Abstract
Coronavirus replicase polyproteins are translated from the genomic positive-strand RNA and are proteolytically processed by three viral proteases to yield 16 mature nonstructural proteins (nsp1 to nsp16). nsp4 contains four predicted transmembrane-spanning regions (TM1, -2, -3, and -4), demonstrates characteristics of an integral membrane protein, and is thought to be essential for the formation and function of viral replication complexes on cellular membranes. To determine the requirement of nsp4 for murine hepatitis virus (MHV) infection in culture, engineered deletions and mutations in TMs and intervening soluble regions were analyzed for effects on virus recovery, growth, RNA synthesis, protein expression, and intracellular membrane modifications. In-frame partial or complete deletions of nsp4; deletions of TM1, -2, and -3; and alanine substitutions of multiple conserved, clustered, charged residues in nsp4 resulted in viruses that were nonrecoverable, viruses highly impaired in growth and RNA synthesis, and viruses that were nearly wild type in replication. The results indicate that nsp4 is required for MHV replication and that while putative TM1, -2, and -3 and specific charged residues may be essential for productive virus infection, putative TM4 and the carboxy-terminal amino acids K398 through T492 of nsp4 are dispensable. Together, the experiments identify important residues and regions for studies of nsp4 topology, function, and interactions.
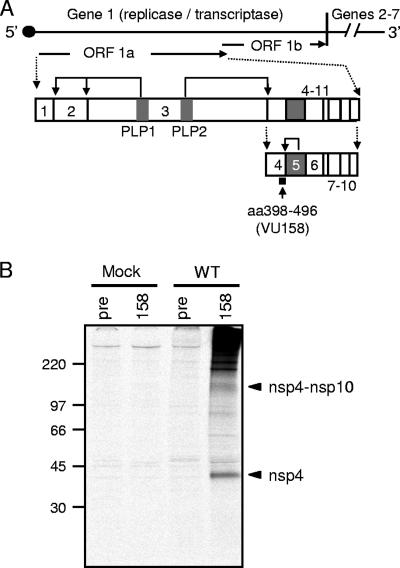
Coronavirus nonstructural proteins (nsp) are hypothesized to function during replication complex formation and viral RNA synthesis; however, the specific functions of many nsp during virus replication remain unknown. To examine the function of these viral proteins and thus better understand the basic replication and pathogenesis strategies of coronaviruses, we use the well-established model coronavirus murine hepatitis virus (MHV). Following the entry of MHV into a cell, the 32-kb positive-strand RNA genome is translated by host cell ribosomes from open reading frame 1 (ORF1), which constitutes approximately 22 kb of the genome and encodes nsp1 to -16. ORF1, which encodes the replicase gene, is comprised of two ORFs (ORF1a and ORF1b) that are connected by a −1 ribosomal frameshift element (Fig. 1A) (2, 5, 6, 26, 29). Translation of ORF1a or the ORF1a-ORF1b fusion results in possible 495-kDa and 803-kDa polyproteins that are processed by three virus-encoded proteases, including two papain-like protease domains (PLP1 and PLP2) in nsp3 and the cysteine protease nsp5 (3CLpro), to yield intermediate precursor products and up to 16 mature nsp (1, 2, 12, 21, 28). nsp4 is processed at its amino terminus by PLP2, and after this initial processing event, nsp4 is detectable as a 150-kDa precursor that includes nsp4 to nsp10 (Fig. 1A and B) (14, 17, 21, 22, 24). 3CLpro (nsp5) mediates processing at the carboxy terminus of nsp4, releasing a mature nsp4 protein with a predicted molecular mass of 56 kDa but an apparent molecular mass of 44 kDa as detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (22, 27). The reason for the difference observed between the predicted and apparent molecular masses of nsp4 has not yet been determined.
FIG. 1.
nsp4 processing and detection in MHV-infected cells using VU158 antiserum. (A) Schematic of nsp4 expression and processing. The approximately 32-kb MHV genome is shown as a line, and the locations of gene 1 (∼22 kb) and genes 2 to 7 (∼9.5 kb) are indicated. Gene 1 is composed of two ORFs (arrows labeled ORF 1a and ORF 1b) connected by a −1 ribosomal frameshift element. The translation of ORF1a yields an nsp1-to-nsp11 polyprotein, shown as connected boxes with each nsp numbered. Viral proteases (PLP1, PLP2, and nsp5) are shown as gray boxes. Bent arrows show cleavage events mediated by the indicated protease. Cleavage by PLP2 at the amino terminus of nsp4 generates an nsp4-to-nsp10 precursor with an approximate molecular mass of 150 kDa. Cleavage at the carboxyl terminus of nsp4 is mediated by nsp5. The black square below nsp4 indicates the region of nsp4 that is recognized by VU158 antiserum. (B) Detection of nsp4 in MHV-infected cells using VU158 antiserum. Radiolabeled lysates were prepared from mock-infected (Mock) or wild-type-MHV-infected (WT) DBT-9 cells and immunoprecipitated with nsp4 rabbit polyclonal antiserum (158) or preimmune serum (pre) from the same rabbit. Molecular mass markers (in kilodaltons) are to the left, and arrowheads indicate the mobilities of the nsp4-to-nsp10 precursor (∼150 kDa) and nsp4 (44 kDa).
All nsp tested colocalize at sites of active viral RNA synthesis, referred to as replication complexes and located on virus-induced double membrane vesicles (DMVs), where they likely mediate virus genome replication and subgenomic RNA synthesis (3, 7, 14, 39). nsp4, which is conserved across all coronaviruses, has been proposed to be involved in both the formation of DMVs in infected cells and the organization and function of virus replication complexes (14). This prediction is based in part on evidence from studies of poliovirus and equine arteritis virus (EAV), two positive-strand RNA viruses that induce the formation of DMVs during infection. It has been demonstrated that exogenous expression of the poliovirus transmembrane replicase proteins 2BC and 3A results in the formation of DMVs that are indistinguishable from those detected during wild-type poliovirus infection (32, 37). Additionally, the expression of EAV transmembrane replicase proteins nsp2 and nsp3 has been shown to be necessary and sufficient for the induction of DMVs identical to those seen in wild-type EAV infection (30, 34). The latter evidence is particularly strong due to the fact that EAV, which like coronaviruses belongs to the order Nidovirales, expresses its nsp as polyproteins in a manner analogous to that of coronaviruses.
Sequence analysis of MHV ORF1a predicted regions of hydrophobicity at the carboxy terminus of nsp3 and within nsp4 and nsp6; subsequently, nsp4 and nsp6 were referred to as membrane protein 1 (MP1) and MP2, respectively (26). Recently, bioinformatic and in vitro analyses determined that nsp3 contains four transmembrane-spanning regions (TMs) that mediate the interaction of nsp3 with membranes (23). Equivalent studies to identify transmembrane regions responsible for mediating nsp4 or nsp6 association with membranes have not yet been reported. nsp4 has been shown to associate with the membrane fraction of infected cell lysates and to be resistant to extraction from membranes following Triton X-114 treatment, suggesting that nsp4 is an integral membrane protein (14). Additionally, in vitro analyses showed that, whereas the expression of nsp5 alone results in an active 3CLpro enzyme, the expression of nsp5 with nsp4 and nsp6 confers a requirement for the presence of membranes for protease activity, supporting the hypothesis that nsp4 and nsp6 are integral membrane proteins important for the organization of nsp at replication complexes (27, 31). However, in spite of an increasing understanding of various aspects of nsp4 expression and processing, no biochemical or enzymatic function has been predicted for nsp4, nor has the requirement for nsp4 in virus replication complex activity been tested.
In this study, we examined the requirement for nsp4 in MHV replication. The MHV-A59 reverse-genetics system (40) was used to generate MHV genomic RNAs with truncations, in-frame deletions, and charge-to-alanine amino acid substitutions across the nsp4 coding region. Mutant viruses were tested for their capacity to grow, synthesize viral RNA, and process the ORF1a polyprotein in cultured cells. The results demonstrate that the carboxy-terminal amino acids K398 through T492 of nsp4 are dispensable for virus replication in culture, allowing for replication at a level similar to that of the wild-type virus. In-frame deletions of the predicted TMs show that TM1, -2, and -3 are required for replication in culture but that TM4 is dispensable. Alanine substitution of conserved, charged residues distributed throughout the amino terminus of nsp4 defines critical residues as well as some that are dispensable for replication. Together, the results demonstrate that nsp4 is required for the rescue of infectious virus and likely contains specific regions or residues amino terminal to TM4 that are essential for virus replication. Additionally, these studies show that mutations and deletions in nsp4 result in altered virus growth, RNA synthesis, and protein processing, suggesting that nsp4 functions at several stages in the virus life cycle.
MATERIALS AND METHODS
Wild-type virus and cells.
Recombinant wild-type MHV strain A59 (wild-type; GenBank accession no. AY910861 [11, 36]) was used as the wild-type control for all experiments. Delayed brain tumor cells selected for high-level expression of the MHV receptor carcinoembryonic antigen cell adhesion molecule-1 (DBT-9) (10, 19, 40) and baby hamster kidney-21 cells expressing the MHV receptor (BHK-MHVR) (9, 10, 40) were grown in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS) for all experiments. Medium for BHK-MHVR cells was supplemented with G418 (0.8 mg/ml) to maintain selection for cells expressing the MHVR.
Antisera.
Rabbit polyclonal MHV nsp4 antiserum (VU158) was generated by Cocalico, Inc., using a recombinant partial nsp4 protein as an antigen. Nucleotides (nt) 9918 to 10208 (gene 1 protein amino acids [aa] 3236 to 3333; nsp4 aa 398 to 496) were amplified by reverse transcription (RT)-PCR using purified MHV-A59 genomic RNA as a template. Primer-generated restriction sites (5′ NcoI and 3′ XhoI) were used to subclone the PCR fragment into the pET23d bacterial expression vector (Novagen). An 11-kDa six-histidine-tagged protein was expressed in Escherichia coli BL21 cells, isolated using nickel resin chromatography according to the manufacturer's protocol, and further purified by SDS-PAGE and electroelution (Bio-Rad) as previously described (3). All other polyclonal antisera used for biochemical experiments have previously been described. These antisera include rabbit anti-nsp2 antisera (VU153) (33), rabbit anti-nsp3 antisera (VU164) (15), and rabbit anti-nsp8 antisera (VU123) (4).
Deletion and charge-to-alanine mutagenesis of nsp4.
To delete portions or introduce charge-to-alanine substitutions in the nsp4 coding sequence (ORF1a nt 8721 to 10208), PCR was performed using the MHV-A59 infectious clone fragment B and C plasmids (pCR-XL-pSMART B and pCR-XL-pSMART C) as templates. Fragment B plasmid contains MHV nsp4 nt 8721 to 9555, and fragment C plasmid contains MHV nsp4 nt 9556 to 10208 (40). Deletions were introduced into fragment B and C plasmids using the ExSite/Quick Change mutagenesis kit (Stratagene) and primers that flanked the region to be deleted. Changes to the manufacturer's protocol include the use of Pfu Turbo instead of the ExSite DNA polymerase blend and the following PCR cycle: initial denaturation at 95°C (once), denaturation at 95°C for 30 seconds, annealing at various temperatures depending on the primers for 1 min, and extension at 68°C for 1 minute per kb of the template, repeating the denaturing/annealing/extension steps for a total of 25 cycles. Primers for deletion of all or portions of the nsp4 coding sequence are detailed in Table 1. Primers for charge-to-alanine mutagenesis within the nsp4 coding sequence are detailed in Table 2. All ligated PCR products, which were plasmids B or C lacking nsp4 sequences, were sequenced across the MHV genome-containing regions to ensure that PCR amplification did not introduce any unintended changes into the plasmid.
TABLE 1.
Deletion mutagenesis of MHV nsp4
| Primer name | Sequence | Template | Purpose |
|---|---|---|---|
| Δnsp4CS3b sense | 5′-CGG GGG AAA TTC TCT CTT GCC ACA TCA ATT-3′ | B | Mutagenesis for Δnsp4CS3 |
| Δnsp4CS3b antisense | 5′-TCT GGC TCT GCC TCA AGT CAT TCC CGC TTA-3′ | B | Mutagenesis for Δnsp4CS3 |
| Δnsp4CS3c sense | 5′-AAA GAA ATC AAT AGG CCG CAC TAA TCC TCC-3′ | C | Mutagenesis for Δnsp4CS3 |
| Δnsp4CS3c antisense | 5′-GCA GAG CTC TGG TAT AGT GAA GAT GGT GTC-3′ | C | Mutagenesis for Δnsp4CS3 |
| Δnsp4CS4b sense | 5′-ACA ACT AAA TTC TCT CTT GCC ACA TCA-3′ | B | Mutagenesis for Δnsp4CS4 |
| Δnsp4CS4b antisense | 5′-AAG AGC TCT GCC TCA AGT CAT TCC CGC TTA-3′ | B | Mutagenesis for Δnsp4CS4 |
| Δnsp4CS4c sense | 5′-AAA GAA ATC AAT AGG CCG CAC TAA TCC TCC-3′ | C | Mutagenesis for Δnsp4CS4 |
| Δnsp4CS4c antisense | 5′-GCA GAG CTT CTT ACA GTC TGG TAT AGT GAA-3′ | C | Mutagenesis for Δnsp4CS4 |
| nsp4ΔN1 sense | 5′-CGG GGG AAA TTC TCT CTT GCC ACA TCA ATT-3′ | B | Mutagenesis for nsp4ΔN1 |
| nsp4ΔN1 antisense | 5′-TTA TTT AAT GTT CCT ACC ACA GTT TTA AGA-3′ | B | Mutagenesis for nsp4ΔN1 |
| nsp4ΔN2 sense | 5′-CGG GGG AAA TTC TCT CTT GCC ACA TCA ATT-3′ | B | Mutagenesis for nsp4ΔN2 |
| nsp4ΔN2 antisense | 5′-GAA GTG GTT AGT GAA GGC ATT GTG CGT GTT-3′ | B | Mutagenesis for nsp4ΔN2 |
| nsp4ΔC1 sense | 5′-GCG GCA GTA AGA GAA CAA CCA CAA TGC ATG-3′ | C | Mutagenesis for nsp4ΔC1 |
| nsp4ΔC1 antisense | 5′-TCT GGT ATA GTG AAG ATG GTG TCG GC-3′ | C | Mutagenesis for nsp4ΔC1 |
| nsp4ΔC2 sense | 5′-AGC ATA TAA ACA AGA CAA TGT GGG ATA AAC-3′ | C | Mutagenesis for nsp4ΔC2 |
| nsp4ΔC2 antisense | 5′-TCT GGT ATA GTG AAG ATG GTG TCG GC-3′ | C | Mutagenesis for nsp4ΔC2 |
| nsp4ΔTM1 sense | 5′-CAC AAA TCG GAT ATG CAG TTG CCT TTA-3′ | B | Mutagenesis for nsp4ΔTM1 |
| nsp4ΔTM1 antisense | 5′-CAA CCA TTG TAA CAT TCT ACT AAA AAC-3′ | B | Mutagenesis for nsp4ΔTM1 |
| nsp4ΔTM2 sense | 5′-CGC CGT TAA GGC AAA GAA ATC AAT AGG-3′ | C | Mutagenesis for nsp4ΔTM2 |
| nsp4ΔTM2 antisense | 5′-AAG CTT AAA CGT GCC TTT GGT GAC TAC-3′ | C | Mutagenesis for nsp4ΔTM2 |
| nsp4ΔTM3 sense | 5′-ACT AGT GTA GTC ACC AAA GGC ACG TTT-3′ | C | Mutagenesis for nsp4ΔTM3 |
| nsp4ΔTM3 antisense | 5′-ACA TTG TCT TGT TTA TAT GCT TGT TTT-3′ | C | Mutagenesis for nsp4ΔTM3 |
| nsp4ΔTM4 sense | 5′-CCA TTG CAA ATG CAT AAC AAC ACT TAT-3′ | C | Mutagenesis for nsp4ΔTM4 |
| nsp4ΔTM4 antisense | 5′-GTT TCA AAC CAT GCA TTG TGT TTG TTC-3′ | C | Mutagenesis for nsp4ΔTM4 |
TABLE 2.
Charge-to-alanine mutagenesis of MHV nsp4
| Primer name | Sequencea | Template | Purpose |
|---|---|---|---|
| VUJS11 sense | 5′-ATA GCC AAT GGT GTG CTA AGG GAT GTG-3′ | B | Mutagenesis for VUJS11 |
| VUJS11 antisense | 5′-AAC GGC AAA ACT GGC ATA TAA AGG CAA-3′ | B | Mutagenesis for VUJS11 |
| VUJS12 sense | 5′-GCC GTG TCT GTT ACT GAC GCA TGC TTC-3′ | B | Mutagenesis for VUJS12 |
| VUJS12 antisense | 5′-GGC TAG CAC ACC ATT ATC TAT AAC TTT-3′ | B | Mutagenesis for VUJS12 |
| VUJS13 sense | 5′-CAA GCC AAT GGC CAT ACC TTA TTT AAT-3′ | B | Mutagenesis for VUJS13 |
| VUJS13 antisense | 5′-GGC TAT TAC AGC AAC CAC AAC AGG ACA-3′ | B | Mutagenesis for VUJS13 |
| VUJS14 sense | 5′-TTG GCC TTT ATA ACC CAT GCA TTT GCT-3′ | B | Mutagenesis for VUJS14 |
| VUJS14 antisense | 5′-CAC GGC AAA TCC ATA TCT TAA AAC TGT-3′ | B | Mutagenesis for VUJS14 |
| VUJS15 sense | 5′-GTC GCC TATAACCTGGCTAGTTCAAAC-3′ | B | Mutagenesis for VUJS15 |
| VUJS15 antisense | 5′-GGC AGG AGC CAA AGA ACT ATA CAG AGA-3′ | B | Mutagenesis for VUJS15 |
| VUJS16 sense | 5′-GTG GCC ACT CGC TCT ATG ACC TAC TGC-3′ | B | Mutagenesis for VUJS16 |
| VUJS16 antisense | 5′-AAC GGC CAC AAT GCC TTC ACT AAC CAC-3′ | B | Mutagenesis for VUJS16 |
| VUJS17 sense | 5′-GCC GCC GAG GAG GGT ATC TGC TTT AAT-3′ | B | Mutagenesis for VUJS17 |
| VUJS17 antisense | 5′-GGC ACA TAA ACC AAC CCT GCA GTA GGT-3′ | B | Mutagenesis for VUJS17 |
| VUJS18 sense | 5′-AAG GGC TAT TAA ATA ATA GAA AGC CAA-3′ | C | Mutagenesis for VUJS18 |
| VUJS18 antisense | 5′-GCC GCC GCC TTT GGT GAC TAC ACT AGT-3′ | C | Mutagenesis for VUJS18 |
Nucleotides used to introduce alanine codons are underlined.
Generation of MHV nsp4 mutant viruses.
Viruses containing the engineered mutations within nsp4 were produced using the infectious cDNA assembly strategy for MHV-A59 described by Yount et al. (40) and modified by Denison et al. (11). Briefly, plasmids containing the seven cDNA cassettes of the MHV genome were digested using the appropriate restriction enzymes. Gel-purified restriction fragments were ligated together in a total reaction volume of 50 μl overnight at 16°C. Following chloroform extraction and isopropanol precipitation of ligated DNA, full-length transcripts of MHV cDNA were generated in vitro using the mMessage mMachine T7 transcription kit (Ambion) according to the manufacturer's protocol with modifications. Fifty-microliter reaction mixtures were supplemented with 7.5 μl of 30 mM GTP, and transcription was performed at 40.5°C for 30 min, 37°C for 60 min, 40.5°C for 30 min, 37°C for 30 min, and 40.5°C for 30 min. In parallel, transcripts encoding the MHV N protein were generated from N cDNA. To prepare for electroporation, BHK-MHVR cells were grown to subconfluence, trypsinized, and then washed twice with phosphate-buffered saline (PBS) and resuspended in PBS at a concentration of 1 × 107 cells/ml. From the resuspended cell solution, a 600-μl aliquot of cells was added to N and MHV transcripts in a 4-mm-gap electroporation cuvette. Three pulses of 850 V at 25 μF were delivered to the cuvette with a Bio-Rad Gene Pulser II electroporator. Electroporated cells were then laid over a layer of 1 × 106 uninfected DBT-9 cells in a 75-cm2 flask and incubated at 37°C. Virus viability was determined by cytopathic effect (CPE) (syncytium formation) in the electroporated cell culture. Progeny virus in the culture medium of electroporated cells (passage 0 [P0]) was passaged onto uninfected DBT-9 cells (P1 cells), and the virus released in the culture medium was designated P1 stock. All constructs that did not yield CPE in the first trial were analyzed two additional times, for a total of three independent trials. For each recoverable virus, RNA harvested from P1 cells was RT-PCR amplified and sequenced to confirm the retention of engineered mutations and the lack of additional mutations within the nsp4 coding sequence and the 500 nt flanking nsp4.
RT-PCR, sequencing, and plaque isolation.
Total intracellular RNA was harvested from P1 cells using TRIzol (Invitrogen) according to the manufacturer's protocol and used as a template for RT-PCR. RT was performed using Superscript III reverse transcriptase (Invitrogen) and random hexamers. The nsp4 coding region was then amplified using PCR with primers complementary to nt 7896 to 7913 (sense) and nt 10344 to 10360 (antisense). The resulting amplicons were directly sequenced to confirm the retention of the engineered mutations and the absence of additional mutations within the nsp4 coding region. To isolate mutant clones for analyses, DBT-9 cells in 60-mm plates were infected with P1 stock virus and overlaid with agar. Following a 16-h incubation at 37°C, three independent plaque clones (P2C1, P2C2, and P2C3) were isolated and sequenced across the nsp4 coding region. All experiments described used P2C1 unless otherwise stated.
Viral growth assays.
For viral growth determination, wild-type and nsp4 mutant viruses were analyzed for replication using growth assays as previously described (11). Briefly, DBT-9 cells were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell with wild-type or nsp4 mutant viruses (P2C1 stock). Following a 30-min adsorption at room temperature, the inoculum was removed and the cells were washed three times with PBS. Cells were supplied with warm 10% FCS-DMEM and incubated at 37°C. Samples of medium were collected from 1 to 30 h postinfection (p.i.), and virus titers were determined by plaque assay as described previously (25).
Radiolabeling of viral proteins.
Confluent monolayers of DBT-9 cells in 60-mm cell culture dishes were either infected with virus at an MOI of 0.01 PFU/cell or mock infected with 10% FCS-DMEM. At 5 h p.i., the medium was replaced with 10% FCS-DMEM lacking methionine and cysteine and supplemented with 5 μg/ml actinomycin D (act D). Cells were incubated from 6 to 9 h p.i. with 100 μCi/ml of [35S]methionine-cysteine (Translabel; ICN). At 9 h p.i., cells were lysed in 500 μl of lysis buffer (150 mM sodium chloride [NaCl], 1% NP-40, 0.5% sodium deoxycholate, 50 mM Tris, pH 8.0). Lysates were then centrifuged at 3,500 × g for 5 min to remove cell nuclei, and the supernatant was retained. Immunoprecipitations were performed in a final volume of 1 ml, using protein A-Sepharose beads (Sigma), 50 μl of radiolabeled lysate (derived from approximately 1 × 106 cells), and a 1:500 dilution of polyclonal antisera in lysis buffer. Following an overnight incubation at 4°C, protein-bead conjugates were pelleted by centrifugation at 12,000 × g for 1 min and washed in low-salt lysis buffer (lysis buffer with 150 mM NaCl) followed by high-salt lysis buffer (lysis buffer with 1 M NaCl) and a final low-salt wash. After being rinsed, proteins were eluted by the addition of 2× lithium dodecyl sulfate (LDS) buffer with 1× dichloro-diphenyl-trichloroethan (NuPage; Invitrogen) to the beads, which were heated at 70°C for 10 min prior to electrophoresis. Proteins were resolved by SDS-PAGE in 4 to 12% polyacrylamide gradient Bis-Tris gels (NuPage; Invitrogen) and analyzed by fluorography. A 14C-labeled high-molecular-weight standard (NEB) and full-range rainbow marker (RPN 800; Invitrogen) were used as molecular weight standards.
Metabolic labeling of viral RNA.
DBT-9 cells were either mock infected or infected at an MOI of 0.01 PFU/cell with wild-type or nsp4 mutant viruses. Following a 30-min absorption at 37°C, virus inoculum was removed and cells were washed twice with PBS. Cells were then incubated for various times in prewarmed 10% FCS-DMEM at 37°C. At 30 min prior to the addition of the radiolabel, the medium was replaced with 10% FCS-DMEM containing act D (5 μg/ml). Viral RNA was metabolically labeled in the presence of act D for 4-h intervals using 100 μCi/ml [3H]uridine. To harvest viral RNA, cells were washed twice with PBS and then lysed with 500 μl of lysis buffer. Lysates were centrifuged at 3,500 × g to remove nuclei, and RNA in 50 μl of cytoplasmic extract was precipitated using 5% trichloroacetic acid (TCA). Precipitated RNA was dried onto glass microfiber filters (Whatman) using vacuum filtration, and radioactivity was measured in a liquid scintillation counter (Beckman). Duplicate samples were collected for each virus at each time point, and duplicate TCA precipitations were performed for each sample. Measurements of total viral RNA synthesis were calculated by measuring the area under the curve using ImageJ 1.38j (http://rsb.info.nih.gov/ij).
RESULTS
Detection of nsp4 in MHV-infected cells.
To detect nsp4 (amino acid residues 2838 to 3333 in the MHV ORF1a polyprotein but renumbered 1 to 496 hereinafter for simplicity) during MHV infection, we generated a rabbit polyclonal nsp4 antiserum (VU158) directed against the carboxy terminus of nsp4 (K398 through Q496). VU158 antiserum was used to probe mock-infected and MHV-infected DBT-9 cells by immunoprecipitation (Fig. 1B). Lysates from radiolabeled, infected cells were immunoprecipitated with VU158, and proteins were resolved by SDS-PAGE. A 44-kDa protein consistent with the apparent molecular mass of nsp4 was detected in infected cell lysates (Fig. 1B) but not in lysates from mock-infected cells with VU158 or after immunoprecipitation of lysates from infected cells with preimmune serum from the same animal (Fig. 1B). Additionally, a protein of approximately 150 kDa, corresponding to the estimated size of an nsp4-to-nsp10 precursor product, was detected by VU158 in MHV-infected cells but not in mock-infected cells or MHV-infected cells probed with preimmune serum. These results show that nsp4 antiserum VU158 specifically recognizes nsp4 and nsp4-containing precursors in MHV-infected cells.
Bioinformatic analyses of nsp4 predicts four TMs.
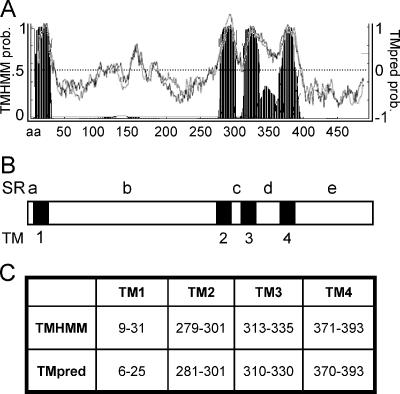
It has been reported that nsp4 exhibits characteristics of an integral membrane protein (14). To predict the location of TMs within nsp4, we analyzed the amino acid sequence of MHV-A59 nsp4 for the probability of TMs using two different programs, TMHMM and TMpred, each designed to search for membrane-spanning sequences (Fig. 2A and B). To predict the location and orientation of TMs within proteins, TMHMM uses a hidden Markov model-based statistical analysis (35) and TMpred uses TMbase, a database of naturally occurring transmembrane proteins, to predict the statistical probability of TMs (20). Using these different algorithms, each program predicted four TMs (TM1, -2, -3, and -4) within MHV nsp4 (Fig. 2A and C).
FIG. 2.
Prediction of TMs in MHV nsp4. (A) TMHMM analysis (www.cbs.dtu.dk/services/TMHMM/) (filled peaks) and TMpred analysis (www.ch.embnet.org/software/TMPRED_form.html) (black and gray graph lines) each predict four transmembrane helices in nsp4. TMpred results show a black line representing the probability (prob.) of TMs if the amino terminus of the protein is cytoplasmic and a lighter, gray line representing the probability of TMs if the amino terminus of the protein is lumenal. TMHMM probability is on the left, TMpred probability is on the right, and the nsp4 amino acid number is on the x axis. The TMHMM threshold is set at 0.5, and the TMpred threshold is set at 0. (B) Schematic of nsp4 indicating the location of predicted TMs (TM1 to -4 [black]) and predicted SRs (SRa to -e [white]). (C) The table shows nsp4 amino acid residues predicted to be involved in TMs by TMHMM and TMpred analyses.
Though the overall numbers and locations of predicted TMs within nsp4 were similar for both the TMHMM and TMpred analyses, the individual predictions generated unique patterns for amino acid residues likely involved in TMs. TMHMM results showed four clear regions within nsp4 that likely span membranes (TM1, -2, -3, and -4) and also indicated a region between predicted TM3 and TM4 that exhibited enough hydrophobic character to generate a peak on the graph but not enough to cross the threshold set for membrane-spanning sequences (Fig. 2A). TMpred results generated two predictions, the first showing the probability of TMs if the amino terminus of the protein is cytoplasmic and the second depicting TM probability if the amino terminus of the protein is lumenal (Fig. 2A). Although both results from TMpred predict the same locations for TMs, the program does not predict a preferred membrane orientation for nsp4. Analysis of TMpred results suggests that more than four regions within nsp4 are potential TMs, crossing the threshold set at 0. However, TMpred results confirm TMHMM results, showing four distinct predicted TMs with amino acid residues that significantly overlap those predicted by TMHMM (Fig. 2C) and a region between TM3 and TM4 that demonstrates high probability of TM character (Fig. 2A).
The prediction of four transmembrane regions within nsp4 would result in five flanking regions that we will refer to as soluble regions (SRa to -e) (Fig. 2C). It is thought that transmembrane helices mediate the membrane association of nsp4, and we predict that the SRs mediate protein interactions or are involved in aspects of virus replication. Bioinformatic analyses of coronaviruses from groups 1, 2, and 3 using the same programs predicted a similar pattern of membrane-spanning regions within the nsp4 coding sequences (data not shown). Though there is limited sequence identity between the individual predicted TMs within nsp4 of different coronavirus groups, the predicted TMs are located at similar positions in the primary sequence, suggesting overall conservation of organization and function.
The carboxy terminus and TM4 of nsp4 are dispensable for MHV replication.
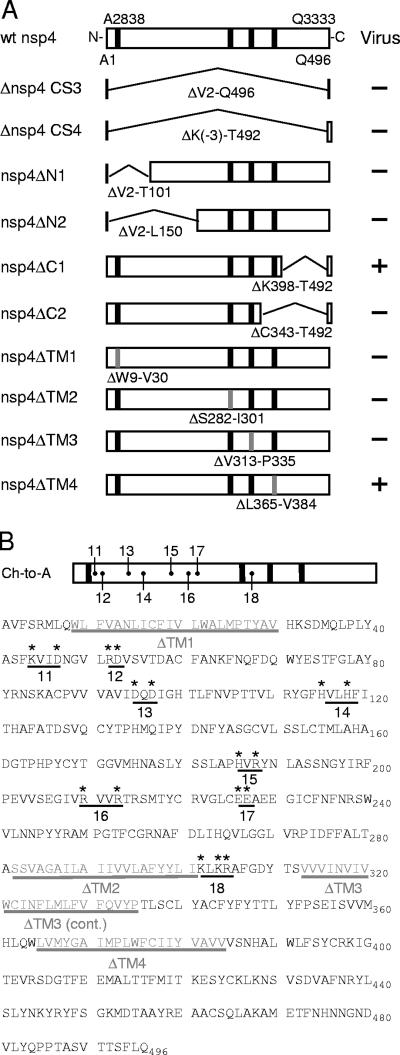
To determine whether nsp4 is essential for virus replication, the nsp4 coding sequence was altered, resulting in in-frame deletions and truncations. Viruses lacking nsp4 were engineered either to remove nsp4 residues V2 through Q496 (Δnsp4 CS3), which retains an nsp3-nsp4 cleavage site, or to remove residues K−3 through T492 (Δnsp4 CS4), which retains an nsp4-nsp5 cleavage site (Fig. 3A). Other viruses were engineered to express truncated forms of nsp4 lacking either V2 through T101 (nsp4ΔN1), V2 through L150 (nsp4ΔN2), K398 through T492 (nsp4ΔC1), or C343 through T492 (nsp4ΔC2) (Fig. 3A). All deletion constructs were designed to maintain the minimal residues proposed to be required for cleavage of nsp4 from the polyprotein by PLP2 and 3CLpro (24, 27). Viral CPE, which for MHV is the formation of syncytia, was detected at 24 h postelectroporation (p.e.) in cells electroporated with recombinant wild-type genomic RNA and at 72 h p.e. in cells electroporated with genomic RNA from the carboxy-terminal truncation mutant nsp4ΔC1.
FIG. 3.
Engineered nsp4 mutants. (A) nsp4 in-frame deletion mutagenesis. The top schematic shows wild-type (wt) nsp4 as a box, with its ORF1a polyprotein amino acid residue and number shown above the box and its nsp4 amino acid residue and number shown below the box. Vertical black bars depict TM1 to -4. Large, internal in-frame deletions of nsp4 amino acids (below the bent lines) are preceded by Δ. Gray vertical bars represent deletions of TMs, with specific amino acid deletions shown below them. The plus sign indicates that infectious virus was recovered, and the minus sign indicates that there was no recovery of infectious virus after multiple attempts. (B) nsp4 charge-to-alanine (Ch-to-A) mutagenesis. The Ch-to-A schematic at the top shows where Ch-to-A substitutions were generated within nsp4. The sequence below the schematic shows nsp4 amino acid residues A1 through Q496, spaced in blocks of 10 aa. Amino acids in gray represent residues deleted from each predicted TM; TMs are underlined in gray and labeled beneath the underlines. Asterisks are shown above each residue replaced with alanine in the nsp4 Ch-to-A mutants. Individual Ch-to-A mutants are indicated by a black line under the clustered, charged residues, with the corresponding numbers below the line representing viruses VUJS11 to -18.
Additionally, to determine if predicted nsp4 TM1, -2, -3, and -4 are critical for virus replication, we engineered viruses lacking individual predicted nsp4 TMs, taking into account the consensus predictions of both bioinformatic analyses (see Fig. 2C). The engineered viruses include nsp4ΔTM1 (ΔW9 to V30), nsp4ΔTM2 (ΔS282 to I301), nsp4ΔTM3 (ΔV313 to P335), and nsp4ΔTM4 (ΔL365 to V384) (Fig. 3A). Cells electroporated with nsp4ΔTM4 genome RNA exhibited virus-induced syncytia. In contrast, cells electroporated with nsp4ΔTM1, nsp4ΔTM2, and nsp4ΔTM3 genome RNA did not exhibit CPE even after 14 days p.e. in three independent experiments. These results demonstrate that residues K398 through T492 (nsp4ΔC1) and, specifically, L365 to V384 (TM4) of nsp4 are dispensable for MHV replication in culture; however, deletions of putative TM1, -2, and -3 were not compatible with productive infection.
Recovery of mutant viruses containing alanine substitutions of conserved, charged residues in the amino-terminal half of MHV nsp4.
It was possible that the deletion of the entire nsp4 coding sequence had unintended effects on the polyprotein or RNA secondary structure critical for replication. To test this possibility, viruses were engineered to contain charge-to-alanine substitutions across the amino-terminal region of nsp4 (Fig. 3B). Residues selected for substitution were situated within a window of four amino acids in the primary sequence and were conserved among group 2 coronaviruses. Eight nsp4 charge-to-alanine mutant viruses were engineered (VUJS11, -12, -13, -14, -15, -16, -17, and -18) (Fig. 3B). Cells electroporated with RNA for VUJS11, -12, -14, -16, and -18 exhibited CPE by 48 h p.e., similar to that of wild-type virus. Cells electroporated with RNA for VUJS17 first exhibited evidence of CPE at 5 days p.e.
Cells electroporated with VUJS13 or VUJS15 RNA did not produce CPE or infectious virus. It has been reported that charge-to-alanine mutagenesis of viral proteins may generate temperature-sensitive mutant viruses (16, 18, 38). nsp4 mutant viruses VUJS13 and VUJS15 could not be recovered at either 30°C or 37°C. Furthermore, no leader-containing subgenomic transcripts could be detected by RT-PCR amplification of RNA harvested from cells electroporated with constructs that did not produce infectious virus (VUJS13, VUJS15, Δnsp4 CS3, Δnsp4 CS4, nsp4ΔN1, nsp4ΔN2, and nsp4ΔC2), providing further evidence that these mutations were profoundly debilitating or lethal for viral replication (data not shown).
nsp4 mutant viruses exhibit defects in viral replication.
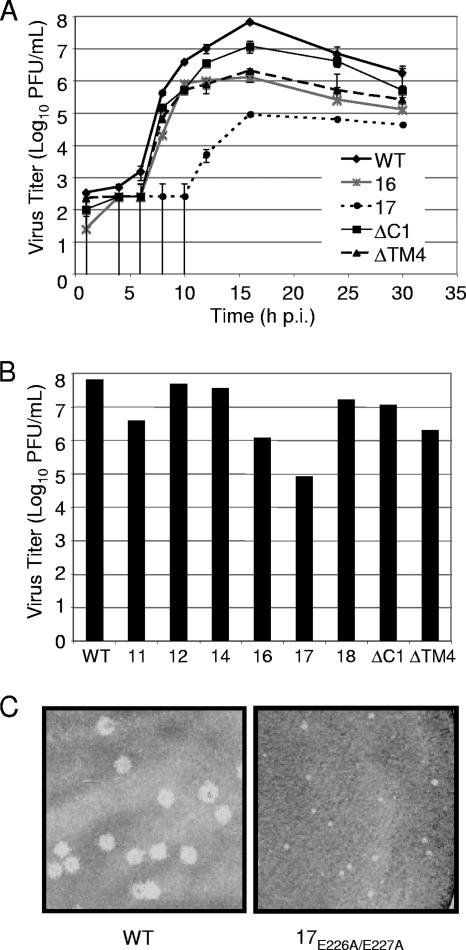
To determine whether nsp4 mutant viruses exhibit replication defects, DBT-9 cells were infected with wild-type virus or nsp4 mutant viruses (VUJS11, -12, -14, -16, -17, and -18, nsp4ΔC1, and nsp4ΔTM4) at an MOI of 0.01 PFU per cell, samples of infected cell culture medium were taken at various times from 1 to 30 h p.i., and virus titers in each sample were determined by plaque assay (Fig. 4A and B). The nsp4 mutant viruses, with the exception of VUJS17, exhibited viral growth kinetics similar to those of wild-type virus, with peak virus release occurring at approximately 16 h p.i. (Fig. 4B). At this time p.i., VUJS12, -14, and -18 and nsp4ΔC1 all produced virus titers within 10-fold of wild-type virus titers (Fig. 4A and B). VUJS11, VUJS16, and nsp4ΔTM4 exhibited a reduction in peak virus titers between 10- and 100-fold the titer of wild-type virus (Fig. 4A and B). VUJS17 demonstrated the greatest impairment, with a prolonged eclipse phase and an approximate 1,000-fold decrease in peak virus titers (Fig. 4A and B). Comparison of plaque phenotypes showed that VUJS11, -12, -14, -16, and -18, nsp4ΔC1, and nsp4ΔTM4 generated plaques similar in size to those of wild-type virus but that VUJS17 exhibited a consistent pinpoint plaque phenotype (Fig. 4C and data not shown). Together, these results indicate that nsp4 mutant viruses have replication defects that range from profound impairment in growth to growth similar to that of the wild type.
FIG. 4.
Growth analysis and plaque morphology of nsp4 mutant viruses. (A) Growth of nsp4 mutant viruses. DBT-9 cells were infected with the indicated viruses at an MOI of 0.01 PFU/cell. Samples of virus supernatants were obtained at 0.5, 4, 6, 8, 10, 12, 16, 24, and 30 h p.i., and virus titers were determined by plaque assay with DBT-9 cells. The x axis shows time (h p.i.), and the y axis shows virus titers (log10 number of PFU/cell). Error bars represent standard deviations between samples. (B) Peak titers of nsp4 mutant viruses. The x axis shows each virus examined, and the y axis shows virus titers (log10 numbers of PFU/cell) at 16 h p.i., the time of peak virus production. WT, wild type; 11, VUJS11; 12, VUJS12; 14, VUJS14; 16, VUJS16; 17, VUJS17; 18, VUJS18; ΔC1, nsp4ΔC1; ΔTM4, nsp4ΔTM4. (C) Plaque morphology of nsp4 mutant viruses. Stocks of the indicated viruses were used to infect DBT-9 cells in six-well plates under 1% agar. At 24 h p.i., cells were fixed. Representative wells were scanned, and images were prepared using Adobe Photoshop CS. An identical region from each well was cropped and magnified to allow better visualization of the plaques. (Left) Plaques from cells infected with wild-type virus; (right) plaques from cells infected with VUJS17 virus.
nsp4 mutant viruses display defects in viral RNA synthesis.
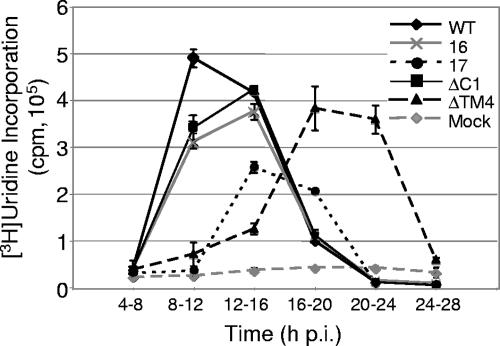
To investigate whether the nsp4 mutant viruses exhibit changes in the timing or levels of viral RNA synthesis, DBT-9 cells were mock infected or infected with wild-type virus, VUJS16, VUJS17, nsp4ΔTM4, or nsp4ΔC1 at an MOI of 0.01 PFU per cell, and viral RNA was radiolabeled from 4 to 28 h p.i. for 4-hour intervals using [3H]uridine in the presence of act D (Fig. 5). RNA was precipitated using TCA, and [3H]uridine incorporation was quantitated using liquid scintillation counting. Analysis of the timing of viral RNA synthesis showed that VUJS16, VUJS17, nsp4ΔTM4, and nsp4ΔC1 each exhibited a delay in reaching peak RNA levels (Fig. 5). Wild-type virus reached peak levels of viral RNA synthesis between 8 and 12 h p.i., whereas VUJS16, VUJS17, and nsp4ΔC1 reached peak levels between 12 and 16 h p.i. nsp4ΔTM4 demonstrated a prolonged delay in exponential growth, reaching peak levels of viral RNA synthesis between 16 and 20 h p.i.
FIG. 5.
RNA synthesis of nsp4 mutant viruses. Cells in six-well plates were either mock infected or infected with the wild type (WT), VUJS16 (16), VUJS17 (17), nsp4ΔC1 (ΔC1), or nsp4ΔTM4 (ΔTM4) at an MOI of 0.01 PFU/cell. Cells were treated with act D for 30 min prior to being labeled. RNA was labeled for the times indicated, cells were lysed, and [3H]uridine incorporation was quantitated by scintillation counting of TCA-precipitable RNA. The x axis shows time (h p.i.), and the y axis shows [3H]uridine incorporation. Data points represent the mean cpm from two individual experiments, and error bars indicate standard deviations between the two experiments.
Delays in the timing of viral RNA synthesis displayed by VUJS16, VUJS17, nsp4ΔTM4, and nsp4ΔC1 were accompanied by decreases in the amount of viral RNA synthesized by each virus throughout the course of infection. For comparison, total levels of viral RNA synthesis (calculated by measuring the area under the curve) for wild-type virus represent 100% of viral RNA synthesized by MHV. No nsp4 mutant virus analyzed reached levels of RNA synthesis equivalent to that of the wild-type virus. Instead, nsp4ΔTM4 synthesized 92.0%, nsp4ΔC1 synthesized 84.6%, VUJS16 synthesized 75.8%, and VUJS17 synthesized 41.1% of wild-type levels of viral RNA (Fig. 5). Though nsp4ΔTM4, nsp4ΔC1, and VUJS16 reached similar levels of viral RNA synthesis, the timing of maximum and total viral RNA synthesis differed between the mutant viruses. These results demonstrate that alterations of different regions or residues within nsp4 have distinct impacts on the timing and levels of viral RNA synthesis.
Mutations in nsp4 result in altered protein expression and processing.
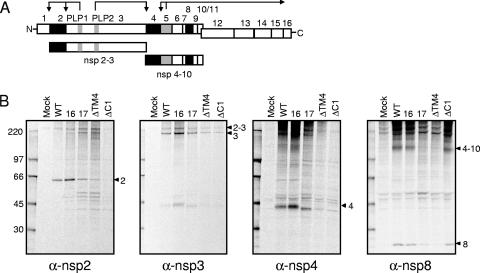
It has been shown that mutations in the ORF1a polyprotein that alter polyprotein processing also affect virus growth and RNA synthesis (8, 10, 15). Recently, a temperature-sensitive virus with a mutation in nsp10, the protein at the carboxy terminus of the nsp4-to-nsp10 precursor, was shown to affect the processing activity of 3CLpro (nsp5) (13). To determine if mutations in nsp4 affected ORF1a-ORF1b polyprotein processing or the activity of PLP1, PLP2, or 3CLpro (nsp5), DBT-9 cells were infected with wild-type or nsp4 mutant viruses, radiolabeled from 6 to 9 h p.i. with [35S]Met/Cys, and cell lysates were immunoprecipitated with antibodies specific for nsp2, -3, -4, or -8. Antiserum specific for nsp2 (anti-nsp2) immunoprecipitated the 65-kDa nsp2 from lysates of cells infected with wild-type virus and VUJS16, VUJS17, nsp4ΔTM4, and nsp4ΔC1 but not from mock-infected cell lysates (Fig. 6B), confirming the intact function of PLP1 in all mutant viruses tested. Immunoprecipitation using antiserum specific for nsp3 (anti-nsp3) detected both nsp3 (210-kDa) and the nsp2-3 precursor (275-kDa) in wild-type and mutant-virus-infected cell lysates but not in mock-infected cell lysates, indicating that PLP2 is active and that processing occurs by PLP2 between nsp3 and nsp4 in the mutant viruses (Fig. 6B). Similarly, antiserum specific for nsp8 (anti-nsp8) detected the 22-kDa nsp8 protein in wild-type and mutant-virus-infected cell lysates but not in mock-infected cell lysates, demonstrating that 3CLpro (nsp5) is active and mediates processing at both the amino and carboxy termini of nsp8. Thus, these results show that PLP1, PLP2, and 3CLpro (nsp5) are functional in the VUJS16, VUJS17, nsp4ΔTM4, and nsp4ΔC1 viruses.
FIG. 6.
Replicase protein expression and processing of nsp4 mutant viruses. (A) The schematic depicts MHV ORF1a-ORF1b nsp1 to nsp16 as boxes. PLP1 and PLP2 in nsp3 and 3CLpro (nsp5) are shown as gray boxes. The arrows above PLP1, PLP2, and 3CLpro depict cleavage events mediated by the indicated protease. nsp2, nsp4, and nsp8 (which are immunoprecipitated in panel B) are shown as black boxes. The nsp2-nsp3 (210-kDa) and nsp4-nsp10 (150-kDa) precursors are depicted below the ORF1a-ORF1b schematic. (B) Cytoplasmic lysates were generated from radiolabeled DBT-9 cells that were either mock infected or infected with wild-type (WT), VUJS16 (16), VUJS17 (17), nsp4ΔTM4 (ΔTM4), or nsp4ΔC1 (ΔC1) virus. Labeled proteins were immunoprecipitated with antiserum specific for nsp2, nsp3, nsp4, and nsp8, as indicated below each gel. α, anti. Proteins were resolved by SDS-PAGE in 4 to 12% polyacrylamide gradient gels and visualized using fluorography. The first (far left) lane of each gel shows molecular mass markers (in kilodaltons), and the arrowheads to the right of each gel points to the indicated protein.
Although PLP1, PLP2, and 3CLpro (nsp5) appear to be active in all nsp4 mutant viruses examined, differences were observed in the patterns of detection of several viral precursor and mature replicase proteins in infected cell lysates (Fig. 6B). The SRb mutant VUJS16 exhibited processing of nsp2, nsp3, nsp2 to nsp3 (nsp2-nsp3), nsp4, nsp4-nsp10, and nsp8 that was identical to that of wild-type virus. Interestingly, the carboxy-terminal truncation mutant, nsp4ΔC1, also demonstrated wild-type processing of nsp2, nsp3, nsp2-nsp3, nsp4-nsp10, and nsp8. As expected, a 44-kDa nsp4 protein was not detected in nsp4ΔC1-infected cell lysates, as the mutant protein lacks the region of nsp4 used to generate anti-nsp4 (Fig. 1A). The SRb point mutant VUJS17 and nsp4ΔTM4 displayed identical patterns of protein detection that differed from those of wild-type virus and the other nsp4 mutant viruses, showing immunoprecipitation of a novel protein of ∼50 kDa by anti-nsp2, the lack of detection of a 44-kDa nsp4 protein by anti-nsp4, minimal detection of the 150-kDa nsp4-to-nsp10 precursor by anti-nsp8, and decreased detection of nsp8 by anti-nsp8. Because both of these mutant viruses contain the anti-nsp4 epitope, the lack of detection of nsp4 and the nsp4-to-nsp10 precursor is likely due to failure of the antibody to recognize this epitope. Both nsp3 and nsp2-nsp3 are detected in VUJS17- and nsp4ΔTM4-infected cell lysates, indicating that the amino terminus of nsp4 is also processed from the ORF1a polyprotein. Thus, the alterations in protein processing observed for VUJS17 and nsp4ΔTM4 are likely due to altered folding or processing of the nsp4-to-nsp10 precursor. Together, the results indicate that removal of the carboxy terminus of nsp4 allows 3CLpro (nsp5) activity and wild-type processing of the nsp4-to-nsp10 precursor, while deletion of TM4 or the SRb mutation of VUJS17 appears to alter the processing of nsp4 to nsp10.
DISCUSSION
Prior to this study, no biochemical or enzymatic function was predicted for coronavirus nsp4, nor was the requirement for nsp4 in virus replication known. In this report, we have shown that MHV nsp4 is required for virus replication, as the nsp4 coding region cannot be deleted from the ORF1a polyprotein and allow for the recovery of infectious virus. Additionally, we demonstrated that the carboxy terminus of nsp4 (K398 through T492) is dispensable for virus replication in culture, whereas deletions of regions within nsp4 containing residues amino terminal to P335 prevented recovery of infectious virus following multiple attempts. These results suggest that nsp4 contains regions or residues in the amino terminus of the protein that are essential for virus replication. To investigate the requirement for individual predicted TMs within nsp4, mutant viruses were generated containing internal in-frame deletions of each predicted TM (TM1, -2, -3, and -4). Results demonstrated that three of the four predicted TMs (TM1, -2, and -3) are essential for recovery of infectious virus, whereas amino acids within TM4 (L365 through V384) are dispensable for virus replication in cell culture. Furthermore, charge-to-alanine substitution of residues within the nsp4 amino terminus identified residues in nsp4 critical for virus replication (D95 and D97 in VUJS13 and H186 and H188 in VUJS15) and allowed for the recovery of several mutant viruses with altered growth and RNA synthesis.
nsp4 membrane topology alterations in nsp4 mutant viruses.
Bioinformatic analysis of MHV nsp4 predicts four TMs and five SRs (SRa to -e) (Fig. 2B). It is hypothesized that the TMs mediate the tight association of nsp4 with membranes and that SRb and SRe, the two largest SRs, mediate protein-protein interactions required for virus replication. Additionally, it is thought that in order for nsp4 to be cleaved from the polyprotein, the amino terminus of the protein must be accessible by PLP2 and the carboxy terminus must be accessible by 3CLpro (nsp5). Taking these considerations into account, along with our experimental results, it is likely that the carboxy-terminal amino acids K398 through T492 of nsp4 are not involved in critical protein-protein interactions during virus replication. Additionally, we conclude that deletion of the carboxy terminus of nsp4 does not significantly alter the function of the amino portion of the protein, as nsp4ΔC1 grows with kinetics and titers similar to those of wild-type virus. Overall, the results suggest that nsp4 contains at least two structurally and functionally distinct domains.
The capacity to remove residues within the putative TM4 in the context of a recoverable virus suggests that either these residues are not required to maintain the proper membrane topology of nsp4 or other residues compensate and span the lipid bilayer. This hypothesis is supported by the bioinformatic prediction that residues between predicted TM3 and TM4 have a high membrane-spanning probability (Fig. 2A). To determine whether other residues might be predicted to compensate for TM4 in nsp4 of nsp4ΔTM4, we analyzed the protein by both the TMHMM and TMpred programs (data not shown). Interestingly, a fourth peak of transmembrane potential is still predicted across the newly juxtaposed residues flanking the deletion of TM4. Though this peak does not cross the threshold of either program to classify a region as membrane-spanning, it does reach up to 75% of the threshold value. Thus, it is possible that juxtaposition of the amino acids flanking those deleted from nsp4ΔTM4 may result in a transmembrane region capable of maintaining interactions with host membranes. This possibility could account for the viability of these mutant viruses in the absence of TM4.
The altered topology of nsp4 in the mutant virus nsp4ΔTM4 has implications not only for nsp4 but also for other nsp downstream of nsp4. It is unknown whether nsp4 insertion in membranes occurs co- or posttranslationally. Because nsp4 is part of a 150-kDa precursor product, nsp4-nsp10, alterations in the membrane topology of nsp4, determining which part of the protein is cytosolic and which part is lumenal, would result in the altered orientation of nsp5 to -10. This change could position the viral proteinase 3CLpro (nsp5) on the opposite side of the membrane relative to its wild-type orientation, affecting both viral protein processing and replication. We did not observe a dramatic decrease in the viral growth of the mutant virus nsp4ΔTM4, suggesting that the basic topology of nsp4 was maintained by amino acids flanking TM4. However, although nsp4ΔTM4 grew with kinetics similar to those of wild-type virus, maximum viral RNA synthesis was delayed longer than any of the other mutant viruses tested, indicating that the deletion of TM4 did alter nsp4 contributions to replication. This change in viral RNA synthesis was associated with a change in the pattern of replicase protein processing, with decreased detection of nsp4, nsp4-nsp10, and nsp8, suggesting that the deletion of TM4 disrupts the normal processing of the nsp4-to-nsp10 precursor. Together, these results raise the possibility that appropriate processing of the nsp4-to-nsp10 precursor serves a role in the timing of viral RNA synthesis.
nsp4 SRb during virus replication.
Mutations in SRb of nsp4 produced mutant viruses with dramatically impaired phenotypes, ranging from viruses that could not be recovered (VUJS13 and VUJS15) to viruses with significant reductions in viral yield and RNA synthesis (VUJS16 and VUJS17). These data support the hypothesis that SRb consists of a functional domain of nsp4 likely involved in protein-protein or protein-RNA interactions. It is possible that the alanine substitutions made within nsp4 of VUJS13 and VUJS15 abolished key protein interactions required for virus replication or altered the structure of nsp4 so that it could no longer accomplish a key function. Alanine substitutions within nsp4 of VUJS16 resulted in the recovery of a mutant virus with slightly decreased growth titers compared to those of wild-type virus and delayed viral RNA synthesis levels that almost reached wild-type levels. These same defects in virus replication were not observed for VUJS17. Instead, VUJS17 exhibited an approximately 1,000-fold decrease in virus yield and a delay in viral RNA synthesis levels that reached only approximately half the synthesis levels of wild-type virus. It was expected that, because the clustered, charged residues targeted for alanine substitution in VUJS16 and VUJS17 were close in proximity in the primary sequence of nsp4, the mutant viruses recovered would have similar phenotypes. It is likely that the differences observed between the two mutant viruses are due to changes in the overall structure of the protein necessary to mediate the function of the SRb domain. Thus, substitutions in VUJS17 likely altered the structure of nsp4 more significantly than substitutions in VUJS16, thereby reducing or inhibiting protein interactions to a greater extent. This conclusion is also supported by our protein processing data (Fig. 6B), in which VUJS16 replicase protein expression was indistinguishable from that in wild-type virus, while VUJS17 displayed decreased expression of nsp4, nsp4-nsp10, and nsp8, similar to that of nspΔTM4.
The experiments in this study represent an in-depth genetic analysis of MHV nsp4, a previously unexplored component of the coronavirus replicase. We have demonstrated a critical requirement for nsp4 in virus replication and have shown that mutations and deletions in nsp4 alter the kinetics and peak growth of MHV, the timing and extent of viral RNA synthesis, and likely the processing of the nsp4-to-nsp10 precursor. Because nsp4 is known to have integral membrane characteristics but no predicted direct enzymatic functions, it is reasonable to propose that its involvement in viral RNA synthesis is via functions in membrane modification, replication complex formation, and protein interactions. While it remains possible that nsp4 may serve a direct role in viral RNA synthesis, we favor the hypothesis that the deletions and mutations in nsp4 ultimately disrupt the optimal formation and function of MHV replication complexes via altered membrane association, protein interactions, or replicase protein processing. The panel of mutant viruses generated in this study will provide powerful tools to further define the precise mechanisms of nsp4 function during coronavirus replication.
Acknowledgments
We thank the Vanderbilt Center for Structural Biology for assistance with bioinformatic analyses. We express our appreciation to Lance Eckerle, Michelle Becker, Rachel Graham, and Adam Sperduto for critical advice and reviews of the manuscript.
This work was supported by Public Health Service award T32 CA009385 (J.S.S.) from the National Institutes of Health, Public Health Service award R01 AI50083 (M.R.D.) from the National Institute of Allergy and Infectious Disease, and the Elizabeth B. Lamb Center for Pediatric Research.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. C. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 67:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., C. J. Pachuk, A. F. H. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. M. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockway, S. M., and M. R. Denison. 2005. Mutagenesis of the murine hepatitis virus nsp1-coding region identifies residues important for protein processing, viral RNA synthesis, and viral replication. Virology 340:209-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W., and R. S. Baric. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, W., V. J. Madden, C. J. Bagnell, and R. S. Baric. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228:318-332. [DOI] [PubMed] [Google Scholar]
- 11.Denison, M. R., B. Yount, S. M. Brockway, R. L. Graham, A. C. Sims, X. Lu, and R. S. Baric. 2004. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 78:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson, E. F., A. C. Sims, R. L. Graham, M. R. Denison, and R. S. Baric. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 81:6356-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 79:13399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanley, K. A., J. J. Lee, J. E. Blaney, Jr., B. R. Murphy, and S. S. Whitehead. 2002. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78:13600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassett, D. E., and R. C. Condit. 1994. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc. Natl. Acad. Sci. USA 91:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2); plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219-225. [PubMed] [Google Scholar]
- 20.Hoffman, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 21.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 74:7911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanjanahaluethai, A., and S. C. Baker. 2001. Processing of the replicase of murine coronavirus: papain-like proteinase 2 (PLP2) acts to generate p150 and p44. Adv. Exp. Med. Biol. 494:267-273. [DOI] [PubMed] [Google Scholar]
- 23.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanjanahaluethai, A., D. Jukneliene, and S. C. Baker. 2003. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 77:7376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. C., R. A. Spence, P. F. Currier, X. T. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, H.-J., C.-K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. LaMonica, J. Tuler, A. Bagdzhadhzyan, and M. M. C. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, X. T., Y. Q. Lu, and M. R. Denison. 1996. Intracellular and in vitro translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, Y., X. Lu, and M. R. Denison. 1995. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J. Virol. 69:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pachuk, C. J., P. J. Bredenbeek, P. W. Zoltick, W. J. Spaan, and S. R. Weiss. 1989. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology 171:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinon, J., R. Mayreddy, J. Turner, F. Khan, P. Bonilla, and S. Weiss. 1997. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology 230:309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 35.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 36.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, R. S., N. Nguyen, H. Zhou, and H. Jin. 2002. Clustered charge-to-alanine mutagenesis of human respiratory syncytial virus L polymerase generates temperature-sensitive viruses. Virology 302:207-216. [DOI] [PubMed] [Google Scholar]
- 39.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]