Abstract
As infection with wild-type (wt) Sendai virus (SeV) normally activates beta interferon (IFN-β) very poorly, two unnatural SeV infections were used to study virus-induced IFN-β activation in mouse embryonic fibroblasts: (i) SeV-DI-H4, which is composed mostly of small, copyback defective interfering (DI) genomes and whose infection overproduces short 5′-triphosphorylated trailer RNAs (pppRNAs) and underproduces viral V and C proteins, and (ii) SeV-GFP(+/−), a coinfection that produces wt amounts of viral gene products but that also produces both green fluorescent protein (GFP) mRNA and its complement, which can form double-stranded RNA (dsRNA) with capped 5′ ends. We found that (i) virus-induced signaling to IFN-β depended predominantly on RIG-I (as opposed to mda-5) for both SeV infections, i.e., that RIG-I senses both pppRNAs and dsRNA without 5′-triphosphorylated ends, and (ii) it is the viral C protein (as opposed to V) that is primarily responsible for countering RIG-I-dependent signaling to IFN-β. Nondefective SeV that cannot specifically express C proteins not only cannot prevent the effects of transfected poly(I-C) or pppRNAs on IFN-β activation but also synergistically enhances these effects. SeV-Vminus infection, in contrast, behaves mostly like wt SeV and counteracts the effects of transfected poly(I-C) or pppRNAs.
All viruses evade the cellular innate immune system in part by expressing gene products that interfere with the ability of the host cell to establish an antiviral state (6). In the case of the Paramyxovirinae, this anti-host-defense activity is due mostly to viral C and V proteins (15, 27, 31). The C and V proteins are encoded by separate alternate open reading frames (ORFs), which both overlap that of the P protein. V and C are also referred to as accessory gene products, as not all members of this virus subfamily express one or the other. More specifically, rubulavirus and avulavirus express V but do not express C proteins, and human parainfluenza virus type 1 (PIV1), a respirovirus most closely related to Sendai virus (SeV), expresses C but does not express a V protein (16, 20).
Paramyxovirus V and C proteins antagonize interferon (IFN) signaling by various mechanisms, and they also target the production of type I IFN (15, 31). Beta IFN (IFN-β) production is one of the earliest events in the cellular innate immune response, which leads to the establishment of an antiviral state. IFN-β production requires the coordinated activation of several transcription factors, including NF-κB and IRF3 (15, 29). For intracellular RNA virus replication, the signaling pathway that leads to IRF3 activation starts with mda-5 and RIG-I, two cytoplasmic DExH/D-box helicases with N-terminal CARD domains. These helicases respond to double-stranded RNA (dsRNA) and, at least for RIG-I, to 5′-triphosphorylated single-stranded RNA (ssRNA) (pppRNA), which are generated in the cytoplasm during RNA virus replication (9, 11, 25). Upon the detection of these viral RNAs, the CARD domains of these helicases interact with IPS-1/Cardif/MAVS/VISA, which is present in the mitochondrial membrane, and this CARD-CARD interaction is thought to lead to the recruitment and activation of TBK1, IKKɛ, and other IKK kinases that activate NF-κB and IRF3, thereby activating the IFN-β promoter (8). The production of these early IFNs initiates autocrine and paracrine signal amplifications via the Jak/Stat pathway to produce a generalized antiviral state and also assists in the subsequent activation of adaptive immune responses.
The role of mda-5 in sensing RNA virus infection was uncovered because mda-5 was found to bind to the PIV5 V protein and other paramyxovirus V proteins, including SeV V. These V-protein-mda-5 interactions, moreover, prevented IFN-β activation in response to transfected poly(I-C) (1). On the other hand, other studies found that RIG-I and not mda-5 acts as the sensor of paramyxovirus infection (13, 28). This paper provides evidence that for SeV infection of mouse embryonic fibroblasts (MEFs), it is the C protein (and not V) that is primarily responsible for this effect and that C acts by countering RIG-I (and not mda-5). Independent expression of C was found to inhibit RIG-I-dependent signaling to the IFN-β promoter induced by either pppRNAs or dsRNAs. Moreover, SeV that cannot specifically express C proteins was unable to counteract poly(I-C)- or pppRNA-induced IFN-β activation, whereas SeV that cannot express V behaved mostly like wild-type (wt) SeV.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
MEFs were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
SeV stocks were grown in the allantoic cavitiesof 9-day-old embryonated chicken eggs for 3 days at 33°C. For nondefective stocks (109 PFU/ml), 0.1 ml of a 105 dilution (ca. 1,000 PFU) was inoculated per egg. In the case of DI stocks, 0.1 ml of a 103 dilution was used. In all cases, the amount of viral proteins present in the resulting allantoic fluid was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining of pelleted virus. Virus titers were determined by plaquing on LLC-MK2 cells.
SeV-GFP(+), which expresses green fluorescent protein (GFP) from a transgene between the M and F genes, and SeV-GFP(−) or SeV-RFP, which expresses antisense GFP mRNA or red fluorescent protein (RFP) (dsRED) from similarly located transgenes, were prepared as previously described (31). DI-H4 stocks were described previously (30).
Primary antibodies used included rabbit anti-RFP (AB3216; Chemicon); anti-actin monoclonal antibody (MAb) (1501; Chemicon); rabbit anti-GFP (632460; BD biosciences); rabbit anti-SeV-P/C/V (homemade); anti-hemagglutinin (HA) MAb (16B12; BABCO), anti-Flag MAb (F1804; Sigma), rabbit anti-mda-5 (J. Tschopp, Lausanne, Switzerland), and rabbit anti-RIG-I (T. Fujita, Kyoto, Japan).
Plasmids, transient transfections, infections, inductions, luciferase assay, and fluorescence-activated cell sorter analysis.
EBS plasmids (3) expressed viral and fluorescent proteins and were constructed by standard methods; precise detail can be obtained from the authors.
NS1 {residues 1 to 77 [NS1(1-77)]} (from Jacques Perrault) and E3L {residues 100 to 190 [E3L(100-190)]} (from Bertram Jacobs), were HA tagged and cloned into pEBS. Flag-tagged RIG-I, RIG-I-C, or RIG-ΔCARDS (dominant negative) and mouse mda-5 were obtained from Jurg Tshopp and Klaus Conzelmann.
pβ-IFN-fl-lucter, which contains the firefly luciferase gene under the control of the human IFN-β promoter, was described previously (14). pTK-rl-lucter, used as a transfection standard, contains the herpes simplex virus TK promoter region upstream of the Renilla luciferase gene (Promega).
For transfections, 100,000 cells were plated into six-well plates 20 h before transfection with 1.5 μg of pβ-IFN-fl-lucter; 0.5 μg of pTK-rl-lucter; 0.5 μg of plasmids expressing RIG-I and MDA-5; 1.5 μg of plasmids expressing V (whose C ORF is closed with a stop codon), C1-204 or C1-23-Tom-C24-204 (or C*), NS1(1-77), wt E3L, mutant E3L(100-190), or RIG-Δ proteins (as indicated); and TransIT-LT1 transfection reagent (Mirus). At 24 h posttransfection, the cells were (or were not) infected with various SeV stocks or transfected with 5 μg of poly(I-C) using TransIT-LT1 transfection reagent. Twenty hours later, cells were harvested and assayed for firefly and Renilla luciferase activity (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of Renilla luciferase.
Immunoblotting.
Cytoplasmic extracts were prepared using 0.5% NP-40 buffer. Equal amounts of total proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto Immobilon-P membranes by semidry transfer. The secondary antibodies used were alkaline phosphatase-conjugated goat anti-rabbit (or mouse) immunoglobulin G (Bio-Rad). The immobilized proteins were detected by light-enhanced chemiluminescence (Pierce) and analyzed in a Bio-Rad light detector using Quantity One software.
In vitro synthesis of RNA, purification, and transfection.
DNA for T7 RNA polymerase synthesis of model RNA1 was prepared by PCR using the following partially complementary primers: 5′-TAATACGACTCACTATA(ggg/gca)ACACACCACAACCAACCCACAAC-3′ (forward) (start sites are in lowercase type) and 5′-GAAAGAAAGGTGTGGTGTTGGTGTGGTTGTTGTGGGTTGGTTGTGG-3′ (reverse). In vitro transcription was performed on 100 pmol of purified PCR product using T7 MEGAshortcript from Ambion according to the manufacturer's instructions. For RNA1 containing the unusual OHGCA start site, RNA was initiated with the dinucleotide 5′ OHGpC24 in a reaction without GTP. For RNA1 containing the usual pppGGG start site, part of the product was treated with 20 units of calf intestinal phosphatase (Roche) for 30 min at 37°C followed by proteinase K treatment (15 min at 37°C), phenol extraction, and ethanol precipitation. The T7 transcripts were purified on NucAway Spin columns from Ambion (to remove unincorporated nucleotides). SeV trailer pppRNAs were synthesized similarly by using specific PCR primers.
For RNA transfection, 1 μg (1×) to 3 μg (3×) of RNA was transfected into MEF cells using TransMessenger transfection reagent (QIAGEN).
RT and real-time PCR via TaqMan.
Confluent MEFs in 10-cm petri dishes (107 cells) were infected with 20 PFU/cell of SeV GFP(+), SeV GFP(−), or both stocks. At 24 h postinfection (hpi), the cells were collected and lysed in 300 μl of NP-40 lysis buffer. Cytoplasmic extracts were then centrifuged in a 20 to 40% (wt/wt) CsCl density gradient (16 h at 35,000 rpm at 12°C). The pellet RNAs were ethanol precipitated and resuspended in 50 μl of Tris-EDTA. Fifteen micrograms of RNA was then mixed with 0.5 μg of the forward or the reverse GFP primer and subjected to a Superscript reverse transcription (RT) reaction, according to instructions provided by the manufacturer (Gibco), in a total volume of 50 μl. Five microliters of each cDNA was then combined with 12 μl MasterMix (Eurogentec), 20 pmol (each) of forward and reverse primers, and 4.4 pmol of TaqMan probe in a total volume of 25 μl. The following primers and probes (Eurogentec or Microsynth) were used for RT and TaqMan analyses of the GFP gene: 5′-CCGACAACCACTACCTGAGCAC-3′ (forward), 5′-GAACTCCAGCAGGACCATGTG-3′ (reverse), and 5′-AAAGACCCCAACGAGAAGCGCGA-3′ (probe). Real-time PCR was carried out in duplicates using a 7700 sequence detector (Applied Biosystems).
RESULTS
Three ways to activate IFN-β.
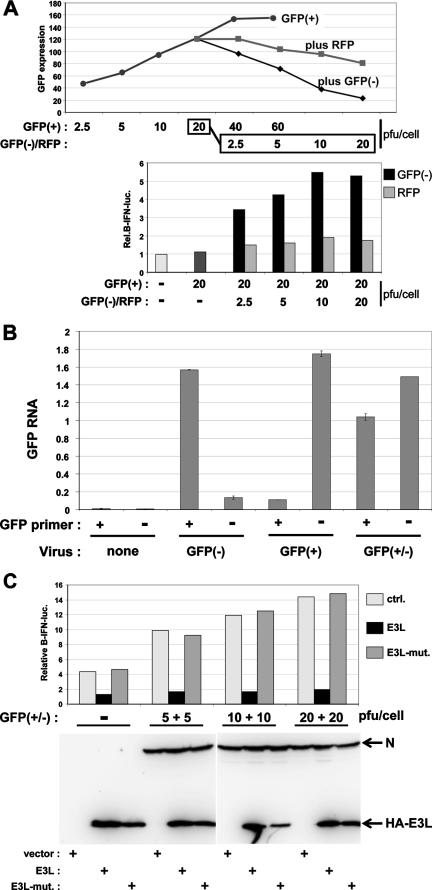
We have used three ways to induce the activation of an IFN-β promoter expressing a luciferase reporter gene in MEFs (see Fig. 2A). The first is to simply transfect a synthetic dsRNA, poly(I):poly(C) [poly(I-C)], into the cells. The second way is to infect the cells with an SeV stock that contains a well-characterized copyback DI genome (H4) (30). The third way is to coinfect cells with SeV-GFP(+), which expresses a GFP transgene, and SeV-GFP(−), which expresses mRNA containing the complement of the GFP ORF, as recently described for vesicular stomatitis virus (VSV) (24). As shown in Fig. 1A, infection with increasing amounts of SeV-GFP(+) alone leads to increasing GFP expression. Coinfection of 20 PFU/cell of SeV-GFP(+) with increasing amounts of SeV-GFP(−) leads to the gradual decrease of GFP expression (top). At 20 PFU/cell of SeV-GFP(−), there are roughly equal amounts of GFP and anti-GFP mRNAs intracellularly (by strand-specific quantitative RT-PCR) (Fig. 1B and see Materials and Methods), and there is a 90% loss of GFP expression (Fig. 1A, top). This loss of GFP expression cannot be accounted for by the reduced level of GFP mRNA (Fig. 1B). In contrast, coinfection with increasing amounts of SeV expressing RFP as a neutral control (SeV-RFP) has a reduced ability to interfere with GFP expression (Fig. 1A). More importantly, whereas infection with SeV-GFP(+) alone or its coinfection with SeV-RFP leads to little or no activation of IFN-β (Fig. 1A), coinfection with SeV-GFP(−) clearly activates the IFN-β promoter (Fig. 1A, bottom). This IFN-β activation is inhibited by the coexpression of the dsRNA-binding domain of the vaccinia virus E3L protein, whereas this activation is unaffected by a mutant form of E3L containing two point mutations that eliminate the binding of dsRNA (10) (Fig. 1C). Taken together, our results show that only SeV coinfections that can form GFP dsRNA induce IFN-β activation.
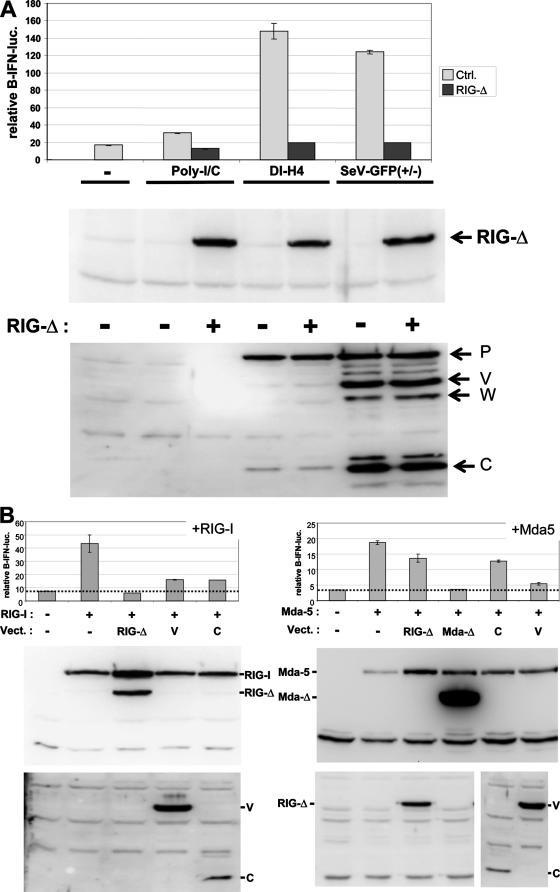
FIG. 2.
Relative contributions of mda-5 and RIG-I in sensing SeV infections in MEFs. (A) Parallel cultures of MEFs were first transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing dominant-negative RIG-I(ΔCARDs) or an empty vector as a negative control. After 24 h, the cells were either transfected with poly(I-C) or infected with either SeV-DI-H4 or SeV-GFP(+/−) (as indicated). Cytoplasmic extracts were prepared after a further 20 h of incubation and used to determine firefly and Renilla luciferase levels and the relative levels of RIG-I(ΔCARDs) (anti-Flag) and viral proteins (anti-P/V/C) by Western blotting (bottom). All transfections were carried out in duplicate, and the range of values obtained is indicated by the error bars. Ctrl., control. (B) Parallel cultures of MEFs were transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing Flag-tagged RIG-I or mda-5 or these helicases plus RIG-I(ΔCARDs), mda-5(ΔCARDs), SeV V (whose overlapping C ORF was closed by a stop codon), SeV C, or an empty plasmid as a negative control, as indicated. Cytoplasmic extracts were prepared after 40 h of incubation and used to determine firefly and Renilla luciferase levels. All transfections were carried out in duplicate, and the range of values obtained is indicated by error bars. The relative levels of the Flag-RIG-I constructs were determined by Western blotting with anti-Flag, those of the Flag-mda-5 constructs were determined with anti-mda-5, and those of the viral V and C proteins were determined with anti-P/V/C serum (bottom). Vect., vector.
FIG. 1.
IFN-β activation induced by SeV-GFP(+/−) infections. (A) Parallel cultures of MEFs were first transfected with pIFNβ-lucff and pTK-lucr and then infected with increasing amounts of either SeV-GFP(+) alone (which expresses a GFP mRNA from a transgene between the M and F genes) or 20 PFU/cell of SeV-GFP(+) plus increasing amounts of either SeV-GFP(−) (which expresses an anti-GFP mRNA from a transgene in the same location) or SeV-RFP (which expresses an RFP mRNA from a transgene in the same location), as indicated. GFP expression was monitored by fluorescence-activated cell sorter analysis at 20 hpi. Cell extracts were prepared at 20 hpi, and equal amounts were used to determine luciferase activities (below). These transfections were carried out three times with independent virus stocks, with similar results. (B) Cytoplasmic extracts were centrifuged on CsCl density gradients to isolate nonencapsidated (pellet) RNAs. The levels of GFP and anti-GFP mRNAs in 15 μg of CsCl pellet RNA were determined using sense- and antisense-specific primers for RT, followed by quantitative PCR (TaqMan) (see Materials and Methods). (C) Parallel cultures of MEFs were first transfected with the luciferase reporter plasmids plus either an empty vector, one expressing wt E3L(100-190), or one expressing mutant E3L(100-190) (E3L-mut.) and then infected with increasing amounts of SeV-GFP(+) and SeV-GFP(−) as indicated. Cell extracts were prepared at 20 hpi, and equal amounts were used to determine luciferase activities. Equal amounts of cell extracts were also Western blotted using anti-N and anti-HA (below).
The two SeV infections that activate IFN-β differ from each other in several respects. First, DI-H4 genomes are of the copyback variety and contain the strong antigenomic replication promoter at their 3′ ends. DI-H4 genomes thus have a strong competitive advantage in replication over nondefective (ND) genomes, and this sometimes leads to less viral structural proteins like N and P being present intracellularly (see, e.g., P protein in Fig. 3A), but sometimes, this difference is minimal (see, e.g., Fig. 2A). However, in either case, viral V and C proteins are almost entirely absent in these DI-H4-infected cells, whereas V and C are found at wild-type levels in GFP(+/−) infections (Fig. 2 and data not shown). Both of these viral proteins are thought to limit IFN-β activation due to virus infection (15, 31). Second, the DI genome replication promoters, like those of the ND genomes, are always active in the presence of viral polymerase, and short 5′-triphosphorylated trailer RNAs (rather than full-length DI genomes) are transcribed by a relatively nonprocessive polymerase, especially when the N protein is limiting. Unlike genome synthesis that is dependent on ongoing (N) protein synthesis, that of trailer RNA actually increases when translation is blocked with cycloheximide (18). SeV trailer RNAs are known to specifically bind to TIAR, a cellular RNA binding protein of the ELAV family (4), and to prevent virus-induced apoptosis (7, 12). DI-H4 infections are expected to overproduce trailer RNAs, which may stimulate RIG-I (11, 25), similar to measles virus leader RNA (26). Lastly, DI-H4-infected cells also contain small amounts of unencapsidated H4 genome RNA that can self-anneal in a concentration-independent manner to form dsRNA panhandles with 5′-triphosphorylated ends. SeV-GFP (+/−)-infected cells, on the other hand, can form dsRNAs with capped ends.
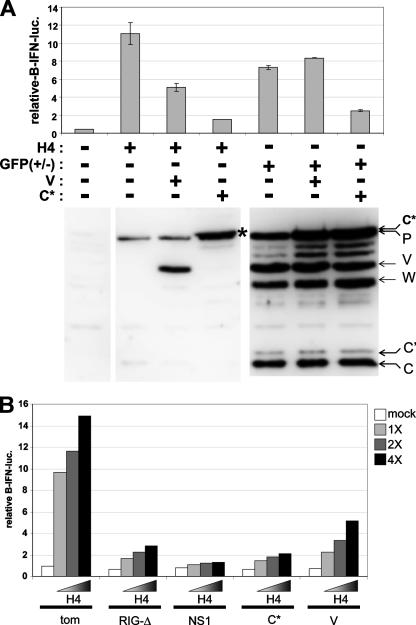
FIG. 3.
SeV V and C inhibition of IFN-β activation induced by SeV infections. (A) Parallel cultures of MEFs were transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing the SeV V protein, the SeV C protein (actually C1-23-Tom-C24-204), or unmodified Tom as a negative control. After 24 h, the cells were infected with either SeV-DI-H4 or SeV-GFP(+/−) (as indicated). Cytoplasmic extracts were prepared after a further 24 h of incubation and used to determine firefly and Renilla luciferase levels. The relative levels of viral P, V, and C proteins were determined by Western blotting with anti-P/C/V serum (bottom). All transfections were carried out in duplicate, and the range of values obtained is indicated by error bars. (B) Parallel cultures of MEFs were transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing either Tom, RIG-I(ΔCARDs), IAV NS1(1-73), C1-23-Tom-C24-204 (C*), or V, as indicated. After 24 h, the cells were infected with increasing amounts of SeV-DI-H4 (1×, 2×, and 4×). Cytoplasmic extracts were prepared after a further 24 h of incubation and used to determine luciferase levels.
Relative contributions of mda-5 and RIG-I in sensing SeV infections in MEFs.
mda-5 signaling to IFN-β was discovered because the PIV5 V protein was found to bind this helicase and thereby prevent poly(I-C)-induced IFN-β activation. Further work showed that the V-mda-5 interaction is a general property of paramyxovirus V proteins, including that of SeV (1, 5). Nevertheless, several groups have now found that SeV infection is sensed by RIG-I (and not mda-5) (13). To determine whether RIG-I was also responsible for signaling to the IFN-β promoter in our MEFs, we examined the effect of expressing a dominant-negative form of RIG-I [RIG-I(ΔCARDs), whose N-terminal CARD domains are deleted] (Fig. 2A). MEFs were first transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing either RIG-I(ΔCARDs) or an empty vector as a negative control. After 24 h, the cells were either transfected with poly(I-C) or infected with either SeV-DI-H4 or SeV-GFP(+/−), and luciferase levels were determined 24 h later. As shown in Fig. 2A, both SeV infections strongly activated the IFN-β promoter, whereas transfected poly(I-C) had a more modest effect in this experiment. RIG-I(ΔCARDs) coexpression did not affect the levels of P, V, and C proteins found intracellularly (bottom), but this coexpression reduced IFN-β activation to background levels in all three cases.
To determine whether the loss of IFN-β activation by RIG-I(ΔCARDs) coexpression was due to its ability to also inhibit mda-5 signaling, e.g., by sequestering cytoplasmic viral RNAs, we examined whether RIG-I(ΔCARDs) could inhibit mda-5 signaling. As mda-5 and RIG-I can be activated by simple overexpression, we examined the effect of RIG-I(ΔCARDs) expression on IFN-β activation due to the overexpression of these two helicases. As shown in Fig. 2B, IFN-β activation clearly occurred upon exogenous mda-5 or RIG-I expression. Moreover, whereas RIG-I(ΔCARDs) coexpression completely inhibited activation due to exogenous RIG-I, RIG-I(ΔCARDs) coexpression had little effect in countering IFN-β activation due to exogenous mda-5. In contrast, the coexpression of a dominant-negative form of mda-5 completely inhibited IFN-β activation due to mda-5 overexpression (Fig. 2B). Taken together, these results suggest that IFN-β activation in our MEFs in response to these SeV infections is predominantly, if not exclusively, due to the action of RIG-I.
The coexpression of either SeV V or C proteins strongly inhibited IFN-β activation due to RIG-I overexpression, whereas only the V protein strongly inhibited IFN-β activation due to mda-5 overexpression (Fig. 2B). The finding that SeV V inhibits RIG-I signaling as well as that of mda-5 is consistent with data from Childs et al. (5), who reported that SeV V was a possible exception to the rule that all V proteins inhibited mda-5 but not RIG-I. They reported that SeV V did in fact modestly inhibit RIG-I (35%), whereas all other V proteins had no effect.
SeV V and C inhibition of SeV-DI-H4 and SeV-GFP(+/−) induced IFN-β activation.
We next examined the abilities of the SeV V and C proteins to inhibit IFN-β activation induced by SeV-DI-H4 and SeV-GFP(+/−) infections. As shown in Fig. 3A, exogenous expression of the SeV V protein did not affect the level of viral P, V, and C proteins in SeV infections, but it did reduce IFN-β activation due to DI-H4 infection (by ∼60%). Remarkably, SeV V overexpression did not inhibit IFN-β activation due to SeV-GFP(+/−) infection. The coexpression of exogenous SeV C protein (actually C1-23-Tom-C24-204, which migrates just slightly slower than the viral P protein) similarly did not affect the level of viral P, V, and C proteins in SeV infections. C overexpression, however, more strongly inhibited IFN-β activation due to either SeV infection [DI-H4-induced activation was reduced by ∼90%, and GFP(+/−)-induced activation was reduced by ∼75%]. Coexpression of the unmodified C protein produced similar results (Fig. 2B and data not shown). The ability of the GFP(+/−) infection to activate IFN-β, despite normal levels of expression of the V and C proteins, is presumably due to the early formation of GFP dsRNA. In this case, the SeV V protein is considerably less potent than C in preventing the response to this dsRNA.
As the DI-H4 infections accumulate so few V and C proteins, we compared the abilities of these proteins (expressed from plasmids) to inhibit IFN-β activation relative to RIG-I(ΔCARDs) and the dsRNA binding domain of the influenza A virus (IAV) NS1(1-73), another viral protein that inhibits RIG-I signaling (22, 25). As shown in Fig. 3B, the SeV C protein was as active as RIG-I(ΔCARDs) in combating an increasing dose of DI-H4 infection and almost as active as NS1. Consistent with above-described results (Fig. 3A), the SeV V protein was less active than C but was still able to inhibit most of the DI-H4-induced IFN-β activation.
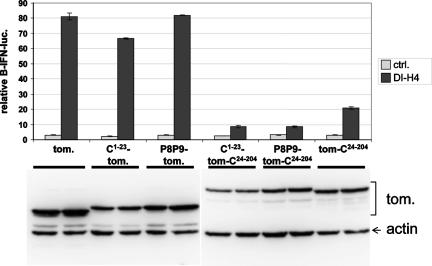
The SeV C1-204 protein is composed of two domains: the N-terminal 23 amino acids (C1-23) which act as a plasma membrane (PM) targeting signal (19) and which is present in the longer (C′/C) but not in the shorter (Y1/Y2) “C” proteins, and C24-204, or the Y1 protein, which acts as a protein interaction domain. Whereas C24-204 (or Y1) is naturally expressed during infection, C1-23 is only found fused to Y1. In order to study the different contributions of these two domains to C-protein function, we have used tomato red fluorescent protein (Tom) in which C1-23 is fused to the N terminus of Tom and C24-204 is fused to its carboxy terminus as a carrier. The interposition of Tom between these two domains of C remarkably does not appear to affect any of the activities of C1-204 (19). MEFs were transfected with the luciferase reporter plasmids along with plasmids expressing various Tom constructs as indicated (Fig. 4). After 24 h, half of the cultures were infected with SeV-DI-H4, and luciferase levels were determined after a further 24 h. The expression of C1-23-Tom, which carries the wt PM anchor and is localized at the cell surface, or P8P9-Tom, which carries a mutant PM anchor and is distributed throughout the cytoplasm (19), had little or no effect on the DI-H4-induced IFN-β activation. In contrast, both C1-23-Tom-C24-204 and P8P9-Tom-C24-204 reduced IFN-β activation to near-background levels. C24-204 alone (Tom-C24-204), moreover, was still quite active in this respect (Fig. 4). Thus, the C24-204 or Y1 protein interaction domain appears to be responsible for inhibiting RIG-I-dependent IFN-β activation, and this inhibition is largely independent of whether C24-204 is localized at the PM.
FIG. 4.
SeV C24-204 (or Y1) protein inhibits IFN-β activation induced by DI-H4 infection. Parallel cultures of MEFs were transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing tomato constructs carrying either the wt (C1-23) or mutant (P8P9) C1-23 fused to their N termini, with and without C24-204 (or Y1) fused to their carboxy termini, C24-204 fused to the carboxy terminus alone (Tom-C24-204), or unmodified Tom as a negative control (ctrl.), as indicated. After 24 h, the cells were infected with SeV-DI-H4 (as indicated). Cytoplasmic extracts were prepared after a further 24 h of incubation and used to determine luciferase levels. The relative levels of the various tomato constructs were determined by Western blotting with anti-dsRED (bottom). All transfections were carried out in duplicate, and the range of values obtained is indicated by error bars.
SeV C protein inhibits IFN-β activation induced by transfected pppRNA.
A general property of nonsegmented negative-strand RNA viruses is that short, promoter-proximal pppRNAs (leader and trailer RNAs) are transcribed from their replication promoters, especially when unassembled N protein is limiting (17, 18). The ability of SeV infections to induce IFN is essentially due to the presence of DI genomes that are present in their egg-grown stocks, especially those of the copyback variety (30). As mentioned above, copyback DI genomes have a strong replicative advantage because they contain strong replication promoters at the DI genome and antigenome 3′ ends. Copyback DI genome replication thus generates short trailer RNAs that are unmodified at either end and can be considered as unstable, abortive replication products (see Discussion).
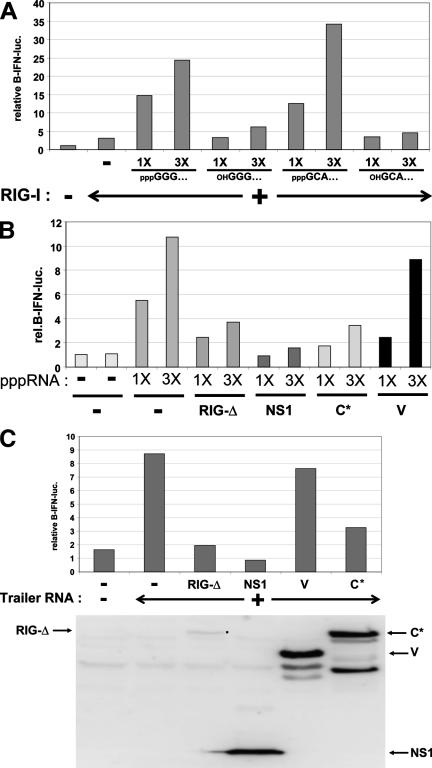
To examine whether trailer RNAs act as pathogen-associated molecular patterns (PAMPs), we transfected trailer RNA made by T7 RNA polymerase in vitro into our MEFs and monitored the activation of IFN-β. As the ability of pppRNAs to induce IFN-β activation is not sequence dependent (11), we also examined model RNAs that were initiated with GTP but then treated with phosphatase or those initiated with the dinucleotide GpC rather than pppG (23). The transfections of all three 5′-triphosphorylated ssRNAs clearly led to IFN-β activation, whereas both RNAs that contained 5′-hydroxyl ends had essentially lost their ability to activate IFN-β in parallel transfections (Fig. 5A and C), confirming previously reported results (11, 25, 26). We then examined the ability of the SeV C and V proteins to inhibit 5′-pppRNA-dependent activation of IFN-β compared to RIG-I(ΔCARDs) and IAV NS1(1-73). MEFs were first transfected with plasmids expressing various viral inhibitory proteins or an empty plasmid as a negative control and then transfected with increasing amounts of pppRNAs. IFN-β activation was monitored after a further 18 h. As shown in Fig. 5B, expression of the SeV C protein was as active as RIG-I(ΔCARDs) in inhibiting IFN-β activation at all amounts of pppRNAs transfected although not quite as active as NS1(1-73). Expression of SeV V was again the least inhibitory; in fact, significant inhibition occurred only at the lowest amount of pppRNA. Thus, short 5′-triphosphorylated ssRNAs such as trailer RNA are potent stimulators of IFN-β when transfected into our MEFs, and expression of the SeV C protein (but not the SeV V protein) can effectively inhibit this stimulation (Fig. 5B and C).
FIG. 5.
pppRNA-induced activation of IFN-β. (A) Parallel cultures of MEFs were transfected with pIFNβ-lucff and pTK-lucr, and pRIG-I was also transfected in some cultures, as indicated. After 24 h, the cells were transfected for 3 h with increasing amounts (1 or 3 μg) of either pppGGG/RNA1, phosphatase-treated GGG/RNA1, pppGCA/RNA1, or OHGCA/RNA1, as indicated. Cytoplasmic extracts were prepared 18 h post-RNA transfection and used to determine luciferase levels. (B) Parallel cultures of MEFs were transfected with pIFNβ-lucff, pTK-lucr, and plasmids expressing Tom, RIG-I(ΔCARDs), IAV NS1(1-73), C1-23-Tom-C24-204 (C*), or V, as indicated. After 24 h, the cells were transfected with increasing amounts (1 μg and 3 μg) of pppGGG/RNA1, as indicated. Cytoplasmic extracts were prepared after 3 h of RNA transfection and used to determine luciferase levels. Rel., relative. (C) Same as above (B), except that the cells were transfected with 3 μg of ppptrailer RNA.
Relative importance of C and V in inhibiting RIG-I-dependent signaling to IFN-β.
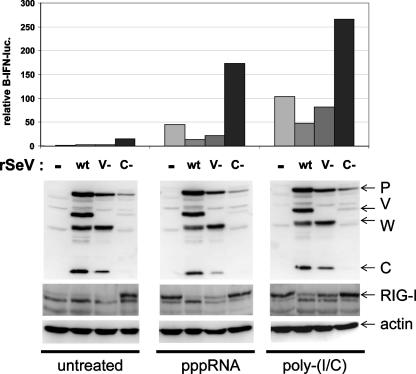
Another way to investigate the relative importance of the C and V proteins in inhibiting RIG-I-dependent signaling to IFN-β is to compare the relative abilities of SeV infections that cannot specifically express the C or V proteins to affect pppRNA- or poly(I-C)-induced IFN-β activation. MEFs were therefore first infected with 20 PFU/ml of either wt SeV, SeV-Vminus (containing a stop codon in the V ORF just downstream of the mRNA editing site, which produces a W-like protein instead of V), or SeV-Cminus (containing three stop codons in the C ORF downstream of the Y2 initiation codon). The infected cells were then transfected (at 24 hpi) with pIFN-β-luc plus either pppRNA, poly(I-C), or no RNA and then harvested after 18 h to determine reporter gene activity. As shown in Fig. 6, these three SeVs replicate to clearly different levels in our highly IFN-competent MEFs (even though they replicate similarly in BSR T7, 293T, and Vero cells), highlighting the essential functions that these accessory proteins play in countering the innate immune response. Nevertheless, in the absence of transfected RNA, only SeV-Cminus infection activates IFN-β to any extent or increases RIG-I levels; RIG-I is an IFN-stimulated gene, and its level reflects that of the antiviral state (Fig. 6, bottom). Transfections of either pppRNA or poly(I-C) strongly activated the reporter gene and increased RIG-I levels. Prior infection with either SeV wt or SeV-Vminus reduced transfected RNA stimulation of the reporter gene and prevented the increase in RIG-I levels (Fig. 6). SeV-Vminus was only slightly less effective than wt SeV in this respect. In sharp contrast, prior infection with SeV-Cminus not only did not prevent the increase in RIG-I levels but also acted synergistically with either pppRNA or poly(I-C) transfection to increase reporter gene activity by increasing the level of RIG-I. These results reinforce the view that it is primarily the SeV C protein (and not V) that inhibits pppRNA- and dsRNA-induced signaling to the IFN-β promoter via RIG-I during SeV infection.
FIG. 6.
RNA-induced activation of IFN-β in cells infected with SeV that cannot express either V or C. Parallel cultures of MEFs were either mock infected or infected with 20 PFU/ml of wt SeV, SeV-Vminus, or SeV-Cminus, as indicated. After 24 h, the cells were transfected with luciferase reporter plasmids and either pppGGG/RNA1, poly(I-C), or no RNA (untreated). Cytoplasmic extracts were prepared after 18 h of RNA transfection, used to determine luciferase levels (above), and Western blotted to determine the levels of P, V, and C proteins as well as endogenous RIG-I and actin as a loading control. rSeV, recombinant SeV.
DISCUSSION
SeV has been one of the most extensively used model viruses to investigate IFN induction in infected cells. Most of this work has used the commercially available Cantell strain of SeV, whose ability to induce IFN, like that of other SeV strains, is due to the presence of DI genomes in egg-grown stocks. Nondefective SeVs that are plaque purified from these stocks, including that of the Cantell strain, do not induce IFN unless cellular RIG-I levels are artificially increased (21, 30). For nondefective SeV infection, the expression of the C and V proteins is apparently sufficient to prevent IFN-β activation under normal conditions. Measles virus infection, in contrast, can apparently induce IFN in the absence of DI genomes, and evidence that this induction is due to the synthesis of leader pppRNAs has recently been provided (26). Leader and trailer RNAs, which are unmodified at either end, are unstable in infected cells unless they are encapsidated with the N protein (presumably after their synthesis as free RNAs) (2). Free leader and trailer RNAs are more easily detected in VSV infections, which synthesize larger amounts of viral RNAs over a shorter period of time. For nondefective VSV infections, eight times as many trailer RNAs/antigenome template are found as leader RNAs/genome template, consistent with the relative strengths of their respective replication promoters. For VSV copyback DI infections, there are 40 times as many trailer RNAs/template (nondefective antigenome plus DI genome) as leader RNAs/genome template, presumably reflecting the increased strength of the copyback DI replication promoters. The VSV polymerase clearly has a strong preference for initiating RNA synthesis at the 3′ ends of copyback DI genomes over both ND genomes and antigenomes (17).
We previously noted that not all SeV stocks that contain DI genomes strongly induce IFN; this ability appears to be restricted to stocks containing relatively small copyback DI genomes (the smaller the DI genomes, the more moles of ends are present for a given weight). The commercially available Cantell strain contains a copyback DI genome of only 546 nucleotides in length, the smallest SeV DI genome described to date (30). SeV copyback DI-H4 genomes (1,410 nucleotides) have the same strong replicative advantage as their VSV counterparts because they also contain strong replication promoters at both their genome and antigenome 3′ ends. Thus, SeV copyback DI infections presumably synthesize considerably more pppRNAs than standard virus infections. We also previously noted that when cytoplasmic extracts of DI-H4-infected cells are centrifuged on CsCl density gradients, small amounts of DI genome RNA are found in the pellet fraction (30). This indicates that this RNA is not encapsidated with the N protein and therefore forms dsRNA panhandles in a concentration-independent manner. Thus, copyback DI-H4 infections apparently produce considerably more of both known PAMPs of RNA virus infection than do standard virus infections. Coupled with their strongly reduced accumulation of the viral C and V proteins, it is easy to see why these copyback DI infections are such potent inducers of IFN-β.
All paramyxoviruses express either C or V proteins, and many viruses express both. In viruses that express only C or V, we presume that either viral protein alone counteracts the innate immune response of the host to aid virus replication. The C and V proteins, which bear no sequence similarity, likely target different key elements of the host response. Viruses that express both C and V presumably have more diverse ways of countering innate immunity. In support of this notion, SeV infections that cannot specifically express either the C or V protein contain increased levels of IFN-β and interleukin-8 mRNAs relative to wt SeV infections (31), and the independent expression of the C or V protein will inhibit poly(I-C)- or Newcastle disease virus-dependent activation of IRF-3 (15). Previous work has identified mda-5 as being a key target of paramyxovirus V proteins in countering the innate immune response (1). This paper provides evidence that for SeV infection of MEFs, it is the C protein (and not V) that is primarily responsible for this effect and that C acts by countering RIG-I-dependent signaling to IFN-β. For example, the independent expression of either C or V inhibited IFN-β activation due to RIG-I overexpression (Fig. 2B). Also, both proteins inhibited IFN-β activation due to DI-H4 infection, although C was always more effective here than V (Fig. 3). However, only C expression effectively inhibited IFN-β activation due to GFP(+/−) infection (Fig. 3) or transfected poly(I-C) or pppRNAs (Fig. 5). Perhaps the strongest indication that the C proteins are primarily responsible for countering the innate immune response comes from experiments with SeV that cannot specifically express the C or V proteins. SeV-Cminus infection not only cannot prevent the effects of transfected poly(I-C) or pppRNAs on IFN-β activation but also synergistically enhances these effects. SeV-Vminus infection, in contrast, behaves mostly like wt SeV infection and counteracts the effects of transfected poly(I-C) or pppRNAs (Fig. 6).
Finally, we note that poly(I-C) (made with polynucleotide phosphorylase that generates 5′ diphosphate ends and which is transfected into cells) and the presumed GFP dsRNA (that is directly generated in the cytoplasm via the viral transcriptase and which contains capped 5′ ends) both activate IFN-β via RIG-I in MEFs. Thus, the ability of dsRNA to induce RIG-I signaling does not depend on the manner in which it is introduced into this cell compartment, nor is it peculiar to the presence of 5′-diphosphorylated ends that are not normally found in cells and could theoretically act as PAMPs. Moreover, in either case, the activation of IFN-β by these dsRNAs is inhibited by the SeV C protein and not V, presumably because this signaling passes through RIG-I and not mda-5. It appears that our MEFs contain insufficient amounts of mda-5 to sense SeV infection, as these MEFs respond well to the expression of plasmid-derived mda-5 (Fig. 2B). This conclusion is also consistent with our finding that three different rubulavirus V proteins that are known to counteract poly(I-C)-induced mda-5 signaling were unable to inhibit IFN-β activation in response to SeV-DI-H4 infection (data not shown). We expect that the SeV V protein will be more important in countering the innate immune response in other cells in which mda-5 functions as a PAMP recognition receptor.
Acknowledgments
We thank J. Tschopp (Lausanne), T. Fujita (Kyoto), J. Perrault (San Diego), B. Jacobs (Arizona), K. Conzelmann (Munich), and R. Randall (St. Andrews) for sharing materials.
This work was supported by the Swiss National Science Fund.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837-845. [DOI] [PubMed] [Google Scholar]
- 3.Bontron, S., C. Ucla, B. Mach, and V. Steimle. 1997. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol. 17:4249-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 2006. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190-200. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 7.Garcin, D., G. Taylor, K. Tanebayashi, R. Compans, and D. Kolakofsky. 1998. The short Sendai virus leader region controls induction of programmed cell death. Virology 243:340-353. [DOI] [PubMed] [Google Scholar]
- 8.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 9.Hiscott, J., T. L. Nguyen, M. Arguello, P. Nakhaei, and S. Paz. 2006. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene 25:6844-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, C. K., and S. Shuman. 1996. Mutational analysis of the vaccinia virus E3 protein defines amino acid residues involved in E3 binding to double-stranded RNA. J. Virol. 70:2611-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 12.Iseni, F., D. Garcin, M. Nishio, N. Kedersha, P. Anderson, and D. Kolakofsky. 2002. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 14.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 15.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137-148. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 17.Leppert, M., and D. Kolakofsky. 1980. Effect of defective interfering particles on plus- and minus-strand leader RNAs in vesicular stomatitis virus-infected cells. J. Virol. 35:704-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leppert, M., L. Rittenhouse, J. Perrault, D. F. Summers, and D. Kolakofsky. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18:735-747. [DOI] [PubMed] [Google Scholar]
- 19.Marq, J. B., A. Brini, D. Kolakofsky, and D. Garcin. 2007. Targeting of the Sendai virus C protein to the plasma membrane via a peptide-only membrane anchor. J. Virol. 81:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuoka, Y., J. Curran, T. Pelet, D. Kolakofsky, R. Ray, and R. W. Compans. 1991. The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J. Virol. 65:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 22.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milligan, J. F., and O. C. Uhlenbeck. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51-62. [DOI] [PubMed] [Google Scholar]
- 24.Ostertag, D., T. M. Hoblitzell-Ostertag, and J. Perrault. 2007. Overproduction of double-stranded RNA in vesicular stomatitis virus-infected cells activates a constitutive cell-type-specific antiviral response. J. Virol. 81:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 26.Plumet, S., F. Herschke, J. M. Bourhis, H. Valentin, S. Longhi, and D. Gerlier. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE 2:e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 28.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 175:5260-5268. [DOI] [PubMed] [Google Scholar]
- 29.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 30.Strahle, L., D. Garcin, and D. Kolakofsky. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101-111. [DOI] [PubMed] [Google Scholar]
- 31.Strahle, L., D. Garcin, P. Le Mercier, J. F. Schlaak, and D. Kolakofsky. 2003. Sendai virus targets inflammatory responses, as well as the interferon-induced antiviral state, in a multifaceted manner. J. Virol. 77:7903-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]