresponding dG-containing DNA sequences. These 8-oxodG-binding RNAs will be used to monitor levels of 8-oxodG in DNA from biological sources and should provide a unique method for evaluating oxygen-mediated DNA damage. This approach should be applicable for the creation of RNA molecules that can bind to and identify the different modifications of DNA produced by a variety of environmental agents.
Molecular recognition is an essential component of all biological processes, with the primary role being assigned to proteins. In recent years, the molecular recognition repertoire has been expanded to include, in addition to proteins, both RNA and DNA molecules evolved to possess high affinity and specificity for small ligands as well as for macromolecular targets (1–7). The in vitro evolution technique systematic evolution of ligands by exponential enrichment (SELEX) (8) produces nucleic acid polymers with the attributes of high specificity and high affinity traditionally associated with antibodies and makes both evolved DNA and RNA attractive candidates for use in diagnostic and therapeutic applications (9).
We asked whether RNA molecules can be evolved that specifically recognize altered nucleotides in DNA, in particular, alterations produced by oxygen free radicals. The genome of every living organism is continuously assaulted by reactive oxygen species. Steady-state levels of oxidative DNA lesions have been estimated by Ames and Shigenaga (10) at 2 × 104 lesions per human cell per day. Many of these adducts in DNA frequently base-pair with noncomplementary nucleotides during DNA synthesis, causing mutations, and are postulated to contribute to the multiple mutations found in human cancers (11, 12). One of the most common mutagenic lesions produced in cellular DNA by oxidative damage is 7,8-dihydro-8-hydroxy-2′-deoxyguanosine (8-oxodG), the presence of which is known to generate G to T transversions (13–16). The levels of 8-oxodG have been shown to increase with age and oxidative stress (17–19), making this lesion a sentinel marker for monitoring diseases associated with oxidative DNA damage.
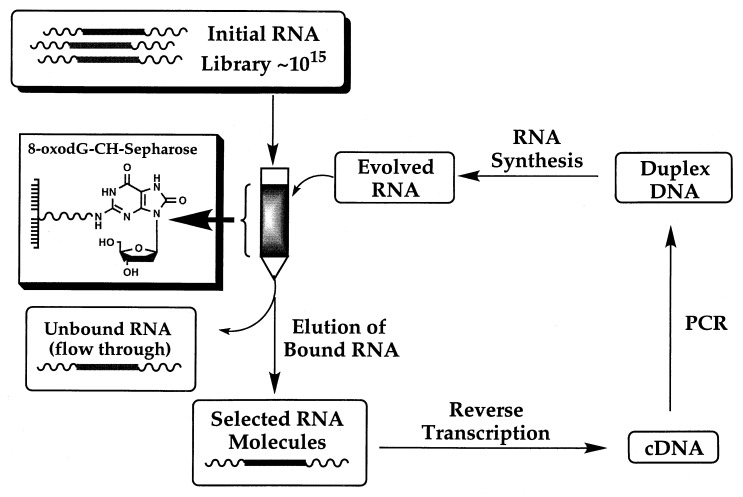
We therefore used the nucleoside 8-oxodG as a target for SELEX to generate RNA ligands that bind 8-oxodG in DNA. An 8-oxodG nucleoside bound to CH-Sepharose (Fig. 1) was used as the target for selection, rather than 8-oxodG embedded in DNA, to avoid the complexities associated with selection against single- and double-stranded nucleic acid targets (20–22). RNA molecules were obtained that distinguish the subtle molecular differences between dG and 8-oxodG that are located in the imidazole portion of the purine ring (see Fig. 1), with the level of specificity exceeding one 8-oxodG in greater than 10,000 dG residues. The high degree of specificity achieved in the evolved molecules meets and exceeds the specificity found thus far in antibodies raised against the free nucleoside 8-oxodG (23–26). This report reveals that RNA aptamers can, in fact, be developed to bind to specific damaged sites present in DNA after in vitro evolution against the nucleoside form of the damage.
Figure 1.
Structures of the 8-oxodG-CH-Sepharose affinity matrix, dG, and 8-oxodG.
MATERIALS AND METHODS
Source of Materials.
The reverse primer (5′-GGGCCAAGCTTCTGCAGAAAAAT-3′) and forward primer (5′-TAATACGACTCACTATAGGGAGGAATTCCCGAG-3′) were synthesized and HPLC-purified by Operon Technologies (Alameda, CA). The 101-nt DNA template (5′-GGGCCAAGCTTCTGCAGAAAAAT(N)45CTCGGGAATTCCTCCCTATAGTGAGTCGTATTA-3′) used to generate the random, double-stranded DNA library was synthesized and HPLCpurified by Keystone Laboratories (Menlo Park, CA). Oligonucleotides 5′-AATCCATTCCAATACCTA-dG (see Fig. 3B), 5′-AATCCATTCCAATACCTA-8-oxodG (see Fig. 3A), 5′-AATCCATTC-G-CAATACCTA-3′, and 5′-AATCCATTC8-oxodG-CAATACCTA-3′ were provided, in crude form, by Midland Certified Reagent (Midland, TX). These synthetic oligos were purified by 20% DPAGE and extracted from the gel in acetate elution buffer using standard conditions (27). The DNA was desalted by using a C18-RP Sep-Pak Cartridge (Waters).
Figure 3.
Binding affinity in cycle 10 pool RNA to 8-oxodG and dG using an electrophoretic mobility-shift assay. Unlabeled RNA was denatured at 100°C for 1 min, mixed with binding buffer to a final concentration of 20 mM NH4CH3CO2 (pH 6), 300 mM NaCl, and 5 mM MgCl2, and renatured for 1 h. This RNA was allowed to bind with the indicated 5′-32P-radiolabeled (∗) DNA substrate (1 pmol) for 30 min at rt before the addition of nondenaturing loading buffer (Promega). Samples were loaded onto a running (25 mA), 10% native polyacrylamide gel (29:1 acryl/bis), electrophoresed for 3–4 h at 10–12°C, and subsequently dried on DE-81 DEAE cellulose paper (Whatman). Quantitation of the single-stranded (ss) DNA and the RNA/DNA complex was obtained by using PhosphorImage analysis with ImageQuant software (Molecular Dynamics). (A) Native gel analysis of binding reactions between cycle 10 pool RNA (30–1,000 pmol) and the terminal-8-oxodG-DNA substrate (1 pmol). (B) Native gel analysis of binding reactions between cycle 10 pool RNA (30–1,000 pmol) and the terminal-dG-DNA substrate (1 pmol).
HPLC-grade water (Fisher Scientific) or Milli-Q Plus PF Ultra Pure water was used in the RNA experiments without further treatment. Sources of chemicals were: dG, Sigma; ammonium acetate, NaCl, and MgCl2, J. T. Baker; 20 mg/ml glycogen, Boehringer-Mannheim; 8-oxodG, Chem-Master International (East Setauket, NY); and G-50 Nick Spin Columns, G-25 Micro Spin columns, and activated CH-Sepharose 4B, Pharmacia. Radionucleotides, [γ-32P]ATP (3,000 Ci/mmol) and [α-32P]UTP (3,000 Ci/mmol) were obtained from New England Nuclear.
8-OxodG Affinity Matrix.
The 8-oxodG affinity matrix (Fig. 1) was prepared by coupling 0.4 ml of rehydrated, activated CH-Sepharose 4B (Pharmacia LKB Biotechnology) to the exocyclic amino group of 8-oxodG (Chem-Master International) according to manufacturers’ specifications. The unreacted sites on the matrix were blocked with 0.1 M Tris⋅HCl (pH 8) and 0.5 M NaCl. The final concentration of 8-oxodG covalently linked to the solid support was 1.5 mM.
Preparation of Randomized Oligonucleotide Libraries.
The initial random DNA library (≈1014 unique molecules) was generated by PCR amplification of the 101-nt template in the presence of both forward and reverse primers. The 101-nt template consists of a centrally located 45-nt randomized region (N) flanked on both sides by constant sequences that hybridize to primers required for reverse transcription and PCR amplification. The template also has a T7 promotor sequence for RNA synthesis by T7 RNA polymerase as well as HindIII and EcoRI sites for cloning. An RNA pool of ≈1015 molecules was generated from the double-stranded random DNA library by in vitro transcription with T7 RNA polymerase (New England Biolabs) for 3 h at 37°C in the presence of [α-32P]UTP. The reactions were incubated with RQ1 DNase (Promega) for 1 h at 37°C to facilitate degradation of DNA template and primers. The reaction product then was applied onto a G50-Nick Spin Column, extracted with phenol/chloroform (1:1, vol/vol) followed by chloroform, and then ethanol-precipitated in the presence of 1 μl of glycogen (20 mg/ml).
In Vitro Selection.
The initial RNA pool (≈65 μg in 90 μl of water) of ≈1015 molecules was denatured in a boiling water bath for 1 min, mixed with 10 μl of 200 mM NH4CH3CO2 (pH 6), 3 M NaCl and 50 mM MgCl2, and allowed to renature for 1 h. The sample was loaded onto the 8-oxodG column pre-equilibrated with binding buffer, followed by 100 μl of 1× binding buffer (20 mM NH4CH3CO2, pH 6/300 mM NaCl/5 mM MgCl2). After 1 h at room temperature (rt), the unbound RNA was eluted gravimetrically with 5 ml of 1× binding buffer, after which the column was equilibrated with 0.55 ml of EDTA elution buffer (50 mM EDTA/1× binding buffer) for 1 h at rt. The bound RNA was eluted with 5.5 ml of EDTA elution buffer, and the percent RNA recovered was calculated from the cpm collected relative to the total cpm loaded onto the column.
Reverse transcription (RT) of the recovered RNA was performed with HIV type 1 RT in the presence of the reverse primer. Subsequent PCR amplification followed by RNA synthesis provided the new RNA pool for the next cycle of selection. Ten cycles of selection were performed. Approximately 40 μg of pool RNA was used in each of cycles 2–9 and 10 μg of RNA was used in cycle 10. Counter-SELEX (28) was used in cycles five and six to obtain RNA molecules with high specificity for 8-oxodG. The column-bound RNAs were first equilibrated (30 min at rt) then washed with 2.3 ml of dG elution buffer (4 mM dG/1× binding buffer) to remove those RNAs having affinity for dG. After washing the column with 2.3 ml of 1× binding buffer, the column was equilibrated (30 min at rt) and then washed with 2.3 ml of 8-oxodG elution buffer (4 mM 8-oxodG/1× binding buffer). In the final cycles (nos. 7–10), elution was solely with the 8-oxodG elution buffer. A matrix-only precolumn was run before cycle four to reduce the level of matrix binders present in the RNA pool. Approximately 4% of the RNA molecules remained bound to the precolumn after the first three cycles of selection. The double-stranded DNA pool amplified from the 10th cycle of selection was used in the synthesis of the corresponding RNA pool by T7 RNA polymerase.
Cloning and Sequencing.
Double-stranded DNA products amplified from the last round of selection were digested by HindIII and EcoRI, extracted first with phenol/chloroform (1:1, vol/vol) followed by chloroform, and the digested DNA was recovered by ethanol precipitation. The double-digested DNA pool was ligated into the EcoRI–HindIII cloning site in the plasmid pGEM-3Z (Promega). The ligation products were used directly for transformation of Escherichia coli strains JM109 or NM522 by electroporation. Transformation of E. coli with the ligated plasmid DNA provided individual clones, which were sequenced by using the Thermosequenase Kit (Amersham).
In Vitro Run-Off Transcription.
Nonradiolabeled RNA was prepared from individual clones after digestion of the plasmid DNA harboring insert with HindIII and phenol/chloroform extraction. The digested plasmid was precipitated with ethanol and resuspended in water. Run-off transcription with T7 RNA polymerase (500 units, NEB, Beverly, MA) was performed by using half of the plasmid previously digested with HindIII in a total volume of 100 μl. The reaction mixture contained rRNasin (120 units, Promega), 100 μg/ml of BSA, inorganic pyrophosphatase (5 units, NEB), and 3 mM NTPs (Amersham) in 1× NEB transcription buffer (40 mM Tris⋅HCl/6 mM MgCl2/10 mM DTT/2 mM spermidine, pH 7.9 at 25°C). After incubation overnight at 37°C, the sample was treated with RQ1 DNase (7.5 units) for 1 h at 37°C. Run-off transcripts were run through a G-50 Nick Spin Column, extracted with phenol/chloroform and chloroform, and precipitated with ethanol before their use in binding experiments. The RNA concentrations were determined from UV absorbance at 260 nm. The secondary structure of individual RNA sequences was predicted by using the program mulfold (29, 30) on a Power Macintosh 7200.
Evaluation of Kd by Electrophoretic Mobility-Shift Assay.
RNA used in the electrophoretic mobility-shift assays was obtained by 8% DPAGE purification of the run-off transcripts. In a typical experiment, unlabeled RNA (2–100 pmol) was denatured at 100°C for 1 min, mixed with 1/10 volume binding buffer (10×), and reannealed in the same water bath for 1 h. The reannealed RNA was mixed with the desired DNA substrate, a trace amount of which was radiolabeled, and the mixture was equilibrated at rt for 30 min before the addition of nondenaturing loading buffer (Promega).
One-half of a binding reaction was loaded into one lane of a running (25 mA), nondenaturing 10% acrylamide gel [29:1 acryl/bis, 40 mM tris-acetate (pH 6), 3 mM NaCl, 5 mM MgCl2], electrophoresed for 3–4 h at 10–12°C, and subsequently dried on DE-81 DEAE cellulose paper (Whatman) by using a Bio-Rad Gel Drier model 583 run on cycle 1 for 30 min at 80°C. Acrylamide gel images were obtained either by autoradiography (Kodak X-Omat AR or Blue XB-1 film) or by PhosphorImage analysis using phosphor screens (Molecular Dynamics). Quantitation of electrophoretic mobility-shift data was obtained from gels dried on DE-81 DEAE cellulose paper (Whatman) using PhosphorImage analysis with imagequant software (Molecular Dynamics).
Kd was calulated from the PhosphorImage data by using Eq. 1 (31):
 |
1 |
where r represents the number of mols of DNA complexed with RNA divided by the total number of mols RNA added; [A] is the molar concentration of unbound DNA; n is the number of DNA binding sites per RNA molecule; Kd is the dissociation constant of the complex. The Kd value of a given complex was first calculated from the negative reciprocal slope of a Scatchard plot of r/[A] as a function of r using the program kaleidagraph. The value of n was subsequently calculated from Eq. 1. The reported Kd values were determined from the average of four independent experiments.
Competition Experiments.
The RNA used in the competition assays was obtained directly from run-off transcription reactions. In a typical experiment, 1 pmol of radiolabeled terminal-8-oxdG-DNA (or centrally located 8-oxodG-DNA) was mixed with 10–10,000 pmol unlabeled terminal-dG-DNA (or centrally located dG-DNA), and the substrates were allowed to compete for binding to R10-B35 RNA (100 pmol, 30 min at rt). The samples were analyzed on a 10% native gel as previously described.
RESULTS
In Vitro Selection.
The steps used in our selection protocol are summarized in Scheme S1. To evolve RNA molecules with affinity for 8-oxodG, we generated a pool of ≈1015 RNA molecules containing a stretch of 45 random nucleotides. The RNA library was loaded onto an 8-oxodG-CH-Sepharose column, and the bound RNA ligands were recovered and quantified. Reverse transcription of the recovered RNA molecules, followed by PCR amplification and subsequent RNA synthesis constitutes one cycle of selection.
Scheme 1.
In vitro evolution of RNA molecules using an 8-oxodG nucleoside affinity matrix.
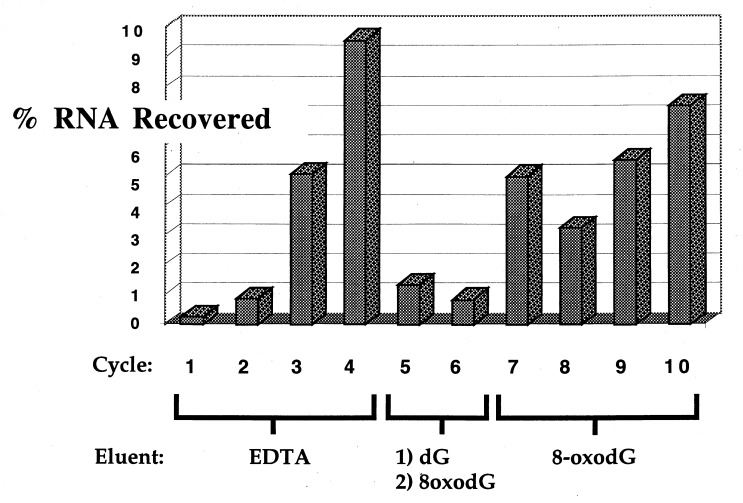
Ten cycles of SELEX were performed, and the percentage of bound RNA recovered from each cycle of selection is presented in Fig. 2. During cycles 1–4, bound RNA molecules were eluted from the matrix with 50 mM EDTA, which resulted in an increasing recovery of bound RNA species. Counter-SELEX (28) was used in cycles five and six, eluting bound RNA first with dG followed by 8-oxodG. These stringent conditions resulted in a substantial decrease in the amount of recovered RNA. Bound RNA in cycles 7–10 were eluted from the column with buffer containing 8-oxodG, which afforded a steady increase in the recovery of binding RNA molecules.
Figure 2.
Percent RNA recovery for each cycle of in vitro evolution.
Evaluation of Round 10 Pool RNA for Binding to 8-OxodG-Containing DNA.
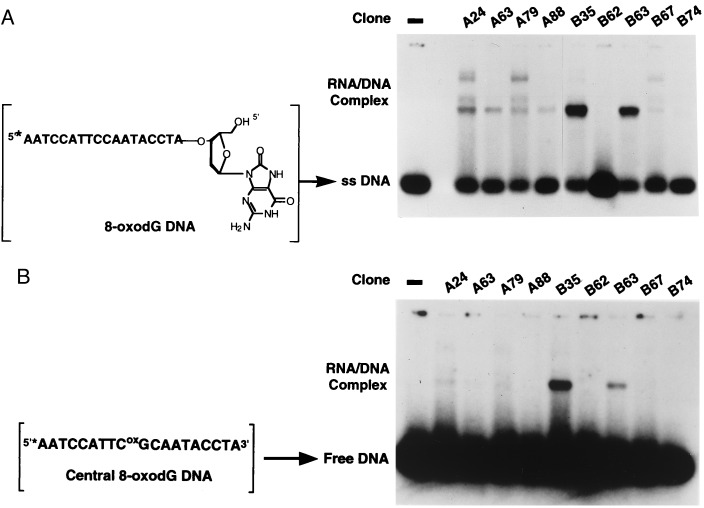
To test the affinity and specificity of the evolved RNA pool for 8-oxodG, single-stranded DNA was used that terminated with either a single 3′-to-3′ linked 8-oxodG or dG residue. The oligodeoxyribonucleotide serves as an arm to present the 3′-terminal residue for binding by the RNA. The RNA/DNA complex was separated from the unbound, single-stranded DNA by using an electrophoretic mobility-shift assay. The pool of RNA molecules obtained after the 10th round of selection gave rise to two bands after incubation with the terminal-8-oxodG-DNA substrate and subsequent native gel analysis (Fig. 3). The faster migrating species represents the unbound single-stranded DNA, while the slower mobility band is indicative of an RNA/DNA complex. The amount of the RNA/DNA complex formed between the RNA molecules and the terminal-8-oxodG-DNA increases as a function of the amount of pool RNA. Kd determined from the data in Fig. 3A was estimated at ≈5 μM, confirming the attainment of a substantial affinity of RNA to 8-oxodG by the selection protocol. In contrast, formation of an RNA/dG-DNA complex is undetectable by using the pool RNA and the terminal-dG-DNA target (Fig. 3B). These results illustrate that a high degree of specificity exists in these RNA molecules for binding 8-oxodG but not dG.
Evaluation of Individual Clones for Binding to 8-OxodG in DNA.
The nucleotide sequences of the 10th selection cycle were determined, and the corresponding RNA was produced by run-off transcription and used without further purification in the binding experiments. RNA molecules derived from individual clones were independently evaluated for their binding properties with the terminal-8-oxodG-DNA substrate by using the electrophoretic mobility-shift assay. In a representative experiment (Fig. 4A), the individual RNA molecules display varying ability to bind to the terminal-8-oxodG target, with some RNA molecules, such as B35, having more pronounced affinities for 8-oxodG relative to other RNA clones.
Figure 4.
Evaluation of 8-oxodG binding by cloned RNA molecules. Individual RNA molecules (100 pmol) were denatured, renatured, and then incubated with the 5′-32P-radiolabeled (∗) terminal-8-oxodG-DNA substrate (1 pmol) as described in Fig. 3. The samples were analyzed on a 10% native polyacrylamide gel, and the single-stranded (ss) DNA and RNA/DNA complex was quantified as previously described. (A) Binding of individual RNA clones to the terminal-8-oxodG-DNA substrate. (B) Binding of individual RNA clones to the centrally located 8-oxodG-DNA substrate.
We next examined the binding affinity of the evolved RNA molecules with 8-oxodG residing at the center of a single-stranded DNA polymer (Fig. 4B), a mimicry of the potential situation in genomic DNA. Results of this experiment show that the RNA molecules having highest affinity for the terminal-8-oxodG-DNA can, in fact, recognize and bind to the centrally located 8-oxodG residue, albeit with a reduced affinity relative to the target with a terminal 8-oxodG residue. None of the RNA molecules evaluated displayed any detectable affinity for the centrally located dG-containing DNA substrate (data not shown).
Affinity and Specificity for Binding 8-OxodG in Single-Stranded DNA by Clone R10-B35.
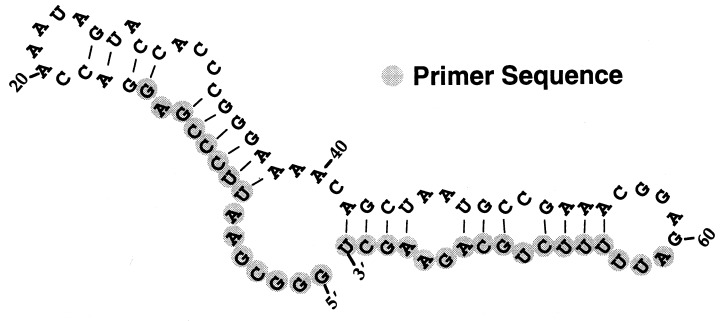
The RNA molecule exhibiting the most robust interaction with the 8-oxdG-DNA substrates was designated R10-B35 (10th selection cycle, clone B35) (Fig. 5) and was evaluated for its specific binding properties. Kd values were determined from Scatchard analysis of electrophoretic mobility-shift data as described in Materials and Methods. The analysis revealed an apparent Kd of ≤ 270 nM for the binding of the terminal-8-oxodG-DNA and a Kd ≤ 2.8 μM for binding to the centrally located 8-oxodG residue. The observed difference in affinity of RNA R10-B35 for the two 8-oxodG targets is not unexpected, given that the RNA molecule was selected by virtue of its binding to the free nucleoside.
Figure 5.
Computer-generated secondary structure of RNA R10-B35 predicted by using the program mulfold (29, 30). The highlighted nucleotides correspond to fixed primer sequence.
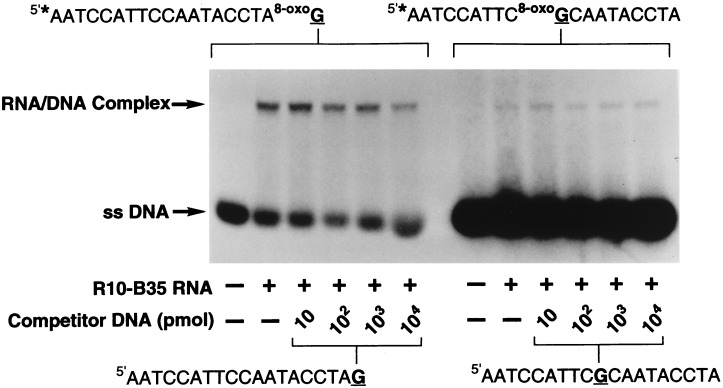
To more directly test the specificity and sensitivity of binding, competition experiments were performed in which an invariable quantity of 32P-radiolabeled terminal-8-oxodG-DNA was incubated with an increasing quantity of unlabeled terminal-dG-DNA (Fig. 6). Under the conditions tested, increasing the amount of terminal-dG-DNA had very little effect on the quantity of terminal-8-oxodG bound by this RNA, consistent with a very specific interaction between the RNA and the 8-oxodG residue. The same result was obtained for the centrally located 8-oxodG DNA in competition with increasing quantities of dG-containing competitor (Fig. 6). The specificity of 8-oxodG detection by RNA R10-B35 can be stated as a ratio of 8-oxodG residues to the total number of dG residues or to the total number of nucleotides present in DNA. Our results demonstrate that the evolved RNA can detect one 8-oxodG in a background of more than 104 dG residues or one 8-oxodG residue in a background of more than 1.9 × 105 nucleotide residues.
Figure 6.
Affinity and specificity of RNA R10-B35 to 8-oxodG. 5′-32P-Radiolabeled (∗) terminal-8-oxodG-DNA (1 pmol) was mixed with unlabeled dG-DNA (10–10,000 pmol), and the substrates were allowed to compete for binding to R10-B35 RNA (100 pmol) for 30 min at rt. The samples were analyzed, and the data were quantified as described.
DISCUSSION
In this report we demonstrate that nucleic acids can be evolved to recognize the subtle differences between 8-oxodG and dG in DNA. The fact that these molecules are able to bind to a modified base located within a DNA strand extends the repertoire of binding nucleic acids to encompass alterations in a DNA polymer. The RNA molecules have been evolved to specifically bind to the modified DNA in a manner that does not depend on sequence context or DNA/RNA hydrogen bonding. To date, most of the reported 8-oxodG antibodies that recognize sites of DNA damage are of limited specificity, and thus the immunoassays developed have quantified the 8-oxodG nucleoside released enzymatically from cellular DNA samples or have been used to visualize the distribution of 8-oxodG in tissue (23–26). The RNA molecules reported herein bind to 8-oxodG in DNA with affinities that rival those reported for antibodies raised against the lesion, and with specificity for 8-oxodG that exceeds the specificity reported thus far for antibodies.
Jenison et al. (28) isolated the first RNA capable of binding to theophylline and not caffeine, two relatively small molecules that differ in their structures by the presence of a hydrogen atom or a methyl group at the N7 position. This RNA molecule was shown to bind theophylline with greater specificity than antibodies. We have shown that RNA molecules can be used to detect small molecular entities, specifically modified nucleosides, within a DNA biopolymer, and can recognize the subtle structural differences between the modified nucleoside target and the normal nucleosides present throughout the genome. These results hold promise for the development of new, simplified methods for evaluating the levels of 8-oxodG present in a variety of biological sources of DNA, including individual cells and samples of tissue. Moreover, this methodology could be extended to target other types of DNA damage, such as UV-induced thymine dimers and bulky alkylation adducts, and may lead to the development of diagnostics for specific diseases associated with DNA damage, deficits in DNA repair, or environmental exposure to genotoxic agents.
Acknowledgments
We thank Dr. Baek Kim for the generous gift of HIV type 1 reverse transcriptase, Katie Lindberg for technical assistance, and Drs. Michael Fry, Terry Newcomb, and Aimee Jackson for their critical advice. This work was supported by grants from the National Institutes of Health (NRSA F2CA73141A to S.M.R. and OIG-R35-CA3990s and AG-01 751 to L.A.L.).
ABBREVIATIONS
- 8-oxodG
7,8-dihydro-8-oxo-2′-deoxyguanosine
- rt
room temperature
- SELEX
systematic evolution of ligands by exponential enrichment
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF083271).
References
- 1. Keene J D. Chem Biol. 1996;3:505–513. doi: 10.1016/s1074-5521(96)90139-8. [DOI] [PubMed] [Google Scholar]
- 2.Tsang J, Joyce G F. Methods Enzymol. 1996;267:410–426. doi: 10.1016/s0076-6879(96)67025-6. [DOI] [PubMed] [Google Scholar]
- 3.Conrad R C, Baskerville S, Ellington A D. Mol Divers. 1995;1:69–78. doi: 10.1007/BF01715810. [DOI] [PubMed] [Google Scholar]
- 4.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 5.Klug S J, Famulok M. Mol Biol Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 6.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 7.Haller A A, Sarnow P. Proc Natl Acad Sci USA. 1997;94:8521–8526. doi: 10.1073/pnas.94.16.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Gold L. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 10.Ames B N, Shigenaga M K. Ann NY Acad Sci. 1992;663:85–96. doi: 10.1111/j.1749-6632.1992.tb38652.x. [DOI] [PubMed] [Google Scholar]
- 11.Loeb L A. Cancer Res. 1989;49:5489–5496. [PubMed] [Google Scholar]
- 12.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 13.Shibutani S, Takeshita M, Grollman A O. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M, Ou C, Bodeudi V, Johnson F, Takeshita M, Grollman A P. Mutat Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 15.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 16.Wood M L, Esteve A, Morningstar M L, Kuziemko G M, Essigmann J M. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Floyd R A, Watson J J, Harris J, West M, Wong P K. Biochem Biophys Res Commun. 1986;137:841–846. doi: 10.1016/0006-291x(86)91156-3. [DOI] [PubMed] [Google Scholar]
- 18.Kasai H, Crain P F, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 19.Shigenaga M K, Cimeno C J, Ames B N. Proc Natl Acad Sci USA. 1989;86:9697–9701. doi: 10.1073/pnas.86.24.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pei D H, Ulrich H D, Schultz P G. Science. 1991;253:1408–1411. doi: 10.1126/science.1716784. [DOI] [PubMed] [Google Scholar]
- 21.Soukup G A, Ellington A D, Maher III L J. J Mol Biol. 1996;259:216–228. doi: 10.1006/jmbi.1996.0314. [DOI] [PubMed] [Google Scholar]
- 22.Mishra R K, Tinevez R L, Toulme J-J. Proc Natl Acad Sci USA. 1996;93:10679–10684. doi: 10.1073/pnas.93.20.10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park E-M, Shigenaga M K, Degan P, Korn T S, Kitzler J W, Wehr C M, Kolachana P, Ames B N. Proc Natl Acad Sci USA. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ide H, Kow Y W, Chen B-X, Erlanger B F, Wallace S S. Cell Biol Toxicol. 1997;13:405–417. doi: 10.1023/a:1007467726635. [DOI] [PubMed] [Google Scholar]
- 25.Yin B, Whyatt R M, Perera F P, Randall M C, Cooper T B, Santella R M. Free Radical Biol Med. 1995;18:1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- 26.Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Yoshida A, Uchida K, Hiai H, Ochi H, Osawa T. Lab Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch D G, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 30.Jaeger J A, Turner D H, Zuker M. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freifelder D. In: Physical Biochemistry. 2nd Ed. Wilson J, Steeds D, editors. New York: Freeman; 1982. pp. 655–684. [Google Scholar]