Abstract
Replication fidelity of RNA virus genomes is constrained by the opposing necessities of generating sufficient diversity for adaptation and maintaining genetic stability, but it is unclear how the largest viral RNA genomes have evolved and are maintained under these constraints. A coronavirus (CoV) nonstructural protein, nsp14, contains conserved active-site motifs of cellular exonucleases, including DNA proofreading enzymes, and the severe acute respiratory syndrome CoV (SARS-CoV) nsp14 has 3′-to-5′ exoribonuclease (ExoN) activity in vitro. Here, we show that nsp14 ExoN remarkably increases replication fidelity of the CoV murine hepatitis virus (MHV). Replacement of conserved MHV ExoN active-site residues with alanines resulted in viable mutant viruses with growth and RNA synthesis defects that during passage accumulated 15-fold more mutations than wild-type virus without changes in growth fitness. The estimated mutation rate for ExoN mutants was similar to that reported for other RNA viruses, whereas that of wild-type MHV was less than the established rates for RNA viruses in general, suggesting that CoVs with intact ExoN replicate with unusually high fidelity. Our results indicate that nsp14 ExoN plays a critical role in prevention or repair of nucleotide incorporation errors during genome replication. The established mutants are unique tools to test the hypothesis that high replication fidelity is required for the evolution and stability of large RNA genomes.
Viruses possessing RNA genomes have been proposed to replicate and evolve near the maximum limit of genetic variability (9, 10, 12, 17). The replication of RNA virus genomes occurs within constraints imposed by a requirement to generate quasispecies diversity important for adaptation to changing environments (27, 41), while simultaneously preventing population extinction due to excessive mutation (5). This precarious balance is thought to be maintained by interplay between RNA polymerase fidelity and error repair by RNA recombination since proofreading activities have not been identified in RNA viruses. Recent studies with poliovirus indicate the “window” of replication fidelity that maintains quasispecies diversity while preventing lethal mutagenesis may be as little as twofold (27, 40). In that background, it is unknown how the largest viral RNA genomes, such as those of coronaviruses (CoVs), are faithfully maintained. At 27 to 32 kb, the positive-sense RNA genomes of CoVs (Fig. 1A), including murine hepatitis virus (MHV) and severe acute respiratory syndrome CoV (SARS-CoV), are the largest known RNA genomes of any virus (14, 15). With the exception of toroviruses and roniviruses, which are classified with CoVs in the order Nidovirales, the RNA genomes of CoVs are up to twice as large as the next-largest nonsegmented RNA genomes of any virus family (15). In addition, CoVs use RNA recombination to generate subgenomic mRNAs (sg mRNAs) (30, 31) and possibly as a genome repair mechanism (15, 34). Thus, a paramount question is whether a specific mechanism is responsible for the evolution and maintenance of the large RNA genomes of CoVs.
FIG. 1.
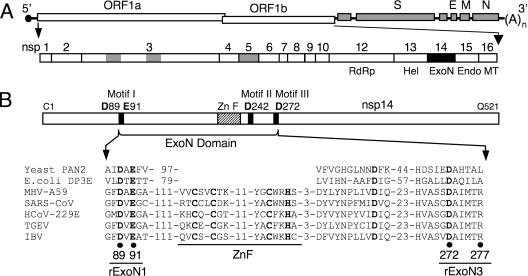
MHV genome organization and nsp14 exoribonuclease motifs. (A) MHV genome organization and ORF1a/b replicase polyprotein expression. The MHV genome is a 31.3-kb positive-sense RNA molecule that is capped (dark circle) and polyadenylated. The genes are indicated for the replicase (ORF1a and ORF1b), spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. ORF1b is accessed by ribosomal frameshift. The ORF1a/b polyprotein is translated from input genome RNA and processed into 16 mature nsps by three virus-encoded proteinases (gray). nsps 12 to 16 have predicted or demonstrated activities as described in the text. Hel, helicase; Endo, endoribonuclease; MT, methyltransferase; (A)n, polyadenylate tract. (B) Organization of nsp14 and partial sequence alignment of representative CoV nsp14 sequences with the sequence of Saccharomyces cerevisiae PAN2 (SwissProt accession number P53010), a poly(A)-specific ExoN, and Escherichia coli DNA polymerase III epsilon subunit (DP3E), the proofreading exonuclease subunit of the replicative DNA polymerase (SwissProt P03007). GenBank accession numbers for the full-length CoV genomes are as follows: MHV-A59, AY910861; SARS-CoV (Urbani strain), AY278741; HCoV-229E, NC_002645; transmissible gastroenteritis virus (TGEV), NC_002306; infectious bronchitis virus (IBV)-Beaudette, NC_001451. Sequence alignment was adapted from Snijder et al. (34). Active-site residues of conserved motifs I to III of the DEDD superfamily (45) are indicated by bold text and amino acid position in MHV nsp14. A predicted zinc finger domain (ZnF) is located between motifs I and II in the viral sequences. Residues replaced with alanine are indicated by black circles for MHV mutants rExoN1 (ExoN motif I) and rExoN3 (ExoN motif III) (see Table 1 for details).
CoVs express and process up to 16 nonstructural proteins (nsps 1 to 16) directly from the input positive-sense RNA genome (Fig. 1A). Among these nsps are experimentally confirmed or predicted proteinases, an RNA primase (18), an RNA helicase (nsp13) (21, 33), and an RNA-dependent RNA polymerase ([RdRp] nsp12) (7). Additionally, novel functions for an RNA virus have been demonstrated in vitro for nsp14 (exoribonuclease [ExoN]) (24) and nsp15 (endoribonuclease) (4, 20) and predicted for nsp16 (RNA methyltransferase) (34). The roles of nsp14 to nsp16 in infection remain to be determined, but these proteins have been predicted to function coordinately to modify viral RNA (34).
Homologs of nsp14 are encoded by all CoVs, and each has 3′-to-5′ exonuclease motifs I (DE residues), II (D), and III (D) within the amino-terminal ∼280 amino acids of the 521-amino-acid protein (Fig. 1B) (25, 34, 45). Bacterially expressed SARS-CoV nsp14 has been shown to have 3′-to-5′ ExoN activity in vitro, and alanine substitutions for the DE-D-D residues profoundly impaired or abolished this activity (24). Further, alanine substitutions for the ExoN active-site residues blocked recovery of recombinant human CoV 229E (HCoV-229E) and resulted in limited viral RNA synthesis in cells electroporated with genome RNA (24). Based on these results, it was concluded that nsp14 is a 3′-to-5′ ExoN requiring the conserved DE-D-D residues for activity and that nsp14 ExoN activity is required for productive CoV replication.
Using genetically engineered mutants of MHV, we demonstrate that nsp14 ExoN is not required for virus replication but is required for efficient RNA synthesis and for faithful replication of the viral RNA genome. The nsp14 mutants described in this report provide a model to experimentally investigate the impact of replication fidelity on the evolution of large viral RNA genomes.
MATERIALS AND METHODS
Cells.
Murine delayed brain tumor (astrocytoma) cells sorted for high-level expression of the MHV receptor carcino-embryonic cell adhesion molecule-1 (DBT-9) (6, 16, 44) and transgenic baby hamster kidney-21 cells constitutively expressing the MHV receptor (BHK-MHVR cells) (6, 44) were grown and maintained as described previously (35).
Mutagenesis of infectious cDNA fragment F plasmids.
The entire MHV-A59 genome was cloned as seven cDNA fragments, A to G, each in a separate plasmid (44). The F fragment plasmid designed for recovery of the recombinant wild-type (WT) virus VUSS3 (35) was used as a template for site-directed PCR-based mutagenesis to replace charged residues of the nsp14 ExoN domain with alanines as detailed in Table 1. For each construct, a 2.1-kbp BssHII-HpaI restriction fragment containing the mutation was substituted into the parental plasmid, and the sequence of the PCR-generated fragment was verified.
TABLE 1.
Engineered mutations in ExoN mutant viruses
| Virus | Engineered mutationsa | Amino acid substitutionsb |
|---|---|---|
| rExoN1 | A18426C, U18427A | Asp89Ala |
| A18432C, A18433U | Glu91Ala | |
| rExoN3 | A18975C, U18976G | Asp272Ala |
| C18989G, G18990C | Arg277Ala |
Nucleotide positions refer to the MHV-A59 complete genome sequence (GenBank accession no. AY910861).
Amino acid positions refer to nsp14 sequence (ORF1a/b polyprotein residues 5985 to 6505).
Engineering and recovery of viruses.
Mutant recombinant ExoN motif I (rExoN1) and rExoN3 viruses were generated by an infectious cDNA clone assembly strategy as previously described (35, 44). Briefly, mutant cDNA fragment F was ligated with the six other WT fragments to generate a full-length infectious cDNA of the MHV-A59 genome that was used as a template for in vitro transcription of capped and polyadenylated genome RNA. These transcripts were mixed with short transcripts for expression of N protein and electroporated into BHK-MHVR cells. Electroporated cells were seeded onto DBT-9 cells and incubated at 37°C until virus-induced syncytia involved more than 20% of the monolayer (48 h postelectroporation for rExoN1 and rExoN3). Clarified supernatants obtained after pelleting cell debris at 700 × g for 10 min were used as passage zero (p0) virus stocks. The WT virulent recombinant virus VUSS3 (35) was generated using the same approach and was included as a control in all assays.
Plaque isolation and passage of viruses.
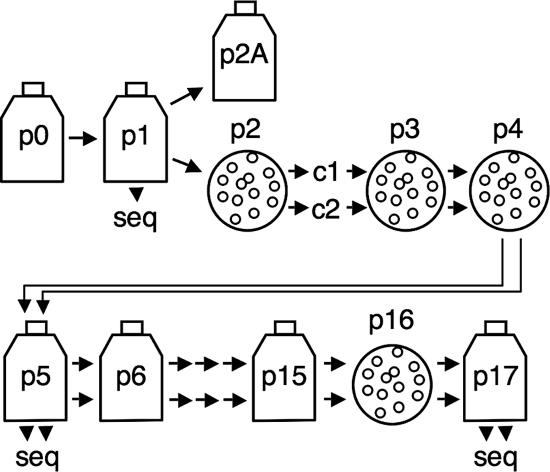
WT and mutant viruses were passaged and plaques were isolated as illustrated in Fig. 3. For plaque isolation, virus stocks were serially diluted in gel saline (0.3% gelatin in phosphate-buffered saline), and DBT-9 cells in six-well plates were infected by application of 200 μl/well. Virus was adsorbed for 30 min at room temperature. Cells were overlaid with 1.0% agar in growth medium and incubated at 37°C for 22 to 28 h. Gel plugs above individual, well-isolated plaques were transferred to 200 μl of gel saline and vortexed for 30 s. The homogenate was diluted and used to infect cells for subsequent rounds of plaque isolation. Alternatively, 30 to 100 μl was used to infect cells in 25- to 150-cm2 flasks to amplify virus.
FIG. 3.
Virus isolation and passage strategy. Flasks indicate population passage, and circles indicate plaque isolation. From p2 forward, two plaque clones were isolated and passed in parallel, indicated by double arrows. Passages where RNA from infected cells was purified and used for RT-PCR and determination of viral sequences are denoted by seq. Passages between p0 and p6 were conducted at an MOI of ≤0.1 PFU/cell. Passages between p6 and 15 were performed at unknown MOIs.
Growth analysis.
DBT-9 cells in 60-mm dishes were infected with WT or mutant viruses in growth medium at the multiplicity of infection (MOI) indicated in the figure legends. After a 30-min adsorption at room temperature, inoculum was removed, cells were washed twice in Dulbecco's modified Eagle medium, and growth medium was added. The first sample was obtained immediately after the addition of growth medium. Cells were then incubated at 37°C, and additional samples of culture medium were collected at the times indicated in the figure legends. Virus titers were determined as described below.
Determination of virus titers and analysis of plaque phenotype.
Virus titers were determined by plaque assay on DBT-9 cells as previously described (16, 22) with the following modifications to enhance visualization of plaques. At 22 to 24 h postinfection (hpi) for WT or 28 to 30 hpi for mutants, cells under an agar-growth medium overlay were fixed with 4% formaldehyde at room temperature. The overlay was then discarded, and the fixed cell monolayer was air dried before plaques were counted. For imaging of relative plaque size, representative wells were scanned using an Epson Perfection 4870 photo scanner, and images were prepared using Adobe Photoshop CS.
Kinetic analyses of viral RNA synthesis.
To monitor the kinetics of viral RNA synthesis, DBT-9 cells in 60-mm dishes were mock infected or infected at an MOI of 0.1 or 3. Virus was absorbed and washed as for viral growth analysis. Cells were then incubated in growth medium at 37°C until 30 min prior to labeling, at which time the medium was replaced with growth medium supplemented with 10 μg/ml actinomycin D (ActD), and cells were incubated for 30 min at 37°C. After this pretreatment period, [3H]uridine was added to a final concentration of 20 μCi/ml, cells were incubated at 37°C for intervals of 4 h (MOI of 0.1) or 2 h (MOI of 3), beginning at the times indicated in the figure legends. At the end of each labeling period, cells were lysed in cell lysis buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and 50 mM Tris, pH 8.0). Nuclei were removed by centrifugation at 14,000 × g for 3 min. RNA in 10% of each lysate was precipitated with cold 5% trichloroacetic acid (TCA) onto glass microfiber filters (Whatman) that were then washed twice in fresh 5% TCA and twice in 95% ethanol and dried using vacuum filtration. Incorporation of radiolabel was quantitated by liquid scintillation counting.
Electrophoretic analysis of viral RNA.
For electrophoretic analysis of RNA, cells in 60-mm dishes were mock infected or infected at an MOI of 3 using adsorption and wash conditions as for RNA kinetics analyses. Cells were pretreated with ActD from 10.5 to 11 hpi, after which [3H]uridine was added to a final concentration of 50 μCi/ml, and cells were incubated for 2 h. At 13 hpi, total intracellular RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. From the total volume of each RNA sample, 2.5% was denatured using glyoxal loading dye (Ambion) at 50°C for 30 min and resolved by electrophoresis in 1% agarose gels. Fivefold less of the RNA from VUSS3-infected cells was also analyzed. After electrophoresis, gels were incubated in 100% methanol for 1 h, in 1% 2,5-diphenyloxazole in methanol for 1 h, and in water for 2 h. Gels were then dried by vacuum filtration at 50°C and exposed to X-ray film.
Reverse transcription-PCR (RT-PCR) analysis.
To detect MHV RNA2 and cellular β-actin mRNA, 1% of each radiolabeled RNA sample from mock-infected cells and cells infected with mutants or 0.25% of the sample from VUSS3-infected cells was reverse transcribed using Superscript III reverse transcriptase (Invitrogen) and random hexamers at 50°C for 60 min. To amplify RNA2, 10% of each cDNA was denatured at 95°C for 2 min and then amplified by PCR using Easy-A High-Fidelity PCR cloning enzyme (Stratagene), a positive-sense leader-specific oligodeoxynucleotide primer (24CGTACCCTCTCAACTCTAAAAC45), and a negative-sense body-specific primer (22144GGAATCCAGTGTCTGGAT22127) for 40 cycles at 95°C for 45 s, 50°C for 30 s, and 72°C for 30 s. Mouse β-actin mRNA was amplified using the same protocol except for the following modifications: 5% of each cDNA was used; the sense and antisense primers were TAAAACGCAGCTCAGTAACAGTCCG and TGGAATCCTGTGGCATCCATGAAAC, respectively; the annealing temperature was 60°C; and 30 cycles were used. A total of 15% (RNA2) or 5% (actin) of each PCR mixture was resolved adjacent to a 100-bp DNA ladder (New England BioLabs) in a 1.2% or 1.5% agarose gel in Tris-acetate-EDTA buffer. DNA was visualized by ethidium bromide staining and photographed using an Eagle Eye II imaging system (Stratagene).
Sequence analysis of viral RNA.
To obtain viral RNA for sequence analysis, cells in 25-cm2 flasks were infected at an MOI of ≤0.1 PFU/cell. When >10% of the cells were involved in syncytia, total intracellular RNA was extracted using TRIzol reagent according to the manufacturer's protocol. Reverse transcription was performed using Superscript III reverse transcriptase and random hexamers at 50 to 55°C for 30 to 60 min. The three regions of the MHV genome shown in Fig. 5 were amplified by 30 to 40 cycles from first-strand cDNA as multiple subregions using Easy-A High-Fidelity PCR cloning enzyme (Stratagene) or Elongase (Invitrogen) DNA polymerases and oligodeoxynucleotides specific for the MHV-A59 genome. Amplicons were gel purified and directly analyzed by automated DNA sequencing. Sequences of PCR and sequencing primers are available upon request.
FIG. 5.
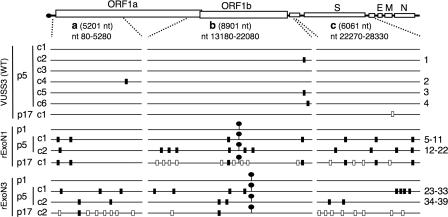
Distribution of mutations in WT, rExoN1, and rExoN3 mutants. Schematic of MHV RNA genome shows the regions a, b, and c that were amplified by RT-PCR and sequenced for each WT and ExoN mutant virus. The 5′ and 3′ terminal nucleotides of the regions are indicated, as is the length in nucleotides. Lollipops indicate locations of engineered ExoN mutations. Black rectangles on each genome indicate additional mutations identified in p5 viruses. White rectangles indicate mutations identified in p17 but not in p5 viruses. Numbers at right indicate mutations for which details are shown in Table 3. VUSS3p17c1, rExoN1p17c1, and rExoN3p17c2 were derived from VUSS3p5c1, rExoN1p5c1, and rExoN3p5c2, respectively. S, spike; E, envelope; M, membrane; N, nucleocapsid protein.
Statistical analyses.
The mutation counts from the 18 sequenced fragments of WT (120,978 nucleotides [nt] total) and 12 sequenced fragments of rExoN1 and rExoN3 combined (80,652 nt total) were normalized as mutation counts per 10,000 nt and analyzed using Wilcoxon rank-sum or Kruskal-Wallis tests, as indicated below. All tests were two-tailed, and a P value of <0.05 was considered significant. Statistical analyses were performed using STATA 9.1 software (StataCorp, College Station, TX).
Mutation rate estimates.
The mutation rate (μ) was estimated using the following formula: μ = number of mutations/number of nucleotides sequenced/number of replication cycles. Based on growth analyses, we estimated that one replication cycle was equivalent to 8 h for WT or 10 h for rExoN3. The total number of cycles between p0 and p5 was approximately 13 for each virus. As an example, for rExoN3, the mutation rate was calculated as follows: μ = 17 mutations/40,326 nt/13 cycles = 3.2 × 10−5. Our values of μ are estimates of neutral mutation rates since highly deleterious or lethal mutations were selected against, and no changes in growth were apparent during virus passage.
RESULTS
MHV nsp14 ExoN mutants are viable and have stable growth defects.
We sought to determine whether mutants in MHV nsp14 ExoN DE-D-D motifs could be recovered from cDNA and, if so, to determine the effect on virus growth, RNA synthesis, and replication fidelity of genome RNA. The nsp14 coding sequence was engineered to introduce alanine substitutions at conserved active-site residues in ExoN motif I (D89A and E91A in virus rExoN1) or motif III (D272A and R277A in rExoN3) (Fig. 1B and Table 1). All mutations were engineered into the in vitro assembled full-length cDNA background of recombinant WT MHV clone VUSS3 (35), followed by electroporation of DBT-9 cells with in vitro transcribed genome RNA. Characteristic MHV-induced syncytia were visible by 24 h in cells electroporated with rExoN1 or rExoN3 mutant RNA, and clarified culture medium contained infectious virus as determined by plaque assay (Fig. 2C). Virus from the rExoN1 and rExoN3 electroporated cell culture medium (p0) was passaged once (p1) followed by isolation of two independent plaque clones each of rExoN1 and rExoN3 (p2c1 and p2c2, where c1 and c2 are clone 1 and clone 2, respectively). These p2 clones were plaque purified two additional times (p3 and p4) and amplified (p5) in cells (Fig. 3). Sequencing of bulk RT-PCR products amplified from viral RNA from cells infected with p1 and p5 viruses confirmed the presence of the engineered mutations in all rExoN1 and rExoN3 viruses. Together, these results demonstrate that intact ExoN active-site residues are not required for MHV replication in cell culture.
FIG. 2.
Replication and plaque morphology of rExoN1 and rExoN3 mutants. (A) Multicycle growth analysis of ExoN mutants. DBT-9 cells were infected with the indicated viruses at an MOI of 0.01 PFU/cell. Viruses are identified in the text. Supernatant samples were obtained at 0.5, 6, 8, 10, 12, 16, 24, 30, 36, 42, and 48 hpi, and virus titers were determined by plaque assay. Mean titers and standard deviations from duplicate samples are indicated. (B) Single-cycle growth analysis of rExoN1 mutants. DBT-9 cells were infected with the indicated viruses at an MOI of 3. Supernatant samples were obtained at 0.5, 4, 6, 8, 10, 12, 16, and 24 hpi, and virus titers were determined by plaque assay. Mean titers and standard deviations from duplicate samples are indicated. (C) Plaque morphology. p0 and p15 stocks of the indicated viruses were diluted and used to infect DBT-9 cells for 24 to 28 h, followed by fixation with 4% formaldehyde. Representative wells were scanned and images were prepared using Adobe Photoshop CS2.
The growth of the rExoN1 and rExoN3 mutants in culture was compared with recombinant WT VUSS3 that was identically recovered, cloned, and passaged in parallel with the mutant viruses. Infection was performed in DBT-9 cells at an MOI of 0.01 PFU per cell to test for subtle differences during multicycle growth. The rExoN1 p5c1 (rExoN1p5c1) and rExoN1p5c2 stocks were also compared with the WT at an MOI of 3 PFU/cell in single-cycle growth assays. At an MOI of 0.01, the growth kinetics of viruses rExoN1 and rExoN3 were indistinguishable (Fig. 2A). The growth kinetics of the two clones of rExoN1 also were indistinguishable at an MOI of 3 (Fig. 2B). Compared to WT, the mutant viruses exhibited delayed growth as well as peak titer reductions of 1.4 to 2.2 log10 at the low MOI and 0.5 to 0.8 log10 at the high MOI. In addition, there was no detectable difference in the growth of the p0 and p5 viruses (Fig. 2A). These results demonstrated that the rExoN1 and rExoN3 mutants were defective in growth compared to WT and that virus passage and plaque isolation did not alter growth fitness of the mutants.
Plaques of WT virus had a consistent size that was maintained upon passage. In contrast, plaques of rExoN1 and rExoN3 from multiple passages showed extreme heterogeneity, ranging from pinpoints to nearly WT size (Fig. 2C and data not shown). While the proportion of small, medium, and large plaques appeared to change during passage for rExoN1 and rExoN3, plaque passage of virus from small, medium, and large plaques all resulted in heterogeneous plaque size. These results suggested that genetic variants were generated even during multiple rounds of plaque isolation.
ExoN mutants have RNA synthesis defects.
To determine whether RNA synthesis of the ExoN mutant viruses was altered during multicycle and single-cycle infections, cells were infected at an MOI of 0.1 or 3, and viral RNA was metabolically labeled for 4-h or 2-h intervals, respectively, with [3H]uridine in the presence of ActD to inhibit cellular DNA-dependent RNA synthesis. Peak levels of viral RNA synthesis for rExoN1 and rExoN3 were decreased by 90 and 89%, respectively, compared to WT at the low MOI (Fig. 4A) and by 81 and 75% at the high MOI (Fig. 4B). In addition, RNA synthesis from the mutants was delayed by about 2 to 4 h, depending on the MOI. Thus, the ExoN mutants exhibited substantially reduced and delayed RNA synthesis under both growth conditions. The progression of cytopathic effect throughout the entire monolayer for WT and both mutants during these infections suggests that the observed RNA synthesis phenotypes of the mutants are due to intrinsically impaired RNA synthesis.
FIG. 4.
RNA synthesis from rExoN1 and rExoN3 mutant viruses. (A) Kinetics of viral RNA synthesis during multicycle growth. DBT-9 cells were mock infected or infected with the indicated viruses at an MOI of 0.1 PFU/cell. Viral RNA was metabolically labeled with [3H]uridine in the presence of ActD at 4-h intervals that began at 4, 8, 12, 16, 20, 24, 28, 32, 36, and 40 hpi. Total RNA was precipitated with 5% TCA, and incorporation of radiolabel was quantitated by scintillation counting. Mean values and standard deviations from duplicate samples obtained from duplicate infection series are indicated. (B) Kinetics of viral RNA synthesis during single-cycle growth. DBT-9 cells were mock infected or infected with the indicated viruses at an MOI of 3 PFU/cell. Viral RNA was metabolically labeled with [3H]uridine in the presence of ActD at 2-h intervals that began at 3, 5, 7, 9, 11, 13, and 15 hpi. (C) Electrophoretic analysis of RNA replication and transcription products. DBT-9 cells were mock infected (m) or infected with the indicated viruses at an MOI of 3 PFU/cell. Viral RNA was metabolically labeled with [3H]uridine in the presence of ActD from 11 to 13 hpi. Intracellular RNA was isolated, denatured, and resolved by electrophoresis in 1% agarose gels. Labeled RNA species were visualized by fluorography. Genomic RNA (R1) and sg mRNAs (R2 to R7) are indicated. The apparent band migrating between RNA3 and RNA4 in lanes 1, 3, 4, and 5 is most likely due to a physical effect of abundant 28S rRNA. The asterisk indicates that lane 1 received the same RNA sample as lane 5 but fivefold less. Note that all samples were electrophoresed on the same gel, but the image was cropped to remove extraneous lanes between lanes 4 and 5. (D) RT-PCR of MHV RNA2 (top) and cellular β-actin mRNA (bottom). The same RNA samples analyzed in panel C were subjected to RT-PCR, and the products were resolved in 1.2% (top) and 1.5% (bottom) agarose gels in the presence of ethidium bromide. The VUSS3 RNA sample was diluted fourfold prior to reverse transcription. Sizes (in kbp) of bands from a 100-bp DNA ladder are indicated. The predicted sizes of the RNA2-specific and actin-specific amplicons are 437 and 348 bp, respectively. Abbreviations: m, mock; con, negative control PCR for β-actin using water as a template. Images in panels C and D were prepared using Adobe Photoshop CS2.
To compare RNA replication and transcription products from the ExoN mutants and WT, cells were infected at an MOI of 3, and viral RNA was metabolically labeled from 11 to 13 hpi. Electrophoretic analysis of denatured RNA showed that both mutants synthesized genome RNA (RNA1) and sg mRNAs (RNA3 through RNA7), and these had mobilities similar to the corresponding species from WT (Fig. 4C). Synthesis of each RNA species was reduced for the mutants compared to WT (Fig. 4, compare lanes 3 and 4 with lane 5). The largest sg mRNA, RNA2, was not detected from the mutants using this method. However, RNA2 was specifically detected by RT-PCR from cells infected with the ExoN mutants or WT MHV but not from mock-infected cells using multiple different sets of primers specific for the leader-body junction (Fig. 4D, top; also data not shown). As a control, mouse β-actin mRNA was amplified from mock-infected and infected cells (Fig. 4D, bottom). Together, these results confirm that the ExoN mutant viruses synthesized all seven viral RNA species.
ExoN mutants accumulate nonengineered mutations.
The heterogeneity in plaque size of the ExoN mutants suggested genetic variation. To determine whether nonengineered (secondary) mutations were present, we sequenced an 8.9-kb region (Fig. 5, region b) that included open reading frame 1b (ORF1b) from viral RNA for two p5 clones of each ExoN mutant and six p5 clones of WT. ORF1b was selected for initial sequencing because it encodes nsp12 through nsp16, each of which has been demonstrated or predicted to participate in viral RNA synthesis or modification. Analysis of WT virus identified zero mutations in three clones and one mutation in each of three additional clones. In contrast, two to six secondary mutations were identified in ORF1b in each clone of rExoN1 and rExoN3 (Fig. 5 and Table 2). Several mutations in WT, rExoN1, and rExoN3 were verified by sequence analysis of amplicons from independent RT reactions. None of the secondary mutations detected at p5 was detected in the p1 population, with the exception of heterogeneity at specific sites in the nsp13 coding sequence (see below), suggesting that the secondary mutations were generated during passage and fixed by plaque isolation. The observations that each rExoN1p5 and rExoN3p5 clone had multiple mutations that were unique to that clone and that no change in growth fitness was observed over the course of virus passage suggested that the mutations were random and neutral with respect to growth.
TABLE 2.
Mutation counts in WT, rExoN1, and rExoN3 mutant viruses
| Virus and clone | No. of mutationsa
|
|||
|---|---|---|---|---|
| Region a | Region b | Region c | Mean per 10,000 ntb | |
| VUSS3p5 (WT) | ||||
| c1 | 0 | 0 | 0 | |
| c2 | 0 | 1 | 0 | |
| c3 | 0 | 0 | 0 | 0.29 |
| c4 | 1 | 0 | 0 | |
| c5 | 0 | 1 | 0 | |
| c6 | 0 | 1 | 0} | |
| rExoN1p5 | ||||
| c1 | 2 | 2 | 3 | |
| c2 | 1 | 6 | 4 | |
| rExoN3p5 | 4.4 | |||
| c1 | 3 | 4 | 4 | |
| c2 | 2 | 2 | 2} | |
Regions a to c refer to the sequenced regions of the MHV genome shown in Fig. 5. A total of 120,978 nt were sequenced for WT, and 80,652 nt were sequenced for the combined mutant viruses.
Mean values for WT and the combined mutants were obtained by normalizing the mutation count in each region of each clone as mutations per 10,000 nt, summing the normalized values, and then dividing by the number of genome fragments sequenced (18 for WT and 12 for the combined mutants). A significant difference in the means was observed between WT and the combined rExoN1 and rExoN3 mutants (P < 0.001, Wilcoxon rank-sum test).
To test this hypothesis, viral RNA was sequenced across two additional regions of the genome (Fig. 5, regions a and c) that included the 5′ proximal region of ORF1a and the spike glycoprotein gene, resulting in a total of 20,163 nt sequenced for each mutant or WT clone. The number of mutations identified in each region for each virus clone is shown in Fig. 5 and Table 2, and details of each mutation are shown in Table 3. Combining the data from all three regions from the six WT virus clones, four mutations were identified in 120,978 nt. No secondary mutations were identified in p1 populations of rExoN1 or rExoN3. In contrast, 18 and 17 secondary mutations were identified in 40,326 nt for p5 clones of rExoN1 and rExoN3, respectively (Fig. 5 and Table 2). The average mutation count of 4.4 per 10,000 nt in the ExoN mutants was 15-fold greater than in WT (0.29 per 10,000 nt), a difference that is highly statistically significant (P < 0.001, Wilcoxon rank-sum test) (Table 2). In contrast, there is no statistically significant difference in mutation counts between rExoN1 and rExoN3 clones (P = 0.936, Wilcoxon rank-sum test). For rExoN1 and rExoN3 combined, there is no statistically significant difference in mutation counts between the three sequenced regions (P = 0.469, Kruskal-Wallis test). Thus, over five passages, the ExoN mutants accumulated an unexpectedly large number of mutations across the sequenced regions of the genome, and with the exception of clustered mutations 30 to 32 (Table 3 and Fig. 7), the mutations appeared to be evenly distributed (Fig. 5).
TABLE 3.
Secondary mutations identified in WT (VUSS3) and ExoN mutant viruses
| Virus | Mutation no.a | Mutationb | Amino acid changec | Locationd |
|---|---|---|---|---|
| VUSS3p5c2 | 1 | C21149A | Pro102Thr | nsp16 CDS |
| VUSS3p5c4 | 2 | A4608G | Asn635Asp | nsp3 CDS |
| VUSS3p5c5 | 3 | C21149A | Pro102Thr | nsp16 CDS |
| VUSS3p5c6 | 4 | A21515C | Met224Leu | nsp16 CDS |
| rExoN1p5c1 | 5 | U150C | Silent | 5′ UTR |
| 6 | G1092A | Val48Ile | nsp2 CDS | |
| 7 | G17723A | Val455Ile | nsp13 CDS | |
| 8 | A21069U | Asp75Val | nsp16 CDS | |
| 9 | U24030C | None | S gene | |
| 10 | U25845C | None | S gene | |
| 11 | U28207C | None | ORF4b | |
| rExoN1p5c2 | 12 | U694A | Leu162Gln | nsp1 CDS |
| 13 | U14122C | None | nsp12 CDS | |
| 14 | G15133A | None | nsp12 CDS | |
| 15 | A15706G | None | nsp12 CDS | |
| 16 | A17747G | Lys463Glu | nsp13 CDS | |
| 17 | A18802U | None | nsp14 CDS | |
| 18 | G19115A | Ala319Thr | nsp14 CDS | |
| 19 | U23407A | Silent | HE | |
| 20 | U25783A | Phe619Ile | S gene | |
| 21 | U26721C | None | S gene | |
| 22 | C28194U | Thr46Met | ORF4b | |
| rExoN3p5c1 | 23 | G812U | None | nsp1 CDS |
| 24 | U2144A | None | nsp2 CDS | |
| 25 | U4081G | Ile459Ser | nsp3 CDS | |
| 26 | U13481C | None | nsp10 CDS | |
| 27 | G15375A | Arg600His | nsp12 CDS | |
| 28 | G17711A | Val451Met | nsp13 CDS | |
| 29 | U18772C | None | nsp14 CDS | |
| 30 | C27928A | Silent | IGR | |
| 31 | G27930U | Silent | IGR | |
| 32 | A27931C | Silent | IGR | |
| 33 | U insertione | Multiple | ORF4b | |
| rExoN3p5c2 | 34 | A1342G | Glu131Gly | nsp2 CDS |
| 35 | C3337U | Ala211Val | nsp3 CDS | |
| 36 | U17227A | His289Gln | nsp13 CDS | |
| 37 | G17452A | None | nsp13 CDS | |
| 38 | A23139G | Silent | HE | |
| 39 | U24188C | Val87Ala | S gene |
Numbers refer to secondary mutations depicted in Fig. 5.
Nucleotide positions refer to the MHV-A59 complete genome sequence (GenBank accession no. AY910861).
Amino acid positions in nsps refer to residues in mature, processed nsps (i.e., the first residue after each viral proteinase cleavage site is renumbered as 1).
CDS, coding sequence; UTR, untranslated region; ORF4b, putative 11.7-kDa protein; HE, hemagglutinin-esterase pseudogene (not expressed) in MHV-A59; IGR, intergenic region between spike gene and ORF4.
A single U insertion was identified in a run of five consecutive U residues (nt 28104 to 28108) and is predicted to result in a frameshift relative to WT sequence and, consequently, in substitutions of His18Ser and Val19Cys and early termination after amino acid residue 19 in ORF4b.
FIG. 7.
Model for generation of intergenic region mutations in rExoN3p5c1. (A) Single template switch during normal synthesis of sg mRNA4. The WT sequences of a portion of the intergenic region (IGR) between the spike gene and ORF4 (first line) and leader regions (bold text, second line) of genome RNA (RNA1) are shown. The conserved consensus sequence in the RNA4 transcription-regulating sequence and at the 3′ end of leader is boxed. The line arrow indicates the replicase-transcriptase template switch from the intergenic region to the leader region to produce negative-sense (−) RNA4, which is then copied to yield positive-sense RNA4. Lowercase text represents negative-sense sequence. (B) Proposed double template switch to generate the intergenic region mutations in rExoN3p5c1. The three clustered nucleotide substitutions (circles) identified in the intergenic region between the spike gene and ORF4 in rExoN3p5c1 (Mut) could result from a single step in which the replicase-transcriptase switches from copying the intergenic region to copying 4 to 5 nt of leader and then back to copying the intergenic region.
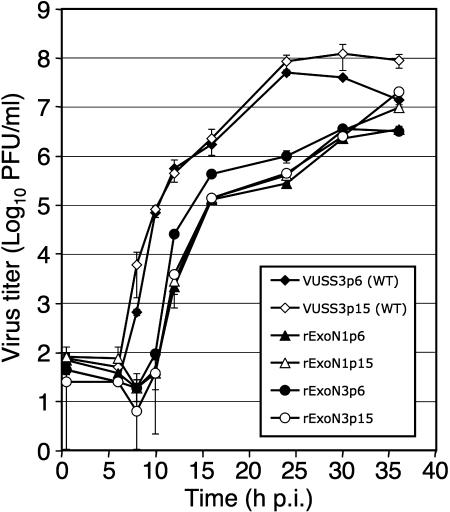
These sequencing results, combined with the absence of changes in growth fitness during virus passage, led us to predict that the ExoN mutants would continue to accumulate neutral mutations during population passage in culture. To test this prediction, we performed an additional 10 passages (p6 through p15) of rExoN1 and rExoN3 (Fig. 3) and compared growth of p15 stocks with identically passaged WT virus. The growth kinetics of WT and mutant viruses at p6 and p15 were identical (Fig. 6), indicating stable growth during passage. rExoN1p15 and rExoN3p15 exhibited similar heterogeneity in plaque size as observed at p0 and p5 (Fig. 2C and data not shown). We selected a single plaque each of rExoN1 and rExoN3 (p16) for expansion (p17) and then sequenced regions a, b, and c as before. For comparison, we sequenced a single p17 clone of VUSS3 (WT). VUSS3p17c1 had 1 mutation, whereas rExoN1p17 contained the engineered rExoN1 mutations, the 7 secondary mutations detected at p5, and 13 additional secondary mutations (Fig. 5). rExoN3p17 contained the engineered rExoN3 mutations, 2 of the 6 secondary mutations detected at p5, and 16 additional secondary mutations (Fig. 5). For rExoN1c1 and rExoN3c2 combined, the number of unique mutations approximately doubled (29 compared with 13) over approximately twice as many passages (12 compared with 5), demonstrating that the mutations accumulated at similar rates from p0 to p5 and from p6 to p17. Finally, sequencing of regions a through c was completed for a single p12 clone of VUSS2. VUSS2 is a mutant containing a Tyr414His substitution in nsp14, downstream of the ExoN motifs, that exhibits growth identical to WT in culture but is attenuated for virulence in mice (35). Analysis of VUSS2p12 identified only a single secondary mutation (data not shown). This result indicates that the accumulation of mutations in rExoN1 and rExoN3 was due to replacement of ExoN active-site residues rather than to nonspecific changes in nsp14.
FIG. 6.
Replication of p6 and p15 viruses. DBT-9 cells were infected with the indicated viruses at an MOI of 0.05 PFU/cell. p6 and p15 viruses of VUSS3, rExoN1, and rExoN3 were derived from VUSS3p5c1, rExoN1p5c1, and rExoN3p5c2, respectively. Supernatant samples were obtained at 0.5, 6, 8, 10, 12, 16, 24, 30, and 36 hpi, and virus titers were determined by plaque assay. Mean titers and standard deviations from duplicate experiments are indicated.
Potential nonrandom mutations in ExoN mutants.
The majority of secondary mutations identified in the ExoN mutants appeared to be randomly distributed across the genome. Two possible exceptions include mutations in nsp13 and mutations adjacent to the RNA4 transcription regulating sequence (TRS). A single nonsynonymous mutation was identified in nsp13 in each ExoN mutant clone at p5 (Fig. 5 and Table 3). These mutations occurred at different residues in each virus, but all were located within the helicase domain of nsp13 (21). These mutations were retained at p17 for those viruses examined. At p5 and p17, the nonsynonymous nsp13 mutations were unambiguous: a single peak was observed on the sequence electropherogram. However, the exact nonsynonymous nsp13 coding mutation found in later passages was also detected in each p1 mutant virus, although as part of a heterogeneous signal that included the WT nucleotide at the same position (data not shown). These mutations (and the remainder of the mutations identified in ExoN mutants) were not detected in WT virus or in the cDNA fragments used to assemble genome cDNA for virus recovery. Consequently, we cannot formally exclude the possibility that nsp13 coding mutations represent early adaptive mutations associated with the recovery of the ExoN mutants.
Three noncoding mutations were identified within a 4-nt region immediately upstream of the TRS for RNA4 in rExoN3p5c1 (Fig. 5 and Table 3). We propose a double template switch model as described in Fig. 7 for the genesis of these three mutations in one round of copying. This model posits that during synthesis of negative-sense RNA, the replicase-transcriptase switches templates once to copy a portion of the leader sequence and then switches templates again to resume copying the intergenic region. The first template switch (to leader sequence) occurs with high frequency as a central step in discontinuous transcription used by CoVs to generate sg mRNAs (30, 31). A template switch from leader back to 3′ proximal regions of the genome is apparently much less frequent but also has precedents (23, 36, 39). The effect of the clustered mutations near the RNA4 TRS on virus replication is unknown, particularly since the ORF4 protein product expressed from RNA4 is dispensable for virus replication (11, 29, 43).
DISCUSSION
Recovery of MHV nsp14 ExoN mutant viruses.
In this study we recovered MHV mutants containing engineered substitutions in ExoN active-site residues that are completely conserved across CoVs and that have been shown to be required for SARS-CoV nsp14 ExoN activity in vitro (24). The recovery of infectious MHV ExoN mutants indicates that these conserved residues are not required for MHV replication in culture. Although amino acid sequences of MHV-A59 and SARS-CoV nsp14 are 57% identical and 72% similar, ExoN activity of MHV nsp14 remains to be demonstrated biochemically. Nonetheless, our results clearly demonstrate the viability of MHV ExoN mutants and raise the question of whether ExoN activity is required for replication of other CoVs in cell culture. Minskaia et al. reported that alanine substitutions for nsp14 ExoN active-site residues in an infectious cDNA clone of HCoV-229E prevented recovery of infectious mutant viruses but allowed severely limited genome replication and sgRNA synthesis in cells electroporated with mutant genomes (24). It is unclear why infectious ExoN mutants were recovered for MHV but not for HCoV-229E. One possibility is that the RNA synthesis defects are greater in magnitude for ExoN mutants of HCoV-229E. Alternatively, it is possible that the RNA synthesis defects of ExoN mutants of both viruses are of similar magnitudes but that the defects exceed the threshold for recovery of only HCoV-229E. Another possibility is that the MHV mutants have residual ExoN activity in cells, or if the HCoV-229E mutants also have residual activity in cells (which has not been investigated), that such activity is greater for the MHV mutants. It will be interesting to determine whether ExoN mutants of other CoVs are viable and to characterize their phenotypes.
RNA synthesis defects of ExoN mutants.
Studies using MHV and other CoVs have suggested that nsp14 ExoN activity is involved in viral RNA synthesis, although the precise role of ExoN in RNA synthesis remains to be determined (1, 24, 32). Minskaia et al. reported severe RNA synthesis defects in ExoN mutants of HCoV-229E, with alterations of ratios and amounts of genomic and sg mRNA species (24). It was concluded that ExoN was critical for correct and efficient RNA synthesis and that the extent of the observed defects was consistent with the inability to recover infectious mutant viruses. Here, we have shown that MHV ExoN mutants have delayed and reduced overall RNA synthesis. Further, the magnitude of the observed RNA synthesis defects may be sufficient to account for the observed growth defects of these mutants. Gel analysis of [3H]uridine-labeled RNA showed bands of genomic mRNAs and sg mRNAs that were qualitatively similar to those of WT, with the exception of RNA2, which was detectable only by RT-PCR for the mutants. In addition, the patterns of RNA species were identical for both rExoN1 and rExoN3. Thus, while all viral RNA species were detected, the level of RNA2 appeared to be reduced. These results are consistent with the change in sg mRNA species observed for HCoV-229E ExoN mutants but differ in that a single sg mRNA species appeared most altered in abundance for the MHV mutants. Quantitative analyses of the molar ratios of individual RNA species will be required to address whether the MHV ExoN mutants have specific defects in genome replication and/or transcription of specific sg mRNAs. The possibility that the mutants have specific defects in positive- or negative-strand RNA synthesis also remains to be investigated.
Although many questions remain to be addressed, the recovery and replication of the MHV rExoN1 and rExoN3 mutants demonstrates that the predicted ExoN active-site residues are dispensable for MHV replication in culture but important for efficient growth and RNA synthesis. The lack of change in growth over 15 passages indicates that the replication defects of the ExoN mutants were not complemented by adaptive mutations, indicating that the functions of nsp14 ExoN are unique in the MHV genome. The stable, reproducible, and similar RNA synthesis defects of the MHV rExoN1 and rExoN3 mutants provide a powerful system to test for specific defects in replication and sg mRNA transcription and define the specific interactions and functions of ExoN in viral RNA synthesis and replication.
ExoN mutants have decreased replication fidelity.
For the MHV ExoN mutants, the large number and continuous accumulation of mutations distributed across the genome over 5 to 17 passages, in the absence of selective pressure or change in growth fitness, indicate that the secondary mutations are neutral for growth in cell culture. Thus, substitutions in different ExoN motifs resulted in significantly decreased replication fidelity and a distinct mutator phenotype. Together with the conservation of exonuclease motifs of known proofreading enzymes of the DEDD superfamily (25, 34, 45) and the demonstrated 3′-to-5′ ExoN activity of the conserved nsp14 of SARS-CoV (24), these results are consistent with the hypothesis that CoV nsp14 mediates RNA proofreading during replication of the viral RNA genome. Such an activity would be unprecedented in RNA viruses and have profound implications for studies of RNA genome replication and evolution. However, distinguishing a direct involvement of nsp14 in proofreading from other models clearly requires additional studies.
Models for a role of nsp14 in replication fidelity.
Although interactions of CoV replicase proteins are poorly understood, it is likely that nsp14 interacts with nsp12 (RdRp), nsp13 (helicase), and other nsps in multisubunit replication/transcription complexes. Thus, there are several possible mechanisms by which nsp14 ExoN could confer increased replication fidelity and which, if disrupted, could result in the observed mutator phenotype. First, nsp14 3′-to-5′ ExoN activity may directly mediate hydrolysis of an incorrect nucleotide from the 3′ end of the nascent RNA chain, similar to the role of DE-D-D ExoN proofreading domains or subunits of DNA polymerases (3, 25). Second, nsp14 may stimulate hydrolysis of a misincorporated nucleotide by an as yet unidentified intrinsic 3′-to-5′ ExoN activity of nsp12. Such a mechanism is used for RNA proofreading during cellular transcription, in which cleavage-stimulatory factors stimulate polymerase-mediated hydrolysis of incorrect nucleotides (28, 37). Third, nsp14 may function through allosteric effects to increase the accuracy of nucleotide incorporation by the RdRp (nsp12). Fourth, nsp14 ExoN may promote error repair by RNA recombination. The available data lead us to favor the first model because there is no obvious reason why conserved ExoN active-site residues would be critical for stimulation of nsp12-mediated nascent-chain cleavage or for allosteric interactions with nsp12. Additionally, although recombination is well described for CoVs, there is no reported role for recombination in correction of nucleotide incorporation errors during CoV replication in the absence of selection. Further studies to distinguish between these possibilities will likely require purification and testing of the membrane-associated replication complexes containing up to 16 nsps.
A direct function of nsp14 in removal of misincorporated nucleotides, as in the first two models presented above, would constitute RNA proofreading, a function that has not been described during genome replication of RNA viruses. For influenza virus, an RNA proofreading function of the RdRp was proposed based on the demonstration that a nuclease activity in purified virions removed excess noncognate residues from 3′ termini of capped primers in the presence of correct substrate (19). While the experiment was an in vitro model for initiation of viral mRNA transcription, there has been no further report of this activity in vitro or during virus infection. The fact that MHV ExoN mutant viruses accumulated mutations throughout the genome suggests that error correction operates during RNA replication and is not limited to initiation. It remains to be tested whether MHV genome replication and sg mRNA transcription have similar fidelities and whether the ExoN mutants exhibit reduced fidelity during transcription.
Estimates of neutral mutation rates reveal atypically high replication fidelity of WT MHV.
A generally established range for mutation rates during replication of RNA viruses is ∼10−3 to 10−5 mutations per nucleotide per replication cycle (12, 17). For comparison, we estimated mutation rates based on the data for p5 viruses in Table 2 for WT and rExoN3 as 2.5 × 10−6 and 3.2 × 10−5 mutations per nucleotide per replication cycle, respectively. Surprisingly, the estimated mutation rate for the rExoN3 viruses was within the range reported for other RNA viruses such as poliovirus (26). In contrast, the estimated mutation rate for WT MHV was well below this range. Sequencing of additional WT and mutant virus clones will refine these estimates. Nonetheless, the low mutation rate of WT MHV is interesting in light of previous experimental evolution experiments with MHV that demonstrate rapid adaptation to growth in cells from different species and recovery of growth fitness following engineered deleterious mutations (2, 23). Thus, CoVs demonstrate a capacity for fast adaptation but maintain high-fidelity replication under stable conditions. SARS-CoV has apparently exhibited both characteristics, with rapid accumulation of mutations early in the 2003 epidemic but with rates of mutation that were lower than expected over the late phase of the epidemic (8, 42) and during passage of a SARS patient isolate in Vero cells (38). It will be important to determine whether other CoVs, and specifically SARS-CoV, also exhibit high-fidelity replication in the absence of selection in cell culture and whether nsp14 ExoN mutants of those viruses also exhibit mutator phenotypes.
We propose that nsp14 ExoN increases the fidelity of a CoV replication machinery that in the absence of ExoN has fidelity comparable to other RNA viruses. Replication fidelity has been proposed as an important constraint on the size of RNA virus genomes (13). Gorbalenya et al. proposed that the last common ancestor of the smallest nidoviruses, the arteriviruses, had sufficient fidelity to achieve genome sizes of ∼15 kb in the absence of ExoN but that acquisition of ExoN-mediated proofreading was critical for genome expansion up to ∼30 kb (15). The experimental results reported here are consistent with this theory, although the exact mechanism by which ExoN increases fidelity remains to be determined.
Acknowledgments
We thank members of the Denison Laboratory for critical reading of the manuscript.
Support for this work was provided by Public Health Service awards AI59443 and AI26603 (M.R.D.) from the National Institute of Allergy and Infectious Diseases and by Training Grant T32AI049824 (L.D.E.) for Cellular and Molecular Microbiology awarded to Vanderbilt University School of Medicine. Additional support was provided by Public Health Service award CA68485 to the Vanderbilt University DNA Sequencing Shared Resource of the Vanderbilt-Ingram Cancer Center.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Almazán, F., M. L. Dediego, C. Galán, D. Escors, E. Álvarez, J. Ortego, I. Sola, S. Zuñiga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80:10900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., B. Yount, L. Hensley, S. A. Peel, and W. Chen. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 71:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, J. J., R. Sanjuan, and C. O. Wilke. 2007. Theory of lethal mutagenesis for viruses. J. Virol. 81:2930-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, W., V. J. Madden, C. J. Bagnell, and R. S. Baric. 1997. Host-derived intracellular immunization against mouse hepatitis virus infection. Virology 228:318-332. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. [DOI] [PubMed] [Google Scholar]
- 9.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 11.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eigen, M. 1993. The origin of genetic information: viruses as models. Gene 135:37-47. [DOI] [PubMed] [Google Scholar]
- 14.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 15.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano, N., K. Fujiwara, and M. Matumoto. 1976. Mouse hepatitis virus (MHV-2): plaque assay and propagation in mouse cell line DBT cells. Jpn. J. Microbiol. 20:219-225. [PubMed] [Google Scholar]
- 17.Holland, J. J., E. Domingo, J. C. de la Torre, and D. A. Steinhauer. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 25:4933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihama, A., K. Mizumoto, K. Kawakami, A. Kato, and A. Honda. 1986. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J. Biol. Chem. 261:10417-10421. [PubMed] [Google Scholar]
- 20.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J. C., R. A. Spence, P. F. Currier, X. T. Lu, and M. R. Denison. 1995. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d. Virology 208:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser, M. J., W. R. Holley, A. Chatterjee, and I. S. Mian. 1997. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 25:5110-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvin, J. D., A. Moscona, W. T. Pan, J. M. Leider, and P. Palese. 1986. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J. Virol. 59:377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeiffer, J. K., and K. Kirkegaard. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLOS Pathog. 1:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole, A. M., and D. T. Logan. 2005. Modern mRNA proofreading and repair: clues that the last universal common ancestor possessed an RNA genome? Mol. Biol. Evol. 22:1444-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarma, J. D., E. Scheen, S. H. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 32.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLOS Pathog. 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taguchi, F., T. Ikeda, S. Makino, and H. Yoshikura. 1994. A murine coronavirus MHV-S isolate from persistently infected cells has a leader and two consensus sequences between the M and N genes. Virology 198:355-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, M. J., A. A. Platas, and D. K. Hawley. 1998. Transcriptional fidelity and proofreading by RNA polymerase II. Cell 93:627-637. [DOI] [PubMed] [Google Scholar]
- 38.Vega, V. B., Y. Ruan, J. Liu, W. H. Lee, C. L. Wei, S. Y. Se-Thoe, K. F. Tang, T. Zhang, P. R. Kolatkar, E. E. Ooi, A. E. Ling, L. W. Stanton, P. M. Long, and E. T. Liu. 2004. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma, S., L. A. Lopez, V. Bednar, and B. G. Hogue. 2007. Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein. J. Virol. 81:5339-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vignuzzi, M., J. K. Stone, and R. Andino. 2005. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107:173-181. [DOI] [PubMed] [Google Scholar]
- 41.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh, S. H., H. Y. Wang, C. Y. Tsai, C. L. Kao, J. Y. Yang, H. W. Liu, I. J. Su, S. F. Tsai, D. S. Chen, and P. J. Chen. 2004. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc. Natl. Acad. Sci. USA 101:2542-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokomori, K., and M. M. Lai. 1991. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J. Virol. 65:5605-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]