Abstract
Hepatitis C virus (HCV) is a global challenge to public health. Several factors have been proven to be critical for HCV entry, including the newly identified claudin-1 (CLDN1). However, the mechanism of HCV entry is still obscure. Presently, among the 20 members of the claudin family identified in humans so far, CLDN1 has been the only member shown to be necessary for HCV entry. Recently, we discovered that Bel7402, an HCV-permissive cell line, does not express CLDN1 but expresses other members of claudin family. Among these claudins, CLDN9 was able to mediate HCV entry just as efficiently as CLDN1. We then examined if other members of the claudin family could mediate entry. We show that CLDN6 and CLDN9, but not CLDN2, CLDN3, CLDN4, CLDN7, CLDN11, CLDN12, CLDN15, CLDN17, and CLDN23, were able to mediate the entry of HCV into target cells. We found that CLDN6 and CLDN9 are expressed in the liver, the primary site of HCV replication. We also showed that CLDN6 and CLDN9, but not CLDN1, are expressed in peripheral blood mononuclear cells, an additional site of HCV replication. Through sequence comparison and mutagenesis studies, we show that residues N38 and V45 in the first extracellular loop (EL1) of CLDN9 are necessary for HCV entry.
Hepatitis C virus (HCV) is the major cause of liver cirrhosis and hepatocellular carcinoma worldwide. Approximately 3% of the global population is infected with HCV, and at least 70% develop chronic hepatitis (13, 17, 32). In patients with chronic HCV infection, about 20% develop liver cirrhosis, about 5% of which go on to develop hepatocellular carcinoma (17). While HCV is generally confined to the liver, there is growing evidence suggesting that HCV can replicate in extrahepatic tissues including peripheral blood mononuclear cells (PBMCs) (4, 15, 23, 24).
HCV is a small, enveloped virus that belongs to the family Flaviviridae (19). Its genome encodes an approximately 3,000-amino-acid precursor polyprotein, which is cleaved into at least 10 mature proteins, including two envelope glycoproteins. These glycoproteins are referred to as E1 and E2 (25). E2 is believed to interact with putative receptors on the surface of target cells (29). CD81 has been shown to interact with the E2 glycoprotein and can serve as a coreceptor for HCV entry of all six genotypes (1, 2, 6, 14, 18, 21, 26, 34, 36). In addition, scavenger receptor class B member I (SR-BI) is another E2 binding protein and was shown to enhance HCV entry in a high-density-lipoprotein-dependent manner (3, 18, 31, 35). However, the overexpression of both CD81 and SR-BI was not able to render nonhepatic cells susceptible to HCV (2, 37). Recently, Evans et al. reported that claudin-1 (CLDN1), an integral membrane protein and a component of tight-junction strands, was able to mediate the entry of HCV in two nonhepatic cell lines, 293T and SW13. In addition, CLDN1 acts after virus binding with CD81 and is most likely important for mediating the fusion between the viral and cellular lipid membranes (7). However, there are several cell lines that are resistant to HCV infection which express CLDN1, CD81, and SR-BI. The existence of such cell lines indicates that other factors are required for HCV entry in addition to the previously identified receptors (7).
The claudin family constitutes a large group of four-transmembrane domain proteins that are essential for the formation of tight junctions (22). Tight junctions are responsible for the control of paracellular transport and maintenance of cell polarity (10). To date, 20 claudins have been identified in humans according to UniGene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene). Most tissues express various claudin proteins, and these proteins interact with each other homologously or heterologously to form the tight-junction strands (9). Variations in tight junctions are most likely determined by combinations of constituent claudins. Members of the claudin family are highly conserved, especially in the first extracellular loop (EL1), which was found to be important for the interaction with HCV (7). It has also been reported that liver expresses multiple claudins (28). This result suggests that other claudins, in addition to CLDN1, might be able to mediate HCV entry.
Here, we show that the human hepatocellular carcinoma cell line Bel7402 is susceptible to HCV but that it does not express CLDN1. CLDN9 expression in Bel7402 cells was found to mediate HCV entry. Furthermore, its closest relative, CLDN6, was shown to function similarly. The expression of CLDN6 and CLDN9 in the liver, where HCV replicates the most, was detected. In addition, PBMCs, the extrahepatic tissue believed to be a site of HCV replication, were shown to express CLDN6 and CLDN9 but not CLDN1. By sequence comparison and mutational analysis, two residues, N38 and V45, were identified as being critical for HCV entry in CLDN9-expressing cells. These data suggest that CLDN6 and CLDN9 can also mediate HCV entry into target cells.
MATERIALS AND METHODS
Cells.
293T cells were used as packaging cell lines for the human immunodeficiency virus (HIV)-backboned pseudotyped particle preparation for the infection assay. 293GP cells were used as packaging cell lines for the pBabepuro-backboned pseudotyped particles. 293T and SW13 cells were used as target cells to construct cell lines stably expressing claudins. Hep3B, Huh7, PLC/PRF/5, Bel7402, SMMC7721, U87, and HeLa human cell lines were used for virus infection assays. All these cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (Gibco). PBMCs were isolated from fresh peripheral blood on lymphocyte separation medium (Ficoll-Paque; Amersham Biosciences) by centrifugation at 400 × g.
Reverse transcription (RT)-PCR.
Total RNA of cells or tissues was isolated with TRIzol reagent (Life Technologies) and reverse transcribed using a Reverse-Transcribe kit (CY-TECH, China). The resulted cDNA were amplified by Ex Taq polymerase (Takara, Japan) using claudin-specific primers with GAPDH as a control. The primer sequences of claudins are available upon request.
Plasmids.
DNA fragments containing a Kozak sequence and the complete open reading frame of claudins were amplified from cDNA from Bel7402 cells, Huh7 cells, or liver and cloned into a BamHI/SalI (New England Biolabs)-digested pBabepuro vector. Flag-tagged claudins were constructed similarly, with the reverse primers containing a Flag tag sequence. Point mutations in CLDN3, CLDN6, and CLDN9 were generated by site-directed mutagenesis using a QuikChange kit (Stratagene).
Generating stable cell lines that express various claudin variants.
A retroviral vector expressing claudin variants was transfected together with a plasmid encoding the vesicular stomatitis virus glycoprotein (VSV-G) into 293GP cells, a cell line that stably expresses a lentiviral gag-pol gene, using a calcium phosphate precipitation protocol. The supernatants were harvested 48 h later and used to infect 293T and SW13 cells. Forty-eight hours after infection, the cells were selected in medium containing puromycin (Sigma) at 5 μg/ml for at least 1 week.
Real-time PCR.
The primes for CLDN1, CLDN6, CLDN9, and GAPDH were designed by using PRIMEREXPRESS software (version 2.0; Applied Biosystems) crossing intron-exon boundaries. For standardization of the amount of mRNA, expression of GAPDH in each sample was also quantified. All PCRs were performed using an ABI Prism 7000 sequence detector (Perkin-Elmer Applied Biosystems) as described previously (27). Briefly, equivalent amounts of cDNA sample, derived from 50 ng of DNase-digested total RNA, were used for each PCR. The threshold cycle values were determined using the AUTOANALYSE features of Sequence Detection System software. The primers were as follows: 5′-GCG CGA TAT TTC TTC TTG CAG-3′ (CLDN1 forward), 5′-GCA GGT TTT GGA TAG GGC CT-3′ (CLDN1 reverse), 5′-AGA AGG ATT CCA AGG CCC G-3′ (CLDN6 forward), 5′-GAT GTT GAG TAG CGG GCC AT-3′ (CLDN6 reverse), 5′-GCG GCT GCA CTG CTT ATG CT-3′ (CLDN9 forward), 5′-GCA GTG GGG AGC AGT GGG CT-3′ (CLDN9 reverse), 5′-CTC AAC TAC ATG GTC TAC AT-3′ (GAPDH forward), and 5′-AGT AGA CTC CAC GAC ATA CT-3′ (GAPDH reverse). Quantitative PCR was performed in duplicate for each sample, and three independent experiments were carried out. The means and standard deviations (SD) were calculated and reported as data from one representative experiment.
Pseudotyped particle generation and infection assays.
HIV/HCV pseudotyped particles (HCVpp) were produced as described previously (14). Briefly, 12 μg pNL4-3.luc.R−E− and 12 μg HCV envelope glycoproteins expressing plasmids were cotransfected into 293T cells using a calcium phosphate precipitation protocol. Empty vector or plasmid expressing VSV-G was used as a control. The medium was replaced 12 h after transfection. Supernatants were harvested 36 h later, cleared by centrifugation, and used for the infection assay. In this paper, HCVpp refers to pseudotyped particles bearing envelope glycoproteins of strain H77 unless otherwise stated.
Target cells (8 × 103 cells per well) were seeded into 96-well plates 24 h before infection. Pseudotyped particles (5 ng HIV-1 p24 for HCVpp and no env and 0.1 ng for VSV-G) plus 8 μg/ml polybrene were incubated with cells for 3 h at 37°C and then replaced with fresh medium. The cells were lysed 48 h after infection, and 30 μl of lysates was tested for luciferase activity by the addition of 50 μl luciferase substrate (Promega) on a Beckman LD400 luminometer (Beckman-Coulter). Infection was performed in triplicate for each sample. Similar results were obtained in three independent experiments.
Cell culture-derived HCV and infection assay.
A plasmid containing the genome of chimeric J6/JFH with a T7 promoter was linearized by XbaI digestion and in vitro transcribed using a MEGAscript T7 kit (Ambion). Huh7.5 cells were electroporated with RNA using an electroporator (Bio-Rad), and the supernatants were collected 72 h later. High-titer stocks were gained by passage on fresh Huh7.5 cells three times. The supernatants were titrated on Huh7.5 cells by a limiting-dilution assay as described previously (19). Huh7.5 cells and naive and claudin-expressing 293T cells were seeded in a four-well plate 24 h before infection. Cells were tested for the presence of J6/JFH RNA by real-time PCR 72 h after infection. The primers were as follows: 5′-CTT CAC GCA GAA AGC GTC T-3′ (forward) and 5′-CAA GCA CCC TAT CAG GCA G-3′ (reverse).
RNA interference.
RNA interference was performed using lentivirus-delivered short hairpin RNAs (shRNAs) as described previously (30). Briefly, vector pLL3.7 expressing shRNAs targeting the CLDN9 reference sequence (GenBank accession number NM_020982) was transfected into 293T cells together with three package plasmids. The resulting supernatants were collected 48 h after transfection and used to infect Bel7402 cells. The expression level of CLDN9 in infected cells was determined by real-time PCR 96 h after infection with GAPDH as a control. The shRNA sequences were designed by siSearch (http://sonnhammer.cgb.ki.se/siSearch/siSearch_1.7.html): 5′-GAC TAC GAG TCT GCT TTG T (342), 5′-GGA AGG TGA CCG CCT TCA T (661), and 5′-TAG CAG CTA AAC ACA TCA A (irrelevant control).
Antibodies and Western blotting.
Mouse anti-Flag M2 monoclonal antibody was purchased from Sigma. Rabbit anti-human CLDN9 polyclonal antibody was purchased from Genway Biotech. Horseradish peroxidase-labeled anti-human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) monoclonal antibody and anti-mouse and anti-rabbit secondary antibodies were purchased from New England Biolabs. fifty micrograms of total protein was separated by a 12% sodium dodecyl sulfate-polyacrylamide gel, which was followed by transfer onto a nitrocellulose membrane (Hybond ECL; GE), and stained with a primary antibody and a horseradish peroxidase-labeled secondary antibody. Naive 293T cells served as a control. The bands were detected by an ECL plus kit (GE).
RESULTS
Identification of CLDN6 and CLDN9 as HCV coreceptors.
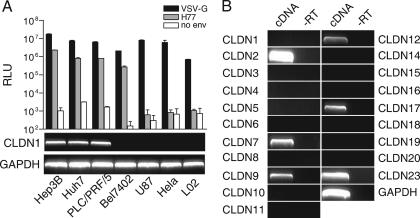
To test whether the newly identified HCV coreceptor CLDN1 is involved in HCV infection of all susceptible cell lines, the association between HCVpp susceptibility and CLDN1 expression was examined in several known HCV-nonpermissive and -permissive cell lines. Three HCV-permissive cell lines, Huh7, Hep3B, and PLC/PRF/5, expressed CLDN1. In contrast, all three cell lines that could not be infected by HCVpp (U87, HeLa, and L02) did not show detectable levels of CLDN1 (Fig. 1A). In addition, we also tested another hepatocellular carcinoma cell line, Bel7402, which was derived from a Chinese hepatocellular carcinoma patient (5, 20, 33, 38). As shown in Fig. 1A, Bel7402 is permissive to HCVpp, but interestingly, no expression of CLDN1 was detected by RT-PCR. This suggests that factors other than CLDN1 could facilitate HCV entry.
FIG. 1.
Lack of expression of CLDN1 in the HCV-susceptible cell line Bel7402. (A) Cell tropism of HCVpp and its correlation to expression of CLDN1. Different cell lines were infected by HCVpp-bearing envelope glycoproteins of strain H77 (gray bar), and luciferase activity was measured as relative light units (RLU) 48 h postinfection. Pseudotyped particles containing VSV-G protein (black bars) or in the absence of envelope protein (white bars) were used as a control (mean of n = 3) (error bars indicate SD). Expression of CLDN1 in these cell lines was analyzed by semiquantitative RT-PCR and is shown under the bar graph, with GAPDH as a control. (B) Expression pattern of known human claudins in Bel7402 cells. The expression of known claudins in Bel7402 cells was analyzed by semiquantitative RT-PCR, using GAPDH as a control. −RT, control reaction of cDNA synthesis in which reverse transcriptase was not added.
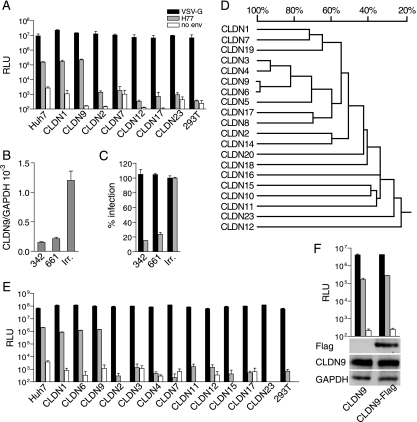
According to the UniGene database for Homo sapiens, at least 20 members of the claudin family have been identified. Therefore, it was reasonable to assume that claudins other than CLDN1 might also serve as coreceptors for HCV. Intriguingly, RT-PCR analysis revealed that among the 20 known claudins examined, CLDN2, CLDN7, CLDN9, CLDN12, CLDN17, and CLDN23 could be detected in Bel7402 cells at various levels (Fig. 1B). This led to the subsequent study of associations between HCV susceptibility and each of these claudins. As described in Materials and Methods, cDNAs of these claudins were cloned into retroviral vectors and used to generate individual stable cell lines, where each would have specific CLDN2, CLDN7, CLDN9, CLDN12, CLDN17, or CLDN23 expression. These cell lines were challenged with HCVpp, and, as shown in Fig. 2A, only CLDN9-expressing 293T cells could be infected by HCVpp as efficiently as cells that expressed CLDN1 (Fig. 2A). Similar results were also obtained in SW13 cells generated to have stable CLND9 expression (data not shown). To further confirm the role of CLDN9 in HCVpp infection, we performed RNA interference to reduce endogenous CLDN9 expression. Two shRNA sequences targeting CLDN9 (referred to as 342 and 661) were delivered into Bel7402 cells using a lentivirus-based system. As shown in Fig. 2B, the expression level of CLDN9 in 342- and 661-treated cells was significantly downregulated compared with the irrelevant shRNA-treated cells. The susceptibility of these CLDN9-silencing cells to HCVpp, but not to VSV-Gpp, was remarkably reduced (Fig. 2C). These data clearly demonstrate that besides CLDN1, CLDN9 was also able to mediate HCV entry into target cells.
FIG. 2.
CLDN6 and CLDN9 mediate HCVpp entry. (A) 293T cells stably expressing claudins that were found in the Bel7402 cell line were infected with pseudotyped particles harboring VSV-G protein (black bar), envelope glycoproteins of HCV strain H77 (gray bars), and no env (white bars), with luciferase as a reporter (RLU, relative light units) (mean of n = 3) (error bars indicate SD). Naive 293T cells were used as a negative control. (B) Expression level of CLDN9 in Bel7402 cells treated with CLDN9-specific or irrelevant (irr.) shRNAs was determined by real-time PCR using CLDN9- and GAPDH-specific primers. The value of GAPDH was initialized to 1 (mean of n = 2) (error bars indicate SD). (C) Bel7402 cells treated with CLDN9-specific shRNAs were infected by pseudotyped particles bearing envelope glycoproteins as indicated in A. Results are presented as percentages of infection in irrelevant shRNA-treated cells (irr.) (mean of n = 3) (error bars indicate SD). (D) High homology between CLDN6 and CLDN9. The phylogenetic tree of EL1 of claudin family members was created by DNAMAN. The homology among EL1 of claudin family members is indicated as a percentage. (E) CLDN6 could also mediate HCVpp entry. Huh7, naive 293T, and 293T cells stably expressing individual CLDN1, CLDN6, CLDN9, CLDN2, CLDN3, CLDN4, CLDN7, CLDN11, CLDN12, CLDN15, CLDN17, and CLDN23 were infected with luciferase reporter viruses pseudotyped with envelope glycoproteins indicated as in A (RLU, relative light units) (mean of n = 3) (error bars indicate SD). (F) Flag-tagged CLDN9 could mediate HCV entry as efficiently as naive CLDN9. CLDN9-expressing and CLDN9-Flag-expressing 293T cells were infected by pseudotyped particles bearing envelope glycoproteins, as indicated in A, with luciferase as a reporter (mean of n = 3) (error bars indicate SD). The expression of CLDN9 and CLDN9-Flag was detected by Western blotting using antibodies against CLDN9 and Flag tag, with GAPDH as a control.
It has been reported that EL1 of CLDN1 was critical for mediating the interaction with HCV envelope glycoproteins (7). To examine if there is any other claudin that might also function as an HCV coreceptor, the amino acid sequences of EL1 of all claudins were aligned to search for any similarities with EL1 of CLDN9. Results showed that CLDN6, followed by CLDN3 and CLDN4, demonstrated the highest order of homology for EL1 compared with CLDN9 (Fig. 2D). In contrast, CLDN12, CLDN23, CLDN11, and CLDN15 showed the least homology with CLDN9. Hence, CLDN3, CLDN4, and CLDN6 were selected for further functional experiments. In addition, two other claudins with the least homology, CLDN11 and CLDN15, were used as controls. The potential of these claudins as HCV coreceptors was investigated using a method similar to the methods described above. As shown in Fig. 2E, the cell lines stably expressing CLDN6 could be efficiently infected by HCVpp, therefore demonstrating that CLDN6 is an additional claudin that is able to mediate the entry of HCV.
To monitor the expression of the claudin proteins in stable cell lines, a Flag tag was fused to the C terminus of all of the claudins studied (8). We show that all of these family members were in fact expressed as expected. Furthermore, 293T cells expressing Flag-tagged CLDN1, CLDN6, and CLDN9 were susceptible to HCVpp, while the other Flag-tagged claudin family members did not support HCV entry (CLDN9 is shown in Fig. 2F, and the others are not shown). These results provide further evidence that CLDN6 and CLDN9 are able to function as HCV coreceptors.
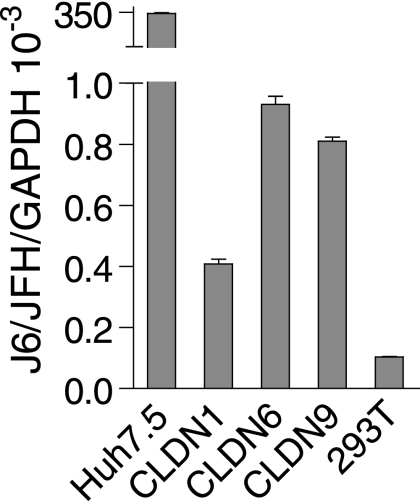
The ability of CLDN6 and CLDN9 as HCV coreceptors was further verified using cell culture-derived virus (HCVcc). Cell culture-derived particles of J6/JFH were produced by the transfection of Huh7.5 cells with in vitro-transcribed J6/JFH RNA (19). To determine if the overexpression of CLDN6 and CLDN9 could confer susceptibility of 293T cells to HCVcc, the supernatants were harvested and used to inoculate 293T cells that were either mock transfected or transfected with CLDN6 or CLDN9. The efficiency of infection was determined using real-time PCR. Both CLDN6- and CLDN9-expressing 293T cells support infection with HCVcc, but the titers of HCVcc on these cells were about 400 times lower than those on Huh7.5 cells (Fig. 3), which might be a result of the inefficient replication of HCV in 293T cells (16).
FIG. 3.
CLDN6 and CLDN9 mediate HCVcc infection. HCVcc infection was analyzed by real-time PCR. 293T cells expressing mock, CLDN1, CLDN6, or CLDN9 were challenged with HCVcc of strain J6/JFH. At 72 h postinfection, total RNAs were isolated from these cells and amplified with primers specific to the corresponding claudins or GAPDH. Huh7.5 cells were used as a positive control. The value of GAPDH was initialized to 1 (mean of n = 2) (error bars indicate SD).
No coreceptor usage preference among CLDN1, CLDN6, and CLDN9 by various genotypes of HCV.
HCV has been classified into six genetically distinct genotypes exhibiting different phenotypic properties (1, 34). To investigate if there is any preference in the coreceptor usage by HCV of various genotypes, we generated a panel of HCVpp-bearing HCV envelope glycoproteins derived from six HCV strains (UKN1A 14.38, UKN2A 2.4, UKN3A 1.28, UKN4 11.1, UKN5 14.4, and UKN6 5.340). These glycoproteins represented genotypes 1 to 6, respectively. Virus particles prepared with these glycoproteins were used to infect 293T cells expressing 12 claudin family members including CLDN1 to CLDN4, CLDN6, CLDN7, CLDN9, CLDN11, CLDN12, CLDN15, CLDN17 and CLDN23. Huh7 and parental 293T cells were used as positive and negative controls. All of these HCVpps were unable to infect parental 293T cells and 293T cells expressing claudins tested, with the exception of CLDN1, CLDN6, and CLDN9 (data not shown). Although six genotypes of HCVpp displayed different levels of infectivity in 293T cells expressing CLDN1, CLDN6, and CLDN9, such differences in the efficiency of infection are similar to that in Huh7 cells (data not shown), suggesting that there is no obvious preference in coreceptor usage by various HCV genotypes.
Expression of CLDN6 and CLDN9 in liver, PBMCs, and human hepatocellular carcinoma cell lines.
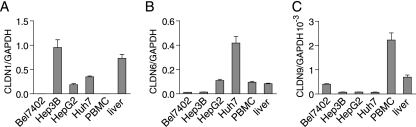
Accumulating evidence suggests that besides the liver, PBMCs are also important target cells for HCV (4, 15, 23, 24). To study the in vivo potential of the involvement of CLDN6 and CLDN9 in HCV infection, the expression patterns of CLDN1, CLDN6, and CLDN9 were examined in liver and PBMCs using real-time PCR. These three claudin family members could be detected in liver, implying that they might be all involved in HCV infection in liver (Fig. 4). In PBMCs, CLDN6 and CLDN9, but not CLDN1, could be detected. This result suggests that they may be involved in mediating HCV entry into PBMCs. Further experiments are needed to reveal their relevance in vivo. The expression pattern of these three claudins was also studied in several HCV-susceptible cell lines. As shown in Fig. 4, CLDN6 was expressed in Huh7 and HepG2 cells, while CLDN9 was expressed in Bel7402 cells.
FIG. 4.
Expression pattern of CLDN1, CLDN6, and CLDN9 in human liver, PBMCs, and human hepatocellular carcinoma cell lines. The cDNA samples of different sources were amplified by real-time PCR using CLDN1 (A)-, CLDN6 (B)-, CLDN9 (C)-, and GAPDH-specific primers. The value of GAPDH was initialized to 1 (mean of n = 2) (error bars indicate SD).
Identification of amino acid residues in CLDN9 critical for HCV entry.
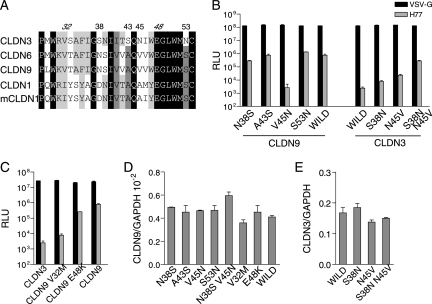
Although CLDN3 shared the highest EL1 homology to CLDN6 and CLDN9, it was unable to serve as an HCV coreceptor. Comparison of EL1 sequences was carried out between human CLDN1, CLDN3, CLDN6, and CLDN9 and murine CLDN1. Four positions in the N terminus of EL1 were selected for mutational analysis, namely, positions 38, 43, 45, and 53 (CLDN9 was used as the reference sequence for numbering of the amino acid positions). At these sites, the non-HCV receptor CLDN3 had residues that were different from those of proposed HCV coreceptors (Fig. 5A). Since positions 32 and 48 have also been reported to be critical for CLDN1-mediated HCV entry (7), they were selected as well. As CLDN6 and CLDN9 are highly conserved in EL1 and differ only at position 29, we first introduced a M29L mutation into CLDN6 and found no difference in levels of infectivity (data not shown). After these mutations were introduced into CLDN9 or CLDN3, they were delivered into 293T cells for functional analysis. First, the expression of wild-type and mutated claudins was confirmed by real-time PCR and showed no obvious difference in expression levels among them (Fig. 5D and E). As shown in Fig. 5B, A43S and S53N had no apparent impact on CLDN9 coreceptor activity, while the N38S mutation alone could mildly decrease CLDN9-mediated HCVpp infection. Most strikingly, a single V45N mutation almost abolished CLDN9-mediated HCVpp infection. Such effects of N38 and V45 were then further verified on CLDN3, in that a single mutation of S38N or V45N in CLDN3 could partially render 293T permissive to HCVpp, while double mutations of S38N and V45N could enhance this effect most significantly (Fig. 5B). V32M and E48K mutations displayed a similar effect on CLDN9 (Fig. 5C) as on CLDN1 (7). Taken together, these data demonstrated that besides V32 and E48, N38 and V45 are critical for HCVpp entry in CLDN9, where V45 might play the most important role during the process.
FIG. 5.
Key residues in EL1 of CLDN9 necessary for HCVpp entry. (A) Alignment of the N-terminal EL1 amino acid sequences of human CLDN1, CLDN3, CLDN6, and CLDN9 and murine CLDN1. Amino acid homology and differences are displayed by different shades of black. Number represents the positions of amino acids in full-length CLDN9 that were selected as candidates for further analysis. The numbers for known critical residues are shown in italics. (B and C) Wild-type and mutated claudin-transduced 293T cells were tested for susceptibility to HCVpp bearing envelope glycoproteins of strain H77 (gray bar) or VSV-Gpp (black bar) with luciferase as a reporter (RLU, relative light units) (mean of n = 3) (error bar indicate SD). (D and E) Expression of wild-type and mutated claudins was verified by real-time PCR. No signal was detected on naive 293T cells. The value of GAPDH was initialized to 1 (mean of n = 2) (error bars indicate SD).
DISCUSSION
In this paper, we demonstrated that in addition to CLDN1, CLDN6 and CLDN9 are able to mediate HCVpp and HCVcc infection of nonhepatic cells, indicating that both claudins have HCV coreceptor activity as well as CLDN1. The expression of CLDN6 and CLDN9 in liver and PBMCs was detected, suggesting their potential role in HCV infection in liver and the extrahepatic compartment. Finally, we found two residues critical for HCV infection in EL1 of CLDN9.
The identification of CLDN6 and CLDN9 as additional HCV coreceptors should assist in future efforts to understand the relationship between claudin proteins and HCV pathogenesis. Representative strains of HCV genotypes 1 and 2 use CLDN1 as a coreceptor, and CLDN1 is expressed at high levels in liver (8), indicating its important role in HCV infection (7). Although the in vivo contribution of CLDN6 and CLDN9 to HCV infection is unresolved, there are several lines of evidence suggesting that CLDN6 and CLDN9 are potential HCV coreceptors. Previous reports have shown that the expression of CLDN6 and CLDN9 in liver is detectable by RT-PCR (11, 12). Our study also detected the expression of CLDN6 and CLDN9 in liver by real-time PCR (Fig. 4). On the other hand, the Huh7 and HepG2 human hepatocellular carcinoma cell lines were found to express both CLDN1 and CLDN6, while Bel7402 expresses CLDN9 (Fig. 4). These data suggest that CLDN6 and CLDN9 may play a role in HCV infection in liver. Furthermore, we found CLDN6 and CLDN9 expressed in PBMCs, which are believed to be an HCV replication site outside the liver, whereas no expression of CLDN1 was detected by real-time PCR (Fig. 4). These results suggest that CLDN6 and CLDN9 may also contribute to extrahepatic HCV infection, especially in PBMCs.
Although HCV displays a high degree of genetic heterogeneity, no coreceptor usage preference was detected among CLDN1, CLDN6, and CLDN9, which may be attributed to the high similarity among their structures. Claudins are a family of integral membrane proteins involved in the formation of tight junctions. Tight junctions are responsible for the formation and maintenance of the epidermal permeability barrier. Claudins have three characteristic functional domains: (i) two extracellular loops responsible for permeability barrier formation and EL1 with specific ion selectivity, (ii) four transmembrane regions, and (iii) a cytoplasmic C tail that functions to anchor to the cytoskeleton (9). Most of the claudin proteins display high levels of homology, especially in CLDN6 and CLDN9. These two claudin family members differ only at amino acid position 29 in EL1. By aligning the amino acid sequences of human CLND3, CLDN6, and CLDN9 and murine CLDN1, we found four candidate residues that could play a role in mediating HCV entry. By mutational analysis, we found that N38 and V45 are important for HCV infection in CLDN9 (Fig. 5B). Residues at positions 32 and 48, critical for HCV infection in cells that express CLDN1 (7), were also shown to be important in CLDN9 in our studies (Fig. 5C). The identification of these residues should be useful for the future development of drugs that inhibit HCV infection.
Acknowledgments
This research was supported by the Ministry of Science and Technology (grant 2005CB522903), Beijing Natural Science Foundation (5051003), National Nature Science Foundation of China for Creative Research Groups (30421004), a Bill & Melinda Gates Foundation grant (37871) and a 111 Project grant to H. Deng.
We thank Jonathan K. Ball for sending plasmids containing HCV envelope glycoproteins of genotypes 1 to 6, Charles M. Rice for providing the plasmid containing the HCV whole genome of strain J6/JFH, Shelia Stewart for the gift of plasmid pBabepuro, and Luk Van Parijs for vector pLL3.7. We also acknowledge Wang Peigang, Matt Stremlau, Joanna W. Y. Ho, and Hong Zhang for critical reading of the manuscript. We also thank Yizhe Zhang for technical assistance in real-time PCR.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Amoroso, P., M. Rapicetta, M. E. Tosti, A. Mele, E. Spada, S. Buonocore, G. Lettieri, P. Pierri, P. Chionne, A. R. Ciccaglione, and L. Sagliocca. 1998. Correlation between virus genotype and chronicity rate in acute hepatitis C. J. Hepatol. 28:939-944. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackard, J. T., N. Kemmer, and K. E. Sherman. 2006. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology 44:15-22. [DOI] [PubMed] [Google Scholar]
- 5.Chen, R. M., D. H. Zhu, and X. Z. Ye. 1975. The establishment and character of human hepacarcinoma cell line (BEL-7402) in vitro. Comm. Sci. 20:434-436. [Google Scholar]
- 6.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 8.Furuse, M., K. Fujita, T. Hiiragi, K. Fujimoto, and S. Tsukita. 1998. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Mariscal, L., A. Betanzos, P. Nava, and B. E. Jaramillo. 2003. Tight junction proteins. Prog. Biophys. Mol. Biol. 81:1-44. [DOI] [PubMed] [Google Scholar]
- 10.Gumbiner, B. M. 1993. Breaking through the tight junction barrier. J. Cell Biol. 123:1631-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewitt, K. J., R. Agarwal, and P. J. Morin. 2006. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 6:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, Y. H., D. Hishikawa, H. Miyahara, Y. Nishimura, H. Tsuzuki, C. Gotoh, T. Iga, Y. Suzuki, S. H. Song, K. C. Choi, H. G. Lee, S. Sasaki, and S. G. Roh. 2005. Up-regulation of the claudin-6 gene in adipogenesis. Biosci. Biotechnol. Biochem. 69:2117-2121. [DOI] [PubMed] [Google Scholar]
- 13.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser, P., B. Niederost, B. Joos, V. von Wyl, M. Opravil, R. Weber, H. F. Gunthard, and M. Fischer. 2006. Equal amounts of intracellular and virion-enclosed hepatitis C virus RNA are associated with peripheral-blood mononuclear cells in vivo. J. Infect. Dis. 194:1713-1723. [DOI] [PubMed] [Google Scholar]
- 16.Kato, T., T. Date, M. Miyamoto, Z. Zhao, M. Mizokami, and T. Wakita. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 18.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 19.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 20.Ling, C. Q., B. Li, C. Zhang, D. Z. Zhu, X. Q. Huang, W. Gu, and S. X. Li. 2005. Inhibitory effect of recombinant adenovirus carrying melittin gene on hepatocellular carcinoma. Ann. Oncol. 16:109-115. [DOI] [PubMed] [Google Scholar]
- 21.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita, K., M. Furuse, K. Fujimoto, and S. Tsukita. 1999. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. USA 96:511-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morsica, G., G. Tambussi, G. Sitia, R. Novati, A. Lazzarin, L. Lopalco, and S. Mukenge. 1999. Replication of hepatitis C virus in B lymphocytes (CD19+). Blood 94:1138-1139. [PubMed] [Google Scholar]
- 24.Muller, H. M., E. Pfaff, T. Goeser, B. Kallinowski, C. Solbach, and L. Theilmann. 1993. Peripheral blood leukocytes serve as a possible extrahepatic site for hepatitis C virus replication. J. Gen. Virol. 74:669-676. [DOI] [PubMed] [Google Scholar]
- 25.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 26.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 27.Qu, X. X., P. Hao, X. J. Song, S. M. Jiang, Y. X. Liu, P. G. Wang, X. Rao, H. D. Song, S. Y. Wang, Y. Zuo, A. H. Zheng, M. Luo, H. L. Wang, F. Deng, H. Z. Wang, Z. H. Hu, M. X. Ding, G. P. Zhao, and H. K. Deng. 2005. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 280:29588-29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahner, C., L. L. Mitic, and J. M. Anderson. 2001. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology 120:411-422. [DOI] [PubMed] [Google Scholar]
- 29.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. Zhang, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 31.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeff, L. B. 1995. Natural history of viral hepatitis, type C. Semin. Gastrointest. Dis. 6:20-27. [PubMed] [Google Scholar]
- 33.Shao, G. Z., R. L. Zhou, Q. Y. Zhang, Y. Zhang, J. J. Liu, J. A. Rui, X. Wei, and D. X. Ye. 2003. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene 22:5060-5069. [DOI] [PubMed] [Google Scholar]
- 34.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 35.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 36.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, X. F., B. F. Xie, J. M. Zhou, G. K. Feng, Z. C. Liu, X. Y. Wei, F. X. Zhang, M. F. Liu, and Y. X. Zeng. 2005. Blockade of vascular endothelial growth factor receptor signal pathway and antitumor activity of ON-III (2′,4′-dihydroxy-6′-methoxy-3′,5′-dimethylchalcone), a component from Chinese herbal medicine. Mol. Pharmacol. 67:1444-1450. [DOI] [PubMed] [Google Scholar]