Abstract
Poliovirus infection remodels intracellular membranes, creating a large number of membranous vesicles on which viral RNA replication occurs. Poliovirus-induced vesicles display hallmarks of cellular autophagosomes, including delimiting double membranes surrounding the cytosolic lumen, acquisition of the endosomal marker LAMP-1, and recruitment of the 18-kDa host protein LC3. Autophagy results in the covalent lipidation of LC3, conferring the property of membrane association to this previously microtubule-associated protein and providing a biochemical marker for the induction of autophagy. Here, we report that a similar modification of LC3 occurs both during poliovirus infection and following expression of a single viral protein, a stable precursor termed 2BC. Therefore, one of the early steps in cellular autophagy, LC3 modification, can be genetically separated from the induction of double-membraned vesicles that contain the modified LC3, which requires both viral proteins 2BC and 3A. The existence of viral inducers that promote a distinct aspect of the formation of autophagosome-like membranes both facilitates the dissection of this cellular process and supports the hypothesis that this branch of the innate immune response is directly subverted by poliovirus.
All positive-strand RNA viruses of eukaryotes replicate their genomes on cytoplasmic membranes, and infections are often accompanied by dramatic reorganization of those membranes. We and others have argued that one function of membrane localization of viral RNA replication proteins is to create two-dimensional surfaces to promote the oligomerization of viral proteins (6, 31). Yet the detailed cell biology of the targeted membranes and the mechanisms by which they are altered differ greatly from virus to virus (40, 53).
Poliovirus infection rearranges intracellular membranes, creating membranous vesicles, 200 to 400 nm in diameter, that accumulate in the cytoplasm (9, 42, 47). Immunoelectron microscopy revealed that the cytoplasmic surfaces of these membranous vesicles, not the interior lumen, are the sites of viral RNA replication (1, 7). The specific morphology of these membranes has proven somewhat controversial, because they are rich in lipids (17, 33) and their apparent morphology is therefore sensitive to the fixation method used in electron microscopy (43, 46). Using high-pressure cryopreservation methods, we have consistently observed double-membraned vesicles (42, 47), as did Dales et al. using more conventional fixation techniques (9). Double-membraned vesicular structures have also been associated with viral RNA replication complexes in cells infected with several other positive-strand RNA viruses, including equine arterivirus (37), murine hepatitis virus (16), and sudden acute respiratory syndrome-associated virus (15). Therefore, understanding the mechanism(s) by which viral proteins induce the formation of double-membrane vesicles is of interest for understanding the formation of RNA replication complexes of several kinds of RNA viruses.
Several of the features displayed by the vesicles induced during poliovirus infection are shared with cellular autophagosomes, double-membraned organelles that become degradative upon maturation, breaking down cytoplasmic proteins and organelles entrapped within their lumen. Originally identified as a process induced by cellular starvation, autophagy is now appreciated as a cellular response to a variety of stimuli. For example, the estrogen receptor agonist tamoxifen is a potent inducer of autophagy, indicating a role for the hormonal induction of autophagy (2-4). In Saccharomyces cerevisiae, formation and maturation of autophagosomes requires the function of many genes (reviewed in reference 52). Human homologs of several yeast autophagy genes have recently been identified, including LC3, the human homolog of ATG8, and it is likely that much of the autophagic pathway has been evolutionarily conserved (22, 50). Many recent experiments have identified the autophagy pathway as a critical component of the innate immune response of plants and mammals (reviewed in references 10 and 30). Therefore, we sought to discover the similarities and differences between the poliovirus-induced membranes and cellular autophagosomes and to investigate the mechanism of formation of the double-membraned vesicles induced by poliovirus and the role of the autophagy pathway during viral infection. Previous studies show that reduced function of the autophagy pathway resulted in reduced amounts of extracellular and intracellular poliovirus (20), arguing that the autophagic pathway plays a positive role in the growth of both these positive-strand RNA viruses.
The formation and maturation of autophagosomes involve the stepwise acquisition of proteins from disparate cellular compartments (reviewed in references 28 and 39). Nascent autophagosomes form either de novo or from the endoplasmic reticulum and comprise cellular cytoplasm surrounded by two lipid bilayers that fuse from a C-shaped intermediate (14, 32, 36). Although relatively protein poor, they have been shown to contain a modified form of the autophagy protein LC3. Originally characterized as microtubule-associated protein light chain 3, or MAP-LC-3, LC3 protein is cleaved after synthesis by cellular protease Atg4, generating an 18-kDa species termed LC3-I (see Fig. 1A). When autophagy is activated, a series of covalent transfers links LC3 to Atg7, then to Atg3, and finally to phosphatidylethanolamine, generating a lipidated species termed LC3-II (Fig. 1A) (19, 45, 50). This modification allows LC3 to become membrane associated, preferentially associating with the developing and newly formed autophagosomes (23, 24).
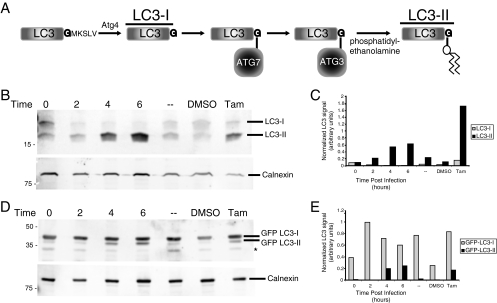
FIG. 1.
Modification of LC3 protein during poliovirus infection. (A) Schematic representation of LC3 processing as it occurs during autophagy activation. LC3-I is the unmodified, cleaved form of LC3, while LC3-II is the lipid-conjugated species. (B and C) Human 293T cells were either infected with poliovirus at a multiplicity of infection of 50 PFU per cell and harvested at 0, 2, 4, or 6 h after infection, mock infected (-), treated with 10 μM tamoxifen, or treated with control solvent (dimethyl sulfoxide [DMSO]) for 48 h prior to harvesting. (B) Cell extracts were subjected to PAGE through a 13.5% gel, and separated proteins were immunoblotted using a rabbit polyclonal anti-LC3 antibody and subsequently detected with goat anti-rabbit alkaline phosphatase and ECF reagent (Amersham Biosciences). The same immunoblots were reprobed with a mouse monoclonal anticalnexin antibody. (C) The amount of signal for both endogenous LC3-I and endogenous LC3-II from panel B is presented graphically. (D) Cells were transfected with a plasmid that expressed GFP-LC3 for 48 h before infection. Cell extracts were subjected to PAGE in a 10% polyacrylamide-SDS gel, and separated proteins were immunoblotted for LC3 and calnexin as for panel B. The asterisk signifies an unidentified cross-reacting band. (E) The amount of signal for GFP-LC3-I and GFP-LC3-II from panel D is presented graphically. The gels are representative of multiple experiments.
The genomes of poliovirus and other picornaviruses consist of a single, positive-stranded RNA. A single encoded polyprotein is processed by viral proteases to produce individual capsid proteins, the viral RNA-dependent RNA polymerase 3D, and several other multifunctional nonstructural proteins. Of the nonstructural proteins, three (2B, 2C, and 3A) associate with membranes when expressed in isolation; mutations in any of these three proteins lead to defects in RNA replication. The expression of stable protein precursor 2BC and protein 3A, in combination but not separately, was found to induce the formation of double-membraned vesicles that display biochemical markers and fractionate similarly to the vesicles induced during poliovirus infection (20, 47). Thus, should these membranes prove to derive directly from the autophagy pathway, these viral proteins will be among the few molecularly characterized inducers of this process.
We showed previously that, during poliovirus infection, a green fluorescent protein (GFP)-fused version of one of the three cellular LC3 proteins, LC3b, becomes associated with cellular membranes that also contain viral RNA replication proteins (20). Here, we investigated the possibility that the targeting of LC3 to membranes results from its covalent modification, as occurs during autophagy. Further, we tested the possibility that expression of an individual viral protein is sufficient to accomplish this modification.
MATERIALS AND METHODS
Cells and viruses.
Human HeLa and monkey kidney COS-7 cells were cultured as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) calf serum, 100 units of penicillin/ml, and 100 units of streptomycin/ml at 37°C and 5% CO2. Human 293T cells were cultured in a similar fashion using 10% (vol/vol) fetal bovine serum. Poliovirus type 1 Mahoney was propagated from an infectious cDNA plasmid as previously described (25, 38).
Plasmids and transfection.
Mutagenesis of a plasmid that encodes GFP-LC3 (20) to create the G120A, G120Stop, and K122A constructs was performed by PCR using primers containing the mutation of interest close to the 3′ EcoRI restriction site used to clone the LC3 gene. Each amplified DNA segment was sequenced in its entirety to ensure that no adventitious mutations were introduced. Plasmids used for expression of polioviral proteins were previously described (12).
Antibodies.
Anti-vesicular stomatitis virus G protein (VSV-G) antibody was purchased from RDI (Concord, MA). Anticalnexin antibody was purchased from Stressgen Bioreagents Corp. (Victoria, B.C., Canada). Mouse monoclonal antibodies directed to human LAMP-1 (H4A3) were purchased from the Developmental Studies Hybridoma Bank (University of Iowa). Anti-2C and anti-2B mouse monoclonal antibodies were the kind gift of K. Bienz (University of Basel). Anti-3A mouse monoclonal antibody was previously described (13). Polyclonal rabbit serum against human LC3 protein was raised against purified LC3b, expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein. The GST-LC3 clone was constructed by excising the LC3b cDNA from the GFP-LC3 construct with EcoRI followed by in-frame ligation into the pGEX-6P-2 vector (Amersham Biosciences, Piscataway, NJ). GST-LC3 protein was purified from bacterial extracts on glutathione Sepharose 4B columns. LC3 protein was released through proteolytic digestion with the PreScission protease (Amersham). Two commercial preparations of antibody (Invitrogen, Carlsbad, CA) were affinity purified (21), depleted of anti-GST antibodies using a GST-Sepharose column, and stored at 4°C.
Protein extraction and immunoblotting.
Cells were seeded at 1 × 106 per 10-cm dish for infection the next day. For transfection, cells were seeded out at 2 × 105 cells per 10-cm dish, followed by transfection with 2 μg of plasmid DNA using Effectene reagent (QIAGEN, Valencia, CA) and incubation for 48 h at 37°C. Cells were harvested with phosphate-buffered saline (PBS) containing 5 mM EDTA (pH 8.0) and collected by centrifugation at 300 × g for 5 min. Cell pellets were washed in the same buffer, pelleted in an Eppendorf microcentrifuge at 6,000 × g, resuspended in RSB-NP-40 (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 1.5 mM MgCl2, 1% NP-40) supplemented with Mini-complete EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), and incubated on ice for 15 min. Nuclei and insoluble debris were pelleted by centrifugation at 6,000 × g for 5 min. Cell extracts were then stored at −20°C or subjected immediately to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For saponin extraction, cell pellets were resuspended in a solution containing 0.5% saponin, 80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8], 1 mM CaCl2, and 5 mM EGTA as described elsewhere (29) and collected by centrifugation prior to the addition of the RSB—NP-40 solution. Cell extracts were mixed with 6× Laemmli loading buffer and heated to 95°C for 5 min prior to electrophoresis at 120 V for 1 to 2 h on either 10% or 13.5% SDS-polyacrylamide gels. For immunoblotting, proteins were transferred to Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) for 75 min at 20 V in a Miniprotean III transfer tank (Bio-Rad, Hercules, CA). Membranes were then air dried before rewetting with 70% methanol followed by blocking with PBS supplemented with 0.1% (vol/vol) Tween-20 and 2% (wt/vol) bovine serum albumin. All primary antibody incubations were done in blocking solution for a minimum of 1 h at room temperature followed by washing with PBS-Tween (PBS supplemented with 0.1% Tween). Alkaline phosphatase-conjugated secondary antibodies were diluted 1:10,000 in PBS-Tween, incubated with the blot for a minimum of 1 h at room temperature, washed with PBS-Tween, and developed with the ECF reagent (Amersham). Exposures to ECF reagent and detection with a Storm phosphorimager were maintained in a linear range to facilitate quantitation.
Immunoprecipitation.
Transfected COS-7 cells were harvested and subjected to homogenization as previously described (47). Briefly, cells were resuspended in a solution containing 250 mM sucrose and 5 mM MOPS (morpholinepropanesulfonic acid) and then syringe filter homogenized with three passes through three 14-μm filters. Homogenates were spun at 6,000 × g for 5 min to pellet nuclei, large membranes, and cell bodies. The postnuclear supernatant was then mixed with anti-2B antibody and rotated for 4 h at 4°C, and NP-40 was added to a final concentration of 1% vol/vol as shown in Fig. 7. Sheep anti-mouse immunoglobulin G magnetic beads (Dynabeads M-280; Invitrogen, Carlsbad, CA) were equilibrated in homogenization buffer with or without 1% NP-40. To each immunoprecipitation, 1.4 × 108 equilibrated beads were added. Samples were rotated overnight at 4°C. Unbound material was separated on a magnetic separation stand. Magnetic beads were washed three times in homogenization buffer with or without 1% NP-40. Beads were resuspended in a small volume of 2× Laemmli sample buffer before boiling. Magnetic beads were separated from sample buffer prior to electrophoresis in polyacrylamide gels.
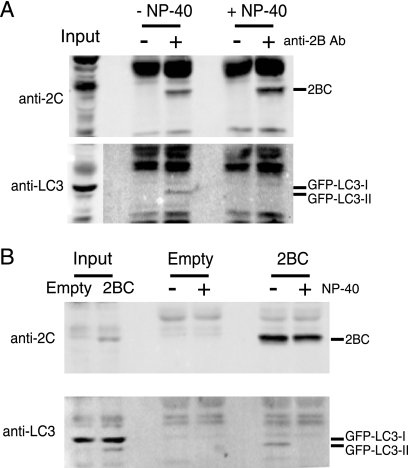
FIG. 7.
Immunoprecipitation of 2BC from cellular homogenates. COS-7 cells were transfected with both GFP-LC3 and 2BC expression plasmids (A and B) or with empty vector (B). Forty-eight hours later, cells were harvested and lysed by filter homogenization. A low-speed-centrifugation supernatant was collected and used as input for immunoprecipitation. (A) Half of the samples were supplemented with NP-40 and immunoprecipitated with antibody as indicated. (B) One sample of each transfection was supplemented with NP-40 prior to antibody addition to all samples. Immunoprecipitates were separated on an 8% Laemmli gel, transferred to PVDF, and blotted with anti-2C and then anti-LC3 antibodies. Exposure of the LC3 Western blot in panel A is lighter for the input to facilitate visualization of bands.
RESULTS
Modification of endogenous LC3 and GFP-LC3 expressed by transfection during poliovirus infection.
To determine whether one of the early hallmarks of cellular autophagy, the conversion of cellular protein LC3 to LC3-II (Fig. 1A), occurs during poliovirus infection, we monitored the electrophoretic mobility of endogenous cellular LC3. Immunoblotting with an anti-LC3 polyclonal antibody (see Materials and Methods) revealed two reactive species of LC3, as can be seen in Fig. 1B: an upper, 18-kDa band corresponding to unmodified LC3-I and a lower band corresponding to previously described species LC3-II. While an increase in mobility for a species to which a phosphatidylethanolamine moiety has been added may seem counterintuitive, its identity has been well documented (23); it is likely that the increased mobility results from increased SDS binding during denaturing gel electrophoresis. The intensity of the LC3-II band increased relative to a control protein, calnexin, after treatment of the cells with tamoxifen to induce autophagy but not after control treatments (Fig. 1B). During a time course of poliovirus infection, a large increase in the amount of endogenous LC3-II could be readily detected by 4 and 6 h after infection. The comigration of this species with bona fide LC3-II in cells that had been treated with tamoxifen made it likely that this species was authentic LC3-II.
To investigate the mechanism of LC3 modification during poliovirus infection and its similarity to the formation of LC3-II during autophagy, we utilized a GFP-LC3 fusion protein expressed by transient transfection (22, 29). The immunoblot in Fig. 1C displays the electrophoretic mobility of GFP-LC3 protein during a time course of poliovirus infection. In the untreated sample, very little GFP-LC3-II was observed. However, tamoxifen treatment caused a readily detectable increase in signal for the GFP-LC3-II band. Infection with poliovirus also caused a large increase in the intensity of a band of identical mobility. Therefore, for both endogenous LC3 and GFP-LC3 expressed by transfection, a clear increase in a species with an altered electrophoretic mobility identical to that of the lipidated LC3-II species observed during bona fide autophagy was observed during poliovirus infection. For both tamoxifen treatment and poliovirus infection, the amounts of GFP-LC3-II clearly increase. However, the ratios of the modified to unmodified species are lower than those observed for the endogenous LC3 protein, and a decrease in the GFP-LC3-I band is not observed. This presumably results from the overexpression of GFP-LC3 and the resulting saturation of some aspect of the lipidation machinery.
Quantitation of these data (Fig. 1C and E) reveals that the apparent increase of the endogenous LC3-II species was larger than any concomitant decrease in the LC3-I signal. If the immunoblotting signals reflected the relative amounts of protein faithfully, this would argue for a net synthesis or stabilization of LC3, present as the LC3-II species, during poliovirus infection. However, it has been reported by others (23) and observed by us that anti-LC3 antibodies can display different affinities for LC3-I and LC3-II species. In support of this possibility, when the modification of GFP-LC3 protein was monitored by immunoblotting with an anti-LC3 antibody (Fig. 2C) and the same blot was probed with an anti-GFP antibody (Fig. 2E), it was clear that the apparent percentages of modification were different for the two probes. Therefore, we cannot conclude that the apparent net increase in LC3-containing species reflects a net synthesis or stabilization, because the LC3-II species are much more reactive with the antibody. However, we can conclude that the amounts of LC3-II and GFP-LC3-II species increase dramatically during infection with poliovirus, as they do during the induction of cellular autophagy.
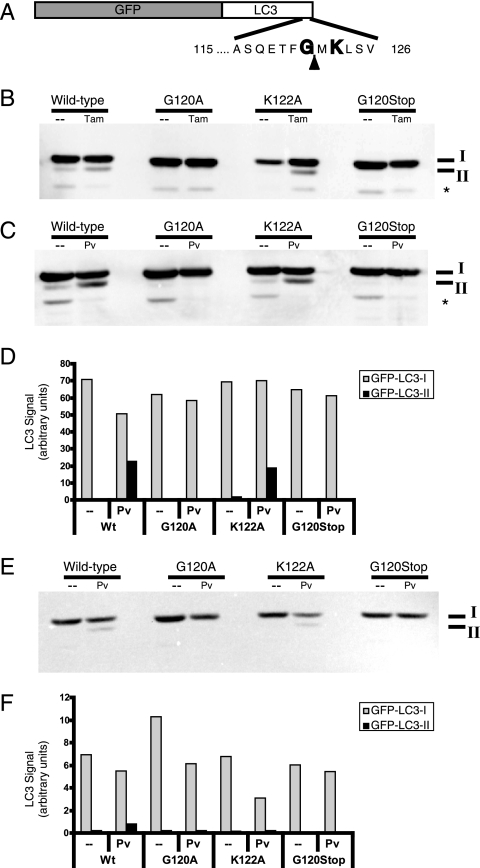
FIG. 2.
Effect of mutations in GFP-LC3 on modification during autophagy induction and poliovirus infection. (A) Schematic depiction of the GFP-LC3 fusion protein sequence. The solid arrowhead indicates the site of cleavage by cellular protease Atg4. Gly120 is the known site of phosphatidylethanolamine addition during autophagy induction. (B) DNA plasmids that encode wild-type GFP-LC3 and variant GFP-LC3 sequences containing G120A, G120Stop, and K122A mutations were transfected into 293T cells. Following transfection, cells were fed with either normal medium (-) or medium that contained 10 μM tamoxifen for 48 h. Cell extracts were displayed on a 10% polyacrylamide-SDS gel and immunoblotted using an anti-LC3 antibody. The asterisk signifies an unidentified cross-reacting band. (C) Wild-type and G120A, K122A, and G120Stop mutant GFP-LC3-encoding plasmids were transfected into 293T cells. After 48 h, cells were infected with poliovirus (50 PFU/cell) and harvested after 6 h. GFP-LC3 protein was detected as for panel B. Gels are representative of multiple experiments. (D) Quantitation of data in panel C is shown. The anti-LC3 immunoblot in panel C was reprobed with an antibody to GFP (E) to test whether the relative immunogenicity of GFP-LC3-I and GFP-LC3-II changed with different antibodies. (F) Relative amounts of GFP-LC3-I and GFP-LC3-II are quantified.
Modification of GFP-LC3 during autophagy and poliovirus infection shows similar dependence on LC3 sequence.
To investigate whether poliovirus-induced modification of LC3 utilizes the same amino acids as autophagy-induced LC3 lipidation, residues of the LC3 protein known to be crucial for lipidation were mutagenized (Fig. 2A). Specifically, LC3 is cleaved C-terminally to Gly120, after which a series of covalent transfers occurs (Fig. 1A) that ultimately lead to the covalent linkage of phosphatidylethanolamine to the carboxyl group of Gly120 (21, 23, 51). As can be seen in Fig. 2B, mutation of this crucial Gly to either an Ala residue (G120A) or to a stop codon (G120Stop) prevented the modification of LC3 observed following treatment with tamoxifen, whereas mutation of residue Lys122, known to be dispensable for LC3 modification during autophagy (51), to Ala (K122A) had no effect. Similarly, the G120A and G120Stop mutations abrogated the poliovirus-induced modification of GFP-LC3, whereas the K122A mutation did not (Fig. 2C and D). These data and the identical electrophoretic mobilities of the modified LC3 species following tamoxifen treatment and poliovirus infection are consistent with the hypothesis that the poliovirus-induced modification is the addition of phosphatidylethanolamine, as occurs during cellular autophagy.
Localization of wild-type and mutant GFP-LC3 during poliovirus infection.
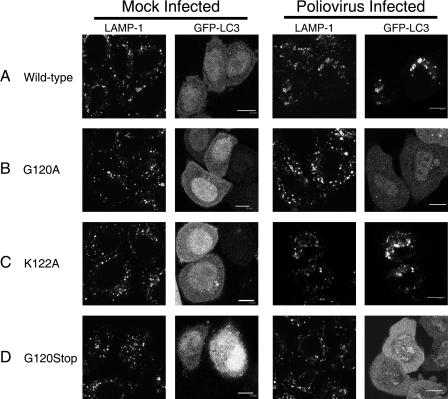
During autophagy, the lipidation of LC3 is necessary and sufficient for its relocalization from cytosolic and microtubule-associated forms to autophagic membranes. To test whether LC3 modification correlated with its relocalization to membranous structures during poliovirus infection as well, we monitored the subcellular localization of the four GFP-LC3 proteins for which data are shown in Fig. 2 in the presence and absence of infection. As shown in Fig. 3, uninfected cells that expressed wild-type GFP-LC3 or any of the mutant GFP-LC3 proteins exhibited diffuse GFP signals that did not colocalize with the punctate staining of the late endosomal marker LAMP-1. Following poliovirus infection, however, wild-type GFP-LC3 was found to colocalize with LAMP-1, as has been shown previously (20). The modifiable mutant protein GFP-LC3 K122A showed a similar pattern of punctate colocalization with LAMP-1 upon poliovirus infection. In contrast, the nonmodifiable GFP-LC3 proteins G120A and G120Stop maintained a diffuse GFP signal upon poliovirus infection, similar to that observed in uninfected cells. Therefore, there was an excellent correlation between the modification of GFP-LC3-I to GFP-LC3-II (Fig. 2) and the localization of all detectable GFP-LC3 to membranous structures during poliovirus infection.
FIG. 3.
Effect of mutations in GFP-LC3 on its colocalization with LAMP-1 during poliovirus infection. The intracellular distributions of cellular LAMP-1, visualized by indirect immunofluorescence, and GFP-LC3, visualized by its intrinsic fluorescence, were studied following poliovirus infection. Localization patterns of LAMP-1 and wild-type GFP-LC3 (A), G120A GFP-LC3 (B), K122A GFP-LC3 (C), or G120Stop GFP-LC3 (D) in the absence (mock infected) and presence of poliovirus infection are shown. HeLa cells were transfected with GFP-LC3-encoding plasmids prior to infection with poliovirus as in Fig. 2. Cells were fixed 4 h postinfection and stained for LAMP-1 protein with subsequent detection with goat anti-mouse antibody labeled with rhodamine. Bars, 10 μm.
The induction of LC3-II formation during poliovirus infection, and its similarity to the process of cellular autophagy, suggests two distinct hypotheses: either poliovirus subverts the autophagic machinery to synthesize subcellular membranes on which to assemble RNA replication complexes, as we have previously suggested (20, 42, 47), or bona fide autophagy is induced by viral infection as part of the innate immune response. We showed previously that viral proteins 2BC and 3A, when expressed together, induce double-membraned vesicles to which both GFP-LC3 and LAMP-1 colocalize (20). Therefore, we tested the ability of individual viral proteins to induce LC3 modification.
Optimizing detection of LC3-II to extend analysis to additional cell types.
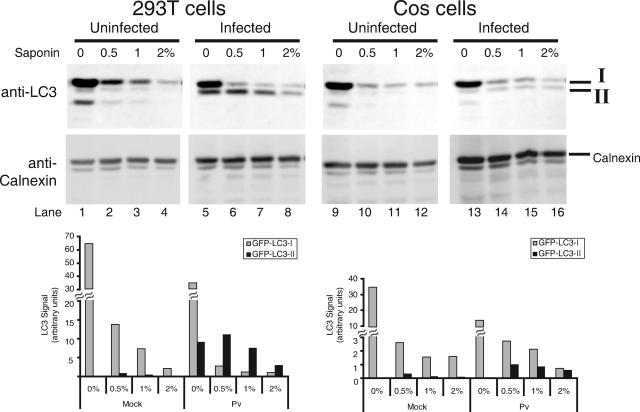
We have demonstrated detectable expression of poliovirus proteins 2B, 2C, 2BC, and 3A after transient transfection only in COS-7 monkey kidney cells (12). Unfortunately, cell lines differ in their abundance of endogenous LC3 and in the extent of its modification during autophagy (49). To test the effect of individual poliovirus proteins on LC3 modification, we needed to establish a detection system in COS-7 cells. As shown in Fig. 1, when total cytoplasmic extracts of uninfected and infected human 293T cells that had been transfected with a GFP-LC3 expression plasmid were displayed by SDS-PAGE and detected using an anti-LC3 antibody, an increase in GFP-LC3-II upon poliovirus infection was readily detected (Fig. 4, compare lanes 1 and 5). However, such a difference was difficult to detect in uninfected and infected COS-7 cells that had been transfected with the same GFP-LC3 expression plasmid (Fig. 4, compare lanes 9 and 13). To enhance the detection of any GFP-LC3-II signal that might be present in infected COS-7 cells, we endeavored to reduce the background from unmodified GFP-LC3-I. To this end, the cell pellets were extracted with the detergent saponin, which has been reported to remove unlipidated LC3 species selectively (29). Extraction of 293T cell pellets with increasing amounts of saponin showed, as reported, that LC3-I was selectively extracted from both the uninfected and infected samples, which improved the detection of the GFP-LC3-II that accumulated during poliovirus infection by reducing the background of GFP-LC3-I. The optimal concentration of saponin for this extraction was determined to be 0.5% (Fig. 4, compare lanes 2 and 6). Importantly, the saponin extraction procedure did not change the recovery of calnexin, a resident endoplasmic reticulum protein, arguing that little membrane disruption occurred during this procedure. Extraction of uninfected and infected COS-7 cells with saponin removed enough GFP-LC3-I that an increased GFP-LC3-II signal could readily be observed in the infected samples (Fig. 4, compare lanes 10 and 14). Therefore, this technique was employed to assess the contribution of individual poliovirus proteins to GFP-LC3 modification in COS-7 cells.
FIG. 4.
Saponin extraction can be used to extract unmodified LC3 species. Human 293T and monkey COS-7 cells were mock infected or infected with poliovirus (50 PFU/cell) for 6 h. Cells were harvested, collected by centrifugation, and subjected to saponin extraction with 0, 0.5, 1, and 2% saponin-containing buffer, as described in Materials and Methods. Proteins were displayed by electrophoresis in 10% acrylamide-SDS gels and visualized by blotting with anti-LC3 or anticalnexin antibodies. The relative amounts of GFP-LC3-I and GFP-LC3-II proteins remaining after saponin extraction were quantified by normalizing the signal in the anti-LC3 immunoblot with the amount of calnexin visualized in the anticalnexin immunoblot.
Effect of individual poliovirus proteins on LC3 modification.
To determine whether any of the individual poliovirus proteins (2B, 2C, 2BC, or 3A) known to affect the structure and function of intracellular membranes (7, 11, 12, 20, 47) was necessary or sufficient to induce the modification of LC3, we expressed these proteins singly and in combination in COS-7 cells as described previously (12). Each individual protein was expressed as a dicistronic mRNA, with the poliovirus protein encoded by the first cistron and the glycoprotein from VSV-G encoded by the second cistron, downstream from the poliovirus internal ribosome entry site. In all the experiments whose results are shown in Fig. 5, the COS-7 cells were transfected with equal amounts of plasmid DNA. In the lanes that show no expression of a poliovirus protein, the transfected DNA was the vector that expressed VSV-G only under the control of the simian virus 40 promoter and the poliovirus internal ribosome entry site. This method was used to ensure that the expression of VSV-G could be used to monitor the relative transfection and transcription efficiencies of the individual poliovirus protein-expressing constructs. Cells were harvested and subjected to saponin extraction before subsequent preparation of cytoplasmic extracts and display of proteins by SDS-PAGE. These gels were then immunoblotted to determine whether the viral protein of interest was expressed in increasing amounts as expected and to determine the amount of VSV-G synthesized (Fig. 5). VSV-G was used as an internal control in this experiment for several reasons. First, as VSV-G is an integral membrane protein, its relatively constant amount served as a way to monitor the gentleness of the saponin extraction conditions. Second, because viral proteins can be toxic, monitoring the amount of VSV-G coexpressed in the same cells served to control for selectively reduced vitality of the transfected cells. For each lane, the amount of GFP-LC3-I and GFP-LC3-II was quantified, normalized to the amount of VSV-G observed, and graphed as a function of the amount of viral protein-expressing plasmid introduced.
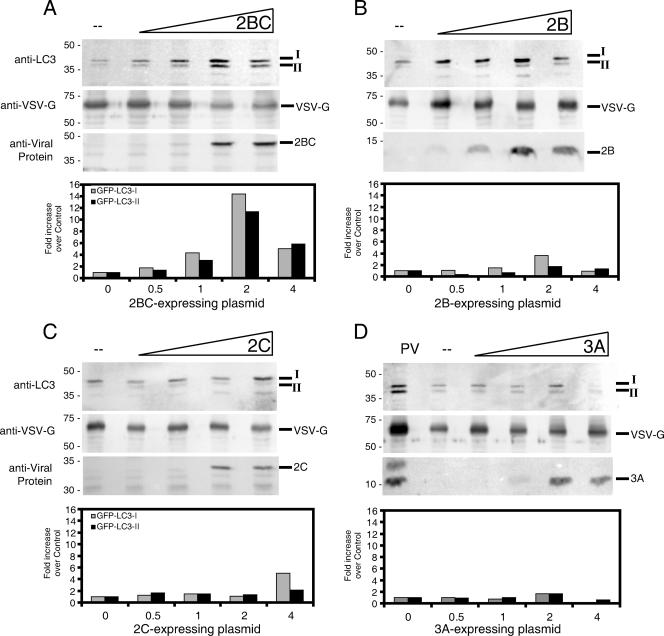
FIG. 5.
Effect of expression of individual viral proteins on the accumulation of saponin-resistant GFP-LC3-I and GFP-LC3-II proteins. COS-7 cells were transfected with identical total amounts of DNA plasmids, in mixtures that contained GFP-LC3-expressing plasmids and increasing amounts of DNA that expressed 2BC (A), 2B (B), 2C (C), or 3A (D). The balance of the DNA samples comprised the VSV-G-expressing vector into which the poliovirus sequences were cloned. Cells were incubated for 48 h, collected by centrifugation, and subjected to extraction with 0.5% saponin. Saponin-resistant proteins were displayed by electrophoresis in 10% polyacrylamide gels, except as indicated, and probed with three different antibodies for each panel: anti-LC3, to detect GFP-LC3-I and GFP-LC3-II; anti-VSV-G, to normalize for transfection efficiency and toxicity; and anti-2B, anti-2C, or anti-3A, to monitor the expression of the viral protein of interest. (A) Effect of increasing amounts of poliovirus 2BC protein expression. (B) Effect of 2B protein expression. The gel used to resolve the 2B protein was 13.5% acrylamide. (C) Effect of 2C protein expression. (D) Effect of 3A protein expression, compared to the amount of GFP-LC3-I and GFP-LC3-II that was saponin resistant following poliovirus infection as for Fig. 1. The lower portion of each panel shows the amount of saponin-resistant GFP-LC3-I and GFP-LC3-II detected divided by the amount of VSV-G observed and normalized to the control transfection in which no poliovirus protein was expressed. Some toxicity was observed for the greatest amounts of DNA transfected.
As can be seen in Fig. 5, the expression of viral protein 2BC caused an increase in the amount of saponin-resistant GFP-LC3-II relative to the amount of VSV-G observed. This approximately 10-fold increase in the amount of GFP-LC3-II was matched by a concomitant increase in saponin-resistant GFP-LC3-I, suggesting that membrane-associated viral protein 2BC increases both the recruitment of unmodified GFP-LC3-I to membranes and its subsequent modification. None of the other expressed proteins, 2B, 2C, or 3A, caused an increase in the amount of saponin-resistant GFP-LC3-II relative to the calnexin control. However, both 2B and 2C caused small increases in the amount of saponin-resistant GFP-LC3-I.
Modification of LC3 by poliovirus protein 2BC can be observed in whole cell extracts as well as following saponin extraction.
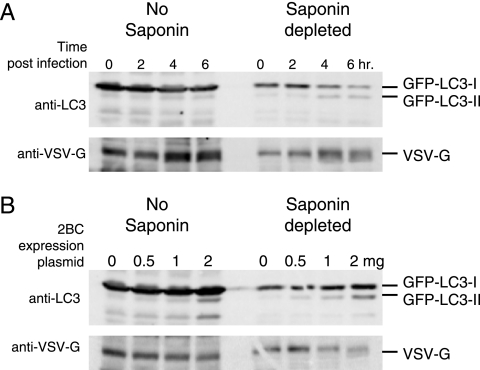
To ensure that the modification of GFP-LC3 by 2BC expression observed in Fig. 5 was not an artifact of saponin extraction, we compared the formation of GFP-LC3-II during poliovirus infection and upon 2BC expression with and without saponin extraction of the cell lysates. As shown in Fig. 6, saponin extraction of the cell lysates was necessary to visualize the increase in GFP-LC3-II formation during poliovirus infection of COS-7 cells. However, 2BC expression in isolation was sufficiently effective at inducing GFP-LC3-II formation that an increase in GFP-LC3-II could be observed with or without saponin extraction to reduce background. Therefore, we conclude that 2BC expression alone is sufficient to induce one of the first steps of cellular autophagy and, perhaps, of membrane vesicle formation during poliovirus infection, the modification of the cellular protein LC3.
FIG. 6.
Comparison of saponin-depleted and nondepleted cell extracts during poliovirus infection and 2BC expression. (A) COS-7 cells were transfected with GFP-LC3 expression plasmid and then 48 h later infected with poliovirus (50 PFU/cell). Cells were then harvested at 0, 2, 4, and 6 h postinfection. (B) COS-7 cells were transfected with a plasmid that expressed GFP-LC3 and increasing proportions of 2BC-expressing plasmids relative to a control vector, followed by harvesting. Recovered cell samples were split in half and either subjected to extraction first with 0.5% saponin and then with NP-40-containing lysis buffer or directly treated with NP-40-containing lysis buffer. Cell extracts were separated by SDS-PAGE on a 10% polyacrylamide gel, transferred to PVDF, and blotted with anti-LC3 and then anti-VSV-G antibodies.
2BC expression recruits GFP-LC3-II to cellular membranes.
To determine whether 2BC expression was sufficient to relocalize GFP-LC3 to cellular membranes, as was observed during poliovirus infection (Fig. 3), we performed immunoprecipitation experiments using anti-2B antibodies, the kind gift of K. Bienz (University of Basel). As can be seen in Fig. 7A, the presence of the anti-2B antibody in immunoprecipitates from cells transfected with 2BC-expressing plasmid led to the recovery of expressed 2BC. In the absence of detergent, GFP-LC3-II was also selectively precipitated by the anti-2B antibody, arguing that this modified protein, and not the unmodified GFP-LC3-I species, was present on the same membranes as 2BC. This result was also observed when the anti-2B antibody was used in all samples and the control cells were transfected with a vector that did not express 2BC but still expressed VSV-G (Fig. 7B). In the absence of the detergent NP-40, the immunoprecipitate from 2BC-expressing cells contained GFP-LC3-II, the modified species, but not unmodified GFP-LC3-I.
To search for a direct interaction between 2BC and GFP-LC3-I, GFP-LC3-II, or both that might provide a clue as to the mechanism by which this viral protein accomplishes this modification, identical immunoprecipitations were performed in the presence of NP-40. While these conditions did not impair the recovery of the expressed 2BC, no GFP-LC3 species were recovered under these conditions (Fig. 7) or any others tested (data not shown). Although these negative results do not preclude the existence of an unstable interaction between 2BC and LC3, it is likely that 2BC protein, and poliovirus infection, accomplishes LC3 modification by less direct means.
DISCUSSION
During poliovirus infection, GFP-LC3 localizes to membranous compartments within the infected cell. Here, we have shown that, as in the process of cellular autophagy, this relocalization correlates with a covalent modification of both endogenous LC3 and GFP-LC3 expressed by transfection. Modification of GFP-LC3 during poliovirus infection was found to require a Gly residue at position 120, the same residue that is essential for autophagy-mediated conjugation of phosphatidylethanolamine to LC3. Further, this modification correlated with the poliovirus-induced relocalization of GFP-LC3 to a LAMP-1-containing structure, arguing that in poliovirus infection, as in autophagy, LC3 modification is essential for membrane localization. This modification of GFP-LC3 observed during poliovirus infection rendered the protein more resistant to extraction from cellular constituents with the mild detergent saponin, as has been observed previously during autophagy (27). This observation both facilitated our experiments and supported the hypothesis that LC3 modification during autophagy and that during poliovirus infection occur by a similar biochemical mechanism.
We suggested previously that the similarities between poliovirus-induced membranes and cellular autophagosomes suggest that poliovirus infection subverts the cellular autophagy pathway to create the membranous vesicles on which RNA replication complexes are assembled (20, 42, 47). This hypothesis was supported by the finding that RNA interference-induced reduction in the expression of either cellular protein LC3 or Atg12, also required for cellular autophagy, reduced viral yield. As reported in the current work, the specific modification of autophagy protein LC3 by an individual poliovirus protein, 2BC, both strengthens this hypothesis and provides insight into the mechanism by which poliovirus might subvert this cellular process. Specifically, viral protein 2BC is sufficient to retain GFP-LC3-II to membranes on which the viral protein is resident, although no evidence for direct interaction between 2BC and LC3-II was obtained from coimmunoprecipitation experiments (Fig. 7). However, the increased resistance of both GFP-LC3-I and GFP-LC3-II to saponin extraction (Fig. 5) suggests direct recruitment of GFP-LC3-I to 2BC-containing membranes, possibly via an intermediate protein or other substituent.
The double-membraned vesicles formed during poliovirus infection resemble those induced during autophagy in their morphology, their sedimentation behavior, and their colocalization of GFP-LC3 and LAMP-1 (20, 47). The poliovirus-induced membranes can be specifically induced by the coexpression of two viral proteins, 2BC and 3A. 2BC and 3A have been shown to interact in two-hybrid assays (8), making their joint participation in a cell biological pathway plausible. Here, however, we show that poliovirus protein 2BC is sufficient to induce LC3 modification (Fig. 5). Coexpression of 3A did not increase the amount of the modification (data not shown). This is, to our knowledge, the first time the mechanistic steps of LC3 modification and double-membrane formation have been decoupled. Therefore, the poliovirus genome has provided not only molecular inducers of double-membraned vesicle formation but also potentially the ability to separate it into two distinct steps: LC3 modification and the wrapping of cytosol required to yield double-membraned vesicles. None of the other known molecular inducers of LC3 modification has been shown to separate these events (Table 1).
TABLE 1.
Molecular inducers of LC3 modification and double-membraned vesicle formation
| Inducer | Site of induction | LC3 modification | Formation of double-membraned vesicles | Potential mechanism | Reference |
|---|---|---|---|---|---|
| Poliovirus 2BC and 3A proteins (coexpressed) | Endoplasmic reticulum | Yes | Yes | Unknown | 20, 47; this report |
| Poliovirus 2BC protein | Endoplasmic reticulum | Yes | No | Unknown | This report |
| Equine arterivirus ns2/ns3 proteins (coexpressed) | Endoplasmic reticulum | NTa | Yes | Unknown | 44 |
| Vibrio cholerae cytolysin-containing supernatants | Plasma membrane | Yes | Yes | V. cholerae toxin known to form pores in plasma membrane | 18 |
| Rapamycin | Cytoplasm | Yes | Yes | Inhibits TOR kinase, affecting the activity of S6 kinase and its targets | 35 |
| Tamoxifen | Plasma membrane | Yes | Yes | Stimulates PI-3 signaling | 4, 41 |
| Concanavalin A in hepatoma cells | Plasma membrane, mitochondria | Yes | Yes | Alters mitochondrial membrane permeability | 5 |
NT, not tested, or no data in the published literature.
If the structures induced by 2BC expression represent temporal or mechanistic intermediates in double-membraned vesicle formation, it is of interest to examine the properties of those structures more closely. It has been shown that 2BC expression leads to the accumulation of large single-membraned vesicles (7, 47) with buoyant densities similar to those of the vesicles induced during poliovirus infection (47). Therefore, 2BC's ability to induce the modification of LC3 leads to two possibilities about the nature of these single-membraned structures. First, they could be the remnants of autophagosomes whose inner contents were fully degraded. Then, 3A would be postulated to retard the maturation of double-membraned structures to single-membraned vesicles. The second possibility is that, when 2BC is expressed in isolation, LC3 is covalently modified and localized to intracellular membranes, but 3A is required to facilitate the completion of a double-membraned vesicle. Further investigation of the ultrastructure and biochemistry of the double-membraned vesicles formed during autophagy and by the expression of individual viral proteins may reveal additional steps to the lipid sequestration, cytosolic wrapping, and maturation of these unique intracellular compartments.
A second hypothesis for the presence of autophagosome-like membranes in poliovirus-infected cells is that these membranes are induced as a component of the innate immune response. Autophagy is increasingly appreciated as a functional pathway in the innate immune response to several viruses, intracellular bacteria, and parasites (reviewed in references 27, 30, and 48). For example, when the autophagy pathway is blocked by genomic knockouts of ATG5, a threefold increase in the growth of group A streptococcus was observed (34). If the induction of double-membraned vesicles during poliovirus infection is part of the cellular innate immune response, it is clearly not effective. Instead, it is possible that one of the reasons that poliovirus induces the membranes on which it replicates its RNA genome from the autophagy pathway is to thwart this newly appreciated component of the innate immune response. Another possibility, that the unique topology of double-membraned vesicles makes possible the nonlytic escape of this nonenveloped cytoplasmic virus from infected cells (20, 26), could provide a benefit to virus spread within infected organisms.
Acknowledgments
We thank William T. Jackson for experimental advice and reagents and Michel Brahic, William T. Jackson, and Peter Sarnow for critical reading of the manuscript.
We are grateful to the National Institutes of Health (training grant GM007276) for support of M.P.T. and to the Ellison Foundation and the National Institutes of Health (grant AI048756) for support of this research.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 2.Bilir, A., M. A. Altinoz, M. Erkan, V. Ozmen, and A. Aydiner. 2001. Autophagy and nuclear changes in FM3A breast tumor cells after epirubicin, medroxyprogesterone and tamoxifen treatment in vitro. Pathobiology 69:120-126. [DOI] [PubMed] [Google Scholar]
- 3.Bursch, W., A. Ellinger, H. Kienzl, L. Torok, S. Pandey, M. Sikorska, R. Walker, and R. S. Hermann. 1996. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis 17:1595-1607. [DOI] [PubMed] [Google Scholar]
- 4.Bursch, W., K. Hochegger, L. Torok, B. Marian, A. Ellinger, and R. S. Hermann. 2000. Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J. Cell Sci. 113:1189-1198. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C. P., M. C. Yang, H. S. Liu, Y. S. Lin, and H. Y. Lei. 2007. Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology 45:286-296. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., A. Noueiry, and P. Ahlquist. 2003. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 77:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 8.Cuconati, A., W. Xiang, F. Lahser, T. Pfister, and E. Wimmer. 1998. A protein linkage map of the P2 nonstructural proteins of poliovirus. J. Virol. 72:1297-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dales, S., H. J. Eggers, I. Tamm, and G. E. Palade. 1965. Electron microscopic study of the formation of poliovirus. Virology 26:379-389. [DOI] [PubMed] [Google Scholar]
- 10.Deretic, V. 2006. Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 18:375-382. [DOI] [PubMed] [Google Scholar]
- 11.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens, J., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn, W. A., Jr. 1990. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 110:1923-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinea, R., and L. Carrasco. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez, M. G., H. A. Saka, I. Chinen, F. C. Zoppino, T. Yoshimori, J. L. Bocco, and M. I. Colombo. 2007. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc. Natl. Acad. Sci. USA 104:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi, N. Ishihara, N. Mizushima, I. Tanida, E. Kominami, M. Ohsumi, T. Noda, and Y. Ohsumi. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408:488-492. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jager, S., C. Bucci, I. Tanida, T. Ueno, E. Kominami, P. Saftig, and E. L. Eskelinen. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837-4848. [DOI] [PubMed] [Google Scholar]
- 22.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabeya, Y., N. Mizushima, A. Yamamoto, S. Oshitani-Okamoto, Y. Ohsumi, and T. Yoshimori. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805-2812. [DOI] [PubMed] [Google Scholar]
- 24.Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi, T. Yoshimori, T. Noda, and Y. Ohsumi. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirkegaard, K., and W. T. Jackson. 2005. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy 1:182-184. [DOI] [PubMed] [Google Scholar]
- 27.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochl, R., X. W. Hu, E. Y. Chan, and S. A. Tooze. 2006. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 7:129-145. [DOI] [PubMed] [Google Scholar]
- 30.Levine, B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159-162. [DOI] [PubMed] [Google Scholar]
- 31.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 32.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosser, A. G., L. A. Caliguiri, A. S. Scheid, and I. Tamm. 1972. Chemical and enzymatic characteristics of cytoplasmic membranes of poliovirus-infected HeLa cells. Virology 47:30-38. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, S. Hamada, and T. Yoshimori. 2004. Autophagy defends cells against invading group A streptococcus. Science 306:1037-1040. [DOI] [PubMed] [Google Scholar]
- 35.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 36.Noda, T., K. Suzuki, and Y. Ohsumi. 2002. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12:231-235. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214:916-919. [DOI] [PubMed] [Google Scholar]
- 39.Reggiori, F., and D. J. Klionsky. 2005. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17:415-422. [DOI] [PubMed] [Google Scholar]
- 40.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scarlatti, F., C. Bauvy, A. Ventruti, G. Sala, F. Cluzeaud, A. Vandewalle, R. Ghidoni, and P. Codogno. 2004. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 279:18384-18391. [DOI] [PubMed] [Google Scholar]
- 42.Schlegel, A., T. Giddings, Jr., M. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 45.Sou, Y.-s., I. Tanida, M. Komatsu, T. Ueno, and E. Kominami. 2006. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 281:3017-3024. [DOI] [PubMed] [Google Scholar]
- 46.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361:304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, M. S. 2006. Autophagy: eating for good health. J. Immunol. 177:4945-4951. [DOI] [PubMed] [Google Scholar]
- 49.Tanida, I., N. Minematsu-Ikeguchi, T. Ueno, and E. Kominami. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84-91. [DOI] [PubMed] [Google Scholar]
- 50.Tanida, I., E. Tanida-Miyake, M. Komatsu, T. Ueno, and E. Kominami. 2002. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 277:13739-13744. [DOI] [PubMed] [Google Scholar]
- 51.Tanida, I., T. Ueno, and E. Kominami. 2004. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 279:47704-47710. [DOI] [PubMed] [Google Scholar]
- 52.Wang, C. W., and D. J. Klionsky. 2003. The molecular mechanism of autophagy. Mol. Med. 9:65-76. [PMC free article] [PubMed] [Google Scholar]
- 53.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]