Abstract
The question of whether retroviruses, including human immunodeficiency virus type 1 (HIV-1), interact with the cellular RNA interference machinery has been controversial. Here, we present data showing that neither HIV-1 nor human T-cell leukemia virus type 1 (HTLV-1) expresses significant levels of either small interfering RNAs or microRNAs in persistently infected T cells. We also demonstrate that the retroviral nuclear transcription factors HIV-1 Tat and HTLV-1 Tax, as well as the Tas transactivator encoded by primate foamy virus, fail to inhibit RNA interference in human cells. Moreover, the stable expression of physiological levels of HIV-1 Tat did not globally inhibit microRNA production or expression in infected human cells. Together, these data argue that HIV-1 and HTLV-1 neither induce the production of viral small interfering RNAs or microRNAs nor repress the cellular RNA interference machinery in infected cells.
MicroRNAs (miRNAs) are small, ∼22-nucleotide (nt)-long regulatory RNAs that are expressed by all metazoan eukaryotes (reviewed in reference 2). The large majority of miRNAs are initially expressed as part of capped, polyadenylated RNA polymerase II (Pol II) transcripts called primary miRNA (pri-miRNA) precursors (7, 40). Within the pri-miRNA, the mature miRNA sequence forms part of one arm of an imperfect RNA stem-loop structure. The first step in miRNA processing involves the recognition of characteristic structural features of pri-miRNA stem-loops by the nuclear RNase III enzyme Drosha, acting in concert with the double-stranded RNA (dsRNA) binding protein DGCR8 (18, 22, 26, 29, 39, 65). Drosha cleaves both strands of the pri-miRNA stem-loop structure to liberate the pre-miRNA precursor, an ∼60-nt RNA hairpin bearing a 2-nt 3′ overhang. The pre-miRNA is exported to the cytoplasm where it is recognized by a second RNase III enzyme called Dicer (23, 29, 34). Dicer binds to the 2-nt 3′ overhang at the base of the pre-miRNA stem and then cleaves both RNA strands ∼22 nt away from its binding site to generate the miRNA duplex intermediate, an ∼20-bp dsRNA bearing two 2-nt 3′ overhangs. The miRNA strand of the duplex intermediate is incorporated into the RNA-induced silencing complex (RISC), where it acts as a guide RNA to target RISC to mRNAs bearing a complementary sequence (25, 45, 53). RISC binding can result in the degradation or translational silencing of target mRNAs (30, 65).
In addition to miRNAs, RISC can also be programmed by a second, related type of short regulatory RNAs called small interfering RNAs (siRNAs). While siRNAs are functionally similar to miRNAs, they differ in that they result from the cytoplasmic processing by Dicer of long dsRNAs, i.e., siRNA production does not require Drosha function (19, 25, 32). Biologically active siRNAs can be readily detected in invertebrate cells that have been transfected with dsRNAs or infected by RNA viruses (20, 42, 61). In contrast, long dsRNAs induce the interferon response in vertebrate cells, which results in a global inhibition of mRNA translation and often leads to cellular apoptosis, and it currently remains unclear whether siRNAs are produced naturally in somatic vertebrate cells (9, 15). However, artificial, biologically active siRNAs can be introduced into vertebrate cells in the form of dsRNA molecules too short to induce the interferon response that structurally mimic intermediates in the miRNA-processing pathway. Specifically, siRNAs can be introduced into vertebrate cells as one strand of an siRNA duplex (19), which mimics the structure of the miRNA duplex intermediate, or as part of a short hairpin RNA (shRNA) (6, 50), which mimics pre-miRNA hairpins. Moreover, shRNAs can be expressed in vivo as RNA Pol III transcripts and will then be exported from the nucleus and processed by Dicer to give functional siRNAs (63).
In principle, one can conceive of at least three ways in which the cellular RNA interference (RNAi) machinery might interact with an infecting virus. One possibility is that viral dsRNAs could be processed by Dicer into siRNAs that could protect the cell against the virus (24, 42). Strong evidence for RNAi as part of the antiviral innate immune response has been presented for both plants and invertebrates, but the question of whether RNAi forms part of the innate immune response in animals has been controversial (15). A second possibility is that the infecting virus could encode viral miRNAs that would program cellular RISCs to downregulate either cellular or viral mRNAs; clear evidence for virally encoded miRNAs has been presented for several DNA tumor viruses (17). Finally, cellular miRNAs might, by chance or due to evolutionary selection, show homology to regions of a viral RNA genome or viral mRNAs. In principle, this could then result in the specific inhibition of virus replication (36).
In this study, we have sought to establish whether human retroviruses, specifically, human immunodeficiency virus type 1 (HIV-1) or human T-cell leukemia virus type 1 (HTLV-1), interact with the cellular RNAi machinery. This question has been controversial. On the one hand, Pfeffer et al. (51) have previously reported that exhaustive cDNA cloning of small RNAs from HIV-1-infected CD4+ HeLa cells failed to detect any viral siRNAs or miRNAs. On the other hand, Bennasser et al. (3) reported the detection of an HIV-1-derived siRNA in HIV-1-infected Jurkat or CEM-SS T cells and argued that this siRNA was inhibitory to HIV-1 replication. Moreover, they also argued that the HIV-1 Tat protein, a potent nuclear transcription factor, was able to specifically block Dicer function and thereby reduce the production of HIV-1-specific siRNAs. In contrast, Omoto et al. (48) have argued that HIV-1 encodes an miRNA that can inhibit the expression of viral, and potentially also cellular, mRNAs. Here, we present data arguing that neither HIV-1 nor HTLV-1 expresses any miRNAs or siRNAs in infected human T cells and demonstrate that HIV-1 Tat is incapable of blocking either cellular miRNA function or RNAi targeted against a cellular gene.
MATERIALS AND METHODS
Cell culture and RNA preparation.
293T, HeLa, and MT-2 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. ACH-2, CEM, and CEM/TART (11) cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 10 mM HEPES. Total RNA was prepared using TRIzol (Invitrogen) according to the manufacturer's directions. Where appropriate, ACH-2 cells were induced with 1 μM phorbol myristate acetate (PMA) (Sigma) for 24 h prior to harvest. The levels of HIV-1 p24 Gag production were assessed by enzyme-linked immunosorbent assay (ELISA) (PerkinElmer) according to the manufacturer's directions.
Peripheral blood mononuclear cells (PBMCs) were prepared and infected as described in Tomaras et al. (56). Briefly, PBMCs were activated for 3 days with OKT3 and anti-CD28 antibodies, depleted of CD8+ cells, and allowed 1 day of rest prior to infection. Cells were infected with HIV-1 at a multiplicity of infection of 0.001 using the NL4-3 or QH0515 isolates or mock infected and then harvested 6 days postinfection. Filtered supernatants were collected for p24 ELISA, and total RNA was harvested with TRIzol for Northern blot analysis.
293T cells were transduced with pNL-SIN-CMV-BLR as described in Lee et al. (37). 293T cells transduced with pNL-BLR were generated by the same method, except that no pcTat was included in the initial transfection. Cells were placed under blasticidin selection 4 days after transduction and cultured for at least 21 days prior to use for experimentation.
Small RNA cloning.
The cloning of siRNAs and miRNAs out of HTLV-1- and HIV-1-infected cells was conducted as outlined by Lau et al. (35), using 750 μg of total RNA from MT-2 or ACH-2 cells. The identification and classification of sequenced small RNA cDNA clones were based on analysis using the GenBank and miRBase databases. The likelihood that we missed potential viral siRNAs in our small RNA cloning was calculated as outlined in Ho et al. (28) for zero-occurrence situations. Briefly, the risk can be estimated with 95% confidence to be no worse than 3/n, where n is the number of trials.
Molecular clones.
Plasmids pcTat, pK41A, pcTas, pcTax, pcRev, pHIT-G, pcβ-ARR2, pSUPER, pCMV/luc, pSUPER-luc, pBC12/CMV, pBC12/HIV/CAT, and pNL-SIN-CMV-BLR have all been previously described (4, 6, 33, 43, 52, 60, 63, 64). pRL is a Renilla luciferase reporter plasmid that was commercially purchased (Promega).
A PCR DNA fragment encoding the 101-amino-acid (aa) form of the HIV-1 Tat cDNA was generated as outlined in Ott et al. (49) and cloned into SalI-XhoI-digested pcTat to generate pcTat101. pNL-BLR was derived from pNL-CD4 (55) by the removal of the CD4 cDNA by NotI-XhoI digestion and its replacement with a PCR-generated blr cDNA. A full-length human TRIM5α cDNA bearing a C-terminal hemagglutinin (HA) tag was generated by reverse transcriptase PCR and cloned into pcDNA3 after AspI-XhoI digestion to generate pcTRIM5α-HA. The pSuper/shTRIM5α constructs were generated by annealing together DNA primers encoding the entire short hairpin and ligating the double-stranded fragments directly into BglII-HindIII-digested pSUPER. Three different shTRIM5α constructs were generated: shTRIM5α-1409 (5′-GATCCCCGCTGAAGAATTGGAAGATGACAATATTCATGTCAGCTTCCAACTCTTCAGCTTTTTGG-3′ and 5′-AGCTCCAAAAAGCTGAAGAGTTG GAAGCTGACATGAATATTGTCATCTTCCAATTCTTCAGCG GG-3′), shTRIM5α-1417 (5′-GATCCCCGCATGCCTCAATGCAAACTACA AGAATATTCATCTTGTGGTTTGCAGTGAGGCATGCTTTTTG-3′ and 5′-AGCTCAAAAAGCATGCCTCACTGCAAACCACAAGATGAATATTCTTGTAGTTTGCATTGAGGCATGCGGG-3′) and shTRIM5α-1422 (5′-GATCCCCGCCTTACGAATTCTGAAATTGAGATATATTCAATCTCAGTTTCAGAGTTCGTAAGGCTTTTTG-3′ and 5′-AGCTCAAAAAGCCTTACGAACTCTGAAACTGAGATTGAATATATCTCAATTTCAGAATTCGTAAGGC GGG-3′).
Northern blots.
MiRNAs were detected by Northern blotting as previously described (7), using 30 μg of total RNA. For Northern blots of HTLV-1 and HIV-1 genomic transcripts, 10 μg of total RNA from MT-2 or ACH-2 cells was separated by electrophoresis through a 0.6% agarose gel. RNA was transferred onto nitrocellulose and fixed by UV irradiation and baking at 80°C under vacuum. The blots were hybridized to probes generated by random priming of a BglII-SphI fragment of the HTLV-1 long terminal repeat (LTR) or a KpnI-HindIII fragment of the HIV-1 LTR. The bands were visualized by exposing the blots to film at −80°C with intensifying screens.
Western blots.
293T cells were transfected with 500 ng of pcTat, pcTas, pcTax, or pBC12/CMV, 500 ng of pcTRIM5α-HA, 500 ng of pβ-ARR2, and 500 ng total of pSuper/shTRIM5α (shTRIM5α-1409, shTRIM5α-1417, and shTRIM5α-1422) or pSUPER filler. The cells were harvested 48 h posttransfection and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Samples were transferred to nitrocellulose and probed with a monoclonal mouse antibody specific for the HA tag (Covance). The bands were visualized with Lumi-Light Western blotting substrate (Roche) according to the manufacturer's directions.
Luciferase assays.
HeLa cells were transfected with 3,000 ng of pSUPER-luc or pSUPER filler, 100 ng of pCMV/luc, 100 ng of pRL, and various amounts of pcTat101, pcTas, or pBC12/CMV as described in the figure. The cells were harvested 48 h posttransfection, and luciferase activities were assayed using a dual-luciferase reporter assay system (Promega) according to the manufacturer's directions.
RESULTS
HIV-1 and HTLV-1 do not express detectable miRNAs or siRNAs in infected T cells.
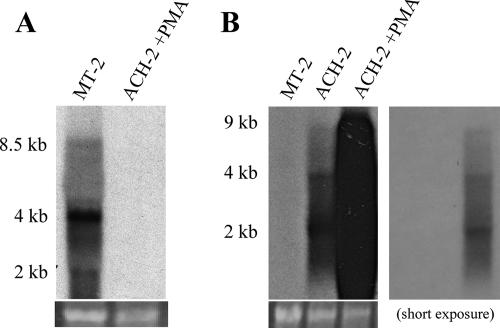
Previously, we and others have cDNA cloned numerous viral miRNAs from human or simian B-cells latently or persistently infected by a range of different DNA tumor viruses (reviewed in reference 17). To examine whether infection of human T cells with either HIV-1 or HTLV-1 would give rise to viral miRNAs or siRNAs, we focused on human T cells persistently infected with either HIV-1 or HTLV-1. The HIV-1-infected T-cell line ACH-2 (13) was initially derived by infection of the CD4+ T-cell line A3.01 using the LAV strain of HIV-1. ACH-2 cells continuously produce low levels of infectious HIV-1 virions in culture and also produce readily detectable levels of the viral structural proteins. Treatment with PMA initially results in the induction of high levels of HIV-1 virion production and then leads to cell death (13). As shown in Fig. 1B, we were able to detect genomic (∼9 kb), singly spliced (∼4 kb), and multiply spliced (∼2 kb) HIV-1 transcripts in both uninduced and PMA-induced ACH-2 cells using Northern blot analysis. In our hands, uninduced ACH-2 cells produced ∼4.1 ng of the HIV-1 p24 capsid protein per ml of supernatant media per day, as determined by ELISA, while treatment with PMA increased p24 production to ∼146 ng/ml.
FIG. 1.
MT-2 and ACH-2 cells express readily detectable levels of viral mRNAs. Total RNA samples were prepared from the HTLV-1-infected MT-2 cell line and the HIV-1-infected ACH-2 cell line, in the latter case either before or after PMA induction. Northern blot analyses were performed using random-primed probes specific for the viral LTRs. All predicted viral mRNAs, including genome-length transcripts, were detected in both MT-2 cells (A) and ACH-2 cells (B). Two exposures are provided for the ACH-2 mRNA Northern blot, to allow the visualization of the level of viral mRNAs produced in both the presence and absence of PMA. The lower panels show photographs of ethidium bromide stains of rRNA bands from the gels used for these Northern blot analyses and represent loading controls.
The HTLV-1-infected T-cell line MT-2 was originally derived by culture of normal human cord leukocytes with HTLV-1-infected adult T-cell leukemia cells (47). MT-2 cells continuously produce infectious HTLV-1 virions and express readily detectable levels of the viral structural proteins. As shown in Fig. 1A, we were able to detect genome-length (∼8.5 kb), singly spliced (∼4 kb), and multiply spliced (∼2 kb) HTLV-1 transcripts in MT-2 cells by Northern blot analysis.
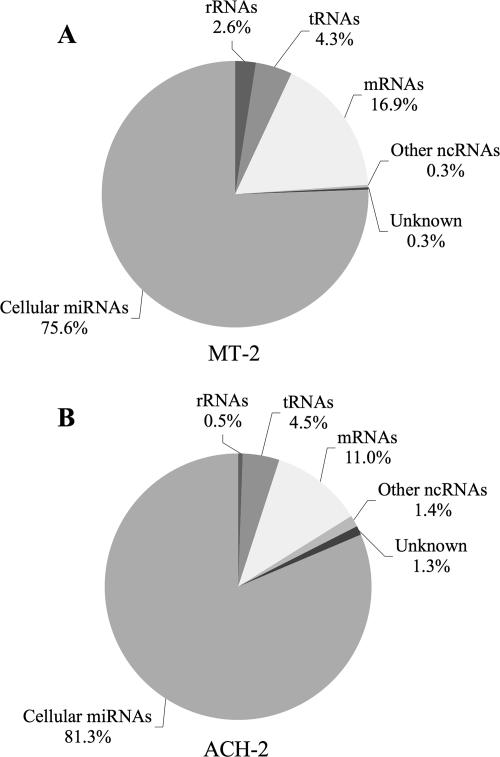
Using previously reported procedures (8, 35), we cDNA cloned 18- to 24-nt-long RNAs from both ACH-2 and MT-2 cells. The resultant cDNA clones were subjected to sequence analysis, and their origin was established by comparison to relevant sequence databases. Totals of 698 MT-2-derived cDNA clones and 625 ACH-2-derived cDNA clones were analyzed. This analysis revealed that cellular miRNAs accounted for the large majority of cDNA sequences derived from both MT-2 cells and ACH-2 cells. As shown in Fig. 2A, cellular miRNAs accounted for 528 (76%) of the 698 MT-2 cDNAs, while the remainder consisted largely of breakdown products of cellular mRNAs (118; 17%), tRNAs (30; 4%), or rRNAs (18; 3%). No cDNA clones of HTLV-1 origin were obtained. Similarly, cellular miRNAs accounted for 508 (81%) of the cDNA clones obtained from ACH-2 cells, with mRNA, tRNA, and rRNA fragments accounting for most of the remaining clones (Fig. 2B). Again, not a single short RNA derived from HIV-1 was obtained. A full listing of all the cellular miRNAs cloned from MT-2 and ACH-2 cells is given in Table 1 at http://dukecullenlab.googlepages.com/cullenlabmain.
FIG. 2.
Summary of the identities of small RNAs recovered from MT-2 or ACH-2 cells. RNAs of 18 to 24 nt in size were cDNA cloned from MT-2 and ACH-2 cells using standard procedures. Totals of 698 MT-2- and 625 ACH-2-derived cDNAs were sequenced and identified by reference to GenBank and miRBase. No small RNAs of viral origin were recovered. ncRNAs, noncoding RNAs.
While it is impossible to prove beyond doubt that HTLV-1 and HIV-1 do not express miRNAs or siRNAs in infected cells, these data, obtained using HTLV-1-infected MT-2 cells and HIV-1-infected ACH-2 cells, do argue that these RNAs must be expressed at a very low level if they do exist. We note that both MT-2 and ACH-2 express all viral mRNA species (Fig. 1), and both produce readily detectable levels of infectious virions, so this result is unlikely to reflect a viral defect specific to these cell lines. Because the protocol used to generate the data summarized in Fig. 2 results entirely in cDNA clones derived from small RNAs bearing a 5′ phosphate moiety, which would be predicted to result from Dicer cleavage, we repeated this analysis using a protocol (38) that can clone all short RNAs, regardless of the presence or absence of a 5′ phosphate. This alternative procedure again failed to detect any virus-derived short RNAs, even though miRNAs and RNA fragments of cellular origin were readily detected (data not shown). Overall, statistical analysis of the data presented in Fig. 2 suggests that, if HIV-1 or HTLV-1 does produce viral miRNAs or siRNAs, these constitute <0.5% of all Dicer-generated small RNAs present in the infected cells analyzed (P < 0.05). This contrasts with our earlier analyses of the viral miRNAs expressed by DNA tumor viruses in latently infected B cells, where the virally derived miRNAs were found to be expressed at approximately the same total level as the various cellular miRNAs (8).
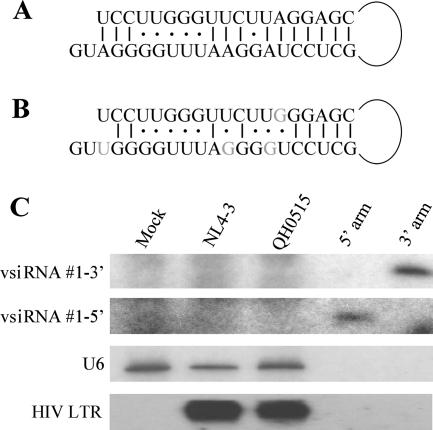
Previously, Bennasser et al. (3) reported the sequence of a hypothetical perfect 19-bp stem in HIV-1 that they argued gave rise to two siRNAs of viral origin. These siRNAs were readily detected in Jurkat or CEM-SS cells infected with the NL4-3 strain of HIV-1 by using Northern blot analysis. However, these proposed siRNAs were not recovered as cDNA clones from our analysis of ACH-2 cells infected with the closely related LAV strain of HIV-1 (Fig. 2). The HIV-1 RNA stem structure proposed by Bennasser et al. (3) is shown in Fig. 3A, while the sequence of this same region in the NL4-3 isolate is shown in Fig. 3B. It should be noted that the terminal loop flanking this stem is actually predicted to be 197 nt in length and that the version of this proposed RNA stem that is found in the NL4-3 isolate (Fig. 3B), unlike the idealized stem sequence proposed in Fig. 3A, is likely to be quite unstable.
FIG. 3.
HIV-1-infected PBMCs fail to express detectable levels of the proposed vsiRNA#1 siRNA. (A) Structure of a hypothetical HIV-1 RNA stem-loop proposed by Bennasser et al. (3). (B) The sequence of the proposed Bennasser stem-loop as found in HIV-1 isolate NL4-3. Differences in sequence are indicated in gray. (C) PBMCs were infected with the NL4-3 laboratory strain of HIV-1 or the primary QH0515 isolate and then cultured for 6 days to allow the virus to spread through the culture. At this point, p24 ELISA assays indicated a viral load of 78 ng/ml of p24 in the NL4-3-infected cultures and 76 ng/ml in the QH0515 culture. Total RNA was isolated from the infected cells and uninfected control PBMCs and analyzed by Northern blotting for expression of the proposed 5′ and 3′ arms of the vsiRNA#1 siRNA, using synthetic probes complementary to the NL4-3 genomic sequence. Synthetic forms of these proposed HIV-1 siRNAs were loaded onto the blot as positive controls at a level equivalent to ∼300 copies per PBMC. The blot was then stripped and rehybridized using the HIV-1 LTR-specific probe described in the Fig. 1 legend. The cellular U6 RNA was used as a loading control.
To more fully address whether HIV-1 indeed gives rise to these two 21-nt-long viral siRNAs, which were named vsiRNA#1-3′ and vsiRNA#1-5′ (3), we purchased synthetic versions of these proposed siRNAs and used these as positive controls for Northern blots to analyze the expression of these RNAs in total RNA samples derived from uninfected primary PBMCs, from PBMCs productively infected with the NL4-3 strain of HIV-1 (3), or with QH0515, a primary HIV-1 isolate (12) (Fig. 3). While the control RNA oligonucleotides were readily detected, we failed to detect either proposed HIV-1 siRNA in PBMCs infected by either the NL4-3 or the QH0515 isolate of HIV-1, even though HIV-1 mRNA transcripts were present at high abundance (Fig. 3C). Of note, the level of the control oligonucleotides loaded on this blot is equivalent to only ∼300 copies per PBMC, a level that is significantly lower than seen for individual miRNAs expressed, for example, by Kaposi's sarcoma-associated herpesvirus in infected B cells (8). Overall, these data again argue that HIV-1 siRNAs, if they do exist, must be expressed at very low levels.
Retroviral transcription factors do not inhibit RNAi.
Many plant and invertebrate viruses encode suppressor of RNA silencing proteins to partially counter the effects of virus-derived siRNAs generated during infection (reviewed in reference 41). Recent reports have suggested that some retroviral transcriptional transactivators might also have suppressor of RNA silencing activity. Specifically, as noted above, it has been proposed that the HIV-1 Tat protein is an inhibitor of Dicer function (3), which would be predicted to result in the inhibition of miRNA production from pre-miRNAs and in the inhibition of siRNA production from shRNAs. In addition, Lecellier et al. (36) have reported that the Tas transcription factor encoded by the retrovirus primate foamy virus (PFV) also inhibits the function of cellular miRNAs, although no mechanism for this effect was proposed. Although PFV is not a human retrovirus, it is a chimpanzee retrovirus that can establish zoonotic infections in humans, and PFV was, in fact, originally cloned out of a human tissue sample (46). Although Tat and Tas show no sequence homology, both proteins are potent transcriptional activators of their cognate retroviral LTR promoter elements (27, 33). Similarly, HTLV-1 also encodes a transcriptional transactivator, termed Tax, that activates the HTLV-1 LTR promoter (21).
It has previously been reported that HIV-1 Tat overexpression can block the function of an shRNA (3), as would indeed be predicted for an inhibitor of Dicer, and we therefore asked whether Tat, Tax, or Tas would be able to block shRNA function. Because the earlier report arguing that Tat is an inhibitor of shRNA function (3) used very high levels of Tat expression, far higher than is seen in HIV-1-infected T cells, this experiment also used a high level of expression of Tat, Tax, or Tas.
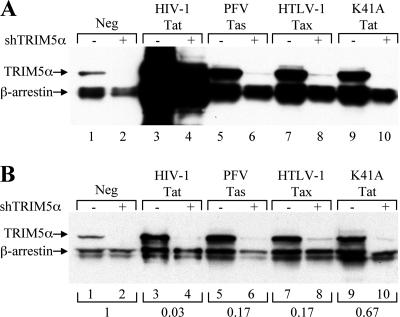
The shRNAs used targeted cellular mRNAs encoding TRIM5α and were expressed by using a Pol III-based expression plasmid called pSuper, as previously described (6). In this experiment, 293T cells were transfected with expression plasmids encoding two influenza virus HA-tagged cellular proteins, i.e., TRIM5α and a second protein, β-arrestin, as an internal control. The cells were also cotransfected with either the parental pSuper vector or pSuper vectors encoding shRNAs specific for the TRIM5α mRNA. Both TRIM5α and β-arrestin were readily detected in transfected cells by Western blot analysis, using an HA-specific monoclonal antibody, when no shRNAs were expressed (Fig. 4A, lane 1). In contrast, when pSuper vectors expressing TRIM5α-specific shRNAs were cotransfected, TRIM5α expression became undetectable while β-arrestin expression was essentially unchanged (Fig. 4A, lane 2).
FIG. 4.
Retroviral transcriptional transactivators fail to inhibit RNAi in human cells. 293T cells were cotransfected with vectors expressing HA-tagged forms of the cellular proteins TRIM5α and β-arrestin. The cells were also cotransfected with pSuper-based expression plasmids that produce shRNAs specific for TRIM-5α (+) or with pSuper as a control (−) and with a vector expressing the indicated wild-type or mutant retroviral transcription factors or with the parental pBC12/CMV as a control (Neg). At 48 h posttransfection, the cells were harvested and lysed and equal amounts of the cellular extracts were used for a Western blot analysis using an HA-specific monoclonal antibody (A). Panel B shows the results of analysis of the same cell extracts but with the amounts of samples loaded normalized to yield equivalent signal intensities, as indicated below the panel.
Cotransfection of the HIV-1 Tat expression plasmid pcTat (Fig. 4A, lanes 3 and 4), the PFV Tas expression plasmid pcTas (Fig. 4A, lanes 5 and 6), or the Tax expression plasmid pcTax (Fig. 4A, lanes 7 and 8) resulted in a nonspecific activation of the expression of both TRIM5α and β-arrestin that was particularly marked in the case of HIV-1 Tat. It has, in fact, been previously reported that Tat, Tas, and Tax can all activate heterologous promoters when overexpressed (1, 31, 32, 62), so this was not unexpected. Although both Tas and Tax clearly enhanced the level of TRIM5α expression seen in the absence of the TRIM5α shRNA expression plasmid (Fig. 4A, compare lane 1 with lanes 5 and 7), TRIM5α protein expression was nevertheless reduced to an essentially undetectable level when TRIM5α shRNAs were present (Fig. 4A, lane 6 and 8). However, the nonspecific activating effect of Tat was so marked that it was not possible to determine whether the shRNAs remained functional (Fig. 4A, lanes 3 and 4). To address this problem, we reduced the amount of cell extract loaded per lane to a level that equalized the signal obtained from the β-arrestin internal control (Fig. 4B). This allowed us to clearly see that not only Tas (Fig. 4B, lanes 5 and 6) and Tax (Fig. 4B, lanes 7 and 8) but also Tat (Fig. 4B, lanes 3 and 4) had no effect on the reduction in TRIM5α protein expression induced by the TRIM5α-specific shRNAs.
Previously, Bennasser et al. (3) reported that a stable missense mutant of Tat called K41A, which has lost the ability to interact with its cellular cofactor cyclin T1 (5) and, hence, its function as a transcription activator (54), nevertheless retained the ability to inhibit shRNA function. As shown in Fig. 4A, lanes 9 and 10, the K41A mutant of Tat had indeed lost the ability to nonspecifically activate the promoter used to drive TRIM5α and β-arrestin expression. However, again, no evidence for a specific inhibition of shRNA function was observed.
As noted above, Bennasser et al. (3) reported that Tat is a potent inhibitor of shRNA function as a result of its ability to inhibit Dicer activity. While the data presented in Fig. 4 disagree with this conclusion, we noted two differences between the experiment whose results are presented in Fig. 4 and the earlier work of Bennasser et al. (3). Specifically, the experiment presented in Fig. 4A was performed in 293T cells and used a plasmid expressing the 86-aa form of Tat, which is fully active as a transcriptional activator of the HIV-1 LTR (16). This form of Tat is seen in many common laboratory isolates of HIV-1, including LAV, NL4-3, and HXB-2, and in some primary isolates. In contrast, Bennasser et al. (3) performed their experiments in HeLa cells and used a plasmid expressing a 101-aa form of Tat.
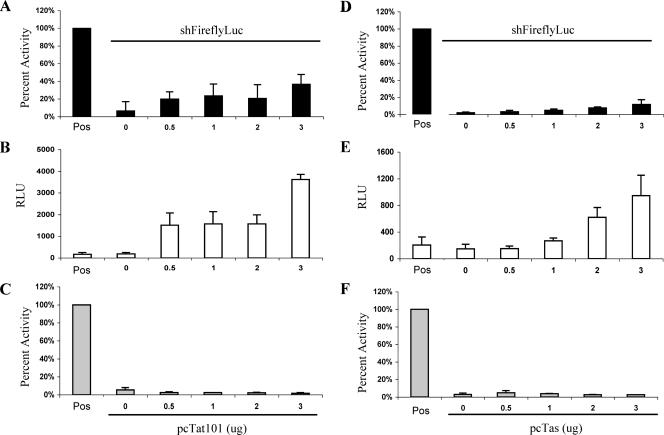
To address whether these differences could explain the different conclusions reached in these two studies, we analyzed the effect of the 101-aa form of Tat on the ability of a pSuper-based plasmid expressing an shRNA specific for firefly luciferase (Fluc) to inhibit Fluc expression in HeLa cells. These experiments were designed to closely mimic the earlier work of Bennasser et al. (3), including transfecting the same levels of the 101-aa Tat expression plasmid, but did differ in that we included a Renilla luciferase (Rluc) expression plasmid as an internal control. Importantly, prior to this analysis, we confirmed that the expression of the 101-aa form of HIV-1 Tat, like the expression of the 86-aa form of Tat, results in the potent transactivation of an indicator construct containing the HIV-1 LTR promoter (data not shown).
As shown in Fig. 5A, cotransfection of increasing amounts of an expression plasmid encoding the 101-aa form of Tat partially rescued the expression of Fluc that had been knocked down using an Fluc-specific shRNA. These data closely parallel the data reported by Bennasser et al. (3). However, analysis of the level of the Rluc internal control enzyme (Fig. 5B) showed that Rluc activity was also increased by the 101-aa form of Tat, suggesting that the expression of the 101-aa form of Tat in HeLa cells, like the expression of the 86-aa form of Tat in 293T cells (Fig. 4A), results in a nonspecific increase in the activity of cotransfected expression plasmids. If the raw Fluc data shown in Fig. 5A are corrected for the internal control Rluc data presented in Fig. 5B, then it becomes obvious that the 101-aa form of Tat in fact has no ability to inhibit the function of an shRNA targeted to Fluc (Fig. 5C).
FIG. 5.
HIV-1 Tat and PFV Tas do not inhibit RNAi mediated by an shRNA. HeLa cells were cotransfected with plasmids expressing Fluc and Rluc together with either pSuper or a pSuper-based plasmid expressing an Fluc-specific shRNA (shFireflyLuc). The HeLa cells were also cotransfected with the indicated levels of a plasmid expressing either a 101-aa form of HIV-1 Tat (pcTat101) (A to C) or PFV Tas (pcTas) (D to F). Panels A and D show the relative levels of Fluc protein expression seen in the absence (Pos) or presence of the pSuper/shFluc expression plasmid. These data are normalized to the level of Fluc expression seen in the absence of both pSuper/shFluc and pcTat101 or pcTas, which was set at 100%. Panels B and E show the raw Rluc expression data derived from the same cultures analyzed in panels A and D. Finally, panels C and F show the Fluc activity data from panels A and D after normalization to the internal control Rluc data presented in panels B and E. The positive control again represents HeLa cells transfected with the Fluc and Rluc expression vectors in the absence of both pSuper/shFluc and pcTat101 or pcTas. The averages and standard deviations of the results of three independent experiments are indicated. RLU, relative light units.
To extend these data, we also asked whether the PFV Tas protein would be able to rescue the inhibition of Fluc expression induced by an Fluc-specific shRNA in HeLa cells. In fact, the expression of Tas in HeLa cells by cotransfection of the pcTas expression plasmid only modestly rescued Fluc expression that had been knocked down by RNAi (Fig. 5D). Moreover, this rescue again reflected a nonspecific activation of gene expression, as visualized by data obtained using the Rluc internal control (Fig. 5E). Once the raw Fluc data were normalized using the Rluc internal control (Fig. 5F), we again saw no evidence for a specific inhibition of RNAi, in this case by the PFV Tas protein.
The HIV-1 Tat protein does not inhibit cellular miRNA biogenesis.
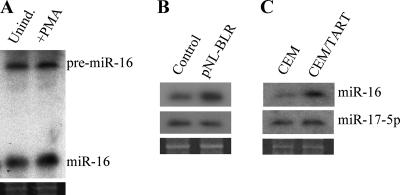
If the HIV-1 Tat protein indeed functions as a potent inhibitor of Dicer function, then Tat expression should inhibit Dicer processing of cellular pre-miRNAs (2), resulting in a drop in miRNA expression. To examine this question, we first asked whether cellular miRNAs would be detectable in ACH-2 cells, which produce viral RNAs and structural proteins and, hence, must express some level of Tat function. As shown in Fig. 6A, we readily detected the cellular miR-16 miRNA in both uninduced and PMA-induced ACH-2 cells. These data, which are consistent with the cDNA cloning of numerous cellular miRNAs from ACH-2 cells (Fig. 2B), therefore argue that productive HIV-1 infection of T cells does not block cellular miRNA expression.
FIG. 6.
The HIV-1 Tat protein does not globally affect cellular miRNA expression. (A) Northern blot analysis of endogenous miR-16 expression in uninduced or PMA-induced ACH-2 cells. Unind, uninduced cells; +PMA, PMA-induced cells. (B) Northern blot analysis of endogenous miR-16 and miR-17-5p expression in 293T cells stably transduced with an HIV-1-based vector, pNL-BLR, that expresses normal levels of HIV-1 Tat, as well as the blr drug resistance gene. The transduced, Tat-expressing cells were cultured for 21 days after selection to allow any pre-existing cellular miRNAs to turn over. 293T cells transduced with the Tat-defective pNL-SIN-CMV-BLR lentiviral vector served as controls. The pre-miRNA forms of miR-16 and miR-17-5p were not detected in either the pNL-BLR- or pNL-SIN-CMV-BLR-transduced 293T cells (data not shown), implying efficient Dicer processing of pre-miRNAs in both the presence and absence of Tat. (C) Northern blot analysis similar to that described for panel B, except that this panel compared cellular miRNA expression in wild-type CEM T cells with that seen in a transduced CEM cell line (CEM/TART) that stably expresses HIV-1 Tat and Rev. Ethidium bromide staining of the gels used in these analyses (lower panels) was performed to confirm equal loading.
To examine whether the expression of physiological levels of HIV-1 Tat would inhibit miRNA biogenesis, we generated an HIV-1 proviral vector, pNL-BLR, that is defective for the expression of Vpr, Nef, Rev, and Env but retains all five other HIV-1 genes, including Tat, in a fully functional form. This vector contains a blastocidin resistance (blr) gene in place of nef. The pNL-BLR vector was produced in an infectious form by cotransfecting 293T cells with plasmids that express the vesicular stomatitis virus glycoprotein (pHIT/G) and the HIV-1 Rev protein (pcRev). The resultant pseudotyped viral particles were then used to infect other 293T cells. After the infection of these target cells, the only gene products that should be expressed from the integrated HIV-1 provirus are Tat and Blr. Because pNL-BLR is defective for Rev function, no viral structural proteins should be expressed, while Nef, the third HIV-1 early protein, has been replaced with the Blr open reading frame. As a control, we also generated transduced, blastocidin-resistant 293T cells using the previously described pNL-SIN-CMV-BLR lentiviral vector (37). This vector is self inactivating, due to a deletion in the U3 region of the 3′ LTR, and also lacks a functional tat gene. After transduction, cells containing this lentiviral vector are, however, resistant to blastocidin, due to the presence of an internal CMV promoter linked to the blr gene.
After selection for resistance to blastocidin, these cells were analyzed for functional expression of the HIV-1 Tat protein from the integrated HIV-1 provirus. This analysis showed that the level of Tat expressed in the pNL-BLR-transduced cells was sufficient to specifically activate the HIV-1 LTR reporter present in a transfected indicator construct (data not shown). Consistent with the absence of a functional rev gene in the pNL-BLR lentiviral vector, these transduced cells did not express detectable levels of the HIV-1 p24 capsid protein (<1 pg/ml). However, transfection of the pNL-BLR-transduced 293T cells with the pcRev expression vector resulted in the detection, by ELISA, of 1.7 ng of p24 per ml of supernatant media. Together, these data argue that the NL-BLR-transduced 293T cells are producing physiological levels of active Tat protein from the integrated NL-BLR provirus.
The pNL-BLR- and pNL-SIN-CMV-BLR-transduced 293T cells were maintained in culture for 21 days after blastocidin selection to permit any pre-existing cellular miRNAs to turn over. At this time, we analyzed the expression levels of the cellular miRNAs miR-16 and miR-17-5p. As shown in Fig. 6B, we saw no drop in the level of expression of either miR-16 or miR-17-5p in the NL-BLR-transduced, Tat-expressing 293T cells compared to their levels of expression in the NL-SIN-CMV-BLR-transduced control cells.
In order to extend these data to T cells, we also analyzed a previously described T-cell line derived from CEM, called CEM/TART, that expresses Tat and Rev at levels sufficient to fully support the replication of Tat- and Rev-deficient HIV-1 mutant proviruses (11). By transfection of an HIV-1 LTR-based Fluc indicator plasmid and an Rluc control plasmid, we first confirmed that the Tat protein expressed in CEM/TART cells was indeed fully capable of transcriptionally activating the HIV-1 LTR promoter (data not shown). Using Northern blot analysis, we then examined the expression levels of the cellular miRNAs miR-16 and miR-17-5p in CEM/TART and control CEM cells. As shown in Fig. 6C, we again saw no evidence of a reduction in the level of miR-16 or miR-17-5p in the Tat-expressing CEM/TART cells in comparison to their levels in control CEM cells.
DISCUSSION
The question of whether viruses in general, and HIV-1 in particular, can induce the expression of virus-specific siRNAs in infected cells has been a controversial one (reviewed in reference 15). In the case of HIV-1, work by Pfeffer et al. (51) failed to identify any HIV-1-specific siRNAs or miRNAs in productively infected CD4+ HeLa cells. In contrast, Bennasser et al. (3) have reported that HIV-1 gives rise to an siRNA that can inhibit HIV-1 replication and have proposed that the HIV-1 Tat protein relieves this inhibition by blocking the function of the cellular RNase III enzyme Dicer, which plays a key role in both siRNA and miRNA biogenesis. Moreover, Omoto et al. (48) have proposed that HIV-1 encodes a viral miRNA that they have suggested plays an important role in regulating viral gene expression. Finally, Lecellier et al. (36), working with the unrelated primate retrovirus PFV, have presented evidence suggesting that PFV replication can be inhibited by cellular miRNAs and that this inhibition is alleviated by the PFV Tas protein, which they proposed can block miRNA and siRNA function by an undefined mechanism.
In the manuscript, we address the question of whether the human retroviruses HIV-1 and HTLV-1 and the primate retrovirus PFV indeed interact with the cellular RNAi machinery. We have performed extensive cDNA cloning analyses of the small RNAs expressed in the persistently HIV-1-infected T-cell line ACH-2 and the persistently HTLV-1-infected T-cell line MT2 and report that there are no detectable viral siRNAs or miRNAs expressed in these cells, although numerous cellular miRNAs were recovered (Fig. 2). We also report that the retroviral nuclear transcription factors HIV-1 Tat, HTLV-1 Tax, and PFV Tas are all incapable of blocking the function of shRNAs in human cells, although all three proteins did reveal an ability to nonspecifically activate heterologous promoters when overexpressed (Fig. 4 and 5). Finally, we report that the stable expression of physiological levels of the HIV-1 Tat protein does not reduce cellular miRNA expression or induce the accumulation of pre-miRNA precursors as would be predicted for an inhibitor of Dicer function (Fig. 6). Our data therefore argue that human retroviruses neither induce the cellular RNAi response by causing the production of viral miRNAs or siRNAs nor inhibit the cellular RNAi response by blocking Dicer or RISC function.
The consideration of earlier work proposing that HIV-1 does encode viral siRNAs or miRNAs and that HIV-1 Tat inhibits Dicer function (3, 48) suggests a few possible reasons for this discrepancy. For example, the potential of retroviral transcriptional transactivators to nonspecifically activate heterologous promoters may not previously have been fully controlled. Moreover, we note that the proposed HIV-1 siRNA and miRNA are not well conserved. The proposed vsiRNA#1 and the proposed RNA stem from which it derives are in fact quite poorly conserved (Fig. 3A and B) and, indeed, the “idealized” RNA stem originally proposed by Bennasser et al. (3) (Fig. 3A) does not appear to be derived from an actual HIV-1 isolate. Nevertheless, and despite the poor predicted stability of this same RNA stem in the NL4-3 laboratory isolate of HIV-1 (Fig. 3B), Bennasser et al. (3) reported that they could detect this viral siRNA in NL4-3-infected T cells. We have not been able to reproduce this result using either cDNA cloning or Northern blot analysis (Fig. 2 and 3C).
In addition to the concerns with the proposed vsiRNA#1 stated above, it is also worth noting that the vsiRNA#1 sequence actually maps to the HIV-1 Rev response element and has been reported by several groups (10, 44) to form part of the Rev response element RNA secondary structure by base pairing to a region of the HIV-1 genome distinct from that proposed by Bennasser et al. (3) (Fig. 3A). The question of whether this RNA stem structure ever forms during the HIV-1 replication cycle clearly requires additional investigation.
The HIV-1 miRNA proposed by Omoto et al. (48) was also not detected in either this or a previous miRNA cloning effort (51) using HIV-1-infected cells. It is worth noting that the excision of a pre-miRNA from a pri-miRNA transcript (in this case the HIV-1 genome) by Drosha would result in the nuclear cleavage and destruction of that transcript (7). This suggests that any HIV-1 miRNA would need to play an important role in the HIV-1 life cycle in order to compensate for the nuclear destruction of viral genomic RNAs by Drosha cleavage. In fact, however, the HIV-1 miRNA proposed by Omoto et al. (48) is not well conserved between different subgroups of HIV-1, including in the critical “seed” region of the miRNA (data not shown). In any event, neither we nor Pfeffer et al. (51) were able to detect this proposed miRNA in HIV-1-infected cells and, if it exists, it must therefore be expressed at a very low level.
If HIV-1 does not make any siRNAs, then there is no obvious benefit from the inhibition of Dicer function (note that the inhibition of Dicer would also block the expression of any HIV-1 miRNAs). In fact, we were unable to obtain evidence in support of the hypothesis that either the 86-aa or 101-aa form of HIV-1 Tat can inhibit either shRNA function, as proposed by Bennasser et al. (3), or miRNA production, as would be expected if Tat indeed inhibited Dicer function (Fig. 4, 5, and 6). This result is perhaps not surprising, given that Tat is a nuclear transcription factor (27), while Dicer is a cytoplasmic RNA-processing enzyme (reviewed in reference 2). Similarly, we also failed to detect any inhibition of shRNA function by either the HTLV-1 Tax or PFV Tas protein, two other potent viral nuclear transcription factors (21, 33). Together, these data argue that human retroviral transcription factors do not inhibit RNAi responses in infected cells.
While the manuscript was being prepared, Triboulet et al. (57) reported that HIV-1 infection can inhibit the expression of some cellular miRNAs in infected cells, including miR-17, although the expression of other cellular miRNAs, e.g., miR-122a and miR-20b, was activated by up to 30-fold. This result is not surprising, as miRNAs are derived from RNA Pol II transcripts (7, 40) and HIV-1 infection has previously been shown to up- or downregulate the expression of numerous cellular mRNAs (14, 58, 59). We note that the finding that HIV-1 infection can upregulate the expression of specific cellular miRNAs is inconsistent with the hypothesis that Tat, an early HIV-1 gene product, acts to block pre-miRNA processing by Dicer. In any event, we did not in fact see any effect of Tat expression on the level of the miR-17 miRNA in infected cells (Fig. 6). The reason for this is not presently clear, but one hypothesis is that miR-17 repression may be mediated by HIV-1 proteins other than Tat, which are not expressed from the Rev- and Nef-defective NL-BLR lentiviral vector the results of whose analysis are shown in Fig. 6B.
Acknowledgments
We thank Georgia Tomaras, Glen Overman, and the Duke Center for AIDS Research (CFAR) Molecular Virology Core for assistance with the infection and culture of primary human T cells.
This work was sponsored by National Institutes of Health grant GM071408.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Ballard, D. W., E. Bohnlein, J. W. Lowenthal, Y. Wano, B. R. Franza, and W. C. Greene. 1988. HTLV-1 Tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science 241:1652-1655. [DOI] [PubMed] [Google Scholar]
- 2.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 3.Bennasser, Y., S. Y. Le, M. Benkirane, and K. T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. [DOI] [PubMed] [Google Scholar]
- 4.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplen, N. J., J. Fleenor, A. Fire, and R. A. Morgan. 2000. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252:95-105. [DOI] [PubMed] [Google Scholar]
- 10.Charpentier, B., F. Stutz, and M. Rosbash. 1997. A dynamic in vivo view of the HIV-I Rev-RRE interaction. J. Mol. Biol. 266:950-962. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H., T. J. Boyle, M. H. Malim, B. R. Cullen, and H. K. Lyerly. 1992. Derivation of a biologically contained replication system for human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:7678-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cleghorn, F. R., N. Jack, J. K. Carr, J. Edwards, B. Mahabir, A. Sill, C. B. McDanal, S. M. Connolly, D. Goodman, R. Q. Bennetts, T. R. O'Brien, K. J. Weinhold, C. Bartholomew, W. A. Blattner, and M. L. Greenberg. 2000. A distinctive clade B HIV type 1 is heterosexually transmitted in Trinidad and Tobago. Proc. Natl. Acad. Sci. USA 97:10532-10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clouse, K. A., D. Powell, I. Washington, G. Poli, K. Strebel, W. Farrar, P. Barstad, J. Kovacs, A. S. Fauci, and T. M. Folks. 1989. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 142:431-438. [PubMed] [Google Scholar]
- 14.Corbell, J., D. Sheeter, D. Genini, S. Rought, L. Leoni, P. Du, M. Ferguson, D. R. Masys, J. B. Welsh, J. L. Fink, R. Sasik, D. Huang, J. Drenkow, D. D. Richman, and T. Gineras. 2001. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 11:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cullen, B. R. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 7:563-567. [DOI] [PubMed] [Google Scholar]
- 16.Cullen, B. R. 1986. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell 46:973-982. [DOI] [PubMed] [Google Scholar]
- 17.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38(Suppl.):S25-S30. [DOI] [PubMed] [Google Scholar]
- 18.Denli, A. M., B. B. Tops, R. H. Plasterk, R. F. Ketting, and G. J. Hannon. 2004. Processing of primary microRNAs by the microprocessor complex. Nature 432:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felber, B. K., H. Paskalis, C. Kleinman-Ewing, F. Wong-Staal, and G. N. Pavlakis. 1985. The pX protein of HTLV-1 is a transcriptional activator of its long terminal repeats. Science 229:675-679. [DOI] [PubMed] [Google Scholar]
- 22.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The microprocessor complex mediates the genesis of microRNAs. Nature 432:235-240. [DOI] [PubMed] [Google Scholar]
- 23.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 25.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 26.Han, J., Y. Lee, K. H. Yeom, J. W. Nam, I. Heo, J. K. Rhee, S. Y. Sohn, Y. Cho, B. T. Zhang, and V. N. Kim. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887-901. [DOI] [PubMed] [Google Scholar]
- 27.Hauber, J., A. Perkins, E. P. Heimer, and B. R. Cullen. 1987. Trans-activation of human immunodeficiency virus gene expression is mediated by nuclear events. Proc. Natl. Acad. Sci. USA 84:6364-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho, A. M., P. W. Dion, M. K. Karmakar, and A. Lee. 2002. Estimating with confidence the risk of rare adverse events, including those with observed rates of zero. Reg. Anesth. Pain Med. 27:207-210. [DOI] [PubMed] [Google Scholar]
- 29.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 30.Hutvagner, G., and P. D. Zamore. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297:2056-2060. [DOI] [PubMed] [Google Scholar]
- 31.Imai, K., K. Nakata, K. Kawai, T. Hamano, N. Mei, H. Kasai, and T. Okamoto. 2005. Induction of OGG1 gene expression by HIV-1 Tat. J. Biol. Chem. 280:26701-26713. [DOI] [PubMed] [Google Scholar]
- 32.Keller, A., E. D. Garrett, and B. R. Cullen. 1992. The Bel-1 protein of human foamy virus activates human immunodeficiency virus type 1 gene expression via a novel DNA target site. J. Virol. 66:3946-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller, A., K. M. Partin, M. Lochelt, H. Bannert, R. M. Flugel, and B. R. Cullen. 1991. Characterization of the transcriptional trans activator of human foamy retrovirus. J. Virol. 65:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau, N. C., L. P. Lim, E. G. Weinstein, and D. P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858-862. [DOI] [PubMed] [Google Scholar]
- 36.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saib, and O. Voinnet. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557-560. [DOI] [PubMed] [Google Scholar]
- 37.Lee, M. T., G. A. Coburn, M. O. McClure, and B. R. Cullen. 2003. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 77:11964-11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, R. C., and V. Ambros. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294:862-864. [DOI] [PubMed] [Google Scholar]
- 39.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 40.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 43.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 44.Mann, D. A., I. Mikaelian, R. W. Zemmel, S. M. Green, A. D. Lowe, T. Kimura, M. Singh, P. J. Butler, M. J. Gait, and J. Karn. 1994. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 241:193-207. [DOI] [PubMed] [Google Scholar]
- 45.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 46.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyoshi, I., I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata, and Y. Hinuma. 1981. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature 294:770-771. [DOI] [PubMed] [Google Scholar]
- 48.Omoto, S., M. Ito, Y. Tsutsumi, Y. Ichikawa, H. Okuyama, E. A. Brisibe, N. K. Saksena, and Y. R. Fujii. 2004. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ott, M., S. Emiliani, C. Van Lint, G. Herbein, J. Lovett, N. Chirmule, T. McCloskey, S. Pahwa, and E. Verdin. 1997. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science 275:1481-1485. [DOI] [PubMed] [Google Scholar]
- 50.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 52.Rimsky, L., J. Hauber, M. Dukovich, M. H. Malim, A. Langlois, B. R. Cullen, and W. C. Greene. 1988. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature 335:738-740. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz, D. S., G. Hutvagner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 54.Tiley, L. S., P. H. Brown, and B. R. Cullen. 1990. Does the human immunodeficiency virus Tat trans-activator contain a discrete activation domain? Virology 178:560-567. [DOI] [PubMed] [Google Scholar]
- 55.Tokunaga, K., M. L. Greenberg, M. A. Morse, R. I. Cumming, H. K. Lyerly, and B. R. Cullen. 2001. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 75:6776-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomaras, G. D., S. F. Lacey, C. B. McDanal, G. Ferrari, K. J. Weinhold, and M. L. Greenberg. 2000. CD8+ T cell-mediated suppressive activity inhibits HIV-1 after virus entry with kinetics indicating effects on virus gene expression. Proc. Natl. Acad. Sci. USA 97:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Triboulet, R., B. Mari, Y. L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K. T. Jeang, and M. Benkirane. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579-1582. [DOI] [PubMed] [Google Scholar]
- 58.van't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen, W., S. Chen, Y. Cao, Y. Zhu, and Y. Yamamoto. 2005. HIV-1 infection initiates changes in the expression of a wide array of genes in U937 promonocytes and HUT78 T cells. Virus Res. 113:26-35. [DOI] [PubMed] [Google Scholar]
- 60.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkins, C., R. Dishongh, S. C. Moore, M. A. Whitt, M. Chow, and K. Machaca. 2005. RNA interference is an antiviral defense mechanism in Caenorhabditis elegans. Nature 436:1044-1047. [DOI] [PubMed] [Google Scholar]
- 62.Williams, C. A., D. Mondal, and K. C. Agrawal. 2005. The HIV-1 Tat protein enhances megakaryocytic commitment of K562 cells by facilitating CREB transcription factor coactivation by CBP. Exp. Biol. Med. (Maywood) 230:872-884. [DOI] [PubMed] [Google Scholar]
- 63.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng, Y., and B. R. Cullen. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng, Y., R. Yi, and B. R. Cullen. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]