Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi's sarcoma. The open reading frame (K9) of KSHV encodes viral interferon regulatory factor 1 (vIRF1), which functions as a repressor of interferon-mediated signal transduction. The amino-terminal region of vIRF1 displays significant homology to the DNA-binding domain of cellular interferon regulatory factors, supporting the theory that the protein interacts with specific DNA sequences. Here, we identify the consensus sequence of vIRF1-binding sites from a pool of random oligonucleotides. Moreover, our data show that vIRF1 interacts with the K3:viral dihydrofolate reductase:viral interleukin 6 promoter region in the KSHV genome.
Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8, a recently discovered human DNA tumor virus, is strongly associated with the development of Kaposi's sarcoma, primary effusion lymphoma, and some cases of multicentric Castleman's disease (3, 4, 24). Analysis of the nucleotide sequence reveals that KSHV belongs to the rhadinovirus subgroup of herpesviruses. Interestingly, the KSHV genome contains a number of open reading frames (ORFs) with unusual homology to known cellular proteins (13, 18). Viral infection triggers a potent antiviral response mediated by interferons (IFNs), which participates in host innate immunity. IFNs perform a variety of biological activities via inducing the expression of over 300 genes encoding proteins with antiviral, antiproliferative, and immunomodulatory functions. IFN signaling is mediated by IFN regulatory factors (IRFs), a family of DNA-binding proteins that serve as activators or repressors (1, 14). The KSHV genome contains four ORFs encoding proteins homologous to IRF families, designated viral IRFs (vIRFs) (15, 18). Among these proteins, vIRF1 of KSHV has been characterized extensively. The vIRF1 protein comprises 449 amino acids. The amino-terminal region contains a conserved tryptophan-rich DNA-binding sequence and displays 70% identity to the IFN consensus sequence-binding protein (18). Several groups report that vIRF1 acts as a negative regulator in cellular IFN signaling (5, 9, 25). vIRF1 expression leads to the transformation of rodent fibroblasts and consequent induction of malignant fibrosarcoma in nude mice, suggestive of a role as a potent oncoprotein (5, 9). A number of studies show that vIRF1 interacts with p300/CREB-binding protein (CBP), inhibiting the transactivation of CBP, the histone acetyltransferase activity of p300, and the formation of transcriptionally active IRF3-p300/CBP complexes (2, 8, 10, 19). Moreover, vIRF1 associates with the tumor suppressor p53, leading to the repression of p53-dependent transcription and apoptosis (12, 22), and inhibits IFN/retinoic acid-induced cell death though interactions with a cell death regulator, GRIM19 (7, 20). vIRF1 additionally suppresses transforming growth factor β-mediated transcription and growth arrest via targeting of Smad proteins (21). Another recent report shows that vIRF1 inhibits ataxia telangiectasia-mutated kinase activity, leading to reduced p53 serine 15 phosphorylation and increased p53 ubiquitination and degradation (23).
IRF transcription factors play an important role in the induction of genes that encode type I IFNs (1, 6, 14). IRFs are characterized by a well-conserved amino-terminal DNA-binding domain (DBD) with five tryptophan repeats, which displays homology to DBD sequences of myb transcription factors. The DBD regions of IRFs form a helix-turn-helix motif and bind similar DNA sequences, designated IFN-stimulated response element, IFN consensus sequence, and IFN regulatory element (6, 11). In view of the significant homology between the amino-terminal region of vIRF1 and DBD sequences of cellular IRFs, it is proposed that vIRF1 interacts with specific DNA sequences (18).
Identification of the DNA sequence interacting with vIRF1.
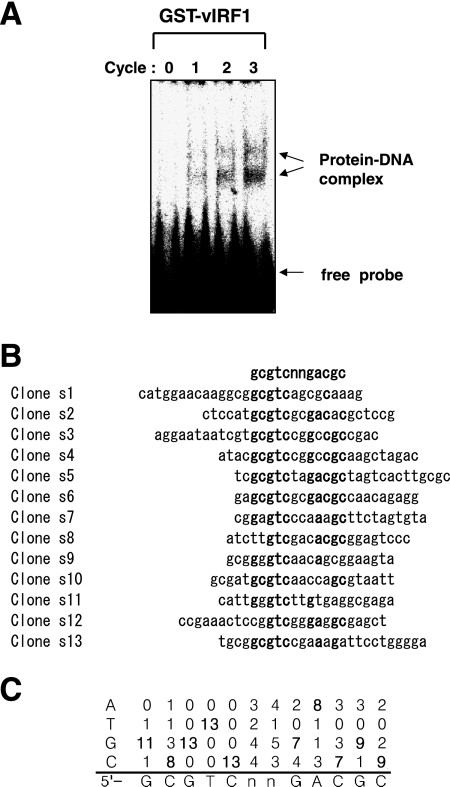
To identify the interacting DNA sequence, we performed a binding site selection assay using the method of Pollock and Treisman (16), with slight modifications. Recombinant glutathione S-transferase (GST)-vIRF1 protein was employed for the assay. Cloning and recombinant GST-vIRF1 protein preparation methods are described in an earlier report (19). The random oligonucleotide pool, N24 (5′CAGGTCAGTAGATCTTGTCG(A/T/G/C)24GAGGCGAGCTCAACTGGAGC3′), contains a synthetic single-stranded random 24-base sequence flanked by two PCR primer sequences (primer 1, 5′CAGGTCAGTAGATCTTGTCG3′; primer 2, 5′GCTCCAGTTGAGCTCGCCTC3′). The flanking sequences contain BglII and SacI restriction sites to facilitate subsequent cloning. To generate the labeled random probe, oligonucleotides were converted to double-stranded DNA by five rounds of PCR with 32P-labeled dCTP and unlabeled dCTP, dATP, dTTP, and dGTP. The binding reaction was performed at room temperature for 30 min in a total volume of 25 μl reaction mixture comprising 20 μl buffer E (16), 0.2 μg poly(deoxyinosine-deoxycytosine), 1 μg GST-vIRF1 protein, and the labeled random probe (0.4 ng). We analyzed binding complexes by electrophoresis on a 5% native gel with 0.5× Tris-borate buffer. Bands were visualized using autoradiography. Oligonucleotides in the complexes were recovered by cutting the appropriate part of the dried gel corresponding to the shifted bands and grinding to powder. DNA was eluted at 50°C with 200 μl of elution buffer (5 mM EDTA, 0.5% sodium dodecyl sulfate, 100 mM sodium acetate, 50 mM Tris-HCl [pH 8.0]), recovered by phenol extraction and ethanol precipitation, and amplified by PCR. Amplified DNA was relabeled and the gel shift assay performed, followed by DNA recovery and amplification, as described above. In total, three cycles of this process were performed. As binding site selection was repeated, the oligonucleotide pools became enriched in sequences forming vIRF1-DNA complexes. We performed gel shift assays using the probe in each cycle. After the first cycle of binding site selection, two vIRF1-DNA complexes were observed. The intensity of the shifted bands increased after each cycle. Following the third cycle, the two shifted bands were clearly detectable (Fig. 1A). DNA was amplified, digested with BglII and SacI, inserted in the pUC vector, and used to transform Escherichia coli JM109 cells. Plasmids were prepared and sequenced to identify the vIRF1-binding sites. The consensus sequence, derived from 13 different clones (Fig. 1B and C), was identified as GCGTCnnGACGC (Fig. 1C).
FIG. 1.
Identification of the vIRF1-binding sequence. (A) Gel shift analysis of selected vIRF1-binding sites. Gel shift assays were performed with DNA selected at each round as probes, together with GST-vIRF1 protein. The number of selection cycles is shown above each lane. Shifted bands are indicated with arrows. (B) Selected vIRF1-binding sequences. Each cloned sequence after three rounds of selection is presented. Sequences were aligned for maximum identity. (C) Statistical results of each base match.
Binding properties of recovered sequences.
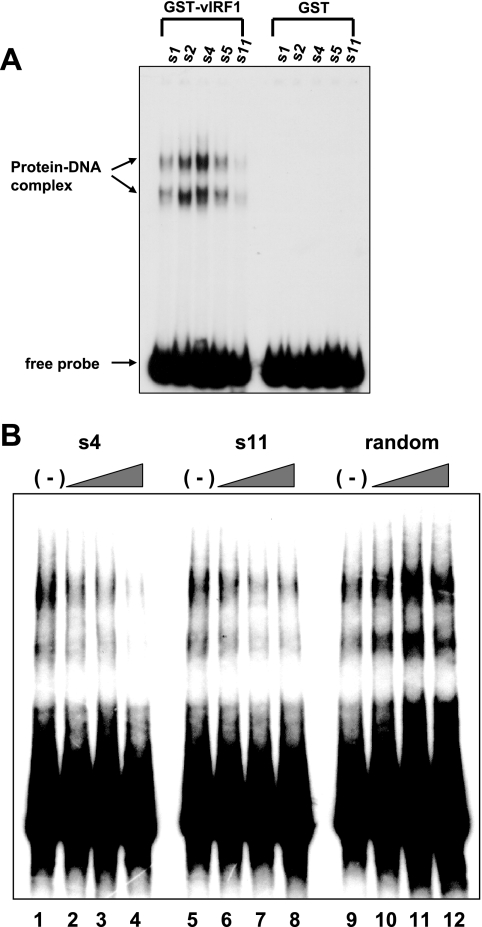
To determine whether the recovered DNA sequences actually bind vIRF1, we performed the gel shift assay using GST-vIRF1 recombinant protein (1 μg). DNA recovered from each clone was labeled with 32P-labeled dCTP by five rounds of PCR and the gel shift assay performed as described above. The GST protein alone did not interact with any sequences, whereas GST-vIRF1 was associated with the recovered s1, s2, s4, s5, and s11 sequences (Fig. 2A). Similar to data from Fig. 1A, two shifted bands were observed, indicating that vIRF1 forms two types of complexes with DNA (Fig. 2A). The s4 sequence displayed the strongest binding affinity for GST-vIRF1, whereas s11 displayed relatively low affinity (Fig. 2A, lanes 3 and 5). To confirm that the DNA-protein complexes are specific for the consensus sequence, we performed competition binding assays with labeled sequenced s5. Preincubation with unlabeled s4 and s11 oligonucleotides led to a dose-dependent decrease in the amount of DNA-protein complexes, indicating that both oligonucleotides are functional in a competition assay (Fig. 2B, lanes 1 to 8). However, unlabeled random oligonucleotides did not inhibit complex formation (Fig. 2B, lanes 9 to 12). The data collectively suggest that vIRF1 binding is highly specific for the consensus sequence.
FIG. 2.
Characterization of vIRF1-binding sites. (A) Gel shift assays with the selected vIRF1-binding sites. Gel shift assays were performed with selected DNA (s1, s2, s4, s5, and s11) sequences and GST-vIRF1 (lanes 1 to 5) or GST protein (lanes 6 to 10). Protein-DNA complexes are specified using arrows. (B) Competition assays were performed by preincubation with selected unlabeled vIRF1-binding sites. Labeled s5 probe was used in each gel shift assay. Unlabeled s4 (lanes 1 to 4), s11 (lanes 5 to 8), and random oligonucleotides (lanes 9 to 12) were used. Twofold (lanes 2, 6, and 11), fivefold (lanes 3, 7, and 12), and 10-fold (lanes 4, 8, and 12) molar excesses of unlabeled probes were used, respectively.
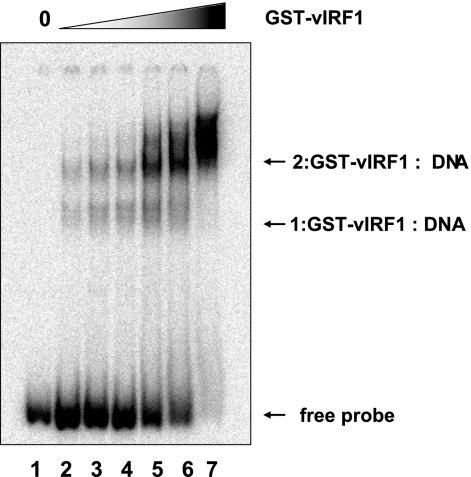
Next, we examined the effects of the levels of GST-vIRF1 proteins with a gel shift assay. Consistent with the above-described data (Fig. 3), two shifted bands were evident in assays with GST-vIRF1 and the consensus sequence s5. Upon the addition of excess GST-vIRF1 protein, the upper band became stronger, whereas the lower band almost disappeared, suggestive of the presence of two binding sites within the consensus sequence (Fig. 3).
FIG. 3.
Effects of the amounts of vIRF1 protein. Gel shift assays were performed with increasing amounts of GST-vIRF1 protein and s5 sequence. Shifted bands and free probe are indicated with arrows.
vIRF1 binds to the promoter region of K3, vDHFR, and vIL6 in the KSHV genome.
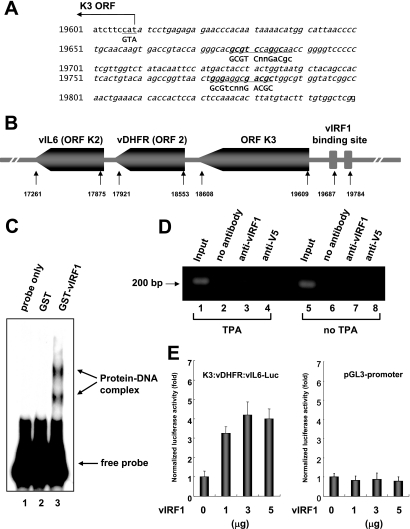
The next step was to determine whether the KSHV genome contains the vIRF1-binding consensus sequence. Interestingly, the promoter region of K3, vDHFR (ORF2), and viral interleukin 6 (vIL6) contains a similar but not identical sequence (Fig. 4A). K3, viral dihydrofolate reductase (vDHFR), and vIL6 are clustered in the KSHV genome (Fig. 4B). To establish whether vIRF1 binds the promoter, gel shift assays were performed using the promoter as a probe. GST-vIRF1 specifically associated with the promoter, in contrast to GST (Fig. 4C), implying specific binding of vIRF1 to this region of the KSHV genome. To confirm that vIRF1 binds to the promoter region in vivo, we performed a chromatin immunoprecipitation (ChIP) assay. BCBL-1 cells (2 × 107) were cultured in the presence or absence of 12-O-tetradecanoylphorbol-13-acetate (TPA) for 48 h. A ChIP assay was performed with a ChIP assay kit (Upstate, Charlottesville, VA) as described in the manufacturer's instructions. The eluted DNA was amplified by PCR using primers for the K3 promoter. The forward and reverse primer sequences were 5′ATCTTCCATATCCTGAGAG3′ and 5′CCGAGCCACAAAGTACATAA3′, respectively, and yielded a product of 200 bp. The K3:vDHFR:vIL6 promoter was found to be associated with vIRF1 when the cells were treated with TPA (Fig. 4D, lane 3), whereas neither of the negative controls with no antibody or anti-V5 antibody precipitated any significant amount of the promoter (Fig. 4D, lanes 2 and 4), indicating that vIRF1 associates with the K3:vDHFR:vIL6 promoter in KSHV infected-cells.
FIG. 4.
vIRF1 binds to the K3:vDHFR:vIL6 promoter region. (A) The KSHV K3:vDHFR:vIL6 promoter sequence is presented. Sequences similar to the vIRF1 consensus sequence are underlined; matched sequences are in boldface type. (B) Schematic diagram of the cluster of K3, vDHFR, and vIL6 genes in the KSHV genome. (C) vIRF1 interacts with the K3:vDHFR:vIL6 promoter. Gel shift assays were performed using the promoter region as a probe, together with no protein, GST, or GST-vIRF1 (s1, s2, s4, s5, and s11 sequences). (D) In vivo association of vIRF1 with the K3:vDHFR:vIL6 promoter. ChIP assays were performed with BCBL-1 cells induced or not induced with TPA. Input DNA (before immunoprecipitation, diluted 1:1,000) and ChIP DNA were amplified by using the K3:vDHFR:vIL6 promoter primer. PCR products were resolved by agarose gel electrophoresis. (E) Activation of the K3:vDHFR:vIL6 promoter by vIRF1. 293T cells were cotransfected with Rous sarcoma β-galactosidase expression plasmid (pRSV-β-gal) (0.5 μg) and increasing amounts of vIRF1 expression plasmid together with the K3:vDHFR:vIL6-Luc reporter plasmid (1 μg) or the blank reporter (pGL3 promoter) (1 μg). Transcriptional changes were measured using the luciferase activity and are presented as severalfold activation. Luciferase activity was derived from three independent experiments and normalized with β-galactosidase expression levels.
To determine whether vIRF1 influences the promoter activity, we performed a transient promoter assay. 293T cells were transiently cotransfected with a vIRF1 expression plasmid plus either a K3:vDHFR:vIL6-pGL3-promoter-Luc plasmid or a pGL3-promoter blank vector. Forty-eight hours after transfection, cells were harvested and assayed for luciferase activity as a measure of the promoter activity. vIRF1 activated the K3:vDHFR:vIL6 promoter in a dose-dependent manner, whereas vIRF1 did not affect the blank promoter (Fig. 4E). These results demonstrate that vIRF1 interacts with the K3:vDHFR:vIL6 promoter and activates the promoter.
Our results suggest that vIRF1 is a DNA-binding protein, analogous to the cellular IRF family. The finding that vIRF1 interacts with the K3 promoter of KSHV supports its role as a viral transcriptional factor that regulates KSHV gene expression. Consistent with this theory, Li et al. (9) reported that vIRF1 antisense expression results in vIL6 downregulation. Since vIRF1 is an activator (17), we speculate that the protein stimulates vIL6 expression through binding to the promoter region, which may explain why vIL6 expression decreases with the antisense of vIRF1.
In gel shift assays using the vIRF1 protein, two different DNA-protein complexes were evident. One possible reason is that the consensus sequence contains two vIRF1-binding sites. When vIRF1 expression is low, all binding sites are not fully occupied. However, at sufficiently high vIRF1 expression, all binding sites are fully occupied. Consequently, two bands (upper band, fully occupied; lower band, partly occupied) are present in the gel shift assay. This hypothesis is supported by the results shown in Fig. 3. Following an increase in vIRF1 expression, the lower band disappeared and the upper band became stronger (Fig. 3), supporting the existence of two vIRF1-binding sites within the consensus sequence. An interesting feature of the consensus sequence is its palindromic structure (GCGTCnnGACGC), resulting in two identical DNA sequences. Another possibility is that vIRF1 forms a dimer. However, further studies are required to examine these hypotheses.
We and other groups have reported that vIRF1 acts as a negative regulator via direct interactions with transcriptional activators, such as p300/CBP, p53, and Smads (8, 10, 12, 19, 21, 22). Most vIRF1 functions are mediated by protein-protein interactions. In this study, we demonstrate that vIRF1 is a DNA-binding protein that binds to a specific consensus sequence. Thus, vIRF1 appears to form both protein-DNA and protein-protein complexes. Studies to establish the importance of vIRF1-DNA interactions in transcriptional regulation and viral gene expression are under way.
Acknowledgments
This work was supported by a Korea Research Foundation grant funded by the Korean Government (MOEHRD) (KRF-2006-521-C00133) (T. Seo) and a Science Research Center (Molecular Therapy Research Center) grant from the Korea Science and Engineering Foundation and a Korea Basic Science Institute grant (KBSI-GJ-D27023) (J. Park).
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 2.Burysek, L., W. S. Yeow, B. Lubyova, M. Kellum, S. L. Schafer, Y. Q. Huang, and P. M. Pitha. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 5.Gao, S. J., C. Boshoff, S. Jayachandra, R. A. Weiss, Y. Chang, and P. S. Moore. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979-1985. [DOI] [PubMed] [Google Scholar]
- 6.Honda, K., A. Takaoka, and T. Taniguchi. 2006. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349-360. [DOI] [PubMed] [Google Scholar]
- 7.Hu, J., J. E. Angell, J. Zhang, X. Ma, T. Seo, A. Raha, J. Hayashi, J. Choe, and D. V. Kalvakolanu. 2002. Characterization of monoclonal antibodies against GRIM-19, a novel IFN-beta and retinoic acid-activated regulator of cell death. J. Interferon Cytokine Res. 22:1017-1026. [DOI] [PubMed] [Google Scholar]
- 8.Li, M., B. Damania, X. Alvarez, V. Ogryzko, K. Ozato, and J. U. Jung. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 20:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, M., H. Lee, J. Guo, F. Neipel, B. Fleckenstein, K. Ozato, and J. U. Jung. 1998. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor. J. Virol. 72:5433-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 11.Mamane, Y., C. Heylbroeck, P. Genin, M. Algarte, M. J. Servant, C. LePage, C. DeLuca, H. Kwon, R. Lin, and J. Hiscott. 1999. Interferon regulatory factors: the next generation. Gene 237:1-14. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura, H., M. Li, J. Zarycki, and J. U. Jung. 2001. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J. Virol. 75:7572-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 15.Offermann, M. K. 2007. Kaposi sarcoma herpesvirus-encoded interferon regulator factors. Curr. Top. Microbiol. Immunol. 312:185-209. [DOI] [PubMed] [Google Scholar]
- 16.Pollock, R., and R. Treisman. 1990. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 18:6197-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roan, F., J. C. Zimring, S. Goodbourn, and M. K. Offermann. 1999. Transcriptional activation by the human herpesvirus-8-encoded interferon regulatory factor. J. Gen. Virol. 80:2205-2209. [DOI] [PubMed] [Google Scholar]
- 18.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo, T., D. Lee, B. Lee, J. H. Chung, and J. Choe. 2000. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) binds to, and inhibits transactivation of, CREB-binding protein. Biochem. Biophys. Res. Commun. 270:23-27. [DOI] [PubMed] [Google Scholar]
- 20.Seo, T., D. Lee, Y. S. Shim, J. E. Angell, N. V. Chidambaram, D. V. Kalvakolanu, and J. Choe. 2002. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus interacts with a cell death regulator, GRIM19, and inhibits interferon/retinoic acid-induced cell death. J. Virol. 76:8797-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo, T., J. Park, and J. Choe. 2005. Kaposi's sarcoma-associated herpesvirus viral IFN regulatory factor 1 inhibits transforming growth factor-beta signaling. Cancer Res. 65:1738-1747. [DOI] [PubMed] [Google Scholar]
- 22.Seo, T., J. Park, D. Lee, S. G. Hwang, and J. Choe. 2001. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. J. Virol. 75:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, Y. C., H. Nakamura, X. Liang, P. Feng, H. Chang, T. F. Kowalik, and J. U. Jung. 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 80:2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 25.Zimring, J. C., S. Goodbourn, and M. K. Offermann. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J. Virol. 72:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]