Abstract
Unique among the retroviruses, mouse mammary tumor virus (MMTV) carries, in addition to the usual long terminal repeat (LTR) promoter, another promoter, P2, which is located in the central part of the proviral U3 sequence, within the LTR open reading frame (ORF). Using an in vitro reporter system based on a sensitive luciferase expression assay, we investigated the regulation of the P2 promoter in the context of the Mtv-2 and Mtv-8 genomes. Irrespective of the genomic source, the activity of the P2 promoter is regulated by a downstream-located enhancer and an upstream-located negative regulatory element (NRE), the activity of which overrides the activator. During this study, we unexpectedly detected another independent neighboring promoter that we called P3. The novel P3 promoter does not seem to be controlled by any NRE but is influenced by the same enhancer that modulates the P2 promoter. The respective transcription starts of the two promoters located in this tight cluster are only 61 bases apart. The transcripts originating from this promoter complex carry the same first intron, which is bound by canonical splice donor and splice acceptor sites located in the LTR. One novel doubly spliced transcript carrying a 459-nucleotide-long ORF was detected in several MMTV-carrying murine cells and could be successfully expressed in murine cells as a His-tagged fusion product. The novel viral protein, the function of which remains to be elucidated, has an apparent molecular mass of 20 kDa.
In common with all other retroviruses, mouse mammary tumor virus (MMTV) contains a major promoter in the U3 region of the long terminal repeat (LTR) upstream of the R region (10, 17, 21). However, in addition to the usual LTR promoter, another promoter, P2, is located in the central part of the proviral LTR within the open reading frame (ORF) region (14, 33, 36). This region is well conserved among the different viral strains and endogenous variants of MMTV and contains several elements that variously repress viral transcription (4, 11, 20, 27, 40), stimulate it (12), are implicated in the response of the LTR to steroid hormones (6, 8, 12, 13, 19, 22, 34), and may control the tissue specificity of expression of the virus (40, 41). The importance of this region in transcriptional control and specificity is underscored by the finding that MMTV proviruses associated with T-cell lymphoma in mice show deletions or mutations in this area, resulting in the genetic reorganization of its sequence (1, 16, 26, 28, 35, 38, 39). Thus, the presence of the P2 promoter in this crucial regulatory region of the MMTV provirus suggests that its activity might be intimately linked to these effects. We first investigated, therefore, if some of the sequences reported previously as acting on the global transcriptional activity of the MMTV LTR influence the promoter activity of the central LTR P2 promoter. An unexpected outcome of these studies on the regulation of the P2 promoter was the detection of another independent promoter in close proximity, thus revealing the presence of a promoter complex in the central part of the MMTV U3 region. Subsequently, we were able to detect a novel viral transcript that originates from this promoter complex in MMTV-carrying murine cells.
MATERIALS AND METHODS
Plasmid construction.
The fragments of the MMTV LTR tested in the luciferase reporter system were obtained by PCR amplification. pdGR102 Cla was used as a template for all Mtv-2 LTR sequences (31), and pdGR102Afl was used as a template for all Mtv-8 LTR sequences. pdGR102Afl was obtained by the religation of pGR102ES (18) previously digested with AflII. MMTV-specific oligonucleotides carrying at the 5′ end the respective restriction site (BamHI or HindIII) were used as primers. After digestion with the relevant enzyme, gel-purified PCR fragments were cloned into the unique BamHI and HindIII restriction sites of the promoterless expression plasmid pZluc (5, 25, 31). The Mtv-2 or Mtv-8 LTR region carried by each resulting plasmid was sequenced to confirm its integrity.
The construction of both pGR102 ES and of pMtv-2pAneoES have been previously reported (see references 18 and 31, respectively).
Construction of pcDNA3.1ORFeHis.
PCR amplification was performed using cDNA isolated from GR producer cells as a template, with 781N (5′-ATATATGCTAGCCTTGGTGTATGCTAACTGAG) as a forward primer and ORFeHT (5′-TTTGCTCTAGATTAGTGGTGGTGGTGG TGGTGAGAACCTCCTCCGCTTCGGAGA) as a reverse primer. The specific DNA fragment thus obtained was gel purified, digested with NheI and XbaI, and then ligated into the vector pcDNA3.1 Hyg+ (Invitrogen) previously digested with NheI and XbaI. The insert of interest carried by the resulting plasmid, pcDNA3.1ORFeHis, was verified by sequencing.
Cell culture.
CrFK cells (9) and GR mouse mammary carcinoma cells (30) productively infected with MMTV (32) were maintained in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen) containing 10% heat-inactivated fetal bovine serum. Whenever needed, cultivated cells were stimulated with 1 μM dexamethasone for 8 h. Two MMTV producer cell lines were established by the stable transfection of CrFK cells (C2AnE15) or NMuMG cells (N2AnE5) with pMtv-2pAneoES. The integrity of the proviral sequence was analyzed by Southern analysis for full-length single copies, and viral productivity was measured by supernatant infectivity on MMTV-sensitive CrFK and NMuMG cells. The CΔP1AnE11 cell clone was previously described (31). All three stably transfected cell clones (CΔP1AnE11, N2AnE5, and C2AnE15) were maintained in medium containing 400 μg/ml of G418.
Transfection and luciferase assays.
For each construct, two independent clones were grown for plasmid preparation and independently tested upon transfection into cells. Transfection experiments were performed using 2 μg of DNA for 4 × 105 cells in six-well plates, each point in duplicate, using the calcium phosphate protocol (Pharmacia Biotech). Transfection efficiencies were determined by cotransfecting the plasmid of interest with 0.5 μg of pEGFPc1 in parallel and by submitting the transfected cells to fluorescence-activated cell sorter analysis 48 h after transfection (FACSCalibur; Becton Dickinson). Experiments were repeated at least three times. The transfected cells were rinsed twice after 6 h and incubated again for luciferase assays. When dexamethasone induction was tested, transfected cell medium was removed 10 h before protein extraction and replaced with the same cell culture medium containing 10−6 M dexamethasone. Luciferase assays were performed 48 h after transfection. After lysis of the transfected cells, protein extracts were kept on ice, and the amount of protein was determined immediately using a protein assay kit (Bio-Rad), followed by total protein concentration measurements at 630 nm on an Elx800 spectrophotometer (Bio-Tech Instruments), and 20 μg of protein was used per point for luciferase assays using the Autolumat LB 953.
RNA extraction.
Total RNA from 106 to 107 cells was extracted from cell pellets using the RNeasy Mini kit (QIAGEN), and mRNA was isolated using a QuickPrep Micro mRNA purification kit (Amersham) according to the manufacturer's instructions. To prevent DNA contamination, RNA was treated with RQ1 RNase-free DNase I (Promega). Ten micrograms of total RNA extract was subjected to DNase I treatment using RNase-free DNase I enzyme (Roche Diagnostics) for 1 h at room temperature followed by 15 min of incubation at 65°C. Subsequently, ethanol precipitation of each RNA sample was performed. The lack of any DNA contamination was determined by performing a control PCR on an aliquot of each RNA sample using a primer pair that is specific for the pol region (oligonucleotides 5′-GCTTGTTCACGAGTGATGTG and 5′- AGTCATTACAGCCTTTGAGGAA) (data not shown). Total RNA and mRNA were stored at −80°C for further analysis.
5′ RACE.
5′ rapid amplification of cDNA ends (RACE) was performed according to the manufacturer's instructions (5′/3′ RACE kit; Roche). Two micrograms of DNase I-treated total RNA extract from the N2AnE5 cell clone (treated or not treated with dexamethasone) was submitted to reverse transcription with oligonucleotide 6890R (5′-CTCCTCCGCTTCGGAGAT), followed by the purification of cDNA thus obtained (Roche). An aliquot of the 5′ poly(A)-tailed, purified cDNA thus obtained was used as a template for the first PCR amplification together with the gag-specific reverse primer −1412 (5′-CTTGTGATGATAGCCAGACAAGA) and the oligo(dT) anchor primer (Roche). One microliter of this first PCR product was used as a template for the next PCR using a nested specific primer, primer −830 (5′-CCTGTTGTTTCATGTCGTC), and the anchor primer. The amplification products of both PCRs were purified from a gel and subcloned into a TOPO vector (TOPO XL; Invitrogen), and 12 independent clones for each PCR fragment were finally analyzed by sequencing. 5′ RACE of transcripts originating from the P1 promoter was performed in a similar way and using the same cDNA preparation, but primer P1mtv-2 (5′-GTGAAGGA TAAGTGACGAGC) was used instead of primer −830.
5′ RLM-RACE.
5′ RNA ligase-mediated rapid amplification of cDNA (RLM-RACE) was performed according to the manufacturer's instructions (GeneRacer kit; Invitrogen) on mRNA extracts from GR or N2AnE5 cells that were not treated with dexamethasone. After dephosphorylation with calf intestine alkaline phosphatase, removal of the cap structures, and ligation into the GeneRacer RNA oligonucleotide, the first-strand cDNA preparation was obtained by reverse transcription using primer −1442 (5′-GTCCGTTCCGCTCTTGTGAT), a specific primer in the gag region. 5′ RLM PCR RACE (35 cycles) was then performed with the GeneRacer 5′ primer and primer −1412. PCR products shown in Fig. 4B were gel purified and subcloned into a TOPO vector (pCR4-TOPO; Invitrogen). Independent clones were analyzed by sequencing.
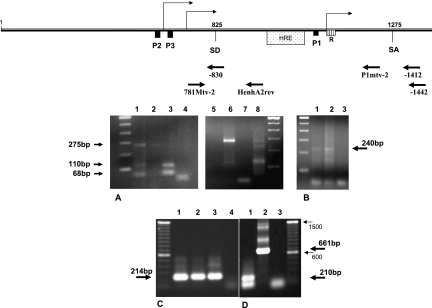
FIG. 4.
Determination of the start of transcription for the novel MMTV P3 promoter. (A) 5′ RACE. DNase I-treated total RNA extract from N2AnE5 or NMuMG cells was reverse transcribed with a primer specific for env, and 5′ poly(A)-tailed, purified cDNA was used as a template for the first PCR amplification together with the gag-specific reverse primer −1412 and the 5′ RACE oligo(dT) anchor primer. Nested PCR was then performed using the anchor primer and primer −830 for the detection of the transcription start from the P3 promoter or using primer P1mtv-2 for the detection of the transcription start from the P1 promoter. PCR products were gel purified and subcloned for sequencing analysis. Lanes 1 to 4 show cells that were not treated with dexamethasone prior to RNA extraction. Lane 1, 5′ RACE PCR products using P1mtv2 from N2AnE5 cDNA; lane 2, 5′ RACE PCR products using P1mtv2 from NMuMG cDNA; lane 3, 5′ RACE PCR products using −830 from N2AnE5 cDNA; lane 4, 5′ RACE PCR products using −830 from NMuMG cDNA. Lanes 5 to 8 show cells that were treated with dexamethasone prior to RNA extraction. Lane 5, 5′ RACE PCR products using P1mtv2 from NMuMG cDNA; lane 6, 5′ RACE PCR products using P1mtv2 from N2AnE5 cDNA; lane 7, 5′ RACE PCR products using −830 from NMuMG cDNA; lane 8, 5′ RACE PCR products using −830 from N2AnE5 cDNA. (B) 5′ RLM-RACE. mRNA was dephosphorylated, the cap structure was removed, RNA was then ligated into the GeneRacer RNA oligonucleotide and reverse transcribed with the gag-specific primer −1442, and nested PCR was performed with primer −1412 and the 5′ GeneRacer primer. Lane 1, GeneRacer PCR products from GR mRNA; lane 2, GeneRacer PCR products from N2AnE5; lane 3, PCR negative control. GeneRacer PCR products were then gel purified before subcloning and sequencing. (C and D) Splicing. Total RNAs from MMTV producer GR cells were first reverse transcribed using specific reverse primer −1442, and RNase H treatment was performed, followed by PCR analysis. (C) Nonspliced transcripts. Primers 781Mtv-2 and HenhA2rev were used. The templates were reverse-transcribed RNAs from GR cells treated with dexamethasone (GR+dex RT RNAs) (lane 1), GR−dex RT RNAs (lane 2), and pGR102 DNA (lane 3). Lane 4, no-template control. (D) Spliced transcripts. PCR was performed with primers 781Mtv-2 and −1412. The templates were GR−dex RT RNAs (lane 1) and pGR102 DNA indicating the genomic size of a nonspliced fragment (lane 2). Lane 3 is a no-template PCR control. The 214-bp- and 210-bp-long DNA products were purified from gels shown in C and D. The sequence of the 210-bp fragment reveals a splicing event that occurred between SD825 and SA1275.
Splicing analysis.
Reverse transcription was performed on 300 ng of total RNA (SuperscriptII reverse transcriptase; Invitrogen) in the presence of 40 units of RNaseOUT (Invitrogen) using oligonucleotide −1442 (5′-GTCCGTTCCGCTCTTGTGAT) as a primer. RNase H treatment was performed prior to PCR analysis. PCR analysis was achieved with oligonucleotide 781Mtv-2 (CCTTGGTGTATGCTAACTGAG) as a forward primer and either primer −1442 or oligonucleotide HenhA2rev (5′-CCCTGGGAACCGCAAGGTTGGGC) as a reverse primer. Aliquots of PCR products were subjected to electrophoresis in 2% agarose gels. DNA from some bands was extracted using NucleoSpin extract (Macherey-Nagel) and sequenced. An aliquot of pGR102 was used as a template for the PCR positive control.
Transcript isolation.
RNA extracts from CΔP1AnE11, N2AnE5, C2AnE15, or GR cells were prepared as described above. cDNAs were obtained by reverse transcription using a poly(dT) primer mix with the terminal 3′ nucleotide being either A or C or G and submitted to RNase H treatment. In the case of the cDNA preparation from CΔP1AnE11, N2AnE5, and C2AnE15 cells, the absence of readthrough originating from the sequence upstream of the provirus was checked by PCR analysis (data not shown). Oligonucleotides used for novel ORF cDNA detection in full-length MMTV-carrying cells were 761mtv2 (5′-GACTCGCCAGAGCTAGACC) and 7383R (5′-CGATCTGGCAGTGAGGATA). Detection of novel ORF from CΔP1AnE11 cells was performed using 781Mtv-2 (5′CCTTGGTGTATGCTAACTGAG) as a forward primer.
Western blot analysis.
Protein extraction from transiently transfected cells was performed 48 h after transfection. The cell pellet was weighed and resuspended in 200 μl phosphate-buffered saline. Proteinase inhibitor (Sigma) was added to the cells at 0.5 μl/0.01 g of pellet (wet weight) before ultrasonication of the cells was performed. The amount of protein was measured using the DC protein assay kit (Bio-Rad), and the absorbance was read at 630 nm (Bio-Tek ELX 800). Twenty micrograms of protein extract was added to protein sample buffer (100 mM Tris HCl [pH 6.8], 5% sodium dodecyl sulfate [SDS], 20% glycerol, 0.02% bromophenol blue, 10% β-mercaptoethanol). Protein lysates were subjected to SDS-polyacrylamide gel electrophoresis, transferred onto a HybondP polyvinylidene difluoride transfer membrane (Amersham), and, after blocking overnight in 5% milk powder-Tris-buffered saline-Tween, probed with polyclonal anti-His rabbit antiserum (Rockland). Anti-rabbit immunoglobulin G, horseradish peroxidase-linked whole antibody, donkey (Amersham), was used as a secondary antibody. Protein detection was performed by using ECL Plus Western blotting detection reagents (Amersham), followed by autoradiography on film for 5 to 15 min (Hyperfilm; Amersham).
RESULTS
Mtv-2 P2 promoter activity is downregulated by an 80-bp-long negative regulatory element (NRE) located immediately upstream of the minimal promoter.
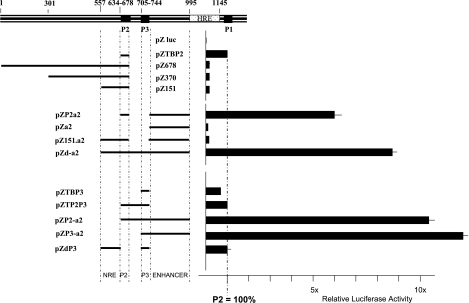
To delineate the regulatory elements that modulate the activity of the P2 promoter, we used an in vitro promoterless reporter system based on luciferase expression (31). Mtv-2 LTR sequences were first tested in this system (Fig. 1). A 40-bp fragment containing the previously described (14) minimal P2 core promoter with the TATA box (pZTBP2), the complete U3 region up to and including the P2 core promoter (pZ678), the region from position 301 (relative to the beginning of the LTR) up to and including the P2 promoter (pZ370), and the region from position 557 up to and including the P2 promoter (pZ151) were individually linked to luciferase coding sequences (Fig. 1).
FIG. 1.
Relative luciferase expression from LTR fragments upon transfection of CrFK cells. The upper schematic is a representation in scale of the LTR U3 region. Numbers indicate nucleotide positions relative to the beginning of the LTR. The filled black boxes represent the P2, P3, and P1 promoter cores, and the dotted box shows the hormone-responsive element (HRE). The left part of the figure shows a schematic summary of the relevant sequences carried by the reporter construct used for transfection. The bold line represents the MMTV sequence, and interruptions in the bold line indicate the site and approximate size of the deletion. Luciferase expression from pZTBP2 is used as a reference and was set to 100%. Luciferase expression was determined in the absence of dexamethasone. Each bar reflects the mean value of at least three independent transfections with standard errors of less than 5% of the mean values.
A number of independent cell lines from different species (feline kidney CrFK cells, mouse mammary tumor GR cells, mouse fibroblast NIH 3T3 cells, mouse lymphocyte A20 cells, and rat hippocampal HiB5 stem cells) were transfected with these four constructs, and transient luciferase activity was measured. The data presented here were obtained after transfection of CrFK cells, but similar patterns of relative luciferase expression were obtained for all cell lines tested (data not shown). In comparison to the construct carrying the minimal P2 promoter (pZTBP2), the activity of which was set to 100%, the other three constructs showed clear down-regulation, with expression levels of around 20 to 25% (pZ678, pZ370, and pZ151) (Fig. 1). This experiment reveals that the smallest region, from positions 557 to 634, is already sufficient to down-regulate the basal activity of the P2 core promoter and is consistent with the presence of a previously described NRE located between position +557 and P2 (27) acting on P2. We observed the inhibitory effect of this NRE in all cell lines tested.
An Mtv-2 P2 promoter enhancer is located downstream of its start of transcription.
To investigate the potential influence of downstream regulatory elements on the P2 promoter, a series of constructs covering the region between positions 744 and 995 with respect to the first base pair of the Mtv-2 LTR was tested. This region has been the subject of some controversy since it has been reported as containing both an enhancer region (12, 23, 29) and several NREs (4, 27, 40). The addition of these sequences (pZP2a2) (Fig. 1) to the minimal P2 promoter (pZTBP2) (Fig. 1) resulted in a clear and consistent sixfold up-regulation of luciferase expression. This up-regulation, however, was totally abrogated if the NRE located between positions 557 and 634 was additionally added (pZ151.a2) (Fig. 1). Since the presence of both the NRE and the positive regulatory element resulted in an expression level that was lower than that of the minimal P2 promoter alone (pZ151a2 versus pZTBP2) (Fig. 1), we can conclude that the NRE overrides the effect of the positive element. The same result was obtained in GR, A20, NIH 3T3, and HiB5 cells (data not shown). To demonstrate that the positive regulatory element (positions 744 to 995) for P2 is really an enhancer and not a region carrying a potential transcription initiator in its own right, we tested the potential transcriptional activity of the positive element in the LTR fragment at positions 744 to 995 in a promoterless reporter system (pZa2) (Fig. 1) and did not observe significant luciferase activity.
The P2 promoter is functionally conserved in Mtv-8.
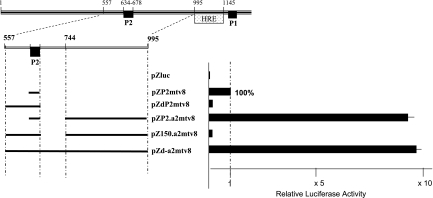
To investigate whether the P2 promoter is conserved and functional in other MMTV isolates, we compared the sequence around the P2 promoters of isolates C3H exo, C3H/HeJ, RIII, Mtv-1, Mtv-8, and Mtv-2 (Fig. 2). The CAAT box and TATA sequence are perfectly conserved among all the isolates investigated (Fig. 2). Exemplarily, we tested the functionality of the P2 promoter using the Mtv-8 isolate (Fig. 3) according to the same strategy as that used for the analysis of Mtv-2 P2 regulation. The activity of the minimal Mtv-8 promoter core was arbitrarily set at 100% (pZP2mtv8) (Fig. 3). The addition of the previously defined NRE (positions 557 to 634) resulted in the expected down-regulation (pZdP2mtv8) (Fig. 3), while the addition of the enhancer element alone (positions 744 to 995) resulted in the expected almost 10-fold up-regulation (pZP2.a2mtv8) (Fig. 3). When both elements were present, the negative effect outweighed the enhancer effect, resulting in a net down-regulation (pZ150.a2mtv8) (Fig. 3). These results clearly demonstrate the same regulation of the P2 promoter in the Mtv-8 MMTV isolate as that observed in the Mtv-2 isolate.
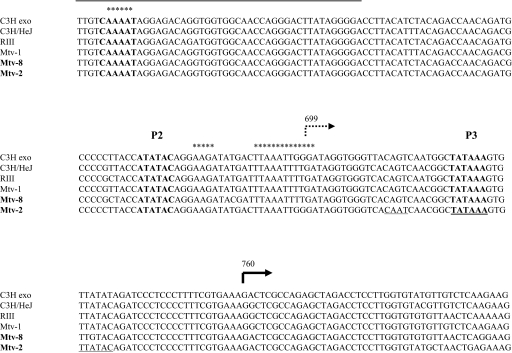
FIG. 2.
Central LTR promoter complex alignment. GenBank accession numbers are as follows: AF228552 for C3Hexo, AF228551 for C3H/HeJ, AF136900 for RIII, and AF228550 for Mtv-1; for Mtv-2, the alignment is according to the 3′ LTR of pGR102 (32), and for Mtv-8, the alignment is according to the 5′ LTR of pGR102. The numbers indicate the position from the first base of the LTR. The promoter elements are shown in boldface type. The transcription start of the P2 promoter is marked by a dotted arrow (14). The region containing the NRE for the P2 promoter is surlined. Stars are placed above the CDP binding sites (41). The transcription start of the P3 promoter is indicated by the thick black arrow at position 760 relative to the 5′ end of the LTR sequence.
FIG. 3.
Mtv-8 P2 promoter regulatory elements. Shown is the relative luciferase expression from Mtv-8 LTR fragments upon transfection of CrFK cells. The symbols and numbers are described in the legend of Fig. 1, with an enlargement of the region of interest also being given. Luciferase expression is here normalized to that obtained from pZP2mtv8, which was set to 100%. Luciferase expression was determined in the absence of dexamethasone. Each bar reflects the mean value of at least three independent transfections with standard errors of less than 5% of the mean values.
A novel promoter (P3) is located in the central part of the MMTV LTR and shares the P2 enhancer but not the P2 NRE.
The relatively high level of luciferase expression from construct pZd-a2 compared to the almost nonexistent luciferase expression from construct pZ151a2 (Fig. 1) suggests that the very short region located between positions 678 and 744 contains an element with a strong positive effect on transcription. Analysis of this 66-bp-long region of the Mtv-2 LTR revealed CAAT and TATA-like sequences that are commonly associated with promoter structures (Fig. 2). We therefore tested the transcriptional potential of this discrete LTR fragment (positions 705 to 743) in the luciferase reporter system (pZTBP3) (Fig. 1). Only marginally lower levels of luciferase expression were observed with pZTBP3 than what was observed with the minimal P2 promoter-carrying luciferase construct (pZTBP2) (Fig. 1), suggesting that this small fragment indeed harbors a promoter element. We propose to call this novel MMTV LTR promoter P3. Similarly to the previously described P2 promoter, the addition of sequences between positions 744 and 995, which contain an enhancer element (pZP3-a2) (Fig. 1), resulted in a greater than 10-fold up-regulation of luciferase expression. However, in contrast to P2, inclusion of sequences between positions 557 and 634 (pZdP3) (Fig. 1) had no effect on luciferase expression, demonstrating that the P3 promoter is not controlled by this NRE.
Constructs containing both P2 and P3 minimal promoter elements (pZTP2P3) (Fig. 1) do not show higher luciferase activity than that observed for the P2 promoter core (pZTBP2) (Fig. 1) or the P3 promoter core (pZTBP3) (Fig. 1) alone, demonstrating that, as expected for promoters in close proximity, the novel P3 promoter does not synergize with the above-described neighboring P2 promoter but rather that they occlude each other. Similarly, a luciferase construct containing both P2 and P3, as well as the enhancer element located between positions 744 and 995 that has been shown to affect both promoters (pZP2-a2) (Fig. 1), does not show significantly different levels of luciferase activity compared to a construct containing only P3 and the enhancer element (pZP3-a2) (Fig. 1).
Determination of the TATA box of the P3 promoter.
One consensus CAAT box and two potential TATA boxes (Fig. 2, underlined elements) were detected in the short Mtv-2 LTR fragment encompassing the minimal P3 promoter sequence. A comparison with the sequence of the Mtv-8 LTR reveals only one of these three elements, a consensus TATA box (Fig. 2). However, this short (66-bp) Mtv-8 LTR sequence located between the P2 promoter and its enhancer is capable of driving high levels of expression even when the P2 promoter is down-regulated by the presence of its NRE (pZd-a2mtv8 compared to pZ150.a2mtv8) (Fig. 3). This is similar to the expression observed in the case of the Mtv-2 LTR (pZ151a2 compared to pZd-a2) (Fig. 1). We conclude, therefore, that the consensus TATA box, which is conserved between Mtv-2 and Mtv-8, is the promoter core of the novel MMTV P3 promoter and that the CAAT box does not influence P3 activity. Thus, the TATA boxes of P2 and P3 appear to be only 47 bp apart (Fig. 2).
Characterization of the transcriptional start of the P3 promoter.
5′ RACE analysis of the mRNAs was performed to determine the start of transcription from the P3 LTR. For this purpose, DNase I-treated total RNA extract from N2AnE5, an MMTV strain carrying the NMuMG cell clone, was subjected to reverse transcription with a reverse primer specific for env. 5′ anchor primer ligation to the cDNA library thus obtained and nested PCR using specific primers were performed according to the 5′ RACE method. Both amplified PCR fragments (Fig. 4A, lane 3) were cloned into a TOPO vector, and 12 independent clones were sequenced for each fragment. All sequences obtained from the larger band were identical and indicated that position 760 was the site of the start of transcription from the novel P3 promoter. The shorter PCR band obtained by 5′ RACE revealed a second signal at position 802 with respect to the beginning of the LTR. When cells were treated with dexamethasone prior to mRNA extraction (Fig. 4A, lane 8), no specific signal could be detected downstream of the P3 TATA box. A 5′ RACE analysis of the transcriptional start site of the P1 promoter using RNA isolated from N2AnE cells grown in the presence of dexamethasone (Fig. 4A, lane 6) confirmed the cap site of the P1 promoter at position 1194 in the presence of dexamethasone. In the absence of dexamethasone (Fig. 4A, lane 1), no unique signal was obtained, but a cluster of signals at the Inr inside the R region as was observed by others previously (7, 31). A 5′ RLM-RACE was also performed on mRNA extracts from both GR and N2AnE5 cells to confirm that the transcription initiation site identified by 5′ RACE is an authentic capped site (Fig. 4B). The 5′ GeneRacer PCR products (about 240 bp long) (Fig. 4B, lanes 1 and 2) were subcloned and sequenced. This analysis confirmed that the stronger signal obtained by 5′ RACE is due to a capped transcript. However the second signal detected by 5′ RACE at position 802 could not be detected by this method, suggesting that it was likely resulting from the presence of a truncated RNA in the extract.
From the independent and identical results obtained by 5′ RACE and 5′ RLM-RACE, we conclude that the start site of the novel P3 promoter is located at position 760 with respect to the beginning of the LTR sequence, 32 bp downstream of the TATA box, and is therefore 61 nucleotides downstream of the transcription start site previously described for P2 (14).
Transcripts originating at the P2/P3 promoter complex utilize the LTR splice donor and acceptor sites.
To determine whether transcripts originating from one or both of the promoters (P2 and P3) constituting the central complex all use the previously described splice donor/splice acceptor sites inside the LTR (splice donor site at position 825 [SD825]/splice acceptor site at position 1275 [SA1275]) (15, 37), we analyzed the occurrence of splicing at these positions using mRNAs from GR cells treated previously or not treated with dexamethasone. Reverse transcription with reverse primer −1442, specific for a sequence located in the gag region downstream of SA1275, was performed and followed by PCR analyses using the cDNAs thus obtained as templates (Fig. 4C and D). Only when primer HenhA2rev, which had been designed to bind inside the intron sequence, was used in combination with primer 781Mtv-2 could a nonspliced message be detected (Fig. 4C, lanes 1 and 2), which corresponds to the product obtained using proviral DNA as a template (Fig. 4C, lane 3). In contrast, when primer −1412, complementary to the splice flanking region, was used in combination with primer 781Mtv-2, only spliced messages could be detected (Fig. 4D, lane 1), whereas the full-length product was obtained when proviral DNA was used as a template (Fig. 4D, lane 2). Sequencing of the spliced fragments obtained after PCR amplification (Fig. 4D, lane 1) confirmed that the splice donor and acceptor at positions 825 and 1275, respectively, are used.
A novel doubly spliced transcript originates at the LTR promoter complex.
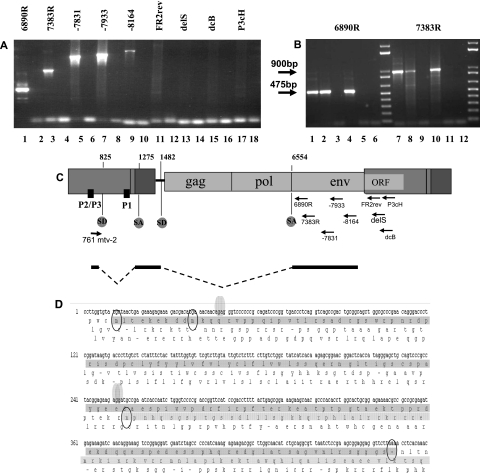
In order to facilitate the detection of potential transcripts originating from the LTR promoter complex, we first analyzed MMTV transcription products from CΔP1AnE11, a CrFK cell clone stably transfected with pmtv2ΔP1pAneoES (31). Basically, pmtv2ΔP1pAneoES carries a full-length MMTV provirus with the exception of a short deletion of the major P1 promoter core in the 5′ LTR. In addition, pmtv2ΔP1pAneoES carries three copies of a polyadenylation signal immediately upstream of the provirus to avoid any readthrough coming either from the bacterial backbone of the plasmid or from the host genome after integration. The integrity of the proviral sequences in the CΔP1AnE11 clone was demonstrated by Southern blotting analyses (31). Total RNA extracted from CΔP1AnE11 was subjected to reverse transcription using poly(dT) primers. Using 781Mtv-2, a forward primer hybridizing 20 nucleotides downstream of the 5′ end of the expected RNA product of the P3 promoter, and a collection of specific reverse primers homologous to sequences situated downstream of the splice acceptor described above (SA1275), a series of PCRs was performed using cDNA from CΔP1AnE11 as a template (Fig. 5A). No specific amplification could be obtained using any reverse primers from the gag, pro, and pol regions (data not shown). However, we observed a strong signal with oligonucleotide 6890R, a primer homologous to the sequence located just downstream of the splice acceptor for Env (SA6554) as well as with oligonucleotides 7383R, −7831, and −7933, all hybridizing downstream of SA6554 within the first half of the env region of the provirus (Fig. 5A). All reverse primers homologous to the 3′ end of the env sequence or to the beginning of the 3′ LTR did not give any PCR signals (Fig. 5A). Since the PCR conditions were accurate for long fragment detection, we cannot rule out alternative polyadenylation in env downstream from primer −8164. All PCR products were sequenced and revealed a doubly spliced message, the sequence of which is shown in Fig. 5D. The complete coding region of the cDNA was readily recovered using primer 7383R as a reverse primer. The first intron is located between SD825 and SA1275, in the U3 and U5 of the 5′ LTR, respectively, and the second intron starts at position 1482 (SD1482) in the gag untranslated region just upstream of the pp21 methionine start codon and is also used as a splicing site for env. This intron ends at the splice acceptor for env (SA6554). The novel, doubly spliced MMTV RNA transcript carries an ORF of 459 nucleotides spanning three exons. Two methionine codons, 25 bp apart, are potential start codons in the first very short exon, which is located upstream of the first splice donor, SD825. The second exon overlaps part of U5 and of the gag nontranslated region, and the third exon ends in the env region but is not in frame with the envelope ORF (Fig. 5D).
FIG. 5.
A doubly spliced message originates from the LTR promoter complex. Total RNA extract from the CΔP1AnE11 cell clone was subjected to reverse transcription using poly(dT) primers. (A) Electrophoresis gel obtained after a series of PCRs using the same forward primer, 781Mtv-2 (hybridizing 20 nucleotides downstream of primer 761mtv2), and different reverse specific primers (arrows in the schematic representation of the provirus given below) of env and LTR sequences is shown here. Each amplification corresponds to two lanes: the left one (odd number) was done in the presence of the cDNA template, and the right one (even number) was done in the absence of cDNA template. Transcripts originating in the central part of the LTR are detectable in lanes 1, 3, 5, 7, and 9. (B) Electrophoresis gel obtained after PCRs using the same forward primer, 761mtv2, and either reverse primer 6890R (lanes 1 to 6) or reverse primer 7383R (lanes 7 to 11). Templates were cDNA from GR cells (lanes 1 and 7), from N2AnE5 cells (lanes 2 and 8), from NMuMG cells (lanes 3 and 9), from C2AnE15 cells (lanes 4 and 10), and from CrFK cells (lanes 5 and 11). Lanes 6 and 12 are no-template PCR controls. Transcripts originating in the central part of the LTR are detectable in cDNAs from the three MMTV producer cell lines GR, N2AnE5, and C2AnE15. (C) Sequencing of these products revealed a doubly spliced message carrying a novel ORF whose schematic representation, in scale with the provirus, is given here. (D) The sequence of the novel ORF is underlined in dark gray. Splicing sites are indicated by dots. The novel ORF revealed by this study is different from the coding frame for the MMTV envelope, which is underlined in light gray.
Using an Mtv-2 forward primer specific for the transcription start in the 5′ LTR as detected by 5′ RACE (primer 761mtv2) in combination with the reverse −6890R primer, or primer 7383R, we could detect the same doubly spliced transcript in cDNA from the MMTV producer GR (cDNA-GR) (Fig. 5B, lanes 1 and 7, respectively), in cDNA from the MMTV producer N2AnE5 (cDNA-N2AnE5) (Fig. 5B, lanes 2 and 8, respectively), and in cDNA from the MMTV producer C2AnE15 (cDNA-C2AnE15) (Fig. 5B, lanes 4 and 10, respectively). Therefore, the novel doubly spliced MMTV transcript is not dependent on the deletion of the P1 promoter core in the 5′ LTR of the provirus since its production is also detected in cells carrying a full-length provirus containing a wild-type 5′ LTR.
The novel doubly spliced MMTV transcript is translated into a 20-kDa protein in mouse cells.
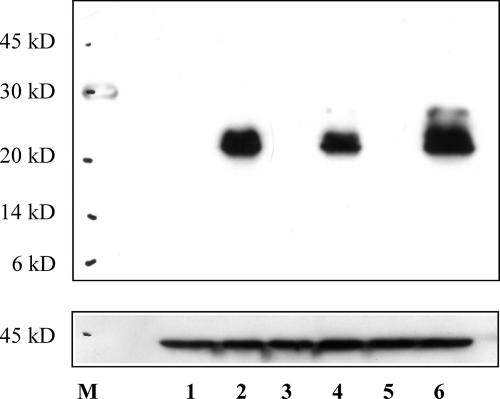
We have shown above that the LTR promoter complex produces, independently of hormone stimulation, a doubly spliced viral mRNA carrying a 459-nucleotide-long ORF (Fig. 5). To analyze whether this novel MMTV transcript obtained from cDNA from GR cells can also give rise to a protein, we cloned the ORF with a 3′ His tag into the mammalian expression vector pcDNA3Hyg. After transient transfections of CrFK, NIH 3T3, and NMuMG cells, protein extracts were assayed by Western blotting using a polyclonal rabbit anti-His antiserum (Fig. 6). The same specific signal was obtained in all three cell lines, indicating the expression of a protein with an apparent molecular mass of around 20 kDa (Fig. 6, lanes 2, 4, and 6). This observation is consistent with the size of this protein of around 17 kDa as calculated from the sequence.
FIG. 6.
The novel doubly spliced LTR transcript can be expressed in murine cells. pcDNA3.1ORFeHis carries a C-terminal His tag fusion of the novel ORF obtained from a GR cell cDNA preparation. After transient transfections of NIH 3T3, NMuMG, and CrFK cells with pcDNA3.1ORFeHis, 20 μg of total protein extract was separated by SDS-polyacrylamide gel electrophoresis, and the novel protein was detected using a polyclonal rabbit anti-His antibody. Lanes 1, 3, and 5 show protein extracts from nontransfected NIH 3T3, NMuMG, and CrFK cells, respectively. Lanes 2, 4, and 6 show protein extracts from transfected NIH 3T3, NMuMG, and CrFK cells, respectively. After stripping, the same blot was assayed with a mouse polyclonal anti-actin antiserum (line below). M, molecular mass marker (in kilodaltons).
DISCUSSION
Identification of a promoter cluster in the center of the MMTV LTR.
Using an improved reporter system for luciferase expression, we have been able to identify and partially characterize a promoter complex located upstream of the classic P1 promoter in the geographic center of both the Mtv-2 and Mtv-8 LTRs. One previously described promoter, P2 (15, 33, 36), is regulated both positively and negatively by adjacent, previously described, elements, while the novel promoter P3, described here, is regulated only by the downstream positive regulatory element. A minimal Mtv-8 P3 promoter, which is lacking two of the three putative promoter elements (TATA and CAAT boxes) that are carried by the equivalent minimal Mtv-2 P3 promoter, is as active as its Mtv-2 homologue, strongly suggesting that the conserved consensus TATA box located at position 721 is the TATA box of the novel P3 promoter.
The P2 and P3 basal promoter elements are highly conserved among MMTV isolates, as are the genetic elements necessary for their activity or regulation, such as the respective cap sites, the splicing sites (37), and the NRE sequences (Fig. 2), although the sequence otherwise presents some variability. The specific conservation of the promoter elements in different MMTV isolates and variants argues for a biological relevance of these promoters.
Efficient splicing at SD825/SA1275.
Two sets of independent and consistent data argue for the importance of the splicing event occurring within the LTR sequences.
In order to follow the fate of transcripts originating in the LTR promoter cluster and especially to test potential splicing events at previously reported sites located within the LTR (15, 37), PCRs were carried out using reverse-transcribed mRNAs from GR cells as a template. A nonspliced message could be detected only when primers designed to bind within the intron sequence were used (Fig. 4C, lanes 1 and 2). When primers complementary to the splice flanking regions were used, only spliced messages could be detected (Fig. 4D, lane 1). Although an advantage in the amplification of shorter over longer fragments cannot be ruled out, this observation suggests that the transcripts originating from the central part of the LTR are very efficiently spliced, since the difference in size between the nonspliced and the spliced products is only 400 nucleotides. The absence of detection of nonspliced messages by reverse transcriptase PCR when using flanking primers suggests that the splice donor site at position 825 is efficiently used and that the splicing event occurs very soon after transcription.
A doubly spliced message giving rise to a 20-kDa protein in mouse cells is one product of this promoter complex.
All the experiments leading to the detection of the novel transcript were performed on mRNA extracts from cells that had not been treated with dexamethasone (Fig. 5). cDNA from a stable clone carrying a full-length MMTV provirus but lacking the P1 promoter core in the 5′ LTR was first used for the detection of transcription products originating in the center of the 5′ U3. A strong and specific PCR signal was obtained using a forward primer identical to the first bases of the expected RNA product of the novel P3 promoter and a specific reverse primer in the proximal env region of the provirus. Cloning and sequencing of the PCR product thus obtained revealed a doubly spliced transcript carrying a 459-nucleotide-long ORF. It could be shown that the second splice acceptor used in the generation of this transcript is identical to the splice acceptor for Env at position 6554 relative to the beginning of the proviral sequence, whereas the third exon of this novel ORF is in the env region but is not in frame with the envelope leader sequence (Fig. 5D). We confirmed the existence of this transcript in MMTV producer GR, N2AnE5, and C2AnE15 cells, all carrying full-length proviral sequences with wild-type 5′ LTRs. We could then show that this novel MMTV transcript isolated from GR cells codes for a protein that can be produced in mouse NIH 3T3 and NMuMG cells as well as in MMTV-permissive CrFK cells. In all cell lines tested, a similar protein with an apparent molecular mass of around 20 kDa was produced, which is consistent with the theoretically estimated 17-kDa size. The function of the MMTV protein detected during this work, the production of which is not dependent on glucocorticoid hormones, remains to be elucidated. We are raising a rabbit polyclonal antiserum directed against a peptide to allow easier characterization. Since the P2 and P3 promoters are only 47 bp apart and the transcripts directed by these promoters seem to follow the same splicing pattern for the first intron within the LTR sequence, it is not yet clear which of these promoters is actually responsible for the production of this novel MMTV protein. This point is also currently under investigation.
Only P2 is negatively regulated by an NRE located upstream.
The P2 and P3 promoters do not function in a synergistic fashion, even in the presence of their common enhancer, and constitute a promoter complex in the central part of the MMTV LTR in between two essential regulatory regions. On the one side, a short upstream flanking region that has been reported due to its inhibitory effects on MMTV transcription (27) correlates exactly with the only NRE that we could identify in this study and appears here to be strictly devoted to the repression of P2 activity. On the other side, a previously reported downstream enhancer (12, 23, 27) affects both P2 and P3 activities.
It has been proposed that the cellular transcription factor CCAAT displacement protein (CDP) is a repressor of MMTV expression and may play a key role in the regulation of MMTV during pregnancy and lactation (24, 40, 41). As can be seen in Fig. 2, the CDP binding sites described previously by Zhu and Dudley (41) that lie in the central promoter complex correlate exactly with the start of transcription of P2 (14), the sequence between the P2 TATA box and its Cap site, and the CAAAT box upstream of the P2 TATA box, which is included in the NRE shown here to downregulate P2 activity. In the luciferase reporter analysis presented here, we observed the repression of P2 promoter activity when these CDP binding sites are present on the LTR fragment controlling luciferase activity, regardless of whether Mtv-2 or Mtv-8 sequences are used (pZTBP2 compared to pZ151 and pZP2mtv8 compared to pZdP2mtv8, respectively) (Fig. 1 and 3). Even though there are additional CDP binding sites in the enhancer region, the enhancer effect is not abrogated by the presence of the CDP sites within the P2 promoter (pZP2a2 compared to pZTBP2 and pZP2a2mtv8 compared to pZP2mtv8, respectively) (Fig. 1 and 3) but is abrogated by the presence of the CDP binding sites in the NRE (pZ151a2 compared to pZP2a2 and pZ150a2mtv8 compared to pZP2a2mtv8, respectively) (Fig. 1 and 3), thus suggesting (i) an essential role of the CDP binding sites, which are located at the CAAAT site of the NRE, in the control of P2 activity and (ii) no involvement of the CDP binding sites in the enhancer in the control of this promoter. We propose here that CDP binding to all sites surrounding P2 may form a superstructure that hides the core promoter and therefore impairs the formation of the transcription complex. This hypothesis remains to be tested and supposes a tight control of the activity of the P2 promoter at a higher level, allowing its activity in the natural proviral context only at a certain stage of the viral replication cycle, when CDP cannot bind to the MMTV LTR.
Since the genetic elements of the promoter complex are extremely conserved among the different exogenous MMTV strains and endogenous Mtv loci, it is likely that they have biological significance. In fact, some of the regulatory elements, which were first described as being modulators of P1 promoter activity (at a time when it was thought that P1 was the only LTR promoter), appear to be located at exactly the same positions as elements that we could show to be essential for the regulation of the promoters constituting the LTR central promoter complex. Therefore, either all three promoters present in the MMTV LTR share some regulatory elements or elements that were first thought to regulate the classical P1 promoter are indeed responsible only for modulating the activity of one or both of the central promoters, the activity of which might in turn modulate the activity of the major P1 promoter or of the R initiator recently shown to be potentially active on its own (31).
Acknowledgments
This work was supported by the Christian Doppler Gesellschaft.
We thank Elzbieta Knapp (Research Institute for Virology and Biomedicine) for help with the fluorescence-activated cell sorter analyses as well as the members of the institute for helpful comments and reagents.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Ball, J. K., H. Diggelmann, G. A. Dekaban, G. F. Grossi, R. Semmler, P. A. Waight, and R. F. Fletcher. 1988. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J. Virol. 62:2985-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Reference deleted.
- 4.Bramblett, D., C. L. Hsu, M. Lozano, K. Earnest, C. Fabritius, and J. Dudley. 1995. A redundant nuclear protein binding site contributes to negative regulation of the mouse mammary tumor virus long terminal repeat. J. Virol. 69:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasier, A. R., J. E. Tate, and J. F. Habener. 1989. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. BioTechniques 7:1116-1122. [PubMed] [Google Scholar]
- 6.Buetti, E., and H. Diggelmann. 1983. Glucocorticoid regulation of mouse mammary tumor virus: identification of a short essential DNA region. EMBO J. 2:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buetti, E., and B. Kühnel. 1986. Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J. Mol. Biol. 190:379-389. [DOI] [PubMed] [Google Scholar]
- 8.Cato, A. C., P. Skroch, J. Weinmann, P. Butkeraitis, and H. Ponta. 1988. DNA sequences outside the receptor-binding sites differently modulate the responsiveness of the mouse mammary tumour virus promoter to various steroid hormones. EMBO J. 7:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crandell, R. A., C. G. Fabricant, and W. A. Nelson-Rees. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176-185. [DOI] [PubMed] [Google Scholar]
- 10.Fasel, N., K. Pearson, E. Buetti, and H. Diggelmann. 1982. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1:3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giffin, W., H. Torrance, H. Saffran, H. L. MacLeod, and R. J. Hache. 1994. Repression of mouse mammary tumor virus transcription by a transcription factor complex. Binding of individual components to separated DNA strands. J. Biol. Chem. 269:1449-1459. [PubMed] [Google Scholar]
- 12.Gouilleux, F., B. Sola, B. Couette, and H. Richard-Foy. 1991. Cooperation between structural elements in hormono-regulated transcription from the mouse mammary tumor virus promoter. Nucleic Acids Res. 19:1563-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groner, B., N. E. Hynes, U. Rahmsdorf, and H. Ponta. 1983. Transcription initiation of transfected mouse mammary tumor virus LTR DNA is regulated by glucocorticoid hormones. Nucleic Acids Res. 11:4713-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Günzburg, W. H., F. Heinemann, S. Wintersperger, T. Miethke, H. Wagner, V. Erfle, and B. Salmons. 1993. Endogenous superantigen expression controlled by a novel promoter in the MMTV long terminal repeat. Nature 364:154-158. [DOI] [PubMed] [Google Scholar]
- 15.Günzburg, W. H., R. M. Saller, and B. Salmons. Retroviral vectors directed to predefined cell types for gene therapy. Biologicals 23:5-12. [DOI] [PubMed]
- 16.Hsu, C. L., C. Fabritius, and J. Dudley. 1988. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J. Virol. 62:4644-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hynes, N., A. J. Van Ooyen, N. Kennedy, P. Herrlich, H. Ponta, H., and B. Groner. 1983. Subfragments of the large terminal repeat cause glucocorticoid-responsive expression of mouse mammary tumor virus and of an adjacent gene. Proc. Natl. Acad. Sci. USA 80:3637-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Indik, S., W. H. Günzburg, B. Salmons, and F. Rouault. 2005. Mouse mammary tumor virus infects human cells. Cancer Res. 65:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Jeanson, L., and J. F. Mouscadet. 2002. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 277:4918-4924. [DOI] [PubMed] [Google Scholar]
- 20.Kang, C. J., and D. O. Peterson. 1999. Identification of a protein that recognizes a distal negative regulatory element within the mouse mammary tumor virus long terminal repeat. Virology 264:211-219. [DOI] [PubMed] [Google Scholar]
- 21.Kusk, P., K. E. Carlson, B. S. Warren, and G. L. Hager. 1995. Role of the TATA box in transcription of the mouse mammary tumor virus long terminal repeat. Mol. Endocrinol. 9:1180-1192. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. W., P. G. Moffitt, K. L. Morley, and D. O. Peterson. 1991. Multipartite structure of a negative regulatory element associated with a steroid hormone-inducible promoter. J. Biol. Chem. 266:24101-24108. [PubMed] [Google Scholar]
- 23.Lefebvre, P., D. S. Berard, M. G. Cordingley, and G. L. Hager. 1991. Two regions of the mouse mammary tumor virus long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol. Cell. Biol. 11:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, J., D. Bramblett, Q. Zhu, M. Lozano, R. Kobayashi, S. R. Ross, and J. P. Dudley. 1997. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol. Cell. Biol. 17:5275-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell, I. H., G. S. Harrison, W. M. Wood, and F. Maxwell. 1989. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques 7:276-280. [PubMed] [Google Scholar]
- 26.Michalides, R., E. Wagenaar, and P. Weijers. 1985. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol. Cell. Biol. 5:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mink, S., H. Ponta, and A. Cato. 1990. The long terminal repeat region of the mouse mammary tumour virus contains multiple regulatory elements. Nucleic Acids Res. 18:2017-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustafa, F., S. Bhadra, D. Johnston, M. Lozano, and J. P. Dudley. 2003. The type B leukemogenic virus truncated superantigen is dispensable for T-cell lymphomagenesis. J. Virol. 77:3866-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuss, F. U., and J. M. Coffin. 2000. The mouse mammary tumor virus transcription enhancers for hematopoietic progenitor and mammary gland cells share functional elements. J. Virol. 74:8183-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ringold, G. M., K. R. Yamamoto, J. M. Bishop, and H. E. Varmus. 1977. Glucocorticoid-stimulated accumulation of mouse mammary tumor virus RNA: increased rate of synthesis of viral RNA. Proc. Natl. Acad. Sci. USA 74:2879-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rungaldier, S., S. B. N. Asl, W. H. Günzburg, B. Salmons, and F. Rouault. 2005. Abundant authentic MMTV-Env production from a provirus lacking the major LTR promoter. Virology 342:201-214. [DOI] [PubMed] [Google Scholar]
- 32.Salmons, B., B. Groner, C. M. Calberg-Bacq, and H. Ponta. 1985. Production of mouse mammary tumour virus upon transfection of a recombinant proviral DNA into cultured cells. Virology 144:101-114. [DOI] [PubMed] [Google Scholar]
- 33.Salmons, B., T. Miethke, S. Wintersperger, M. Müller, G. Brem, and W. H. Günzburg. 2000. Superantigen expression is driven by both MMTV LTR promoters in transgenic mice. J. Virol. 74:2900-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka, H., Y. Dong, Q. Li, S. Okret, and J. A. Gustafsson. 1991. Identification and characterization of a cis-acting element that interferes with glucocorticoid-inducible activation of the mouse mammary tumor virus promoter. Proc. Natl. Acad. Sci. USA 88:5393-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theunissen, H. J., M. Paardekooper, L. J. Maduro, R. J. Michalides, and R. Nusse. 1989. Phorbol ester-inducible T-cell-specific expression of variant mouse mammary tumor virus long terminal repeats. J. Virol. 63:3466-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wintersperger, S., B. Salmons, T. Miethke, V. Erfle, H. Wagner, and W. H. Günzburg. 1995. Negative acting factor and superantigen are separable activities encoded by the mouse mammary tumor virus long terminal repeat. Proc. Natl. Acad. Sci. USA 92:2745-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, L., T. J. Wrona, and J. P. Dudley. 1997. Strain-specific expression of spliced MMTV RNAs containing the superantigen gene. Virology 236: 54-65. [DOI] [PubMed] [Google Scholar]
- 38.Yanagawa, S., A. Murakami, and H. Tanaka. 1990. Extra mouse mammary tumor proviruses in DBA/2 mouse lymphomas acquire a selective advantage in lymphocytes by alteration in the U3 region of the long terminal repeat. J. Virol. 64:2474-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanagawa, S., K. Kakimi, H. Tanaka, A. Murakami, Y. Nakagawa, Y. Kubo, Y. Yamada, H. Hiai, K. Kuribayashi, T. Masuda, and A. Ishimoto. 1993. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J. Virol. 67:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Q., K. Gregg, M. Lozano, J. Liu, and J. P. Dudley. 2000. CDP is a repressor of mouse mammary tumor virus expression in the mammary gland. J. Virol. 74:6348-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, Q., and J. P. Dudley. 2002. CDP binding to multiple sites in the mouse mammary tumor virus long terminal repeat suppresses basal and glucocorticoid-induced transcription. J. Virol. 76:2168-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]