Abstract
Nog1 is a conserved eukaryotic GTPase of the Obg family involved in the biogenesis of 60S ribosomal subunits. Here we report the unique dominant-inhibitory properties of a point mutation in the switch II region of mouse Nog1; this mutation is predicted to restrict conformational mobility of the GTP-binding domain. We show that although the mutation does not significantly affect GTP binding, ectopic expression of the mutant in mouse cells disrupts productive assembly of pre-60S subunits and arrests cell proliferation. The mutant impairs processing of multiple pre-rRNA intermediates, resulting in the degradation of the newly synthesized 5.8S/28S rRNA precursors. Sedimentation analysis of nucleolar preribosomes indicates that defective Nog1 function inhibits the conversion of 32S pre-rRNA-containing complexes to a smaller form, resulting in a drastic accumulation of enlarged pre-60S particles in the nucleolus. These results suggest that conformational changes in the switch II element of Nog1 have a critical importance for the dissociation of preribosome-bound factors during intranucleolar maturation and thereby strongly influence the overall efficiency of the assembly process.
Proteins of the GTPase superfamily are critical control components in many fundamental cellular processes, including translation, cellular transport, and signal transduction. Despite functional diversity, members of this superfamily share a common mechanism of action, based on reversible conformational rearrangements initiated within mobile segments (termed switches I and II) that are driven by GTP binding and hydrolysis (50). In recent years, genomic studies have uncovered several new families of GTPases conserved in a wide range of organisms (31). Many of these GTPases appear to function in connection with the ribosome, although their roles differ from those of canonical translation factors (3). One of the emerging GTPase families found in all domains of life is the Obg family, which includes prototypical Obg/CgtA proteins in eubacteria and several related subfamilies in archaea and eukaryotes (31). Bacterial members of the Obg family have been implicated in diverse cellular processes, including stress response, chromosome replication, sporulation, and differentiation (9, 22, 23, 39, 43, 47, 49, 58). Strong genetic and biochemical data also suggest that Obg family GTPases play important roles in ribosome synthesis. Bacterial Obg proteins associate with and function in the maturation of ribosomal subunits (16, 32, 45, 54). Eukaryotic genomes encode several distinct proteins from the Obg family that are found in association with ribosomes or the ribosome assembly machinery in the nucleolus, cytoplasm, and mitochondria, but the roles of these proteins are not well understood (5, 14, 40).
The synthesis of cytoplasmic ribosomes in eukaryotic cells is a complex multistep process that begins in the nucleolus and involves more than 170 different accessory proteins (10, 25, 55, 57). Several GTPases have been implicated in the assembly of both large and small ribosomal subunits (reviewed in reference 18). Nog1 is a putative eukaryotic GTPase from the Obg family that exhibits high evolutionary conservation of the amino acid sequence along the length of the N-terminal and G domains (40). Mammalian Nog1 was originally described as the product of the chronic renal failure gene CRFG, regulated differentially in renal disease (28); however, the protein has not been functionally characterized. Studies of yeast (Saccharomyces cerevisiae) and trypanosomes have shown that in these organisms, Nog1 is an essential factor in the biogenesis of 60S ribosomal subunits (15, 17, 46). Yeast cells depleted of Nog1p or harboring temperature-sensitive alleles of the gene displayed defects in the processing of pre-rRNA and the release of nascent pre-60S subunits from the nucleolus (17, 46). Proteomic analyses of yeast showed the association of Nog1p with multiple components of pre-60S ribosomes (11, 29). As is the case for most ribosome synthesis factors, the precise molecular mechanisms by which Nog1p performs its function remain to be elucidated.
A challenge of studying the roles of Obg GTPases has been the lack of dominant phenotypes produced by mutations of key residues involved in GTP binding and hydrolysis. Such phenotypes are often observed in other GTPase families and have been used extensively in studies of small Ras-like GTPases and α subunits of heterotrimeric G proteins (7, 30, 36). In this report, we show that a substitution of alanine for the conserved glycine 224 residue located in the switch II segment of mouse Nog1 induces global arrest of 60S ribosomal subunit synthesis upon ectopic expression in mammalian cells. The dominant mutation in Nog1 leads to a marked accumulation of 32S pre-rRNA-containing complexes in the nucleolus and affects multiple pre-rRNA processing steps. The molecular architecture of GTPases requires the presence of glycine at this position to allow proper conformational transitions upon guanine nucleotide binding. These results demonstrate the role of mammalian Nog1 in 60S ribosome synthesis, identify the first dominant-negative mutation in the Obg family of GTPases, and reveal the importance of structural transitions in the switch II element for Nog1 function in ribosome assembly.
MATERIALS AND METHODS
Cloning, plasmids, and antibodies.
The coding sequence of mouse Nog1 was amplified from cDNA and cloned in pX14, a modified IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pX11 vector (41) with an N-terminal hemagglutinin (HA) tag. The sequence of the clone was identical to the database sequence of NOG1_MOUSE (UniProt accession number Q99ME9). Point mutations in Nog1 were introduced using QuikChange reagents (Stratagene) and verified by sequencing the entire coding region. For bacterial expression, the Nog1-CBP fusion protein was cloned into the pCAL-c vector (Stratagene) and purified by binding to calmodulin agarose beads and preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Rabbit polyclonal antibodies were raised against Nog1-CBP, affinity purified, and concentrated on a protein A column. The NG fragments (amino acids 1 to 357) of wild-type and mutant Nog1 were cloned into the BamHI and HindIII sites of the pET-ABC vector that provides an N-terminal tag biotinylated in Escherichia coli by the BirA enzyme coexpressed from the same vector (59).
Cell culture and proliferation assays.
Cell culture and the bromodeoxyuridine-light assay for a reversible cell cycle arrest were performed as described previously (27). Mouse cell lines were obtained by an expansion of individual clones from stably transfected LAP3 cells (41). The following clonal lines were used after verifying induction: D1021 and D1106 (wild-type HA-Nog1) and D1041, D1046, and D1411 (HA-Nog1G224A). Cells were quantified using a CyQUANT kit (Molecular Probes).
RNA, protein analyses, and microscopy.
Protocols for the analysis of pre-rRNA processing, Western blotting, and indirect immunofluorescence have been described previously (27). Cellular RNA was counterstained by consecutive treatment with 2 μg/ml Hoechst 33342 for 10 min and 0.7 μM pyronin Y for 1 min (4), followed by a brief rinse with water before mounting for microscopy. Images were acquired with a Zeiss LSM5 confocal microscope.
Isolation of nucleolar preribosomes.
Nucleoli were isolated following the procedure of Muramatsu and Onishi (37). Cells from six 150-mm plates of D1411 cells were rinsed with phosphate-buffered saline (PBS) and scraped into 10 ml/plate ice-cold PBS, pelleted at 500 × g for 5 min, resuspended in 3 ml cold reticulocyte standard buffer (10 mM Tris-HCl [pH 7.2], 10 mM NaCl, 1.5 mM MgOAc2), and incubated on ice for 10 min. Cells were pelleted as described above (this and subsequent steps were performed at 4°C) and resuspended in 3 ml reticulocyte standard buffer. A total of 99 μl 10% Igepal CA-630 was added, followed by brief vortexing, and 66 μl 10% sodium deoxycholate was added, followed by vigorous vortexing for 30 s. The resulting crude nuclei were pelleted at 1,000 × g for 5 min and resuspended in 1 ml sucrose (0.25 M)-3.3 mM CaCl2 by being passed through a 20G1 needle 10 to 15 times. Nuclei were further purified by centrifugation at 1,200 × g for 10 min through a cushion of 1 ml sucrose (0.88 M)-0.1 mM MgCl2 and resuspended in 0.75 ml sucrose (0.34 M). Nuclei were sonicated three times for 10 s at low power on ice until no nuclei remained intact (observed microscopically), overlaid on top of 0.5 ml sucrose (0.88 M)-0.1 mM MgCl2 in a microcentrifuge tube, and centrifuged in a swinging bucket rotor at 2,000 × g for 15 min. The compact nucleolar pellet was gently resuspended in 15 to 20 μl sucrose (0.34 M), transferred to a new tube, flash frozen in liquid nitrogen, and stored at −80°C. To obtain preribosomes, the frozen nucleoli were resuspended in 300 μl high-salt buffer (27) containing 100 U DNase I (DPRF grade; Worthington Biochemicals) and Complete protease inhibitors (Roche), incubated for 5 min at room temperature with gentle mixing, and precipitated at 16,000 × g for 10 min at 4°C. Preribosomes were extracted from the remaining pellet and separated by sucrose gradient centrifugation as described previously (27).
Bacterial expression and purification of Nog1NG.
Constructs encoding Nog1NG with an N-terminal biotinylation tag were induced in Rosetta(DE3)pLysS cells (Novagen) in LB medium containing 2 μM biotin and 1 mM IPTG for 2 h at 37°C. Bacteria were collected by centrifugation, frozen in pellets, resuspended in 25 mM Tris-HCl (pH 7.4), 100 mM KOAc, 10% glycerol, 1 mM EDTA, 10 mM dithiothreitol, and protease inhibitors (Roche) and lysed by adding Igepal CA-630 to 0.2%. After 20 min at 4°C, 10 mM MgOAc2, 1 mM CaCl2 and DNase I were added to reduce viscosity for 10 min and the lysate was clarified at 10,000 × g for 15 min and filtered through a 0.22-μm filter. KOAc was adjusted to 300 mM, heparin was added to 0.1 mg/ml, and biotinylated proteins were bound to SoftLink monoavidin beads (Promega) by incubation with mixing for 2 h. Beads were extensively washed with B300 buffer (25 mM Tris-HCl [pH 7.4], 10 mM MgOAc2, 10% glycerol, 0.05% Brij 30, 1 mM dithiothreitol, and 300 mM KOAc) and B1000 buffer (same as above but with 1 M KOAc). Proteins were eluted with the B300 buffer containing 5 mM biotin, concentrated and purified from biotin by several rounds of ultrafiltration using Millipore Microcon YM-50, and stored in B300 buffer containing 50% glycerol at −20°C. The protein was estimated to be more than 95% pure by SDS-PAGE.
UV cross-linking assays of GTP binding.
An aliquot of 0.5 to 2 μM protein was incubated with 0.132 μM [α-32P]GTP (Perkin-Elmer) in B300 buffer for 5 min on ice, mixed with 66 μM unlabeled nucleoside triphosphates (Fermentas) when indicated, and irradiated in open Terasaki plates placed on an ice-cold metal block in a Stratalinker for 30 min. The reaction mixture was diluted to 50 μl, and proteins were recovered by binding to Strataclean beads (Stratagene) and analyzed by SDS-PAGE. For the determination of Kd values, proteins were incubated in buffer with optimized concentrations of KOAc (100 mM) and MgOAc2 (5 mM) for 40 min on ice with 0.2 or 0.08 μM [α-32P]GTP in the presence of 10 different concentrations (10−9 to 3 × 10−3 M) of unlabeled GTP, followed by UV cross-linking. The samples were then mixed with 0.5 volume of guanidinium hydrochloride (7.5 M), and the proteins were quantitatively recovered by binding to a SAM2 streptavidin-coated membrane (Promega), owing to the presence of an N-terminal biotinylated tag. The membrane was washed with several changes of 0.5 M NaH2PO4 for 40 min (followed by PBS containing 2 mM EDTA plus 0.1% SDS for 10 min) and water for 5 min, dried, and quantitated by phosphorimaging.
RESULTS
A point mutation in the G domain of Nog1 inhibits 60S subunit synthesis.
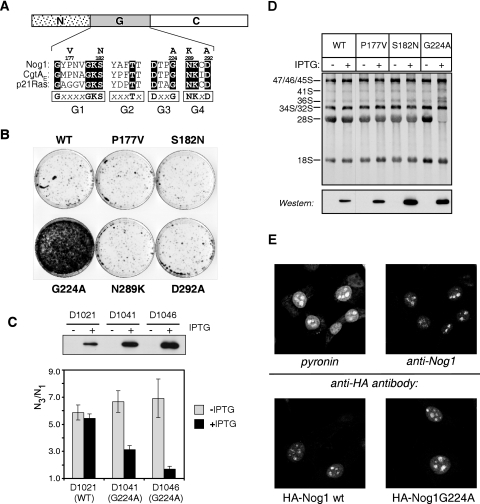
The structurally conserved GTP-binding core (the G domain) of GTPases contains a set of signature sequence motifs designated G1 to G5 that are directly involved in forming contacts with guanine nucleotides and critical for function (Fig. 1A) (1, 50). Based on sequence homology with other GTPases, we generated mutations in conserved residues within the G domain of mouse Nog1 by using site-directed mutagenesis. To search for mutations with strong effects on ribosome synthesis, we took advantage of the observation that mammalian cells with the functional p53 pathway undergo a reversible cell cycle arrest in response to defects in ribosome biogenesis (42). Out of several mutants expressed in mouse cells, the G224A mutant stood out by a pronounced dominant phenotype in a cell cycle inhibition assay (Fig. 1B). We therefore decided to analyze this mutation in more detail.
FIG. 1.
The G224A mutant of Nog1 exerts dominant-inhibitory effects on cell proliferation and rRNA maturation and is localized to the nucleolus. (A) Alignment of the consensus boxes (G1 to G4) within the G domain with sequences from mouse Nog1, the E. coli Obg GTPase CgtAE and p21Ras. Point mutations described in this study are shown above the Nog1 sequence. (B) Reversible cell cycle arrest upon induction of G224A expression. Transfected cell pools were induced in the presence of bromodeoxyuridine to selectively kill proliferating cells (41); arrested cells survive this treatment, resume growth, and form colonies after removing the inducer. (C) Ectopic expression of HA-tagged wild-type Nog1 and Nog1G224A in stable IPTG-inducible clonal cell lines and their effects on cell proliferation. Induction was analyzed by Western blotting with an anti-HA antibody. Growth rates were analyzed by measuring the ratio of cell number on day 3 (N3) to cell number on day 1 (N1) after plating in medium with (+) or without (−) IPTG. Error bars indicate standard deviations from counting triplicate wells. (D) [3H]uridine labeling of newly synthesized rRNA after the induction of wild-type (WT) or mutant Nog1 proteins in stable cell clones uninduced or induced for 16 h. Protein expression was verified with an anti-HA antibody (bottom panel). (E) Immunofluorescent staining of endogenous Nog1 in mouse cells with affinity-purified anti-Nog1 antibodies. Cellular RNA was counterstained with pyronin (top). Ectopically expressed HA-tagged constructs were detected by using an anti-HA antibody (bottom).
Because G224A expression strongly inhibited cell growth, we established cell lines in which the mutant or wild-type protein was expressed using an IPTG-inducible promoter (Fig. 1C). To confirm that the G224A mutant affected ribosome biogenesis, we induced its expression in the cells and performed metabolic labeling of the newly synthesized RNA with [3H]uridine. An analysis of the labeled rRNA revealed that G224A-expressing cells synthesized the primary 47S rRNA transcript, but its processing to mature 28S rRNA was strongly impaired, while 18S continued to be produced normally (Fig. 1D). We estimate that in the cell lines used for this study, the induction of Nog1G224A reduced the total output of newly synthesized 28S rRNA to less than 10% of the normal level. Thus, a single amino acid change in the G domain of Nog1 can effectively shut down the 60S subunit biosynthetic pathway despite the availability of the endogenous wild-type protein in the cell.
We next investigated how the G224A mutation affects subcellular location of Nog1. The indirect immunofluorescence analysis of endogenous Nog1 in mouse cells by using affinity-purified antibodies showed the predominantly nucleolar localization of the protein, with a weaker signal in the nucleoplasm (Fig. 1E, top panel). We expressed HA-tagged wild-type and mutant Nog1 and used antibodies against the tag to differentiate the ectopic proteins from the endogenous Nog1. The G224A mutant showed a nuclear localization pattern, with strong accumulation in the nucleolus similar to that of the wild-type HA-Nog1, although nucleoli after G224A expression often looked somewhat enlarged compared to cells expressing wild-type protein (Fig. 1E, bottom panel). The nucleolar localization of Nog1G224A supports the expectation that the inhibitory effects of the mutant protein result from its direct interaction with the ribosome synthesis machinery in the nucleolus.
Effects of Nog1G224A on pre-rRNA processing.
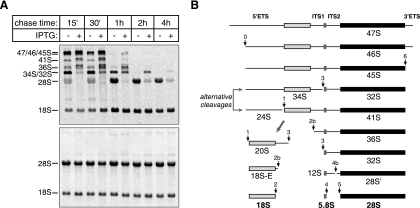
To investigate the changes in rRNA synthesis caused by the G224A mutant, we performed pulse-chase labeling with the radioactive donor of methyl groups, [3H]methyl-methionine. The expression of G224A led to severe abnormalities in the processing of 28S precursors (Fig. 2A). First, these cells exhibited high levels of aberrant precursors appearing as secondary bands above normal 36S and 41S bands. Furthermore, the 32S pre-rRNA in these cells appeared with a significantly slower kinetics, indicating slowly occurring early processing steps, although none of the major cleavages in pre-rRNA (Fig. 2B) was completely blocked. Finally, very little 28S rRNA was formed at the end, and most of the 28S precursors were degraded. This pattern of nonproductive maturation of 28S rRNA was in striking contrast with the nearly normal kinetics of 18S rRNA formation in these cells (Fig. 2A).
FIG. 2.
Effects of Nog1G224A on the kinetics of rRNA processing steps. (A) Cell line D1046 was cultured with IPTG (+) for 18 h to induce Nog1G224A expression or without IPTG (−) for a control. rRNA was pulse-chase labeled with l-[methyl-3H]methionine, separated on a formaldehyde-agarose gel, blotted, and detected by fluorography (top). The same blot stained with methylene blue is shown at the bottom. (B) Major rRNA precursors formed during the processing of the primary rRNA transcript (47S) to mature 18S, 5.8S, and 28S rRNAs in mouse cells. Main processing sites involved in the synthesis of each precursor are indicated. After 47S pre-rRNA is synthesized, cleavages at sites 0 and 6 in the 5′ETS and 3′ETS normally occur first, followed by a variable set of processing events in ITS1, which separates the rRNA destined for 40S subunits from the rest of the transcript. The final maturation of rRNAs in 60S subunits involves processing within ITS2.
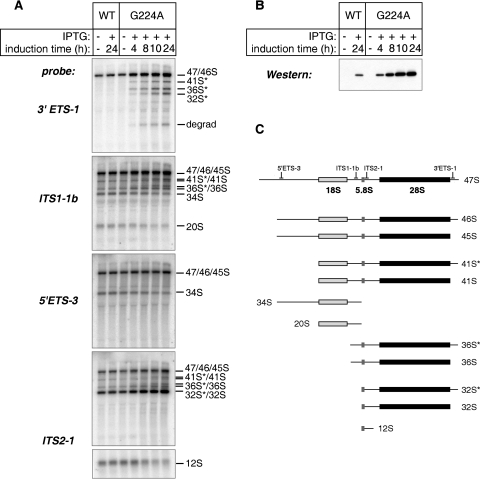
We next examined changes in steady-state levels of pre-rRNAs by Northern hybridization. The G224A mutant was induced for 4 to 24 h and analyzed in parallel with control cells expressing the wild-type protein (Fig. 3A and B). Hybridization with a 3′ external transcribed spacer (3′ETS)-specific probe revealed a dramatic accumulation of precursors with unremoved 3′ETS tails (32S*, 36S*, 41S*, and 46S) (Fig. 3C) after the induction of G224A, explaining the pattern of double bands appearing in [3H]methyl-methionine labeling (Fig. 2A). Of note, hybridization with the 3′ETS probe also showed bands migrating faster than 28S (Fig. 3A), indicating that the aberrant 3′-extended products were degraded through a mechanism that involves an endonucleolytic or 5′-to-3′ exonucleolytic activity. A moderate reduction in 34S and 20S pre-rRNAs, with a concomitant increase in 36S pre-rRNA levels, was observed in hybridizations with internal transcribed spacer 1 (ITS1)- and 5′ETS-specific probes, indicating that processing in ITS1 preferably utilized cleavage at site 2b rather than site 3 (Fig. 2B), although neither of these cleavages was blocked. ITS2 processing was also strongly affected, as indicated by accumulation of 32S pre-rRNA at 24 h and a decline in 12S pre-rRNA levels (ITS2-1 probe [Fig. 3A]). Thus, the presence of the mutant G224A protein reduced the efficiency of multiple pre-rRNA processing events. Furthermore, even though the vast majority of the newly synthesized 28S rRNA precursors were completely degraded within 4 h of synthesis (Fig. 2A), some of the intermediates, particularly the 3′-extended “star” species and 32S pre-rRNA, appeared to have a relatively long half-life, leading to their elevated steady-state levels relative to normal cells.
FIG. 3.
Hybridization analysis of pre-rRNA processing in cells expressing Nog1G224A. (A) D1106 (wild-type [WT]Nog1) and D1411 (mutant) cells were cultured with (+) or without (−) IPTG for the times indicated. Steady-state levels of pre-rRNAs were analyzed by Northern hybridization of total RNA with the oligonucleotide probes indicated on the left. Pre-rRNAs detected with each probe are indicated on the right. (B) Induction of G224A in cells used for the RNA analysis was assessed by detection with anti-HA antibodies in Western blotting of cell lysates normalized for protein content. (C) Relative positions of probes used in the experiment depicted in panel A on the primary pre-rRNA transcript and the structure of processing intermediates detected with these probes.
Stable association of Nog1 with nucleolar preribosomes.
Nog1 was identified as a component of preribosomal complexes in yeast (11, 17, 46) and trypanosomes (15) as well as mammalian cells (12). In yeast, protein complexes containing multiple preribosome-associated factors have been isolated by tandem affinity purification using Nog1p-TAP bait (29, 46). Despite repeated efforts with a variety of affinity tags, we were not able to purify Nog1 from mammalian cells unless we treated samples with low concentrations of SDS, suggesting that the level of accessibility of the mammalian protein to affinity reagents under native conditions is low. Mammalian preribosomes can be isolated from nuclei by centrifugation in sucrose gradients (60); however, these preparations also contain large amounts of contaminating mature subunits. We therefore established a multistep purification protocol to obtain highly enriched preribosomes by exploiting differential sedimentation properties of mammalian nucleoli that allow their selective isolation from cell nuclei (37). The nucleoli prepared by this technique were additionally washed with high-salt buffer (0.5 M NaCl, 50 mM MgCl2) (27), which removed the bulk of soluble nucleolar proteins but did not release preribosomal particles. We then extracted preribosomes from the remaining nucleolar material with EDTA-containing buffer (see Materials and Methods for details). The resulting salt- and EDTA-resistant “core” ribonucleoprotein complexes contained both early (47-41S) and late (36S, 32S, and 12S) 28S/5.8S rRNA precursors (see below). Pre-18S rRNA was virtually absent from these preparations, consistent with the observations of a rapid transfer of newly synthesized pre-40S subunits out of the nucleolus and nucleus (44, 56).
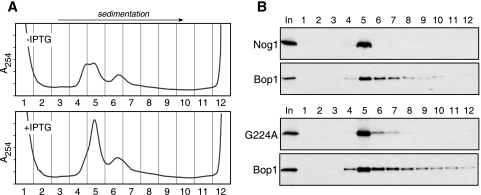
When we analyzed the extracted nucleolar complexes by sucrose gradient centrifigation, we observed a heterogeneous mixture of particles that sedimented with estimated rates of 50S and above (Fig. 4A, upper panel). A significant change in sedimentation properties of the particles was evident after the induction of the G224A mutant. In cells expressing the mutant, a new major peak of absorbance appeared in fraction 5 and a larger amount of particles was observed in fractions 6 to 12 (Fig. 4A, lower panel). To determine whether Nog1 was retained in high molecular complexes under these conditions, we analyzed gradient fractions by immunoblotting. This analysis showed that all Nog1 extracted from nucleoli cosedimented with ∼50S complexes in fraction 5 (Fig. 4B), suggesting that the protein is incorporated into preribosomes. The peak of Nog1G224A was detected in fraction 5 like the wild-type protein was, although we also observed small amounts of the mutant protein in the faster-sedimenting fractions 6 to 7 of the gradient, likely reflecting the increase of particles in these fractions (Fig. 4B).
FIG. 4.
Sucrose gradient centrifugation of nucleolar preribosomes. (A) Changes in sedimentation properties of nucleolar ribonucleoprotein complexes after Nog1G224A expression. Uninduced (−) D1411 cells or cells induced (+) with IPTG for 16 h were used for the isolation of purified nucleoli, followed by the extraction of preribosomes. After separation on a 10 to 30% (wt/wt) sucrose gradient, 12 fractions were collected from the top of the gradient, with continuous measurement of UV absorbance (A254). (B) Proteins precipitated from each gradient fraction depicted in panel A were analyzed by Western blotting. Endogenous Nog1 and Bop1 were detected with affinity-purified antibodies, and Nog1G224A was detected with anti-HA antibodies.
Overall, these data indicate that both wild-type Nog1 and the G224A mutant are present in the nucleolus as part of stable macromolecular complexes with the sedimentation rates of preribosomal particles. One surprising observation from these experiments was that all Nog1 from the nucleolar particles prepared with the use of high-ionic-strength buffer was found in a single fraction of the gradient. By comparison, another tightly bound component of pre-60S ribosomes, Bop1 (51), was broadly distributed on the gradient spanning a range of fractions containing different preribosomes (Fig. 4B). This result indicates that a fraction of pre-60S ribosomes is capable of forming especially tight interactions with Nog1. We speculate that such tight binding may require a specific composition or conformation of complexes that occurs only at a certain point in the assembly process.
Accumulation of 32S pre-rRNA complexes in G224A-expressing cells.
To understand the nature of preribosomes accumulating in G224A-expressing cells, we examined individual gradient fractions by Northern hybridizations using probes specific for different regions of the primary rRNA transcript. Previous studies utilizing different experimental techniques have shown that preribosomes become progressively more compact and retain fewer associated accessory factors in the course of their assembly (35, 38). Consistent with these observations, the largest amount of mature 28S and 5.8S rRNAs was detected in fraction 4 (Fig. 5A). Under normal conditions (without IPTG and no G224A expression), complexes containing the abundant 32S pre-rRNA formed a broad peak spanning fractions 4 and 5, and earlier pre-rRNAs were mostly found in faster-sedimenting complexes in the lower part of the gradient. The expression of the G224A mutant resulted in significant changes in the distribution of pre-rRNA-containing complexes on the gradient. In cells expressing Nog1G224A, the overall amount of 32S-containing complexes was increased (Fig. 5A). In addition, these complexes sedimented faster and now formed a pronounced peak in fraction 5 (Fig. 5C). We conclude that the strong increase in the UV absorbance of fraction 5 in G224A-expressing cells observed above (Fig. 4A) is due largely to the accumulation of complexes containing 32S pre-rRNA.
FIG. 5.
Changes in the pre-rRNA content of preribosomal particles after Nog1G224A expression. (A) RNA isolated from each gradient fraction (Fig. 4A) was separated on a formaldehyde-agarose gel, blotted, stained with methylene blue, and hybridized with the 32P-labeled probes indicated to detect 32S and 12S pre-rRNAs and 5.8S rRNA. (B) The ratio of 12S to 32S in fractions 4 and 5 determined by quantitation of the phosphorimaging data from hybridizations depicted in panel A. (C) Normalized distribution of 32S and 12S pre-rRNAs on the gradient. For comparison purposes, the level of the precursor in each fraction was normalized to the level of the same precursor in fraction 4 (hence, fraction 4 is 1.0). −, absence of; +, presence of.
Hybridization analysis of gradient fractions also revealed the presence of 12S pre-rRNA in the particles (Fig. 5A), indicating that the ITS2 cleavage in mammalian cells occurs within the nucleolus. Consistent with the analysis of steady-state levels of total RNA showing inefficient cleavage in ITS2 (Fig. 3A), the ratio between 12S and its precursor 32S pre-rRNA in nucleolar particles was strongly reduced in G224A cells (Fig. 5B). Interestingly, the distribution of the 12S-containing particles also showed a shift toward faster-sedimenting fractions after the induction of G224A (Fig. 5C), suggesting that ITS2 cleavage might also occur in larger particles, although apparently with a greatly reduced efficiency.
Taken together, these data indicate that the expression of the G224A mutant of Nog1 results in the accumulation of 32S-containing preribosomal complexes within the nucleolus. The 32S-containing assembly intermediates in normal cells are clearly heterogeneous, as shown by their broad sedimentation peak. Moreover, a considerable fraction of nucleolar 32S pre-rRNA-containing complexes in normal cells cosediment with the particles in which 32S has undergone ITS2 cleavage into 28S and 12S pre-rRNAs. These particles are underrepresented in G224A-expressing cells, while the faster-sedimenting 32S-containing complexes that likely contain additional components show strong accumulation, suggesting that the G224A mutant inhibits their conversion to the smaller form.
GTP binding properties of Nog1 and the G224A mutant.
The prominent dominant phenotype puts G224A in a unique category among G domain mutations in Obg proteins. Mutations in many other well-defined residues involved in GTP binding inactivate function of Obg GTPases but have never been observed to elicit dominant effects (6, 33). For instance, the overexpression of mouse Nog1 mutations P177V and S182N, analogous to classic dominant mutations G12V and S17N in Ras, produces no discernible effects on growth or the levels of 60S synthesis when endogenous Nog1 is present (Fig. 1B and D). Corresponding mutations in yeast Nog1p cannot support ribosome biogenesis and significantly impair growth, but this phenotype is also recessive (11). Interestingly, the P177V and S182N mutants of mouse Nog1 can be expressed in cells at high levels (Fig. 1D) and these mutant proteins associate with preribosomal complexes (data not shown). This suggests that the assembly machinery can tolerate the incorporation of inactive Nog1 into preribosomes either because such particles can be efficiently disassembled or because defects in Nog1 function can be corrected in some other way, for instance, through exchange with wild-type Nog1.
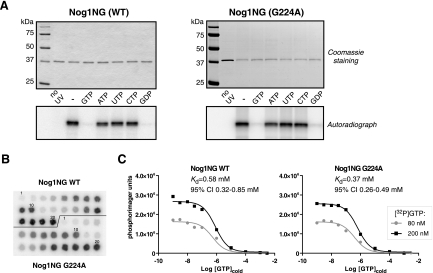
Although the crystal structure of Nog1 has not yet been determined, structural analysis of related bacterial Obg proteins has shown similarities in the overall architecture of the G domains of Obg proteins and Ras-like GTPases (2, 26). Bacterial Obg/CgtA proteins were found to bind GTP; however, one recently characterized member of the Obg family, Ola1/YchF, has the unusual property of binding ATP with higher affinity than GTP (24). Biochemical properties of Nog1 from any organism have not yet been reported. To determine whether Nog1 is indeed a guanine nucleotide binding protein, we sought to obtain a purified protein by expression in bacteria. Consistent with the previously reported lack of success in expressing recombinant Nog1 (11, 40), we found that mouse Nog1 was misfolded in E. coli and inactive in vitro. We discovered, however, that the behavior of recombinant Nog1 could be improved by removing the C-terminal domain, which shows little conservation among eukaryotic Nog1 orthologs and is highly divergent among the rest of the Obg family. We expressed Nog1 lacking the C-terminal domain (Nog1NG) in E. coli by using an N-terminal biotinylated tag for purification and performed UV cross-linking of the purified protein with 32P-labeled GTP. This assay showed that Nog1NG was capable of selectively binding GTP, which was effectively competed out by unlabeled GTP and GDP, but not other nucleotides (Fig. 6A, left panel). Thus, the conserved NG portion of Nog1 is sufficient to provide specific GTP binding, while the divergent C-terminal domain apparently plays other functions. We could not estimate the rate of GTP hydrolysis by Nog1NG due to the presence of a contaminating bacterial nucleotide-hydrolyzing activity in the protein preparation.
FIG. 6.
Guanine nucleotide binding by Nog1 and Nog1G224A. (A) Purified NG fragments of the wild-type (WT) Nog1 and G224A mutant were UV cross-linked with [α-32P]GTP in the absence (−) or presence of a 500-fold excess of unlabeled nucleotides as indicated. Cross-linked products were resolved by SDS-PAGE. Gels were stained with Coomassie for loading control, dried, and analyzed by autoradiography. No UV, non-cross-linked reaction in the absence of competitors. (B) Homologous competition assay to compare binding of GTP to wild-type Nog1NG and G224A. For each protein, samples 1 to 10 were cross-linked with 80 nM [α-32P]GTP and samples 11 to 20 were cross-linked with 200 nM [α-32P]GTP in the presence of various concentrations of unlabeled GTP as detailed in Materials and Methods. Cross-linked samples were loaded onto a streptavidin-coated membrane and detected by phosphorimaging. (C) Data were analyzed in Prism software (GraphPad) by simultaneously fitting two data sets for each protein to a homologous competitive binding model accounting for ligand depletion. The data shown are representative of two independent experiments. CI, confidence interval.
Cross-linking assays with Nog1G224A showed that the mutation did not prevent GTP binding or exchange (Fig. 6A, right panel), implying that the overall structure of the G domain in the mutant was largely intact. To compare affinities of the wild-type and mutant Nog1NG to GTP, we performed cross-linking in the presence of a range of concentrations of the unlabeled nucleotide. The Kd values of Nog1NG proteins calculated from this assay were not significantly different, 0.58 μM for the wild type and 0.37 μM for G224A (Fig. 6B and C). The lack of drastic effects on GTP binding observed in the G224A mutant is consistent with the relatively small changes in nucleotide affinities previously reported for the corresponding glycine mutations in Ras and EF-Tu (19, 53). Crystallographic studies of these GTPases have implicated the G3 box glycine residue as a pivot for the movements occurring within the G domain upon GTP binding and hydrolysis (8, 20, 50). Consequently, the replacement of this glycine with any other residue results in the impairment of conformational changes and protein-protein interactions even though guanine nucleotide binding is not necessarily perturbed. The affinity of Nog1NG to GTP also compares well with the moderate nucleotide binding exhibited by bacterial Obg/CgtA proteins (34, 61), suggesting that the NG part of the eukaryotic Nog1 and bacterial Obg proteins may act in similar ways. The rapid exchange rates previously observed in Obg proteins have led to the proposal that these proteins do not function in their free form but rather require binding to targets to stabilize the bound nucleotide (33). The ability of the G224A mutant to effectively bind GTP might therefore contribute to its unusual dominant properties by allowing interactions with its targets and/or regulatory factors and sequestering them into defective preribosomal particles.
DISCUSSION
In this study, we have found that a single substitution of alanine for a conserved glycine in the G domain of Nog1 produces a remarkably strong dominant-inhibitory effect that virtually shuts down the biosynthesis of 60S ribosomal subunits in mammalian cells. The G224A mutant of mouse Nog1 binds GTP, localizes to the nucleolus and readily associates with ribosome precursor particles like the wild-type protein. The presence of the mutant protein, however, causes severe perturbations of the entire 60S assembly process by stalling the maturation of nucleolar pre-60S particles and inducing aberrant processing and degradation of the newly synthesized 5.8S/28S rRNA precursors. Because dominant-negative phenotypes have not been observed previously for Obg proteins, the mutation described here introduces a useful new tool for the investigation of molecular mechanisms in this family of GTPases.
Our results demonstrate that mammalian Nog1, like its orthologs from lower eukaryotes, is required for ribosome synthesis. Despite the identification of multiple preribosome components associated with Nog1p in yeast (11, 17, 29, 46), the exact role of this protein in ribosome assembly is not known. Our data show that the impairment of a critical pivot point in the G domain of Nog1 allows for the assembly of preribosomal complexes but creates particles from which bound components apparently fail to disengage. In particular, the dramatic changes in the amount and properties of nucleolar preribosomes occurring after G224A expression (Fig. 4 and 5) indicate that Nog1 function is required for the productive maturation of complexes containing 32S pre-rRNA. It is possible that the deficiency of factors due to sequestration by the stalled 32S-containing precursor complexes is also the primary reason for the multiple anomalies in pre-rRNA processing observed in cells expressing this mutant, ultimately leading to the degradation of the newly synthesized 28S/5.8S pre-rRNA (Fig. 2 and 3).
The assembly of each ribosomal subunit in vivo involves the binding and dissociation of dozens of processing factors. How these binding/dissociation events are controlled mechanistically is not well understood. Recently, studies of yeast have implicated two cytoplasmic GTPases, Efl1p/Ria1p and Lsg1p, in facilitating the release of the preribosome-associated shuttling proteins Tif6p and Nmd3p from nascent subunits in the cytoplasm (13, 48). Our data raise an interesting question of whether Nog1 might similarly mediate the release of specific factors bound to preribosomes, but during their earlier, intranucleolar maturation steps. Although we do not yet understand the mammalian ribosome synthesis machinery well enough to determine the identity of factors affected by Nog1 function, this issue may be well worth addressing in better explored systems.
The critical role of the glycine 224 residue in Nog1 suggests that conformational rearrangements of the switch II element are the key factors in Nog1 activity after it binds to pre-60S ribosomes. The essential role of the pivotal glycine residue within the DXXG motif for the mobility of the switch II element is well established, explaining the universal conservation of this residue in GTPases (1, 20, 50). Replacement of the glycine with any other residue impairs conformational changes and results in altered protein interactions in a number of GTPases, including Ras, Gαs, and EF-Tu (19, 21, 36, 52). The crystal structure of Ras with the corresponding G60A mutation has been solved recently (8). This study showed that the substitution of the switch II glycine created a novel open conformation in the G domain that precluded the binding of effectors but at the same time stabilized bound Sos, a guanine nucleotide exchange factor for Ras. The depletion of intracellular pools of guanine nucleotide exchange factors was proposed as the mechanism underlying dominant-negative effects of this mutation with respect to cellular Ras signaling. The failure of G protein subunit dissociation was also observed for the corresponding mutation G226A in Gαs (30, 36). The accumulation of the faster-sedimenting complexes resulting from G224A expression suggests intriguing parallels with the model of the persistent association of RasG60A and Gα subunits with their binding partners. Further investigation will be needed to confirm whether analogous molecular mechanisms account for the dominant effects of the switch II glycine mutations. It is remarkable, however, that the impairment of structural flexibility of the switch II region shows a common propensity for dominant-inhibitory effects in distantly related GTPases, indicating a common “weak link” in their molecular structure, despite functional diversification during evolution.
The finding that a subtle alteration in the G domain of mouse Nog1 is sufficient to cause profound consequences for preribosome assembly suggests a potential approach to specifically inhibit ribosome biogenesis in mammalian cells. Given the high evolutionary conservation of Nog1, analogous mutations may also provide a useful tool for studying ribosome assembly in other eukaryotes. It would be interesting to see whether mutations that limit structural transitions in other Obg GTPases would also lead to informative phenotypes. Obg GTPases are found in all living organisms, where these GTPases participate in processes ranging from the assembly and maintenance of ribonucleoprotein complexes to the replication of chromosomes and stress response, offering the possibility of control over a number of important cellular functions.
Acknowledgments
We thank David C. H. Yang (Georgetown University) for the gift of plasmids for bacterial expression of biotinylated proteins.
Y.R.L. was supported by a postdoctoral fellowship from the American Heart Association. This work was supported by NIH grant GM074091 to D.P.
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117-127. [DOI] [PubMed] [Google Scholar]
- 2.Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10: 1581-1592. [DOI] [PubMed] [Google Scholar]
- 3.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6: 135-139. [DOI] [PubMed] [Google Scholar]
- 4.Darzynkiewicz, Z., J. Kapuscinski, F. Traganos, and H. A. Crissman. 1987. Application of pyronin Y(G) in cytochemistry of nucleic acids. Cytometry 8: 138-145. [DOI] [PubMed] [Google Scholar]
- 5.Datta, K., J. L. Fuentes, and J. R. Maddock. 2005. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell 16: 954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta, K., J. M. Skidmore, K. Pu, and J. R. Maddock. 2004. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol. 54: 1379-1392. [DOI] [PubMed] [Google Scholar]
- 7.Feig, L. A. 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1: E25-E27. [DOI] [PubMed] [Google Scholar]
- 8.Ford, B., K. Skowronek, S. Boykevisch, D. Bar-Sagi, and N. Nassar. 2005. Structure of the G60A mutant of Ras: implications for the dominant negative effect. J. Biol. Chem. 280: 25697-25705. [DOI] [PubMed] [Google Scholar]
- 9.Foti, J. J., J. Schienda, V. A. J. Sutera, and S. T. Lovett. 2005. A bacterial G protein-mediated response to replication arrest. Mol. Cell 17: 549-560. [DOI] [PubMed] [Google Scholar]
- 10.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313: 17-42. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes, J., K. Datta, S. Sullivan, A. Walker, and J. Maddock. 2007. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics 278: 105-123. [DOI] [PubMed] [Google Scholar]
- 12.Fujiyama, S., M. Yanagida, T. Hayano, Y. Miura, T. Isobe, F. Fujimori, T. Uchida, and N. Takahashi. 2002. Isolation and proteomic characterization of human Parvulin-associating preribosomal ribonucleoprotein complexes. J. Biol. Chem. 277: 23773-23780. [DOI] [PubMed] [Google Scholar]
- 13.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24: 567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano, Y., R. L. Ohniwa, C. Wada, S. H. Yoshimura, and K. Takeyasu. 2006. Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells 11: 1295-1304. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, B. C., Q. Wang, C. T. Kifer, and M. Parsons. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60 S ribosomal subunit. J. Biol. Chem. 278: 32204-32211. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, M., K. Datta, A. Walker, J. Strahler, P. Bagamasbad, P. C. Andrews, and J. R. Maddock. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188: 6757-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23: 4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karbstein, K. 2007. The role of GTPases in ribosome assembly. Biopolymers 87: 1-11. [DOI] [PubMed] [Google Scholar]
- 19.Kjaersgard, I. V., C. R. Knudsen, and O. Wiborg. 1995. Mutation of the conserved Gly83 and Gly94 in Escherichia coli elongation factor Tu. Indication of structural pivots. Eur. J. Biochem. 228: 184-190. [DOI] [PubMed] [Google Scholar]
- 20.Kjeldgaard, M., P. Nissen, S. Thirup, and J. Nyborg. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1: 35-50. [DOI] [PubMed] [Google Scholar]
- 21.Knudsen, C., H. J. Wieden, and M. V. Rodnina. 2001. The importance of structural transitions of the switch II region for the functions of elongation factor Tu on the ribosome. J. Biol. Chem. 276: 22183-22190. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, G., S. Moriya, and C. Wada. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 41: 1037-1051. [DOI] [PubMed] [Google Scholar]
- 23.Kok, J., K. A. Trach, and J. A. Hoch. 1994. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol. 176: 7155-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koller-Eichhorn, R., T. Marquardt, R. Gail, A. Wittinghofer, D. Kostrewa, U. Kutay, and C. Kambach. 2007. Human OLA1 defines an ATPase subfamily in the OBG family of GTP-binding proteins. J. Biol. Chem. 282: 19928-19937. [DOI] [PubMed] [Google Scholar]
- 25.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kukimoto-Niino, M., K. Murayama, M. Inoue, T. Terada, J. R. H. Tame, S. Kuramitsu, M. Shirouzu, and S. Yokoyama. 2004. Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J. Mol. Biol. 337: 761-770. [DOI] [PubMed] [Google Scholar]
- 27.Lapik, Y. R., C. J. Fernandes, L. F. Lau, and D. G. Pestov. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell 15: 17-29. [DOI] [PubMed] [Google Scholar]
- 28.Laping, N. J., B. A. Olson, and Y. Zhu. 2001. Identification of a novel nuclear guanosine triphosphate-binding protein differentially expressed in renal disease. J. Am. Soc. Nephrol. 12: 883-890. [DOI] [PubMed] [Google Scholar]
- 29.Lebreton, A., C. Saveanu, L. Decourty, A. Jacquier, and M. Fromont-Racine. 2006. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J. Biol. Chem. 281: 27099-27108. [DOI] [PubMed] [Google Scholar]
- 30.Lee, E., R. Taussig, and A. G. Gilman. 1992. The G226A mutant of Gs alpha highlights the requirement for dissociation of G protein subunits. J. Biol. Chem. 267: 1212-1218. [PubMed] [Google Scholar]
- 31.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317: 41-72. [DOI] [PubMed] [Google Scholar]
- 32.Lin, B., D. A. Thayer, and J. R. Maddock. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186: 481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, B., J. M. Skidmore, A. Bhatt, S. M. Pfeffer, L. Pawloski, and J. R. Maddock. 2001. Alanine scan mutagenesis of the switch I domain of the Caulobacter crescentus CgtA protein reveals critical amino acids required for in vivo function. Mol. Microbiol. 39: 924-934. [DOI] [PubMed] [Google Scholar]
- 34.Lin, B., K. L. Covalle, and J. R. Maddock. 1999. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol. 181: 5825-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuura, S., T. Morimoto, Y. Tashiro, T. Higashinakagawa, and M. Muramatsu. 1974. Ultrastructural and biochemical studies on the precursor ribosomal particles isolated from rat liver nucleoli. J. Cell Biol. 63: 629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, R. T., S. B. Masters, K. A. Sullivan, B. Beiderman, and H. R. Bourne. 1988. A mutation that prevents GTP-dependent activation of the alpha chain of Gs. Nature 334: 712-715. [DOI] [PubMed] [Google Scholar]
- 37.Muramatsu, M., and T. Onishi. 1978. Isolation and purification of nucleoli and nucleolar chromatin from mammalian cells. Methods Cell Biol. 17: 141-161. [DOI] [PubMed] [Google Scholar]
- 38.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21: 5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto, S., and K. Ochi. 1998. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30: 107-119. [DOI] [PubMed] [Google Scholar]
- 40.Park, J. H., B. C. Jensen, C. T. Kifer, and M. Parsons. 2001. A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci. 114: 173-185. [DOI] [PubMed] [Google Scholar]
- 41.Pestov, D. G., and L. F. Lau. 1994. Genetic selection of growth-inhibitory sequences in mammalian cells. Proc. Natl. Acad. Sci. USA 91: 12549-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21: 4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raskin, D. M., N. Judson, and J. J. Mekalanos. 2007. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 104: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouquette, J., V. Choesmel, and P. Gleizes. 2005. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 24: 2862-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10: 393-408. [DOI] [PubMed] [Google Scholar]
- 46.Saveanu, C., A. Namane, P. Gleizes, A. Lebreton, J. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23: 4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, J. M., and W. G. Haldenwang. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181: 4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8: 1363-1373. [DOI] [PubMed] [Google Scholar]
- 49.Słomińska, M., G. Konopa, G. Wegrzyn, and A. Czyz. 2002. Impaired chromosome partitioning and synchronization of DNA replication initiation in an insertional mutant in the Vibrio harveyi cgtA gene coding for a common GTP-binding protein. Biochem. J. 362: 579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sprang, S. R. 1997. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 66: 639-678. [DOI] [PubMed] [Google Scholar]
- 51.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20: 5516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung, Y. J., M. C. Hwang, and Y. W. Hwang. 1996. The dominant negative effects of H-Ras harboring a Gly to Ala mutation at position 60. J. Biol. Chem. 271: 30537-30543. [DOI] [PubMed] [Google Scholar]
- 53.Sung, Y. J., M. Carter, J. M. Zhong, and Y. W. Hwang. 1995. Mutagenesis of the H-ras p21 at glycine-60 residue disrupts GTP-induced conformational change. Biochemistry 34: 3470-3477. [DOI] [PubMed] [Google Scholar]
- 54.Tan, J., U. Jakob, and J. C. A. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184: 2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13: 255-263. [DOI] [PubMed] [Google Scholar]
- 56.Vaughan, M. H., J. R. Warner, and J. E. Darnell. 1967. Ribosomal precursor particles in the HeLa cell nucleus. J. Mol. Biol. 25: 235-251. [DOI] [PubMed] [Google Scholar]
- 57.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261-311. [DOI] [PubMed] [Google Scholar]
- 58.Vidwans, S. J., K. Ireton, and A. D. Grossman. 1995. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177: 3308-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, T., E. Evdokimov, K. Yiadom, Z. Yan, P. B. Chock, and D. C. H. Yang. 2003. Biotin-ubiquitin tagging of mammalian proteins in Escherichia coli. Protein Expr. Purif. 30: 140-149. [DOI] [PubMed] [Google Scholar]
- 60.Warner, J. R., and R. Soeiro. 1967. Nascent ribosomes from HeLa cells. Proc. Natl. Acad. Sci. USA 58: 1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welsh, K. M., K. A. Trach, C. Folger, and J. A. Hoch. 1994. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 176: 7161-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]