Abstract
The ORF49 gene product (ORF49p) of the varicella-zoster virus (VZV) is likely a myristylated tegument protein, and its homologs are conserved across the herpesvirus subfamilies. The UL11 gene of herpes simplex virus type 1 and of pseudorabies virus and the UL99 gene of human cytomegalovirus are the homologs of ORF49 and have been well characterized by using mutant viruses; however, little research on the VZV ORF49 gene has been reported. Here we report on VZV ORF49p expression, subcellular localization, and effect on viral spread in vitro. ORF49p was expressed during the late phase of infection and located in the juxtanuclear region of the cytoplasm, where it colocalized mainly with the trans-Golgi network-associated protein. ORF49p was incorporated into virions and showed a molecular mass of 13 kDa in VZV-infected cells and virions. To elucidate the role of the ORF49 gene, we constructed a mutant virus that lacked a functional ORF49. No differences in plaque size or cell-cell spread were observed in human embryonic fibroblast cells, MRC-5 cells, infected with the wild-type or the mutant virus. However, the mutant virus showed diminished cell-cell infection in a human malignant melanoma cell line, MeWo cells. Therefore, VZV ORF49p is important for virus growth in MeWo cells, but not in MRC-5 cells. VZV may use different mechanisms for virus growth in MeWo and MRC-5 cells. If so, understanding the role of ORF49p should help elucidate how VZV accomplishes cell-cell infections in different cell types.
Varicella-zoster virus (VZV) is a member of the human alphaherpesvirus family and the causative agent of varicella (chickenpox) and zoster (shingles) (46, 47). Varicella is a manifestation of lytic infection that presents as fever and a generalized vesicular rash, usually in childhood; herpes zoster is a localized, painful, vesicular rash caused by VZV reactivation from latency, a characteristic of Herpesviridae infections.
One conundrum in VZV research is that VZV is highly infectious as airborne virions during outbreaks of chickenpox, but once it is grown in vitro, it becomes highly cell associated and is propagated only by cell-cell spread. In vitro, VZV particles are degraded in the late endosome (11, 12). The degradation depends on the mannose 6-phosphate receptor (5) and occurs after nucleocapsids are released from nuclei and become coated with the tegument and envelope at the trans-Golgi network (TGN) (45). Recently, insulin-degrading enzyme was identified as a cellular receptor for infection, which VZV binds via its glycoprotein E (gE). This binding occurs under both cell-free and cell-associated conditions (22), but the mechanism of the cell-cell spread of VZV infection has not been elucidated.
The tegument is a structural component of the herpesvirus virion that lies between the nucleocapsid and the envelope, derived from the cellular cytoplasmic membrane containing the viral glycoproteins. Some tegument proteins have recently been well defined in studies on herpes simplex virus type 1 (HSV-1), pseudorabies virus (PrV), and human cytomegalovirus (HCMV). The homologs of the HSV-1 UL11 gene product are among these tegument proteins, and they are conserved throughout the herpesvirus family (1, 4, 8, 9, 34, 43). HSV-1 UL11 accumulates on nuclear and Golgi-derived membranes (2), and its accumulation on the Golgi-derived membranes occurs in the absence of other viral proteins (23). Interactions between UL11 and UL16 (24, 44) and between UL11 and VP22, gE, and gD have been reported (10).
An HSV-1 UL11 deletion mutant forms only small plaques, and its final titers are reduced 5- to 20-fold compared with the wild type, but it is not essential for viral replication (27). On the other hand, deletion of the PrV UL11 gene has a drastic effect on replication, showing a 30- to 250-fold-reduced final titer (21). The secondary envelopment of the mutant virus is also impaired, as evidenced by the accumulation of naked capsids in the cytoplasm (21). Furthermore, a PrV UL11/gM deletion mutant produces no infectious enveloped virions (20). The homolog of UL11 in HCMV is the UL99-encoded pp28, which accumulates in the cytoplasm of infected cells within the juxtanuclear packaging region, along with other tegument proteins and envelope glycoproteins (37). An HCMV pp28-deficient mutant shows extremely impaired growth in normal fibroblasts and produces no detectable infectious progeny because it does not assemble enveloped virus particles (40). Nonetheless, this mutant does spread from cell to cell via an unknown mechanism that allows tegument-coated capsids to bud through cellular membranes, including the plasma membrane (39). In addition, the phosphorylation of pp28 by UL26 is important for the stability of viral particles and normal viral growth (26, 29). HSV-1 UL11 and HCMV pp28 are N-myristylated proteins and bear a characteristic cluster of acidic amino acids (3, 27, 37). These modifications are necessary for the virus to bind the host cell membrane and to target accurately the appropriate intracellular trafficking pathway (16, 23-25, 38).
The transcript of the open reading frame 49 (ORF49) gene is the VZV homolog of the HSV-1 UL11 gene. It is one of the most abundant viral messages expressed during lytic infection (7), and its gene product (ORF49p) is likely myristylated in infected cells (14). However, its other characteristics and functions have not been investigated. In the present study, to elucidate the functions of VZV ORF49p in infected cells, we raised a monospecific polyclonal antibody to this protein and constructed a mutant virus lacking a functional ORF49 gene. We found that ORF49p was a virion component that was localized mainly to the TGN in infected cells. The growth of the ORF49 mutant virus was reduced in human malignant melanoma cells, MeWo cells, but not in those from human embryonic fibroblasts, MRC-5 cells.
MATERIALS AND METHODS
Cells and viruses.
The human embryonic fibroblast cells (MRC-5; ATCC, Manassas, VA) were maintained as monolayer cultures in modified Eagle's minimal essential medium (Nissui Pharmaceutical, Ueno, Tokyo) supplemented with 10% fetal bovine serum (Gibco-BRL, Grand Island, NY) and used in the 27th to 32nd generations. Human malignant melanoma cells, MeWo cells, were grown in Dulbecco's modified Eagle medium (Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL). The VZV parental Oka (pOka) strain was isolated from a patient with varicella and propagated only in human embryonic lung cells (13, 41).
Recombinant virus (rpOka-BAC) was obtained by the reconstitution of infectious viruses using a bacterial artificial chromosome (BAC) clone termed pOka-BAC, which contained the full-length genomic sequence of pOka and BAC sequence in the intergenic region between ORF11 and ORF12 (30). Cell-free viruses were prepared as follows. The infected cells were treated with 0.1% EDTA in phosphate-buffered saline (PBS; pH 7.4) and sonicated, and the lysates were spun at 500 × g for 5 min at 4°C. The supernatant was transferred to new tubes for use as cell-free viruses.
The purification of virus particles from the infected MRC-5 or MeWo cells was as follows. The cell-free virus solutions were loaded onto a Histogenz gradient (Sigma-Aldrich, St. Louis, MO) and subjected to ultracentrifugation at 27,000 rpm for 1 h at 4°C (P40ST-1689 rotor; Hitachi High-Technologies). The virion fraction obtained from the gradient was diluted in PBS, pelleted by centrifugation, and suspended in TE buffer (10 mM Tris-HCl-1 mM EDTA; pH 7.4). The virion fraction was analyzed by PCR amplification with viral gene-specific primer pairs (VZVgB2239ecoF and VZVgB2604salR, described below in detail) and subjected to Western blotting (WB) and electron microscopic (EM) analyses.
Plasmids.
pGEX-ORF49 was constructed to express an ORF49 gene fragment corresponding to amino acids (aa) 2 to 81 in Escherichia coli, as described elsewhere (35). Briefly, the primer pairs ORF49bamF (5′-ACCGGATCCGGACAATCTTCATCCAGCGGTCGA-3′) and ORF49salR (5′-ACCGTCGACACATTTTGCGCATTTGGAATG-3′) were used to amplify the ORF49 gene from the pOka genome. The PCR products were inserted, in frame, into the pGEX4T-1 bacterial expression vector (GE Healthcare Bio-science, Piscataway, NJ) via the BamHI and SalI sites (underlined). The same procedure was used to produce pMAL-gB-C, in which the cytoplasmic domain of glycoprotein B (a DNA fragment spanning positions 2239 to 2604 of gB) was cloned, in frame, into the pMALC-2 bacterial expression vector (New England Biolabs, Beverly, MA) via the EcoRI and SalI sites. The primer pairs were VZVgB2239ecoF (5′-ACCGAATTCGTGCTTAAACTTAAAACAAGCC-3′) and VZVgB2604salR (5′-ACCGTCGACCACCCCCGTTACATTCTCGG-3′). pMAL-ORF16 was constructed by using the same procedure with pMAL-gB-C. ORF16 DNA fragment corresponding to nucleotides 4 to 1125 was amplified by using the primer pairs of ORF16ecoF4 (5′-ACCGAATTCGATTTGAGGTCGCGTACAGAC-3′) and ORF16xhoR1125 (5′-ACCCTCGAGGATTCCCTCCTGCCATGTGGTTTCG-3′) and cloned into pMALC2 via the EcoRI and SalI sites. The full-length ORF49 gene and the gene fragment encoding the N terminus from aa 1 to 53 were cloned into the eukaryotic expression vector pCAGGS (32) to obtain plasmids pCAGGS/ORF49F and pCAGGS/ORF49N, respectively. ORF49ecoF (5′-ACCGAATTCTTACATCAGCATTGCG-3′) was used as the forward primer for both reactions, ORF49xhoR246 (5′-ACCCTCGAGCAGATCCTCTTCTGAGATGAGTTTTTGTTCACATTTTGCGCATTTGG AATGGG-3′) was the reverse primer for pCAGGS/ORF49F, and ORF49xhoR160 (5′-ACCCTCGAGTGGATTTATCGGCGTCC-3′) was the reverse primer for pCAGGS/ORF49N. MeWo and MRC-5 cells were transfected with each of these expression vectors using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). The pCAGGS plasmid was kindly provided by Jun-ichi Miyazaki, Osaka University, Japan.
RNA extraction and cDNA preparation for RT-PCR.
Total RNA was extracted from virus-infected MRC-5 cells by using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. cDNA was synthesized with the SuperScript II First-Strand cDNA synthesis kit (Invitrogen) using an oligo(dT) primer, in accordance with the manufacturer's instructions. PCR amplification was performed with the primer pairs for each gene described below, using the cDNA as the template. The PCR conditions were an initial denaturation at 94°C for 4 min and 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, followed by 72°C for 5 min. The primer pairs were as follows: for ORF48, ORF48F (5′-ATGGCACGATCGGGATTGGATAGG-3′) and ORF48R (5′-TCAAAGCAACGGTTTCTCCG-3′); for ORF49, ORF49ecoF and ORF49bamR (5′-ACCGGATCCACATTTTGCGCATTTGGAATGGG-3′); for cellular elongation factor (EF), EF-F (5′-GCTCCAGCATGTTGTCACCATTC-3′) and EF-R (5′-GGTGAATTTGAAGCTGGTATCTC-3′). PCR of the extracted RNA was performed without the reverse transcriptase (RT) reaction to rule out DNA contamination.
Antibodies.
To make polyclonal antibodies against ORF49p, a recombinant fusion protein, glutathione-S-transferase (GST)-ORF49, was expressed in E. coli BL21trx, which was transformed with pGEX-ORF49, purified, and used to immunize rabbits (MBL, Nagoya, Japan). The anti-ORF49 polyclonal antibodies (Abs) were purified with GST-conjugated N-hydroxysuccinimide (NHS)-activated Sepharose (GE Healthcare Bio-science) and GST-ORF49-conjugated NHS-activated Sepharose. For the purification procedure, 2 mg GST or GST-ORF49 protein was coupled with 1 ml NHS-activated Sepharose via an amide linkage. The rabbit serum was reacted with the GST-conjugated NHS-activated Sepharose for 2 h at 4°C and centrifuged briefly, and the supernatant was collected. The collected supernatant was allowed to react with GST-ORF49-conjugated NHS-activated Sepharose overnight at 4°C. The flowthrough was discarded, and the specific conjugates were fractionated through 0.2 M glycine-HCl (pH 2.5). Anti-gB polyclonal Abs were raised in rabbits (Sigma Genosys, Hokkaido, Japan) immunized with a maltose-binding protein (MBP)-gB fusion protein, which was expressed in E. coli DH5α transformed with pMAL-gB-C and purified with amylose resin (New England Biolabs). The anti-gB Abs were purified with NHS-activated Sepharose as described above, using purified MBP or MBP-gB fusion protein, and named anti-gB-C. Anti-gM polyclonal Abs were produced in rabbits immunized with a synthesized oligopeptide (CEDELLYERSNSGWE; Sigma Genosys). Anti-ORF16 polyclonal Abs were raised in rabbits immunized (MBL) with an MBP-ORF16 fusion protein, which was expressed in E. coli BL21 transformed with pMAL-ORF16. The following mouse monoclonal Abs (MAbs) for cellular proteins were purchased: anti-LAMP-1 (H4A3; Santa Cruz Biotechnology, Santa Cruz, CA), p230 trans-Golgi (clone 15; BD Biosciences Pharmingen), EEA1 (clone 14; BD Biosciences Pharmingen), and anti-α-tubulin (B-5-1-2; Sigma-Aldrich). Fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G and tetramethyl rhodamine isothiocyanate-conjugated swine anti-rabbit immunoglobulin G (Dako, High Wycombe, United Kingdom) were used as secondary antibodies, and Hoechst 33258 (Sigma-Aldrich) was used for nuclear staining.
VZV-BAC plasmid isolation and construction for BAC mutagenesis.
VZV-BAC DNA was extracted using the Nucleobond BAC 100 kit (Machery-Nagel, Düren, Germany), according to the manufacturer's instructions. A linear fragment for homologous recombination was amplified from pCR2.1-TOPO (Invitrogen) by PCR with primer pairs that also encoded a kanamycin resistance gene (KMr), the flp recognition target (FRT) sequences, and the sequences flanking the pOka ORF49. The primers were as follows: forward primer ORF49FRTKMF160 (5′-tgaagactttgactttgatgagaatgtaacagaggacgccgataaatccaGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCagcaagcgaaccggaattgc-3′) and reverse primer ORF49FRTKMR231 (5′-ttttattagaacgtttatcagggtttatcagggtttaacattttgcgcatGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCctttttcaattca gaagaactc-3′) (the FRT sequences are shown in capital letters).
BAC mutagenesis.
To construct the ORF49 deletion mutant BAC named pOka-BACΔ49, mutagenesis was performed as described previously (48) with a slight modification. The linear recombination fragment mentioned above was prepared by PCR amplification and used in the electroporation of competent E. coli DH10B harboring pOka-BAC (30) and pGETrec. pGETrec, which encodes recombinases E and T (recE/T) under the control of an arabinose-inducible promoter, was a kind gift from Panayiotis A. Ioannou, Murdoch Childrens Research Institute, Australia (31). Electroporated E. coli cells were incubated with shaking at 37°C for 3 h and plated onto agar plates containing ampicillin (50 μg/ml), chloramphenicol (17 μg/ml), and kanamycin (35 μg/ml) to select colonies harboring KMr inserted into the ORF49 locus. pOka-BACΔ49KMr was isolated as described above, and successful homologous recombination was confirmed by PCR amplification and sequencing analysis, using primers for sequences upstream and downstream of ORF49. To remove KMr, E. coli harboring pOka-BACΔ49KMr was transformed with pCP20 by electroporation, incubated with shaking at 28°C for 3 h, plated onto agar plates containing ampicillin and chloramphenicol, and grown overnight at 28°C. Further incubation at 37°C induced expression of the flp recombinase and removed pCP20. The flp recombinase expression plasmid pCP20 was kindly provided by Wilfried Wackernagel, Universitat Oldenburg, Germany (6). The loss of KMr was confirmed by restreaking the colonies in duplicate on chloramphenicol-containing agar plates and chloramphenicol-kanamycin-containing plates and growing them overnight at 37°C. Chloramphenicol-resistant and kanamycin-sensitive colonies were selected, and the mutant BAC was extracted and analyzed by PCR amplification and sequencing analysis with the primers for the upstream and downstream ORF49 sequences, to confirm the loss of KMr. These recombinations were confirmed by Southern blot analysis with specific probes of the ORF49 gene region and KMr. One clone, named pOka-BACΔ49, was chosen for further analysis.
Reconstitution of recombinant VZV.
Recombinant virus (rpOkaΔ49) was obtained as previously described for rpOka (30). Briefly, transfection using Nucleofector technology (Amaxa, Köln, Germany) was performed with 1 μg purified BAC DNA (pOka-BACΔ49) per 1.0 × 106 MRC-5 cells, which were then cultured on six-well plates at 37°C for 3 to 4 days. After typical cytopathic effects (CPEs) were seen in cells expressing green fluorescent protein, the cells were isolated with glass isolation cups and transferred onto AxCANCre-infected MRC-5 cells to remove the BAC sequences containing the guanine phosphoribosyl transferase gene (gpt) and gfp. A recombinant adenovirus, AxCANCre, which expresses the Cre recombinase, was kindly provided by Yasushi Kawaguchi (Tokyo University, Japan) (17, 42). At 3 to 4 days after the superinfection of the Cre recombinase-expressing MRC-5 cells with rpOka-BACΔ49, typical plaques that did not express green fluorescent protein appeared and were isolated. The recombinant virus without the BAC sequence was named rpOkaΔ49. After several propagations in MRC-5 cells, cell-free virus was prepared as described above, titrated on MRC-5 cells, and used in this study. The successful excision of the BAC sequences was verified by PCR amplification and sequencing analysis of the rpOkaΔ49 genome (extracted by proteinase K treatment) with primers corresponding to the upstream and downstream sequences flanking the BAC region between ORF11 and ORF12.
Analysis of virus growth.
To analyze the growth of the recombinant virus, an infectious center assay was performed as described previously (13). The plaques were counted, and the numbers of infectious centers were evaluated as the relative increase rate over the value on day zero. The plaque areas in the same samples used in the infectious center assay were measured by ImageJ software (http://rsb.info.nih.gov/ij/) at the indicated time points, and the values from cells infected with rpOka and rpOkaΔ49 were compared.
Immunohistochemical analysis.
For indirect immunofluorescence assays (IFAs), cells were grown on glass coverslips and infected at a multiplicity of infection (MOI) of 0.1. At 48 h postinfection (hpi), the cells were washed twice with PBS, fixed, and permeabilized with ice-cold acetone and methanol for 20 min. The cells were incubated with primary antibodies for 20 min at 37°C, washed three times with PBS containing 0.02% Tween 20 (PBS-T), incubated with secondary antibodies for 20 min at 37°C, and washed three times with PBS-T. All samples were analyzed on a model DMIRE2 Leica confocal microscope (Leica Microsystems).
Western blotting.
WB was performed as described elsewhere (35).
Electron microscopy.
Cells were scraped and collected by centrifugation at 500 × g for 5 min. The cells were fixed in 0.05 M cacodylate-buffered 2.5% glutaraldehyde (GA) solution for 1 h at room temperature, washed three times with 5% sucrose buffered with 0.1 M cacodylate, postfixed in 0.1 M cacodylate-buffered 1% osmium tetroxide for 1 h at room temperature, and washed three times with distilled water. The samples were block stained with a 0.5% aqueous solution of uranyl acetate for 1 h and dehydrated with a graded series of ethanol and embedded in EPONmix. Ultrathin sections were cut with an ultramicrotome (Reichert-Nissei Ultracuts; Leica Microsystems, Wetzlar, Germany) and stained with uranyl acetate and lead citrate. Viral particles, purified by ultracentrifugation on a Histogenz gradient as described above, were negatively stained using 3% uranyl acetate. The samples were observed with a Hitachi H7100 electron microscope (Hitachi High-Technologies).
RESULTS
The ORF49 gene of VZV encodes an 81-residue, likely myristylated protein, ORF49p. To characterize ORF49p, we produced anti-rabbit monospecific Abs against it. The specificity of the anti-ORF49 antibody was confirmed by IFA and WB of VZV-infected cells, mock-infected cells, and ORF 49p-expressing cells (see Fig. 1C, 2, and 3, below).
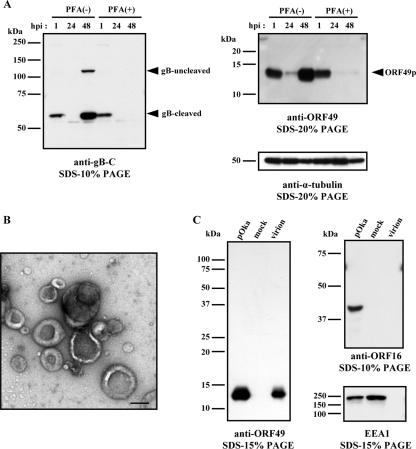
FIG. 1.
Kinetics of protein synthesis and identification of ORF49p in viral particles. (A) MRC-5 cells were infected with pOka cell-free virus at an MOI of 0.1, cultured without (−) or with (+) PFA, and harvested at the indicated time points. The cell lysates were analyzed by WB using Abs to gB and ORF49. An anti-α-tubulin MAb was used as the control. The arrowheads indicate the positions of each protein. (B and C) MRC-5 cells were infected with pOka cell-free virus and cultured until CPE was observed. Histogenz gradient-purified virions were negatively stained with 3% uranyl acetate for electron microscopy (bar, 200 nm) (B) and subjected to WB analysis using Abs to ORF49, ORF16, or EEA1 (C). Molecular mass markers are shown on the left of each panel (A and C).
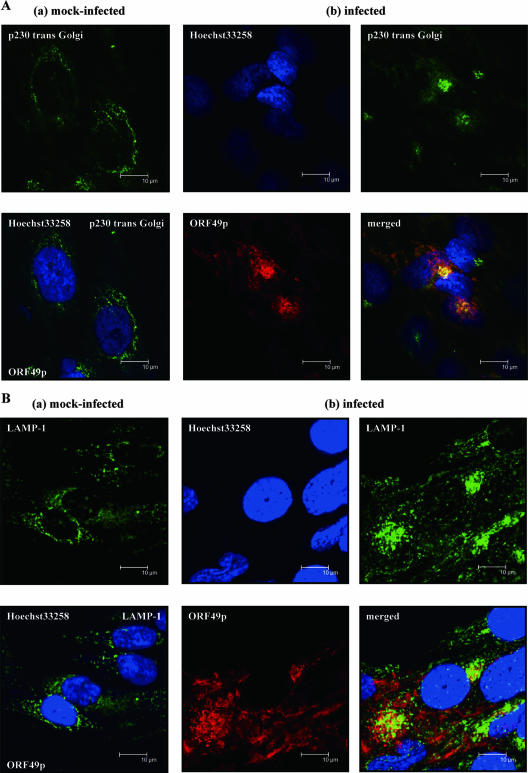
FIG. 2.
Intracellular colocalization of ORF49p in infected cells. MRC-5 cells that were mock infected (a) or pOka infected (b) at an MOI of 0.1 were fixed at 48 hpi and subjected to confocal microscopic analyses. Cells were double labeled for the TGN marker, p230 trans-Golgi (A) or the late endosome marker LAMP-1 (B) and ORF49p; nuclei were stained with Hoechst 33258. Bars (all panels), 10 μm.
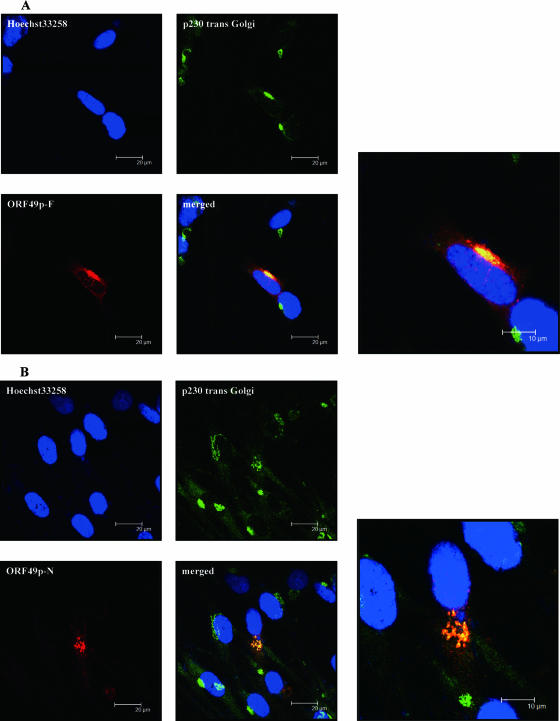
FIG. 3.
Intracellular localization of ORF49p in transiently transfected cells. Confocal images are of MRC-5 cells transfected with expression plasmids for the full-length (A) or N-terminal portion (B) of ORF49p fixed at 48 h posttransfection. ORF49p-F and -N were detected by anti-ORF49, p230 trans-Golgi was used as a TGN marker, and Hoechst 33258 was used to label nuclei. Bars, 10 μm or 20 μm.
ORF49p is expressed during the late phase of infection and is a component of the viral particle.
MRC-5 cells were cultured in medium supplemented with (+) or without (−) 200 μg/ml phosphonoformic acid (PFA), which inhibits viral DNA synthesis, for 1 h. The medium was washed out, pOka cell-free viruses at 0.1 MOI were allowed to adsorb to the cells for 1 h, and the viruses were then washed out. The cells were then cultured with or without PFA until the indicated time points.
We used WB analyses of whole-cell lysates to evaluate the expression of ORF49 and of gB. gB is synthesized as a type I membrane glycoprotein (gB uncleaved) in infected cells. It is then cleaved into 68-kDa and 66-kDa proteins and incorporated into the virion as a heterodimer (18, 19, 28). As shown in Fig. 1A, the only cleaved form of gB was detected at 1 hpi, and both forms of gB were detected in only the PFA (−) cells at 48 hpi, indicating that the cleaved form of gB detected at 1 hpi is derived from viral particles and gB proteins are synthesized at the late phase of infection as reported previously (19, 28, 33). The anti-ORF49 Abs specifically detected a 13-kDa protein in both the PFA (−) and PFA (+) samples at 1 and 24 h hpi, but this band was not present at 48 hpi in the PFA (+) samples (Fig. 1A). The detection of ORF49p and gB in both the PFA (−) and PFA (+) samples at 1 hpi implies that ORF49p may exist in the virion, like gB. Both proteins were scarcely detected at 24 hpi but were detected at 48 hpi abundantly. It appeared that this was due to a low infectious rate. Therefore, ORF49p is expressed at the late phase of infection and may be included in virions.
To confirm that ORF49p is a component of viral particles, VZV viral particles were purified (see Materials and Methods), and the samples were examined by EM using negative staining to confirm the presence of the particles, as shown in Fig. 1B. Because the particles were clearly detected by EM, WB analysis was performed with the same sample. The WB showed ORF49p was present in the virion sample, at a level similar to that in the pOka-infected cell lysates, indicating that ORF49p was abundant in the virions. On the other hand, the ORF16 protein, which is a homolog of the processivity subunit of DNA polymerase and a nonstructural protein, was not detected in the purified virion lysates (Fig. 1C), indicating that the virion preparation was not contaminated with nonstructural viral proteins. In addition, EEA1, which is a 180-kDa hydrophilic peripheral membrane protein that localizes to early endosomes, was not detected in the virion lysates, indicating that the preparation was not contaminated with cytoplasmic membranes (Fig. 1C).
Subcellular localization of ORF49p in infected cells.
To examine the ORF49p localization in VZV-infected cells, an IFA was performed with pOka-infected MRC-5 cells. ORF49p-specific fluorescence was detected in the cytoplasm of the infected cells, most strongly at the juxtanuclear region (Fig. 2). Therefore, the cells were stained for ORF49p and Golgi-associated proteins, to look for colocalization. We found that ORF49p colocalized with p230 trans-Golgi, a peripheral membrane protein associated with the cytoplasmic face of the TGN (Fig. 2A), while it showed similar localization with GM130 (cis-Golgi matrix protein) (data not shown). ORF49p did not colocalize at all with calnexin, an integral membrane protein of the endoplasmic reticulum (data not shown).
Interestingly, although both GM130 (data not shown) and p230 trans-Golgi (Fig. 2A, group a) were diffusely distributed in the cytoplasm of uninfected MRC-5 cells, they accumulated near the nucleus of the pOka-infected MRC-5 cells, where they colocalized with ORF49p (Fig. 2A, group b). This suggests that the viral assembly compartment (36) may form in this juxtanuclear position.
We also analyzed the relative distributions of ORF49p and lysosome-associated membrane protein 1 (LAMP-1; a late endosome/lysosome marker) (Fig. 2B) and CD63/LAMP-3 (a late endosome marker) (data not shown). These proteins were expressed diffusely in the cytoplasm of uninfected MRC-5 cells (Fig. 2B, group a) but they accumulated near the nucleus of pOka-infected MRC-5 cells (Fig. 2B, group b), although their staining patterns were different from those of GM130 or p230 trans-Golgi. Both LAMP-1 (Fig. 2B, group b) and CD63/LAMP-3 (data not shown) showed faintly overlapping expression with ORF49p.
Subcellular localization of ORF49p in transiently transfected cells.
ORF49p's homologs in HSV-1 and HCMV target the Golgi-derived membranes in the absence of other viral proteins (23, 37). Therefore, to examine the localization of ORF49p without other viral gene products, full-length ORF49p (ORF49p-F) or its N-terminal portion (ORF49p-N; corresponding to aa 1 to 53) was expressed in MRC-5 cells (Fig. 3) and MeWo cells (data not shown) by transfecting the cells with pCAGGS/ORF49F or pCAGGS/ORF49N. The expression pattern of the exogenous protein was compared with that of p230 trans-Golgi (Fig. 3A and B). Both ORF49p-F and ORF49p-N colocalized with p230 trans-Golgi at the juxtanuclear region, indicating that ORF49p can target the TGN without the expression of any other viral genes and that the N-terminal portion of ORF49p is sufficient for this localization. The result also indicates that the anti-ORF49 Abs employed in this assay recognize the N-terminal portion of ORF49p.
Construction of an ORF49 gene deletion mutant in E. coli.
To examine the function of the ORF49 gene in virus-infected cells, a deletion mutant of the gene was constructed using the BAC system. For the present study, pOka-BACΔ49KMr, which lacked ORF codons 160 to 231, was generated by BAC mutagenesis in E. coli. The codons were replaced by a linear fragment containing the kanamycin resistance gene (KMr) flanked by FRT sites and 50 nucleotides homologous to ORF49 sequences (for a detailed description, see Materials and Methods) (Fig. 4A). Because ORF48, which encodes the DNase, overlaps with ORF49 nucleotides 1 to 97, only the C-terminal region corresponding to nucleotides 160 to 230 of the ORF49 gene was deleted, to avoid disrupting the ORF48 gene. Successful homologous recombination mediated by recE/T and the removal of the KMr cassette were validated by PCR amplification, sequencing, and Southern blotting of the BAC genomes (data not shown).
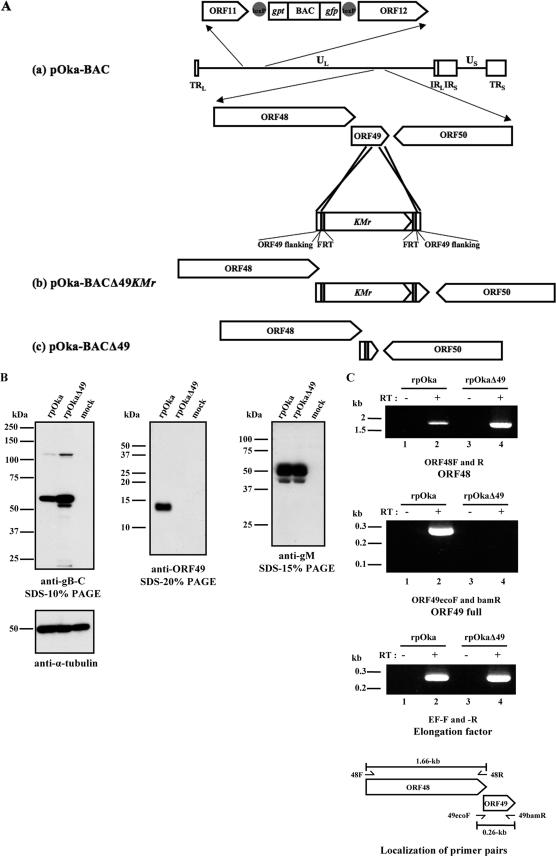
FIG. 4.
Construction of the VZV ORF49 deletion mutant and expression levels of neighboring genes. (A) Schematic map of the VZV genome, showing its two unique regions (UL and US) and inverted repeat sequences, TRL and IRL, which flank the UL region, and TRS and IRS, which flank the US region. (a) A BAC cassette, containing gpt and gfp sequences flanked by loxP sequences (gray circles), was inserted between ORF11 and ORF12. ORFs are drawn as pointed rectangles. A KMr (kanamycin resistance gene) cassette flanked by FRT sites (gray rectangles) and an ORF49 homologous region (from nucleotide positions 86196 to 86245 and 86315 to 86356) was substituted for a substantial portion of the ORF49 coding region from nucleotide positions 86246 to 86314 by the recE/T method. (b) In plasmid pOka-BACΔ49KMr, a 72-nucleotide stretch was replaced by KMr. (c) The KMr cassette was removed by the flp/FRT recombination method, leaving one FRT site behind in the ORF49 region (pOka-BACΔ49). (B) MRC-5 cells were mock infected or infected with rpOka or rpOkaΔ49 at an MOI of 0.01 and harvested at 72 hpi. Equivalent amounts of protein were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes. The blots were reacted with anti-gB-C, anti-ORF49, and anti-gM. An anti-α-tubulin MAb was used as the control, and molecular mass markers are shown at the left of each panel. (C) MRC-5 cells were infected with rpOka or rpOkaΔ49. RNA was extracted when full CPE was observed, and RT-PCRs of ORF48 and ORF49 were performed using specific primer pairs (lanes 2 and 4 of each panel). PCRs without RT were performed as a control for contamination with genomic DNA and are shown in lanes 1 and 3 of each panel. Molecular mass markers are shown at the left of each panel. The RT reaction control used elongation factor-specific primer pairs. The schematic shows the locations of primers used for the PCR amplifications (arrows).
ORF49 protein is not expressed in recombinant virus-infected cells.
Reconstitution of infectious recombinant virus from pOka-BACΔ49 in MRC-5 cells was successful, and ORF49 mutant virus without BAC cassette, named rpOkaΔ49, was obtained. The result indicates that the ORF49 gene is not essential for virus replication in MRC-5 cells. To verify that rpOkaΔ49 did not express ORF49p, cells infected at an MOI of 0.1 were harvested at 72 hpi and lysed. Lysates from MRC-5 cells that were mock infected or infected with rpOka or rpOkaΔ49 were subjected to WB analyses, probing with anti-gB-C, anti-ORF49, anti-gM (ORF50), or anti-α-tubulin Abs (Fig. 4B). The 13-kDa ORF49p could not be detected in the rpOkaΔ49-infected cell lysates, whereas it was abundant in the rpOka-infected cell lysates. The envelope glycoproteins, gB and gM, were detected in the lysates of both rpOka- and rpOkaΔ49-infected cells, proving the expression of the other virus proteins in rpOkaΔ49-infected cells. Regarding gB, a smaller band was seen just below the major band on the blot of rpOkaΔ49-infected cell lysates, but not of rpOka. The band was also seen on the blot of rpOka-infected cell lysates when the film was exposed longer, showing that the smaller band was detected on either blot.
Deletion of the ORF49 gene does not affect the expression of neighboring genes in infected cells.
To examine whether the ORF49 deletion affected the expression of nearby genes, total RNA was extracted from infected cells, and RT-PCR with specific primer pairs was performed in the ORF48 gene, which is adjacent to ORF49 and encodes DNase (Fig. 4C). The ORF48 gene was expressed normally in the rpOkaΔ49-infected cells identical to EF as an internal control, although the ORF49 gene was not expressed in the rpOkaΔ49-infected cells. The expression of the ORF50 (gM) gene was already confirmed by Western blotting (Fig. 4B). Therefore, the deletion of ORF49 did not impair the expression of adjacent genes by the reconstituted virus.
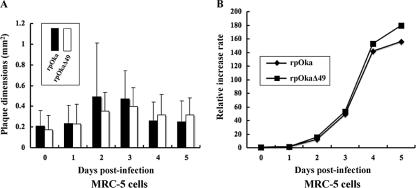
The deletion of ORF49 does not affect plaque formation or virus growth in MRC-5 cells.
Deletion mutant viruses of the ORF49 homologs in HSV-1, HCMV, and PrV all show a smaller plaque size and delayed growth, compared with their wild-type counterparts. In the present study, to examine plaque formation by rpOkaΔ49 and virus growth, MRC-5 cells were infected with rpOka or rpOkaΔ49 cell-free virus at an MOI of 0.005. We found that rpOkaΔ49 formed plaques with similar areas to those formed by the rpOka virus at 7 days pi (data not shown). The mean plaque areas of the two viruses were also similar for cell-cell infection at all the indicated time points (Fig. 5A). An infectious center assay showed that rpOkaΔ49's growth curve was identical to that of rpOka (Fig. 5B) in MRC-5 cells. These results suggest that VZV ORF49p is completely nonessential for the viral life cycle in MRC-5 cells.
FIG. 5.
Growth properties of recombinant viruses in MRC-5 cells. (A) MRC-5 cells were infected with rpOka or rpOkaΔ49 (MOI, 0.005), harvested at the indicated time points, serially diluted, added to newly prepared MRC-5 cells on six-well plates, and cultured for 7 days. Viral plaques were scanned, and the mean areas of single plaques were calculated. (B) Growth kinetics of rpOka and rpOkaΔ49 were compared in an infectious center assay. The infected cells were stained with crystal violet, and the plaques were counted.
EM analysis in MRC-5 cells.
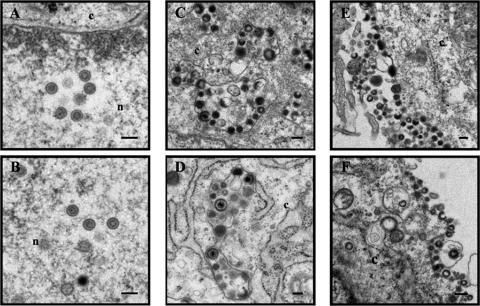
To point out the difference(s) between rpOka and rpOkaΔ49, an ultrastructural analysis was performed. MRC-5 cells were infected with viruses by cell-cell spreading and harvested 48 h later. Numerous nucleocapsids were found in the nucleus of either type of virus-infected cells, and incomplete or complete viral particles were also found in vacuoles of cytoplasm and on cell surfaces of either type of virus-infected cells (Fig. 6). The result suggested that there were no obvious differences between them.
FIG. 6.
Ultrastructural analysis of wild-type and mutant viruses. MRC-5 cells were infected with rpOka (A, C, and E) and rpOkaΔ49 (B, D, and F) by cell-cell spreading and harvested when CPE was observed in almost all cells. The harvested cells were fixed and processed for electron microscopy. Nucleocapsids were observed in the nucleus (n) (A and B), and viral particles were in the cytoplasm (c) (C and D) and on the cell surface (E and F). Bars, 200 nm.
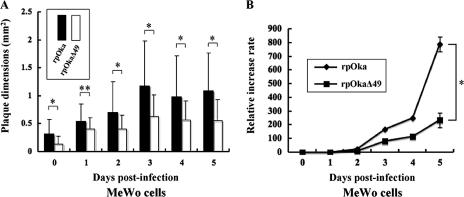
The deletion of ORF49 affects plaque formation and virus growth in MeWo cells.
MeWo cells, in addition to MRC-5 cells, are commonly used to study VZV infection. VZV infection of MeWo cells induces syncytium formation by the cells, a phenotype that is rarely seen in VZV-infected MRC-5 cells. Therefore, we used rpOkaΔ49 reconstituted in MRC-5 cells to infect MeWo cells and observed virus growth in these cells. Interestingly, rpOkaΔ49 growth was slower than that of rpOka in the MeWo cells. Because of this discrepancy in virus growth in the two cell types, we did a direct comparison of plaque size and growth of rpOkaΔ49 and rpOka in MeWo and MRC-5 cells. The plaque areas caused by rpOkaΔ49 were significantly smaller than those caused by rpOka in the MeWo cells (data not shown), but they were essentially the same in the MRC-5 cells (data not shown). Next, we compared cell-cell infection at the indicated time points in both cell lines (Fig. 7A) and again found obviously reduced infection by rpOkaΔ49 in MeWo cells compared with rpOka, while this effect was not seen in the MRC-5 cells (Fig. 5A). In addition, the growth of rpOkaΔ49 was significantly impaired compared with that of rpOka at 5 days pi in the MeWo cells (P < 0.01) (Fig. 7B), but not in the MRC-5 cells (Fig. 5B). These data show that ORF49p is not essential for virus growth in either MRC-5 cells or MeWo cells, but it is required for efficient virus growth in MeWo cells.
FIG. 7.
Growth properties of recombinant viruses in MeWo cells. (A) MeWo cells were infected with rpOka or rpOkaΔ49 (MOI, 0.005), harvested at the indicated time points, serially diluted, added to newly prepared MeWo cells on six-well plates, and cultured for 7 days. Viral plaques were scanned, and the mean areas of single plaques were calculated. *, P < 0.01; **, P < 0.05. (B) Growth kinetics of rpOka and rpOkaΔ49 were compared in an infectious center assay. The infected cells were stained with crystal violet, and the plaques were counted. *, P < 0.01.
DISCUSSION
The VZV ORF49p belongs to a group of highly conserved herpesvirus tegument proteins whose functions have just been elucidated in the alphaherpesviruses HSV-1 (2, 23-25, 27) and PrV (5, 20, 21) and a betaherpesvirus, HCMV, as an important component for secondary viral envelopment (16, 26, 29, 37-40). Here, we showed that VZV ORF49p is synthesized late in the infectious cycle and is a virion component.
We also showed that ORF49p colocalized with TGN marker proteins in both infected and transfected cells. Furthermore, ORF49p colocalized partially with late endosomes, where VZV virions are believed to be sequestered and degraded after packaging events. Because ORF49p is a quite small protein, with only 81 aa, and because it does not contain obvious motifs for targeting the Golgi-related membranes, as glycoproteins do, the mechanism for ORF49p's direct targeting of the Golgi and related membranes is unclear. It is possible that ORF49p myristylated at the N terminus (14) may bind directly to the Golgi-derived membrane via these fatty acids without first binding the endoplasmic reticulum (data not shown); the bound ORF49p might then be transported to the TGN or some more distal compartment for virion maturation and degradation.
VZV infection had an effect on host protein localization. In mock-infected MRC-5 cells, GM130 and p230 trans-Golgi were expressed diffusely in the cytoplasm, but in pOka-infected MRC-5 cells, GM130 and p230 trans-Golgi accumulated in a juxtanuclear region which, according to a study on HCMV, is probably an assembly compartment (36). The same phenomena were also observed in MeWo cells (data not shown). In a transient-expression assay (Fig. 3), ORF49p-N, a truncated protein containing aa 1 to 53 of ORF49p, colocalized with p230 trans-Golgi, indicating that the N-terminal portion of ORF49p is sufficient to direct its targeting to the TGN, even in the absence of other viral factors. This feature is also observed in the HSV-1 homolog of ORF49p, UL11 (25), which targets the membrane normally, even when in a mutant form lacking the C-terminal-third of the protein. HSV-1 UL11 is myristylated and palmitylated at the N terminus and additionally has the acidic cluster and dileucine motif (23, 25), but ORF49 has no obvious acidic cluster or dileucine motif.
To evaluate the roles of ORF49 in the VZV replication cycle in vitro, we generated a mutant VZV that lacked a functional ORF49 gene, rpOkaΔ49, using the BAC mutagenesis method established in our laboratory previously (30). The deleted region of ORF49 was only 23 aa at the C terminus and the first 58 aa of the protein were left in the mutant genome, but the absence of both the full and first 58 aa of ORF49p in rpOkaΔ49-infected MRC-5 cells was confirmed by WB (Fig. 4B). Furthermore, anti-ORF49 was confirmed to recognize the N-terminal portion of ORF49p (aa 1 to 53; ORF49p-N) in transiently expressing cells by both IFA (Fig. 3B) and WB (data not shown), and the region corresponded to the left region in rpOkaΔ49. These facts indicate that the expression of ORF49p (ORF49p-F and -N) was completely defective in rpOkaΔ49-infected cells.
Unexpectedly, unlike deletion mutants for ORF49 homologs in other herpesviruses, HSV-1, PrV, and HCMV, rpOkaΔ49 showed no reductions in plaque size and virus growth compared with the reconstituted virus bearing full-length ORF49, rpOka in MRC-5 cells (Fig. 5). This discrepancy may be due to differences in these viruses’ mechanisms for viral spread. HSV-1, PrV, and HCMV can all infect new cells either as free virus particles secreted into the medium or via cell-cell infection of adjacent cells. In vitro, VZV can spread only through cell-cell infection and not via cell-free virus particles, because VZV-infected cells do not secrete infectious virus particles into the medium. Therefore, ORF49p and its homologs may be dispensable for cell-cell infection, as we observed in MRC-5 cells.
In contrast to our results in MRC-5 cells, we found that MeWo cells were less susceptible to VZV infection (data not shown). When both cell lines were infected with wild-type virus at the same titer, MRC-5 cells produced an approximately 10-fold-higher titer of the progeny viruses than MeWo cells, indicating a potential difference in the ability of these cells to support viral production (data not shown).
In this study, although we found no difference in virus growth between rpOkaΔ49 and rpOka in MRC-5 cells, the plaque size and viral growth were reduced in MeWo cells infected with rpOkaΔ49 compared with rpOka (Fig. 7). The results indicate that VZV ORF49 is important for virus growth in MeWo cells, but not MRC-5 cells, and that VZV may use different mechanisms for virus growth in these two cell lines. Harson et al. reported that the differences in VZV replication in melanoma cells versus MRC-5 cells (15), which showed the entire assembly and envelopment process was more aberrant in MRC-5 cells. To examine this possibility in more detail, we performed EM analyses with MRC-5 and MeWo cells. As reported previously (11, 12, 15), many incomplete virus particles were found in the cytoplasm and plasma membrane of MRC-5 cells infected with either virus type. In this respect, VZV is different from the other alphaherpesviruses. Moreover, there were no noticeable differences at the EM level between rpOkaΔ49 and rpOka in MRC-5 cells. It was even more difficult to find complete viruses in MeWo cells than in the rpOka-infected cells, most likely because the MeWo cells produce a very low titer of infectious progeny. More extensive EM analysis is currently being pursued in our laboratory.
In summary, our results suggest that, like HCMV pp28, VZV ORF49p may play an important role in the production of complete virions (26, 29, 39, 40), and the production of complete virions may be more important for virus spread in MeWo cells than in MRC-5 cells. Further studies are required to elucidate the role of VZV ORF49p in virion production.
Acknowledgments
We thank Panayiotis A. Ioannou (Cell and Gene Therapy Research Group, Murdoch Children Research Institute, Royal Children's Hospital, Australia) for providing pGETrec, Wilfried Wackernagel (Genetics, Department of Biology and Environmental Sciences, Universitat Oldenburg, Germany) for pCP20, Yasushi Kawaguchi (Division of Viral Infection, Department of Infectious Disease Control, International Research Center for Infectious Diseases, The Institute of Medical Science, The University of Tokyo, Japan) for AxCANCre, Jun-ichi Miyazaki (Division of Stem Cell Regulation Research, Osaka University Graduate School of Medicine, Japan) for pCAGGS, and Ulrich H. Koszinowski (Max von Pettenkofer Institut fur Virologie, Ludwig-Maximilians-Universitat Munchen, Germany) for pHA2. We also thank Y. Kawaguchi for helpful suggestions and Eiko Moriishi (Laboratory of Virology and Vaccinology, Division of Biomedical Research, National Institute of Biomedical Innovation, Japan) for technical assistance.
This study was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, and et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. J., Z. Zhu, A. A. Gershon, and M. D. Gershon. 2004. Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell 119:915-926. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 7.Cohrs, R. J., M. P. Hurley, and D. H. Gilden. 2003. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J. Virol. 77:11718-11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.Dijkstra, J. M., W. Fuchs, T. C. Mettenleiter, and B. G. Klupp. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17-35. [DOI] [PubMed] [Google Scholar]
- 10.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabel, C. A., L. Dubey, S. P. Steinberg, D. Sherman, M. D. Gershon, and A. A. Gershon. 1989. Varicella-zoster virus glycoprotein oligosaccharides are phosphorylated during posttranslational maturation. J. Virol. 63:4264-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper, D. R., and H. O. Kangro. 1990. Lipoproteins of varicella-zoster virus. J. Gen. Virol. 71:459-463. [DOI] [PubMed] [Google Scholar]
- 15.Harson, R., and C. Grose. 1995. Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J. Virol. 69:4994-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78:1488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanegae, Y., G. Lee, Y. Sato, M. Tanaka, M. Nakai, T. Sakaki, S. Sugano, and I. Saito. 1995. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 23:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller, P. M., A. J. Davison, R. S. Lowe, C. D. Bennett, and R. W. Ellis. 1986. Identification and structure of the gene encoding gpII, a major glycoprotein of varicella-zoster virus. Virology 152:181-191. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura, K., J. Namazue, H. Campo-Vera, T. Ogino, and K. Yamanishi. 1986. Induction of neutralizing antibody against varicella-zoster virus (VZV) by VZV gp3 and cross-reactivity between VZV gp3 and herpes simplex viruses gB. Virology 149:74-82. [DOI] [PubMed] [Google Scholar]
- 20.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Q., M. A. Ali, and J. I. Cohen. 2006. Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2006. Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 80:10534-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorz, K., H. Hofmann, A. Berndt, N. Tavalai, R. Mueller, U. Schlotzer-Schrehardt, and T. Stamminger. 2006. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. J. Virol. 80:5423-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 28.Montalvo, E. A., and C. Grose. 1987. Assembly and processing of the disulfide-linked varicella-zoster virus glycoprotein gpII(140). J. Virol. 61:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munger, J., D. Yu, and T. Shenk. 2006. UL26-deficient human cytomegalovirus produces virions with hypophosphorylated pp28 tegument protein that is unstable within newly infected cells. J. Virol. 80:3541-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagaike, K., Y. Mori, Y. Gomi, H. Yoshii, M. Takahashi, M. Wagner, U. Koszinowski, and K. Yamanishi. 2004. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 22:4069-4074. [DOI] [PubMed] [Google Scholar]
- 31.Narayanan, K., R. Williamson, Y. Zhang, A. F. Stewart, and P. A. Ioannou. 1999. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 6:442-447. [DOI] [PubMed] [Google Scholar]
- 32.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 33.Okuno, T., K. Yamanishi, K. Shiraki, and M. Takahashi. 1983. Synthesis and processing of glycoproteins of varicella-zoster virus (VZV) as studied with monoclonal antibodies to VZV antigens. Virology 129:357-368. [DOI] [PubMed] [Google Scholar]
- 34.Osterrieder, N. 1999. Sequence and initial characterization of the UL10 (glycoprotein M) and UL11 homologous genes of serotype 1 Marek's disease virus. Arch. Virol. 144:1853-1863. [DOI] [PubMed] [Google Scholar]
- 35.Sadaoka, T., K. Yamanishi, and Y. Mori. 2006. Human herpesvirus 7 U47 gene products are glycoproteins expressed in virions and associate with glycoprotein H. J. Gen. Virol. 87:501-508. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo, J. Y., and W. J. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva, M. C., J. Schroer, and T. Shenk. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. USA 102:2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi, M., T. Otsuka, Y. Okuno, Y. Asano, and T. Yazaki. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288-1290. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 44.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Z. H., M. D. Gershon, O. Lungu, Z. Zhu, S. Mallory, A. M. Arvin, and A. A. Gershon. 2001. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J. Virol. 75:323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weller, T. H. 1983. Varicella and herpes zoster. Changing concepts of the natural history, control, and importance of a not-so-benign virus. N. Engl. J. Med. 309:1362-1368. [DOI] [PubMed] [Google Scholar]
- 47.Weller, T. H. 1996. Varicella: historical perspective and clinical overview. J. Infect. Dis. 174(Suppl. 3):S306-S309. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]