Abstract
Low-virulence Newcastle disease viruses (loNDV) are frequently recovered from wild bird species, but little is known about their distribution, genetic diversity, or potential to cause disease in poultry. NDV isolates recovered from cloacal samples of apparently healthy waterfowl and shorebirds (WS) in the United States during 1986 to 2005 were examined for genomic diversity and their potential for virulence (n = 249). In addition 19 loNDV isolates from U.S. live bird markets (LBMs) were analyzed and found to be genetically distinct from NDV used in live vaccines but related to WS-origin NDV. Phylogenetic analysis of the fusion protein identified nine novel genotypes among the class I NDV, and new genomic subgroups were identified among genotypes I and II of the class II viruses. The WS-origin viruses exhibited broad genetic and antigenic diversity, and some WS genotypes displayed a closer phylogenetic relationship to LBM-origin NDV. All NDV were predicted to be lentogenic based upon sequencing of the fusion cleavage site, intracerebral pathogenicity index, or mean death time in embryo assays. The USDA real-time reverse transcription-PCR assay, which targets the matrix gene, identified nearly all of the class II NDV tested but failed to detect class I viruses from both LBM and WS. The close phylogenetic proximity of some WS and LBM loNDV suggests that viral transmission may occur among wild birds and poultry; however, these events may occur unnoticed due to the broad genetic diversity of loNDV, the lentogenic presentation in birds, and the limitations of current rapid diagnostic tools.
Viruses of the avian paramyxovirus serotype 1 (APMV-1), synonymous with Newcastle disease viruses (NDV), belong to the genus Avulavirus in the family Paramyxoviridae (34) and encompass a diverse group of single-stranded, negative-sense, nonsegmented RNA viruses of approximately 15.2 kb that have a broad host range in bird species. The virulent forms of the viruses, which exhibit an intracerebral pathogenicity index (ICPI) of ≥0.7, are the cause of Newcastle disease, a notifiable avian disease that must be reported to the World Organization for Animal Health. Classically these viruses have been grouped by virulence phenotype, with lentogenic, mesogenic, and velogenic strains reflecting increasing levels of virulence. Those viruses categorized as either mesogens or velogens are currently defined as virulent NDV (vNDV), and lentogens are viruses of low virulence (loNDV). NDV can cause clinical signs ranging from subclinical infections to 100% morbidity and/or mortality, depending on the virulence of the virus and the susceptibility of the host. Lentogenic viruses typically cause subclinical infections or mild respiratory disease. Mesogens are of intermediate virulence, usually resulting in moderate respiratory disease with occasional nervous signs. Velogens, the most virulent viruses, are those that may cause extensive hemorrhagic lesions, particularly in the gastrointestinal tract (viscerotropic), and/or a predominance of nervous signs (neurotropic) (4). vNDV are considered exotic to U.S. poultry; however, the lentogenic strains are common among domestic poultry (27, 32, 36, 45) and wild bird populations (25, 27, 43, 50). The occurrence of vNDV infections in birds is a notifiable event, and when these infections occur in poultry, trade restrictions are imposed. vNDV infections present a significant threat to the U.S. poultry industry, as evidenced by the California outbreak in 2002 to 2003 (41), which resulted in the destruction of 3.3 million birds and cost nearly $200 million dollars to eradicate (53).
NDV have historically been grouped into either genotypes (9) or genetic lineages (2) under one serotype (APMV-1). Recent analysis of the genome sizes and sequences of the F and L genes has revealed two distinct clades within APMV-1: classes I and II (14). While there are few published genomic sequences of class I isolates, which have been recovered primarily from waterfowl of the order Anatidae and samples from U.S. live bird markets (LBMs), the class II viruses comprise the vast majority of sequenced NDV and include isolates recovered from poultry (gallinaceous birds) and from pet and wild birds (2, 45). The class II NDV are further categorized into genotypes I to IX, with the genomic sequences of commonly used vaccine strains resembling vNDV isolated during the 1940s (class II, genotype II).
The natural ecology of NDV is not fully understood, and research has focused mainly on tracing either the origin of specific viruses or the spread of virulent viruses during Newcastle disease outbreaks in poultry (5-7, 12, 15, 21, 41, 44). Of the few studies characterizing NDV infections in wild bird populations, some suggest that waterfowl provide a natural reservoir for NDV, and epidemiological links between outbreak isolates recovered from poultry and those isolates found in wild bird populations have been hypothesized (18, 25-27, 42, 50). For example, phylogenetic analyses have identified vNDV isolates recovered from pigeons and migrating cormorants as the likely source of some NDV outbreaks in poultry (1, 10, 22, 31, 37, 51).
Although recent data have implicated U.S. LBMs as the source of respiratory pathogens such as avian influenza virus (AIV) and NDV that cause disease in domestic poultry (11, 32, 40, 45, 46), there is a paucity of data on the relationship between NDV circulating in wild waterfowl and shorebirds (WS) and NDV causing infections in poultry from LBMs. Earlier reports on the genomic analysis of NDV recovered in U.S. LBMs showed that these loNDV isolates were related to class II NDV strains which have been used for live vaccines in commercial poultry (32), but three U.S. LBM isolates recovered more recently, during 2001 and 2002, are representative of class I viruses (45). Additionally, the recently reported LBM-origin isolates from Hong Kong were also found to be class I viruses (28).
Understanding the epidemiology of loNDV infections in waterfowl and LBM poultry has been hampered by the facts that these viruses typically produce no clinical signs and that class I isolates are often not detected by rapid screening methods such as reverse transcription-PCR (RT-PCR). Sensitive rapid diagnostic assays exist to detect class II NDV, such as the USDA-validated real-time RT-PCR, which targets the matrix gene (M gene assay); however, due to the heterogeneous genetic nature of this virus, class I viruses tested often fail to be detected by the M gene assay (28). Previous evaluation of the nucleotide sequence alignment of the M gene assay probe sites of class I and II viruses revealed a high degree of mismatches between the two clades, and this is likely the reason that the class I viruses escape detection by real-time RT-PCR (28, 29).
Here, NDV from U.S. LBMs (n = 19) that demonstrated NDV-specific hemagglutination inhibition (HI) activity with polyclonal antiserum, but which tested negative for NDV using the M gene assay, were partially sequenced and found to be genetically distinct and phylogenetically distant from vaccine and virulent viruses reported worldwide. In addition, 249 NDV isolates recovered from cloacal samples collected during 1986 to 2005 from apparently healthy WS in the United States were analyzed, and selected isolates were tested using the M gene assay and a panel of monoclonal antibodies (MAbs). We established the phylogenetic relationship between the LBM- and WS-origin NDV isolates, determined the geographic distribution and broad genetic diversity among the WS-origin viruses, and evaluated the potential for virulence of U.S. WS-origin viruses.
MATERIALS AND METHODS
Hemagglutination (HA) and HI assays.
The HA and HI assays were completed by microtiter methods. The HA assay of allantoic fluids harvested from inoculated embryonating eggs was used to identify NDV-positive embryos. Confirmation of NDV-positive fluids and antigenic characterization of virus isolates were conducted by HI using microtiter methods as previously described (30). Four HA units of viral test antigen were used in completing the HI assay with MAbs and polyclonal antiserum.
Isolates and sequence data.
NDV were obtained from the Southeast Poultry Research Laboratory (SEPRL) repository or collected during a multi-institutional cooperative WS virus monitoring project between the University of Georgia Southeastern Cooperative Wildlife Disease Study, the University of Alaska Museum, The Ohio State University Department of Veterinary Preventive Medicine, and USDA-SEPRL (47). Viruses from wild-captured WS were isolated from cloacal swab material by standard virus isolation methods in embryonating chicken eggs (3, 49). Two hundred forty-nine U.S. WS isolates (Table 1) were obtained from monitoring samples for shorebirds of the family Scolopacidae in Delaware (n = 17) and New Jersey (n = 8) and from duck and goose sampling in Maryland (n = 121), Minnesota (n = 43), Ohio (n = 15), Louisiana (n = 16), Texas (n = 26), and Alaska [n = 3; includes one previously published isolate, Northern Pintail/US(AK)/196/1998 (GenBank accession no. EF027165)]. The 19 U.S. LBM environmental and poultry isolates were obtained from surveillance samples submitted to the USDA APHIS National Veterinary Services Laboratory (NVSL) from Massachusetts (n = 1), New Jersey (n = 10), New York (n = 7), and Rhode Island (n = 1). In addition, previously described samples from the Agriculture, Fisheries, and Conservation Department of Hong Kong SAR (n = 21) (28) and Shelduck/France/MC-110/1977 (GenBank accession no. AF003726) were included, totaling 290 isolates (see Table S1 in the supplemental material). For comparisons, all isolates were grouped into four major categories based on the source: Anas platyrhynchos (mallard and Anas rubripes or black duck, n = 169), other Anatidae (other ducks and geese, n = 56), Scolopacidae (shorebirds, n = 25), and LBM (environmental and domestic poultry isolates, n = 40).
TABLE 1.
Distribution of NDV isolates by source group, class, and genotype (n = 290)
| Source group (order) | Species name | Common name or typeb | No. of isolatesa
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I genotype:
|
Class II
|
Total | |||||||||||||||
| 1 | 2 | 3* | 4 | 5 | 6 | 7 | 8 | 9 | Total | I | Ia | IIa | Total | ||||
| WS | |||||||||||||||||
| Anas platyrhynchosc | Anas platyrhynchos | Mallard (MLD) | 1 | 9 | 0 | 10 | 65 | 0 | 30 | 0 | 1 | 116 | 7 | 1 | 45 | 53 | 169 |
| Other Anatidae (Anseriformes) | Anas discors | Blue-winged teal (BWT) | 2 | 1 | 21 | 24 | 1 | 7 | 8 | ||||||||
| Branta canadensis | Canada goose (CGS) | 3 | 3 | 1 | 1 | ||||||||||||
| Anas sp. | Wild duck (DCK) | 1 | 1 | 0 | |||||||||||||
| Anas crecca | Green-winged teal (GWT) | 2 | 2 | 1 | 2 | 7 | 0 | ||||||||||
| Anas fulvigula | Mottled duck (MTD) | 1 | 1 | 2 | 4 | 4 | |||||||||||
| Anas acuta | Northern pintail (NOP) | 1 | 1 | 2 | 2 | 2 | |||||||||||
| Tadorna tadorna | Shelduck (SHD) | 1* | 1* | 0 | |||||||||||||
| Aix sponsa | Wood duck (WDK) | 1 | 1 | 0 | |||||||||||||
| 1* | 1* | ||||||||||||||||
| Total | 7 | 5 | 0 | 1 | 1 | 0 | 3 | 1* | 23 | 40 | 0 | 1 | 14 | 15 | 55 | ||
| Scolopacidae (Charadriiformes) | Calidris alpina | Dunlin (DLN) | 1 | 1 | 2 | 0 | |||||||||||
| Calidris minutilla | Least sandpiper (LSP) | 1 | 1 | 0 | |||||||||||||
| Calidris canutus | Red knot (RKN) | 6 | 6 | 1 | 1 | ||||||||||||
| Arenaria interpres | Ruddy turnstone (RDT) | 4 | 4 | 8 | 6 | 6 | |||||||||||
| Calidris alba | Sanderling (SDL) | 0 | 1 | 1 | |||||||||||||
| Total | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 6 | 0 | 17 | 0 | 8 | 0 | 8 | 25 | ||
| U.S. total | 8 | 14 | 0 | 11 | 66 | 0 | 44 | 6 | 24 | 173 | 7 | 10 | 59 | 76 | 249 | ||
| 14* | |||||||||||||||||
| LBM | Gallus gallus | Chicken (CHK) | 1 | 14* | 2 | 1 | 4* | 0 | |||||||||
| Gallus sp. | Domestic fowl (DFL) | 2* | 2* | 0 | |||||||||||||
| Phasianidae | Mixed poultry (PLT) | 5* | 5* | 0 | |||||||||||||
| Anas sp. | Domestic duck (DDK) | 1 | 1 | 0 | |||||||||||||
| Gallus gallus | Egg (EGG) | 1 | 1 | 0 | |||||||||||||
| NA | Environment (ENV) | 1 | 11 | 1 | 13 | 0 | |||||||||||
| Total | 21* | 21* | |||||||||||||||
| 1 | 0 | 21* | 1 | 15 | 2 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 19 | |||
| U.S. total | 9 | 14 | 0 | 12 | 81 | 2 | 44 | 6 | 24 | 192 | 7 | 10 | 59 | 76 | 268 | ||
| Non-U.S. total | 21* | 1* | 22 | 290 | |||||||||||||
Asterisks indicate non-U.S. isolates.
Abbreviations used in the phylogenetic analysis are in parentheses.
Includes one environmental mallard isolate and two black duck (Anas rubripes) isolates.
RNA was extracted from allantoic fluids using Trizol LS (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 750 μl of Trizol LS reagent was added to 250 μl of allantoic fluid, vortexed, and incubated at room temperature for 7 min. The RNA was separated into the aqueous phase with the addition of 200 μl of chloroform, precipitated with isopropanol, and then centrifuged to pellet the RNA. After one wash with 70% ethanol, RNA was dried and resuspended in RNase-free water. PCR amplification of the RNA was performed using the QIAGEN one-step RT-PCR kit (QIAGEN, Valencia, CA). Amplified products were separated on a 1% agarose gel, the bands excised and eluted using the QIAquick Gel extraction kit (QIAGEN), and the samples quantified using a standard spectrophotometer.
All sequencing reactions were performed with fluorescent dideoxynucleotide terminators in an automated sequencer (ABI 3700 automated sequencer; Applied Biosystems Inc., Foster City, CA). Nucleotide sequence editing and analyses were conducted with the LaserGene sequence analysis software package (LaserGene version 5.07; DNAStar, Inc., Madison, WI). Using the full-length genome positions from the NDV LaSota vaccine strain complete genome (accession no. AF077761), the homologous regions sequenced were as follows: a 374-bp partial F gene (positions 4554 to 4917; n = 206), a 254-bp partial fragment spanning the M and F genes (positions 4826 to 5066; n = 40), and the complete coding region for the F gene (positions 4544 to 6205; n = 21).
Phylogenetic analysis.
Maximum-likelihood (ML) phylogenetic analysis with bootstrap values for n = 100 replicates was performed using Phyml under the general time-reversible model of nucleotide substitutions, ML estimates of base frequencies, estimated transition/transversion ratio, and proportions of invariable sites with four categories of substitution rates (19). For DNAML trees (16) default conditions were used, and distance-based analysis conducted with the neighbor-joining method was done using the Tajima-Nei nucleotide substitution model in the BioEdit sequence alignment editor (19a). The 374-bp region of the F gene, which has commonly been used for phylogenetic analysis of NDV (2), was sequenced to compare the U.S. LBM isolates (n = 19) to 209 WS viruses. For reference, previously published GenBank sequences from known NDV clades (class I, n = 13; class II, n = 47), in addition to 21 class I LBM isolates from Hong Kong (28), were included. The 254-bp region was used to compare the 40 remaining WS isolates (for a total of 249) to 106 LBM and WS viruses previously analyzed in the 374-bp analysis (data not shown). For the purpose of discussion and to maintain consistency with previous naming conventions, the term “genotype” is used here to describe isolates that reproducibly group together on a distinct branch of a phylogenetic tree. The class I genotypes were indicated using arabic numerals for ease of differentiating them from class II genotypes.
Pathogenicity assessment.
The pathogenic potential for selected class I isolates was evaluated using standard assay methods to determine the ICPI in 1-day-old chicks (3), and a modified procedure was used to determine mean death time (MDT) in embryonating eggs. The MDT was determined by inoculating the allantoic cavities of 9- to 11-day-old embryonating specific-pathogen-free eggs with serial 10-fold dilutions of a virus isolate. The eggs were incubated at 37°C and candled twice daily (early morning and late afternoon) for 7 days, and the time of embryo mortality was recorded. The MDT for a minimum lethal dose was interpreted as the mean time in hours for the embryo death in the highest dilution at which all eggs died (3).
Real-time RT-PCR.
Selected LBM and WS NDV isolates (n = 50) were tested using the USDA-validated M gene assay (53).
MAbs and antiserum.
Nine MAbs with different NDV specificities were used for differentiating isolates by the HI assay as previously described (30, 33). The MAbs obtained from USDA, APHIS, National Veterinary Services Laboratories (Ames, IA) included B79, 15C4, and 10D11 (35); AVS (48); and 617/161 (13). Additional MAbs prepared at SEPRL included P15D7, P11C9, P3A11, and P10B8. The reactivities of the MAbs were as follows: the AVS MAb inhibits many lentogens, including B1 and LaSota strains; 10D11 inhibits neurotropic velogens and mesogens such as the Roakin strain; 15C4 inhibits most APMV-1 strains except pigeon paramyxovirus type 1 (PPMV-1); B79 inhibits most APMV-1 strains, including most PPMV-1 strains; P15D7, P11C9, P3A11, and P10B8 identify additional antigenic diversity among APMV-1; and the 617/161 MAb inhibits only PPMV-1 within the APMV-1 group. A positive result was defined as antibody-inhibited HA, and a negative result was defined as no HI. The polyclonal chicken NDV antiserum was prepared at SEPRL by immunization of chickens with inactivated NDV-LaSota.
Nucleotide sequence accession number.
The sequences for 267 U.S. isolates reported here have the following GenBank accession numbers: 374-bp sequence, EF564874 to EF565079; 254-bp sequence, EF564834 to EF564873; and F gene sequence, EF564813 to EF564833 (see Table S1a in the supplemental material). Accession numbers for previously published sequences used in the analyses are in Table S1b in the supplemental material.
RESULTS
NDV isolates from U.S. LBMs were genetically distinct and phylogenetically distant from vaccine and virulent strains of NDV.
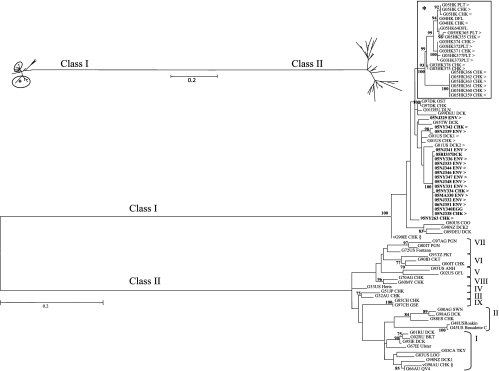
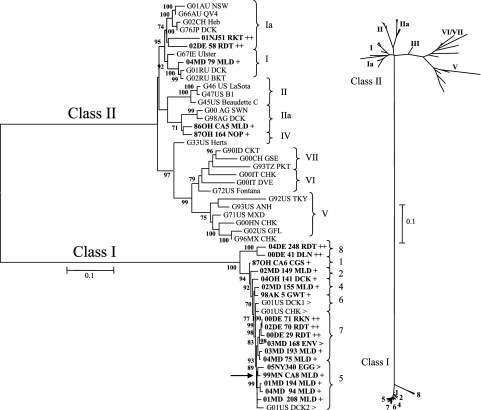
A 374-bp region of the fusion proteins from 19 U.S. LBM samples during 2005 to 2006 was sequenced and phylogenetically compared to the corresponding regions of representative viruses from the NDV class I and II clades (n = 81) (Fig. 1). Based upon analysis of this region, all of the U.S. LBM viruses corresponded to the class I clade and were therefore phylogenetically distant from class II viruses such as the vaccine viruses in genotypes I and II and the frequently observed virulent viruses (genotypes II to IX). The largest cluster of U.S. LBM-origin viruses (Fig. 1) in this study was most closely related to previously reported U.S. LBM-origin viruses from 2001 (G01US DCK1, -DCK2, and -CHK; accession no. AY626266 to -68) (45). Recently identified NDV recovered from Hong Kong LBM samples (Fig. 1) formed a separate cluster which lacked close association with other viruses. One of the U.S. LBM-origin NDV isolates (05NJ329ENV) grouped together with a duck virus isolated in Taiwan during 1995 (G95T DCK; accession no. AY135757) (2), and European isolates from Germany (G01DEU DLN and G99DEU DCK; accession no. DQ096595 and DQ097393, respectively) (14) and Denmark (G97DK CHK and -OST; accession no. AY175733 and AY75744, respectively) (2) formed a nearby cluster. Another cluster near the largest LBM cluster included WS viruses from the United States (G80US COO; accession no. DQ096594) (14), New Zealand (G98NZ DCK2; accession no. AY175774) (2), and Germany (G89DEU DCK; accession no. AY175732) (2). A virulent virus from Ireland that caused an outbreak in poultry during 1990 (vG90IE CHK; accession no. AY972102) (5) was also found to be in the class I clade (Fig. 1).
FIG. 1.
Phylogenetic comparison of a 374-bp region of the fusion gene from HI-positive, matrix real-time RT-PCR-negative U.S. LBM viruses (bold and designated by >; n = 19 [previously published designated with a G, n = 3]) to recent Hong Kong LBM isolates (boxed with asterisk; n = 21) and representative NDV from all known lineages (class I, n = 9; class II, n = 29). The tree was constructed using Phyml ML with 100 bootstrap replicates. The symbol § at isolates in class I and in class II, genotype I, represents atypical virulent viruses. Vaccine viruses are in genotypes I (G67IE Ulster and G66AU QV4) and II. All known class II viruses in genotypes II to IX are virulent viruses. The inset displays the distance between class I and class II viruses, with the large cluster of U.S. LBM isolates circled and the Hong Kong LBM circled with an asterisk. Class II genotypes are denoted in brackets. The scale indicates the branch length based on the number of nucleotide substitutions per site. Virus designations represent an 8- to 10-character name containing the two-digit year of collection, location abbreviation, unique virus identification (one to three characters), and species abbreviation (see Table S1a in the supplemental material). Previously published GenBank sequences are denoted by the letter G, the two-digit year, location abbreviation, and either a species abbreviation or common name (see Table S1b in the supplemental material).
LBM-origin isolates were found to be closely related to WS-origin isolates.
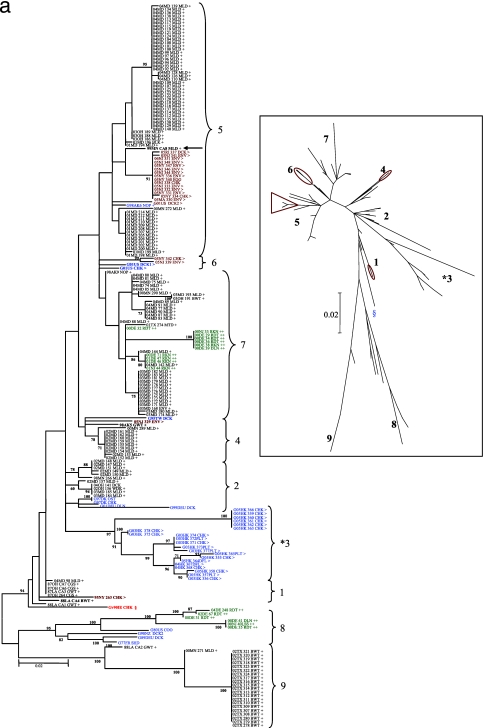
To investigate the source of the U.S. LBM loNDV, they were compared to 249 WS isolates obtained from healthy birds during 1986 to 2005 in eight different states (Table 1; see Table S1a in the supplemental material). Phylogenetic analysis of the 374-bp region demonstrated that U.S. LBM-origin viruses were related to class I WS-origin isolates and that of the nine class I genotypes identified, four were represented by U.S. LBM viruses (Fig. 2a). The largest cluster of U.S. LBM viruses, which were isolated from environmental samples, eggs, chickens, and ducks in several northeastern U.S. states during 2005 to 2006, corresponded to genotype 5 (15/19), which also included a large proportion of the U.S. WS isolates (60/173). The WS isolates in closest association with the U.S. LBM viruses were all from mallards (Anas platyrhynchos), the most notable of which was a 1999 isolate from Minnesota (Fig. 2a, 99MNCA8MLD, genotype 5) that is closely related to LBM viruses isolated in the northeastern United States during 2005 to 2006. Statistical analysis of the branches of this tree using ML confirmed the significant probability (>0.01) for the node connecting the 99MNCA8MLD isolate to the cluster of genotype 5 LBM isolates. Interestingly, genotype 6 was found only among the U.S. LBM isolates including previously described isolates from 2001 (G01US DCK1 and -CHK; accession no. AY626266 and -68, respectively) (45). A single U.S. LBM isolate from New York in 1995 (95NY263CHK) was classified as genotype 1, which comprised many of the oldest class I WS isolates (ca. 1987) identified here, and it was most similar to 88LACA4BWT (Fig. 2a). The only known virulent class I virus appears on a separate branch (vG90IECHK; accession no. AY972102) (5) apparently basal or sister to genotypes 1 to 7 (Fig. 2a, genotype 1), indicating that it represents a separate genotype. The final U.S. LBM isolate (05NJ329ENV) was categorized in genotype 4 and was most closely related to a 1995 Taiwan duck isolate (G95TW DCK; accession no. AY135757) (2) and a U.S. WS isolate from Alaska (98AK5GWT). The LBM viruses from Hong Kong were clearly separated from the other class I viruses and formed genotype 3 (Fig. 2a).
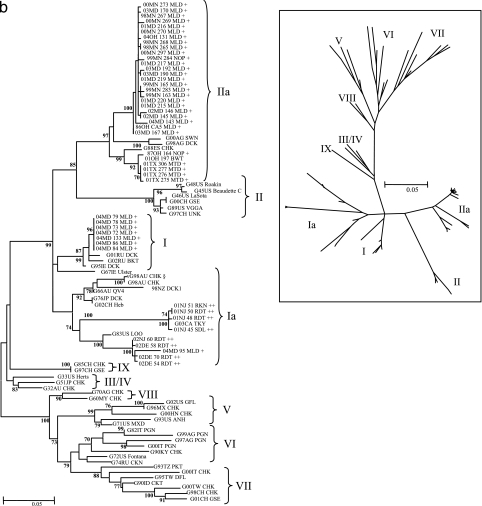
FIG. 2.
(a) Phylogenetic analysis of the 374-bp fusion gene fragment for class I WS (waterfowl designated by + in black typeface and shorebird designated by ++ in green typeface) and LBM (designated by > in brown) isolates with previously published GenBank sequences for reference (designated with a G in blue). The tree was constructed using Phyml ML with 100 bootstrap replicates. The asterisk indicates LBM isolates from Hong Kong; the arrow represents close relation to LBM viruses; and § indicates a virulent isolate, which as a single outlier was not assigned a genotype. The inset displays the relative distance between genotypes, and demarcations show locations of LBM isolates. The scale indicates the branch length based on the number of nucleotide substitutions per site. (b) Phylogenetic analysis of the 374-bp fusion gene fragment for class II waterfowl (designated by +) and shorebird (designated by ++) isolates with previously published GenBank sequences for reference (designated with a G). The tree was constructed using Phyml ML with 100 bootstrap replicates. Vaccine viruses are in genotypes I (G67IE Ulster and G66AU QV4) and II (G46US LaSota and G89US VGGA). The Inset displays the relative distance between genotypes. The scale indicates the branch length based on the number of nucleotide substitutions per site.
Broad phylogenetic diversity was found among the U.S. WS isolates (n = 249) representing class I and II viruses from nine different genotypes.
Seven of the nine novel class I genotypes were represented by U.S. WS isolates (Table 1 and Fig. 2a). For the class I WS viruses, those from Anatidae species (especially those from mallards) were found to be related to the LBM viruses, while the isolates from shorebirds tended to form distinct clusters within genotypes 7 and 8 that were distinct from either the Anatidae or LBM viruses (Fig. 2a). Some temporal and geographic clustering was observed; however, this could be due to the isolation of multiple related samples from the same location, and multiple genotypes were often present in the same year and location (Table 2).
TABLE 2.
Distribution of U.S. WS and LBM NDV isolates by year, state of collection, class, and genotype (n = 268)
| Source | Yr | Statea | No. of isolates
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I genotype:
|
Class II
|
Total | ||||||||||||||
| 1 | 2 | 4 | 5 | 6 | 7 | 8 | 9 | Total | I | Ia | IIa | Total | ||||
| WS | 1986 | OH | 1 | 1 | 1 | |||||||||||
| 1987 | LA | 2 | 1 | 3 | 1 | 6 | 7 | 10 | ||||||||
| OH | 3 | 3 | 3 | 3 | 6 | |||||||||||
| 1988 | LA | 2 | 2 | 2 | 6 | 6 | ||||||||||
| 1998 | AK | 1 | 1 | 1 | 3 | 3 | ||||||||||
| MN | 1 | 3 | 4 | 6 | 6 | 10 | ||||||||||
| 1999 | MN | 4 | 4 | 21 | 21 | 25 | ||||||||||
| 2000 | DE | 7 | 3 | 10 | 10 | |||||||||||
| MN | 1 | 1 | 1 | 1 | 4 | 4 | 4 | 8 | ||||||||
| NJ | 1 | 1 | 2 | 2 | ||||||||||||
| 2001 | DE | 2 | 2 | 2 | ||||||||||||
| MD | 16 | 16 | 5 | 5 | 21 | |||||||||||
| NJ | 1 | 1 | 4 | 4 | 5 | |||||||||||
| OH | 1 | 1 | 1 | |||||||||||||
| TX | 1 | 1 | 4 | 4 | 5 | |||||||||||
| 2002 | DE | 1 | 1 | 3 | 3 | 4 | ||||||||||
| MD | 6 | 9 | 15 | 2 | 2 | 17 | ||||||||||
| NJ | 1 | 1 | 1 | |||||||||||||
| OH | 1 | 1 | 1 | |||||||||||||
| TX | 21 | 21 | 21 | |||||||||||||
| 2003 | MD | 2 | 15 | 17 | 4 | 4 | 21 | |||||||||
| OH | 3 | 1 | 4 | 4 | ||||||||||||
| 2004 | DE | 1 | 1 | 1 | ||||||||||||
| MD | 1 | 38 | 14 | 53 | 7 | 1 | 1 | 9 | 62 | |||||||
| OH | 1 | 1 | 1 | 1 | 2 | |||||||||||
| Total | 8 | 14 | 11 | 66 | 0 | 44 | 6 | 24 | 173 | 7 | 10 | 59 | 76 | 249 | ||
| LBM | 1995 | NY | 1 | 1 | 1 | |||||||||||
| 2005 | MA | 1 | 1 | 1 | ||||||||||||
| NJ | 1 | 7 | 1 | 9 | 9 | |||||||||||
| NY | 5 | 1 | 6 | 6 | ||||||||||||
| RI | 1 | 1 | 1 | |||||||||||||
| 2006 | NJ | 1 | 1 | 1 | ||||||||||||
| Total | 1 | 0 | 1 | 15 | 2 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 19 | ||
| Total | 9 | 14 | 12 | 81 | 2 | 44 | 6 | 24 | 192 | 7 | 10 | 59 | 76 | 268 | ||
AK, Alaska; DE, Delaware; LA, Louisiana; MA, Massachusetts; MD, Maryland; MN, Minnesota; NJ, New Jersey; NY, New York; OH, Ohio; RI, Rhode Island; TX, Texas.
The class II WS viruses formed distinct clusters related to the previously described genotypes I and II (Fig. 2b); for the purpose of discussion, these subgroups will be referred to as Ia and IIa. A cluster of mallard isolates from Maryland during 2004 was closely related to viruses in genotype I which are primarily lentogenic viruses such as the 1967 Ulster strain (G67IE Ulster; accession no. AY562991). However, no viruses closely related to the widely used LaSota and B1 live vaccines were isolated from any of the WS-origin samples. A separate cluster of predominantly shorebird viruses formed a subgroup (genotype Ia) with the viruses endemic to Australia (vG98AU CHK, G98AU CHK, and G66AUQV4; accession no. AY175722, AY175658, and AF217084, respectively), New Zealand (G98NZ DCK1, accession no. AY175730), Asia (G76JP DCK and G02CH Heb; accession no. M24705 and AF217084, respectively), and the United States (G03CA TKY and G83US LOO; accession no. AY175642 and AY175736, respectively). Of particular note was the presence of an isolate from a domestic 12-week-old turkey from a farm in Ontario, Canada (G03CA TKY) within a closely related cluster of shorebird isolates from the U.S. Anatidae viruses related to genotype II. This cluster, which includes a diverse group of lentogens and mesogens in addition to many vaccine strains, formed another subgroup (IIa) with previously published WS viruses from Argentina (G98AG DCK and G00AG SWN; accession no. AY727881 and -82) (14) and a chicken isolate from Spain (G88ES CHK; accession no. AY175642) (2).
Overall, the majority of WS-origin viruses were isolated from mallards, which represented nearly 70% of the WS viruses sequenced (169/249) (Table 1), and the most frequently observed genotype among these viruses was class I genotype 5 (65/249). The Anatidae species alone were host to genotypes 1, 2, 4, 5, 7, and 9 of class I and to I, Ia, and IIa of class II (Table 1), with genotypes 5 and 7 being identified most often. The shorebird (Scolopacidae) isolates were predominantly of genotype 7, with genotype 5 being conspicuously absent among these viruses. Class II genotype IIa represented the third most common genotype (59/249) (Table 1), with mallards representing the most common host.
Diverse WS genotypes were distributed across eight states over the 20-year period.
Multiple genotypes circulated across the United States over time and within a region during a given year (Table 2); for example, three different class I genotypes occurred in Alaska during 1998 (from Anatidae spp.), and four class I genotypes occurred in Minnesota during 2000 (all from mallards). In Maryland during 2004 this held true for both class I and II viruses as well. Multiple genotypes were also found circulating within a specific source group (species) in the Maryland region (Table 3). Mallards during 2001 to 2004 (n = 101) were infected by five of the nine class I genotypes (1, 2, 4, 5, and 7). The data in Table 3 also demonstrated a change in the predominating genotype from year to year. Whereas genotype 5 was most commonly observed in this subset during 2001 (16/16), it was not identified again until 2004 (38/54). Interestingly, genotype 5 was most prevalent in mallards in 2004 and in the U.S. LBM isolates during 2005 to 2006 (15/17).
TABLE 3.
Distribution of U.S. class I NDV isolates from resident, wild mallard ducks (Anas platyrhynchos) collected in Maryland by year and genotype (n = 101)a
| Collection yr | No. of isolates
|
|||||
|---|---|---|---|---|---|---|
| Class I genotype:
|
Total | |||||
| 1 | 2 | 4 | 5 | 7 | ||
| 2001 | 16 | 16 | ||||
| 2002 | 6 | 9 | 15 | |||
| 2003 | 2 | 15 | 17 | |||
| 2004 | 1 | 38 | 14 | 53 | ||
| Total | 1 | 8 | 9 | 54 | 29 | 101 |
Only those genotypes represented by isolates are shown (five of nine genotypes).
The broad phylogenetic diversity found in the 374-bp F gene analysis was confirmed by analysis of the complete coding region of the F gene.
Representative WS viruses and LBM viruses were analyzed using Phyml ML with bootstrap values for n = 100 replicates (Fig. 3) (bootstrap values of ≥70 are shown for each major node), and the results were reproducible using both DNAML and neighbor-joining techniques (data not shown). These results confirmed the phylogenetic diversity observed using the shorter F gene genomic regions, clearly distinguished the LBM viruses from vaccine and virulent viruses, and validated the close phylogenetic relationship between WS and LBM viruses.
FIG. 3.
Phylogenetic comparison of the complete coding regions of the fusion genes from U.S. LBM viruses (designated by >; n = 2) and waterfowl (designated by +) and shorebird (designated by ++) isolates (n = 21) with previously published NDV from other known genotypes as reference (all designated by a G; class I LBM [designated by >, n = 3; class II, n = 25). The tree was constructed using Phyml ML with 100 bootstrap replicates. Genotypes for classes I and II are denoted in brackets; the arrow within genotype 5 represents a close relation to LBM viruses. The inset displays the distance between class I and class II viruses. The scale indicates the branch length based on the number of nucleotide substitutions per site.
WS isolates from the United States possessed a highly attenuated phenotype.
Classical pathotyping using MDT, ICPI, and molecular classification based on the deduced amino acid sequence of the fusion cleavage site were used to determine the pathogenic potentials of representative WS isolates (Tables 4 and 5). Eighty WS isolates representing various genotypes among classes I and II were pathotyped using the ICPI in day-old chicks (Table 4). All isolates tested (80/80) had low ICPI values of <0.2 (on a 0 to 2.0 scale). The result of the MDT assay in eggs was >90 h (data not shown) for 58 of these 80 isolates, which is characteristic of lentogenic viruses and indicates that the viruses tested have a low potential to cause significant disease in poultry (3).
TABLE 4.
Results for ICPI assay of NDV isolates by class and genotype (n = 80)
| ICPIa | No. of isolates
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I genotype:
|
Class II
|
Total | |||||||||
| 1 | 2 | 4 | 5 | 7 | 8 | 9 | I | Ia | IIa | ||
| 0-0.1 | 8 | 5 | 1 | 8 | 7 | 2 | 3 | 1 | 5 | 33 | 73 |
| >0.1-0.2 | 1 | 2 | 4 | 7 | |||||||
| Total no. of isolates | 8 | 6 | 1 | 10 | 7 | 2 | 3 | 1 | 5 | 37 | 80 |
Viruses with an ICPI of <0.7 are considered lentogens.
TABLE 5.
Deduced fusion cleavage site amino acid motifs (positions 110 to 117) for NDV isolates from WS and LBM samples by class and genotype (n = 290)
| Fusion cleavage site motif | No. of isolatesa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I genotype:
|
Class II genotype
|
Total | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | I | Ia | IIa | ||
| EGERQERL | 5* | 5 | |||||||||||
| GGEQQERL | 6* | 6 | |||||||||||
| GGERQDRL | 16 | 1 | 17 | ||||||||||
| GGERQERL | 12 | 11 | 10* | 13 | 59 | 1 | 46 | 6 (1*) | 24 | 183 | |||
| GGERQGRL | 1 | 1 | |||||||||||
| GGVRQERL | 2 | 2 | |||||||||||
| EGGKQGRL | 4 | 4 | |||||||||||
| GGEKQGRL | 46 | 46 | |||||||||||
| GGGKQGRL | 8 | 5 | 8 | 21 | |||||||||
| GVEKQGRL | 1 | 1 | |||||||||||
| RGGKQGRL | 4 | 4 | |||||||||||
| Total | 12 | 11 | 21* | 13 | 75 | 2 | 49 | 6 (1*) | 24 | 8 | 9 | 59 | 290 |
Asterisks denote non-U.S. isolates.
The deduced amino acid sequences confirmed the presence of lentogenic fusion cleavage sites for all WS- and LBM-origin isolates characterized by the limited number of basic amino acids at positions 112 to 116 and the presence of leucine at position 117 (Table 5) (n = 290). The predominant motif among class I isolates, 110-GGERQERL-117, was found in 85.5% of isolates (183/214) and is common among other reported lentogenic strains (45). This motif was also found among U.S. (3/19) and Hong Kong (10/21) LBM isolates; however, the majority of the U.S. LBM isolates had the single-amino-acid substitution of aspartate for glutamate at position 115 (15/19). The remaining LBM isolates from Hong Kong shared two motifs that were unique to that group (11/21). The predominant motif among class II isolates, 110-GGEKQGRL-117, had the amino acid substitutions of lysine for arginine at position 113 and glycine for glutamate at position 115.
Broad phylogenetic diversity may affect the efficacy of rapid detection methods for class I WS and LBM viruses.
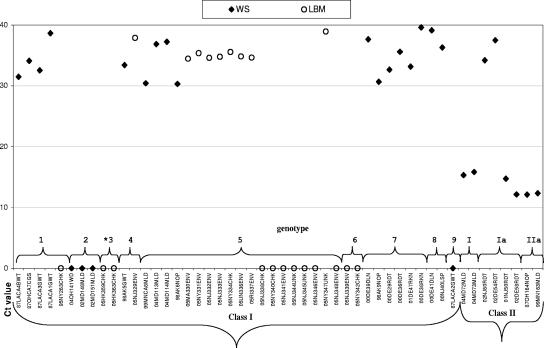
To determine whether the WS isolates would be detected using the USDA-validated M gene assay developed for class II NDV, a panel of 29 WS and 21 LBM viruses (n = 19 U.S. isolates and n = 2 Hong Kong isolates) were tested (class I, n = 42; class II, n = 8) (Fig. 4). Over two-thirds (30/42) of class I viruses were suspect or failed to be detected (cycle threshold [CT] value of ≥35 or zero), and the remainder (10/42) were weakly positive with high CT values (30 to 34). Although the majority of class II viruses (6/8) were accurately identified with the M gene assay, the remaining two viruses from ruddy turnstones in genotype Ia demonstrated suspect CT values (02NJ60RDT and 02DE54RDT; CT values, 34 to 38). All samples contained quantifiable RNA and produced a product of the expected size when amplified using F gene primers in a standard RT-PCR assay. Additionally, selected class I viruses were positive (CT values of <30) with a previously developed L gene-targeted real-time RT-PCR assay (28) (data not shown).
FIG. 4.
Distribution of CT values for 50 NDV isolates from WS (n = 29) and LBMs (n = 21) representing class I (n = 42) and II (n = 8), using the USDA matrix gene-targeted real-time RT-PCR assay. Class I genotype 3 (*) contains only isolates from LBMs in Hong Kong. CT value interpretation: lower values indicate earlier detection, values of ≥35 were considered suspect, and a value of zero is negative for the assay.
The 58 U.S. isolates from Anatidae species isolated during 1986 to 1999 that were used for MDT determination were also evaluated using a MAb binding assay with a novel panel (Table 6). Isolates were classified into seven distinct groups based on the MAb binding patterns, as follows: group 1 (n = 33) was characterized by binding of MAbs AVS, 15C4, B79, P15D7,P11C9, and P3A11; group 1A (n = 3) included binding of MAb P10B8; group 2 exhibited binding of 15C4, B79, P15D7, P11C9, and P3A11 (n = 1), whereas group 2A did not bind MAb P11C9 (n = 1); group 3 (n = 17) bound only to MAb B79; group 4 did not bind to any of the MAbs (n = 2); and group 5 demonstrated binding to MAbs AVS, 15C4, B79, P15D7, and P11C9 (n = 1). MAb groups 2, 3, and 4 correlated with class I viruses, with group 3 representing genotypes 1, 2, 4, 5, 7, and 9, which are found in the Anatidae group. Groups 1, 1A, 2A, and 5 corresponded to the class II viruses, with group I composed primarily of genotypes II and IIa. Although differentiation between class I and II viruses was possible using the MAb assay, only a few of the MAbs recognized class I viruses, and the assay was not capable of distinguishing differences between class I genotypes.
TABLE 6.
MAb patterns for class I (n = 20) and class II (n = 38) NDV isolates obtained from Anatidae species during 1986 to 1999a
| MAb group | MAb patternb
|
No. of isolates
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I genotype:
|
Class II
|
Total | ||||||||||||||||
| AVS | 10D11 | 15C4 | B79 | P15D7 | P11C9 | P3A11 | P10B8 | 617/161 | 1 | 2 | 4 | 5 | 7 | 9 | Ia | IIa | ||
| 2 | 1 | 1 | ||||||||||||||||
| 3 | − | − | − | + | − | − | − | − | − | 7 | 1 | 1 | 1 | 5 | 2 | 17 | ||
| 4 | − | − | − | − | − | − | − | − | − | 2 | 2 | |||||||
| 1 | + | − | + | + | + | + | + | − | − | 1 | 32 | 33 | ||||||
| 1A | + | − | + | + | + | + | + | + | − | 3 | 3 | |||||||
| 2A | − | − | + | + | + | − | + | − | − | 1 | 1 | |||||||
| 5 | + | − | + | + | + | − | − | − | − | 1 | 1 | |||||||
| 10 | 1 | 1 | 1 | 5 | 2 | 1 | 22 | 58 | ||||||||||
Seven different MAb patterns differentiated the viruses by class and distinguished some by genotype.
+, positive binding; −, no binding.
DISCUSSION
The discovery of a large number of class I NDV recovered from LBMs in the northeastern United States and the existence of genetically related viruses found in WS suggest that epidemiological connections may exist between them. Identification of an earlier LBM isolate (Fig. 2a, 95NY263CHK) in class I, genotype 1, which is related to the oldest WS-origin viruses analyzed here and of a class I genotype 5 mallard isolate (99MNCA8MLD) that was closely related to the largest cluster of 2005 to 2006 LBM isolates suggest that transmission between WS and birds in the LBM has occurred. While the mallard isolate (99MNCA8MLD) was geographically removed from where the cluster of LBM viruses was isolated (northeastern United States), the data for the geographic distribution combined with the natural mobility of wild birds indicates that NDV strains are likely highly mobile. Another explanation for the similarity between the WS- and LBM-origin viruses could be the existence of parallel reservoirs that are either evolutionarily static or evolving in similar directions and at comparable rates; however, this would be an evolutionarily implausible scenario based on the rapid capacity for RNA viruses to change, the history of evolution for other NDV (2, 51, 54), and the year-to-year changes in the phylogenetic trees presented here. In this study, there were a few cases where the class I LBM viral genotypes lacked WS counterparts (see, e.g., Fig. 2a, class I genotype 6 from the United States and genotype 3 from Hong Kong), and the data are simply insufficient to determine a viral ancestor. In each of these cases, increased sampling of at-risk domestic and wild birds may potentially reveal reservoirs of loNDV, but the absence of such data does not discard the possibility of viral transmission among these populations.
The identification of a class II genotype Ia virus from a 12-week-old domestic turkey in Ontario during 2003 (Fig. 2b, G03CA TKY) that clustered closely with U.S. shorebird isolates from New Jersey in 2001 (01NJRKN, RDT, and SDL) suggests that class II loNDV may also occasionally be transmitted between wild and domestic populations. It is noteworthy that while billions of doses of live NDV vaccine are used worldwide in poultry, the absence of class II vaccine or vaccine-like viruses in wild birds suggests that transmission from vaccinated domestic poultry into wild birds may not be a frequent occurrence.
The data here support a complex viral ecology for loNDV infections in wild birds, because they present evidence of circulation of more than one genotype within a species and within a geographic location as well as circulation of more than one genotype during the same year. The reoccurrence of genotypes over time presents the possibility of cyclic predominance of various lineages. Cyclic patterns of subtypes have been reported for AIV, where isolation of an HA subtype may be followed 1 to 2 years later by reduced isolation rates for the same subtype (39).
These data also provide substantial insight into the broad phylogenetic diversity of NDV and emphasize the potential for more genotypes yet to be discovered; however, this study is likely not sufficient to establish the precise genotypes that may be encountered. For example, viruses from class I genotypes 1, 2, and 6 were delineated based upon distance and clustering, but they may in fact represent additional genotypes. The ability to discern separate genotypes will improve over time as more viruses are identified and sequenced. The diversity observed at the phylogenetic level would be expected to follow at the antigenic and protein levels as well, as suggested by the MAb results and the differences in the deduced F proteins (up to 17.4% amino acid differences [data not shown]). Here we have explored only avian WS species from the orders Anseriformes and Charadriiformes, and while NDV has been documented to infect at least 241 different bird species (4), other nondomestic avian families in these orders and other nondomestic avian orders remain virtually unexplored for loNDV.
Although class I and II loNDV in this study were predicted to exhibit lentogenic phenotypes, concern exists regarding possible genetic changes to loNDV upon replication in poultry. Virulent viruses with genotypes similar to those found in WS and LBM have previously been recovered (5, 18). The 1990 Ireland outbreak was caused by a virulent class I virus (vG90IE CHK; accession no. AY972102), which in this study corresponds to genotype 1 or the progenitor-type viruses (Fig. 2a), and the Australian outbreak of 1998 to 2000 was caused by the mutation to virulence of a class II genotype I virus, which are typically lentogenic. Genomic analysis of the class II genotype I viruses (genotype Ia in the present study, which included several species of U.S. waterfowl) from the Australian outbreak provided evidence that lentogenic viruses have the potential to become virulent over time (18) and that a change in the cleavage site of the fusion protein of the native virus resulted in increased virulence (17).
The performance of two diagnostic tools commonly used for rapid identification of NDV viruses is also of concern. Approximately 70% of WS-origin isolates identified corresponded to class I viruses (174/249) that are poorly or not detected by the USDA M gene assay (28, 29). The use of MAb assays for rapid characterization may also not be optimal for class I viruses, as these assays have been developed and optimized predominantly to recognize class II viruses. Because MAbs are directed against single epitopes, their ability to detect a broad spectrum of viruses is often limited. Preliminary testing using an L gene-targeted real-time RT-PCR assay detected class I LBM viruses from Hong Kong (28) and other selected WS and LBM isolates (data not shown); however, due to the phylogenetic diversity of loNDV, multiple real-time RT-PCR tests may be needed to detect all WS genotypes.
Active surveillance of LBMs focused on the detection of vNDV is laudable, but many challenges still remain. LBMs promote the commingling of multiple bird species in close quarters and provide opportunities for transmission of disease agents (38, 40, 41, 46). In the United States, LBMs are part of a complex system that provides fresh poultry, typically in larger cities. Because these markets receive birds from multiple sources, confine multiple bird species in close quarters, and provide processing services onsite, they provide an environment in which reservoir species such as waterfowl are closely housed with gallinaceous hosts. While the USDA has been working to control low-pathogenic avian influenza in U.S. LBMs since 1986, the Uniform Standards were released in 2004 and the official program was instituted during 2006, which includes registration of LBM premises, documentation of test-negative status of birds entering the market, and testing of premises and examination of records (52). Over 15,000 samples were submitted to the NVSL specifically for AIV and NDV surveillance in 2003 (48a). In 2002, more than 15,000 specimens were tested at the NVSL for AIV and NDV surveillance; however, few of the surveillance samples originated from backyard poultry. Since the 2002 to 2003 outbreak of vNDV in California, Nevada, and Arizona, which was epidemiologically linked to infections in backyard poultry and game fowl, surveillance among this population has been significantly intensified. For WS populations, NDV monitoring has been sporadic and often occurs in conjunction with other monitoring programs, such as those for AIV (8, 20, 23, 24). Detection of vNDV in wild bird populations often occurs after outbreaks in poultry (18, 25-27, 50). To date, hardly any data exist regarding loNDV circulating in WS populations, and their highly attenuated phenotype makes them difficult to track among poultry because they may circulate undetected due to a lack of clinical signs.
In summary, we have demonstrated the broad genetic diversity and lentogenic presentation of loNDV from WS and have identified a close phylogenetic proximity with LBM-origin loNDV, suggesting that viral transmission may occur among wild birds and poultry. Evidence that NDV monitoring in the United States using the USDA M gene assay may not be adequate to detect all NDV from classes I and II emphasizes the need for new diagnostic assays to identify circulating reservoirs of loNDV in wild bird populations and LBM poultry. Further epidemiologic studies will be needed to clarify the prevalence and origin of loNDV in wild bird populations and LBM poultry, in addition to the ongoing genetic characterization of new isolates.
Supplementary Material
Acknowledgments
We gratefully acknowledge Joan Beck, Scott Lee, Dawn Williams-Coplin, Ginger Goekjian, Britta Hanson, and Page Luttrell for technical assistance; the South Atlantic Area Sequencing Facility for nucleotide sequencing; and Michael Wege for providing samples for screening.
This work was funded by USDA CRIS project numbers 6612-32000-039-00D, 6612-32000-041-01S, 6612-32000-041-04S, 6612-32000-041-06S and 6612-32000-039-03S, and USPEA (SEPEA no. 332).
Mention of trade names or commercial products in this paper is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 12 September 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aldous, E. W., C. M. Fuller, J. K. Mynn, and D. J. Alexander. 2004. A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathol. 33:258-269. [DOI] [PubMed] [Google Scholar]
- 2.Aldous, E. W., J. K. Mynn, J. Banks, and D. J. Alexander. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239-256. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, D. J. 1998. Newcastle disease virus and other avian paramyxoviruses, p. 156-163. In D. E. Swayne (ed.), A laboratory manual for the isolation and identification of avian pathogens. The American Association of Avian Pathologists, Kennett Square, PA.
- 4.Alexander, D. J. 2003. Newcastle disease, p. 64-87. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Disease of poultry. Iowa State Press, Ames, IA.
- 5.Alexander, D. J., G. Campbell, R. J. Manvell, M. S. Collins, G. Parsons, and M. S. McNulty. 1992. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet. Rec. 130:65-68. [DOI] [PubMed] [Google Scholar]
- 6.Alexander, D. J., H. T. Morris, W. J. Pollitt, C. E. Sharpe, R. L. Eckford, R. M. Sainsbury, L. M. Mansley, R. E. Gough, and G. Parsons. 1998. Newcastle disease outbreaks in domestic fowl and turkeys in Great Britain during 1997. Vet. Rec. 143:209-212. [DOI] [PubMed] [Google Scholar]
- 7.Alexander, D. J., G. W. Wilson, P. H. Russell, S. A. Lister, and G. Parsons. 1985. Newcastle disease outbreaks in fowl in Great Britain during 1984. Vet. Rec. 117:429-434. [DOI] [PubMed] [Google Scholar]
- 8.Alfonso, C. P., B. S. Cowen, and H. van Campen. 1995. Influenza A viruses isolated from waterfowl in two wildlife management areas of Pennsylvania. J. Wildl. Dis. 31:179-185. [DOI] [PubMed] [Google Scholar]
- 9.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee, M., W. M. Reed, S. D. Fitzgerald, and B. Panigraphy. 1994. Neurotropic velogenic Newcastle disease in cormorants in Michigan: pathology and virus characterization. Avian Dis. 38:873-878. [PubMed] [Google Scholar]
- 11.Bulaga, L. L., L. Garber, D. A. Senne, T. J. Myers, R. Good, S. Wainwright, S. Trock, and D. L. Suarez. 2003. Epidemiologic and surveillance studies on avian influenza in live-bird markets in New York and New Jersey, 2001. Avian Dis. 47:996-1001. [DOI] [PubMed] [Google Scholar]
- 12.Capua, I., P. M. Dalla, F. Mutinelli, S. Marangon, and C. Terregino. 2002. Newcastle disease outbreaks in Italy during 2000. Vet. Rec. 150:565-568. [DOI] [PubMed] [Google Scholar]
- 13.Collins, M. S., D. J. Alexander, S. Brockman, P. A. Kemp, and R. J. Manvell. 1989. Evaluation of monoclonal antibodies raised against an isolate of the variant avian paramyxovirus type I responsible for the current panzootic in pigeons. Arch. Virol. 104:53-62. [DOI] [PubMed] [Google Scholar]
- 14.Czegledi, A., D. Ujvari, E. Somogyi, E. Wehmann, O. Werner, and B. Lomniczi. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120:36-48. [DOI] [PubMed] [Google Scholar]
- 15.Farley, J. M., C. H. Romero, M. G. Spalding, M. L. Avery, and D. J. Forrester. 2001. Newcastle disease virus in double-crested cormorants in Alabama, Florida, and Mississippi. J. Wildl. Dis. 37:808-812. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 17.Gould, A. R., E. Hansson, K. Selleck, J. A. Kattenbelt, M. Mackenzie, and A. J. Della-Porta. 2003. Newcastle disease virus fusion and haemagglutinin-neuraminidase gene motifs as markers for viral lineage. Avian Pathol. 32:361-373. [DOI] [PubMed] [Google Scholar]
- 18.Gould, A. R., J. A. Kattenbelt, P. Selleck, E. Hansson, A. la-Porta, and H. A. Westbury. 2001. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998-2000. Virus Res. 77:51-60. [DOI] [PubMed] [Google Scholar]
- 19.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Hall, T. A. 1999. Abstr. Nucleic Acids. Symp. Ser. 41:95-98. [Google Scholar]
- 20.Hanson, B. A., D. E. Swayne, D. A. Senne, D. S. Lobpries, J. Hurst, and D. E. Stallknecht. 2005. Avian influenza viruses and paramyxoviruses in wintering and resident ducks in Texas. J. Wildl. Dis. 41:624-628. [DOI] [PubMed] [Google Scholar]
- 21.Haruna, E. S., D. Shamaki, G. O. Echeonwu, K. A. Majiyagbe, Y. Shuaibu, and D. R. Du. 1993. A natural outbreak of Newcastle disease in guinea-fowl (Numida meleagris galeata) in Nigeria. Rev. Sci. Tech. 12:887-893. [DOI] [PubMed] [Google Scholar]
- 22.Heckert, R. A., M. S. Collins, R. J. Manvell, I. Strong, J. E. Pearson, and D. J. Alexander. 1996. Comparison of Newcastle disease viruses isolated from cormorants in Canada and the USA in 1975, 1990 and 1992. Can. J. Vet. Res. 60:50-54. [PMC free article] [PubMed] [Google Scholar]
- 23.Hlinak, A., R. U. Muhle, O. Werner, A. Globig, E. Starick, H. Schirrmeier, B. Hoffmann, A. Engelhardt, D. Hubner, F. J. Conraths, D. Wallschlager, H. Kruckenberg, and T. Muller. 2006. A virological survey in migrating waders and other waterfowl in one of the most important resting sites of Germany. J. Vet. Med. B 53:105-110. [DOI] [PubMed] [Google Scholar]
- 24.Hlinak, A., T. Muller, M. Kramer, R. U. Muhle, H. Liebherr, and K. Ziedler. 1998. Serological survey of viral pathogens in bean and white-fronted geese from Germany. J. Wildl. Dis. 34:479-486. [DOI] [PubMed] [Google Scholar]
- 25.Huovilainen, A., C. Ek-Kommone, R. Manvell, and L. Kinnunen. 2001. Phylogenetic analysis of avian paramyxovirus 1 strains isolated in Finland. Arch. Virol. 146:1775-1785. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen, P. H., K. J. Handberg, P. Ahrens, H. C. Hansen, R. J. Manvell, and D. J. Alexander. 1999. An outbreak of Newcastle disease in free-living pheasants (Phasianus colchicus). Zentbl. Vet. B. 46:381-387. [DOI] [PubMed] [Google Scholar]
- 27.Jorgensen, P. H., K. J. Handberg, P. Ahrens, O. R. Therkildsen, R. J. Manvell, and D. J. Alexander. 2004. Strains of avian paramyxovirus type 1 of low pathogenicity for chickens isolated from poultry and wild birds in Denmark. Vet. Rec. 154:497-500. [DOI] [PubMed] [Google Scholar]
- 28.Kim, L., D. King, D. Suarez, C. Wong, and C. Afonso. 2007. Characterization of class I Newcastle disease virus isolates from Hong Kong bird markets and detection using real-time reverse transcription PCR. J. Clin. Microbiol. 45:1310-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, L., D. Suarez, and C. Afonso. 2007. Use of bioinformatics in improving detection of Newcastle disease virus, p. 227-249. In M. Rogeria de Almeida, M. Pires Moraes, J. Patarroyo, P. Vidigal, and A. Borem (ed.), Biotechnology and Animal Health International Meeting. Federal University of Viscosa, Viscosa, Brazil.
- 30.King, D. J. 1996. Avian paramyxovirus type 1 from pigeons: isolate characterization and pathogenicity after chicken or embryo passage of selected isolates. Avian Dis. 40:707-714. [PubMed] [Google Scholar]
- 31.King, D. J. 1996. Influence of chicken breed on pathogenicity evaluation of velogenic neurotropic Newcastle disease virus isolates from cormorants and turkeys. Avian Dis. 40:210-217. [PubMed] [Google Scholar]
- 32.King, D. J., and B. S. Seal. 1997. Biological and molecular characterization of Newcastle disease virus isolates from surveillance of live bird markets in the northeastern United States. Avian Dis. 41:683-689. [PubMed] [Google Scholar]
- 33.Kommers, G. D., D. J. King, B. S. Seal, and C. C. Brown. 2001. Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens. Avian Dis. 45:906-921. [PubMed] [Google Scholar]
- 34.Lamb, R., P. L. Collins, D. Kolakofsky, J. A. Melero, Y. Nagai, M. B. A. Oldstone, C. R. Pringle, and B. K. Rima. 2005. The negative sense single stranded RNA viruses, p. 607-738. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 35.Lana, D. P., D. B. Snyder, D. J. King, and W. W. Marquardt. 1988. Characterization of a battery of monoclonal antibodies for differentiation of Newcastle disease virus and pigeon paramyxovirus-1 strains. Avian Dis. 32:273-281. [PubMed] [Google Scholar]
- 36.Marin, M. C., P. Villegas, J. D. Bennett, and B. S. Seal. 1996. Virus characterization and sequence of the fusion protein gene cleavage site of recent Newcastle disease virus field isolates from the southeastern United States and Puerto Rico. Avian Dis. 40:382-390. [PubMed] [Google Scholar]
- 37.Meulemans, G., T. P. van den Berg, M. Decaesstecker, and M. Boschmans. 2002. Evolution of pigeon Newcastle disease virus strains. Avian Pathol. 31:515-519. [DOI] [PubMed] [Google Scholar]
- 38.Mullaney, R. 2003. Live-bird market closure activities in the northeastern United States. Avian Dis. 47:1096-1098. [DOI] [PubMed] [Google Scholar]
- 39.Olsen, B., V. J. Munster, A. Wallensten, J. Waldenstrom, A. D. Osterhaus, and R. A. Fouchier. 2006. Global patterns of influenza a virus in wild birds. Science 312:384-388. [DOI] [PubMed] [Google Scholar]
- 40.Panigrahy, B., D. A. Senne, and J. C. Pedersen. 2002. Avian influenza virus subtypes inside and outside the live bird markets, 1993-2000: a spatial and temporal relationship. Avian Dis. 46:298-307. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen, J. C., D. A. Senne, P. R. Woolcock, H. Kinde, D. J. King, M. G. Wise, B. Panigrahy, and B. S. Seal. 2004. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002-2003 outbreak in California and other recent outbreaks in North America. J. Clin. Microbiol. 42:2329-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfitzer, S., D. J. Verwoerd, G. H. Gerdes, A. E. Labuschagne, A. Erasmus, R. J. Manvell, and C. Grund. 2000. Newcastle disease and avian influenza A virus in wild waterfowl in South Africa. Avian Dis. 44:655-660. [PubMed] [Google Scholar]
- 43.Rosenberger, J. K., S. Klopp, and W. C. Krauss. 1975. Characterization of Newcastle disease viruses isolated from migratory waterfowl in the Atlantic flyway. Avian Dis. 19:142-149. [PubMed] [Google Scholar]
- 44.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seal, B. S., M. G. Wise, J. C. Pedersen, D. A. Senne, R. Alvarez, M. S. Scott, D. J. King, Q. Yu, and D. R. Kapczynski. 2005. Genomic sequences of low-virulence avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from live-bird markets in North America not related to commonly utilized commercial vaccine strains. Vet. Microbiol. 106:7-16. [DOI] [PubMed] [Google Scholar]
- 46.Senne, D. A., D. L. Suarez, J. C. Pedersen, and B. Panigrahy. 2003. Molecular and biological characteristics of H5 and H7 avian influenza viruses in live-bird markets of the northeastern United States, 1994-2001. Avian Dis. 47:898-904. [DOI] [PubMed] [Google Scholar]
- 47.Spackman, E., D. E. Stallknecht, R. D. Slemons, K. Winker, D. L. Suarez, M. Scott, and D. E. Swayne. 2005. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Res. 114:89-100. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasappa, G. B., D. B. Snyder, W. W. Marquardt, and D. J. King. 1986. Isolation of a monoclonal antibody with specificity for commonly employed vaccine strains of Newcastle disease virus. Avian Dis. 30:562-567. [PubMed] [Google Scholar]
- 48a.Suarez, D. L. 2004. Abstr. 108th U.S. Animal Health Association Annual Meeting, p. 93. U.S. Animal Health Association, St. Joseph, MO.
- 49.Swayne, D. E., D. A. Senne, and C. W. Beard. 1998. Avian influenza, p. 150-155. In D. E. Swayne (ed.), A laboratory manual for the isolation and identification of avian pathogens. The American Association of Avian Pathologists, Kennett Square, PA.
- 50.Takakuwa, H., T. Ito, A. Takada, K. Okazaki, and H. Kida. 1998. Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations. Jpn. J. Vet. Res. 45:207-215. [PubMed] [Google Scholar]
- 51.Ujvari, D., E. Wehmann, E. F. Kaleta, O. Werner, V. Savic, E. Nagy, G. Czifra, and B. Lomniczi. 2003. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 96:63-73. [DOI] [PubMed] [Google Scholar]
- 52.USDA APHIS. 2004. Prevention and control of H5 and H7 low pathogenicity avian influenza in the live bird marketing system, p. 1-21. USDA APHIS, Riverdale, MD.
- 53.Wise, M. G., D. L. Suarez, B. S. Seal, J. C. Pedersen, D. A. Senne, D. J. King, D. R. Kapczynski, and E. Spackman. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, L., Z. Wang, Y. Jiang, L. Chang, and J. Kwang. 2001. Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.