Abstract
Misregulation of NF-κB signaling leads to infectious, inflammatory, or autoimmune disorders. IκB kinase β (IKKβ) is an essential activator of NF-κB and is known to phosphorylate the NF-κB inhibitor, IκBα, allowing it to undergo ubiquitin-mediated proteasomal degradation. However, beyond IκBα, few additional IKKβ substrates have been identified. Here we utilize a peptide library and bioinformatic approach to predict likely substrates of IKKβ. This approach predicted Ser381 of the K63 deubiquitinase A20 as a likely site of IKKβ phosphorylation. While A20 is a known negative regulator of innate immune signaling pathways, the mechanisms regulating the activity of A20 are poorly understood. We show that IKKβ phosphorylates A20 in vitro and in vivo at serine 381, and we further show that this phosphorylation event increases the ability of A20 to inhibit the NF-κB signaling pathway. Phosphorylation of A20 by IKKβ thus represents part of a novel feedback loop that regulates the duration of NF-κB signaling following activation of innate immune signaling pathways.
NF-κB signaling drives much of the initial acute inflammatory response to innate immune stimulation. Cells of the innate immune system express membrane-bound Toll-like receptors (TLRs) and intracellular pattern recognition receptors, such as the NOD proteins and RIG-I, all of which are responsible for recognizing specific components of bacterial, viral, or fungal organisms (21, 30, 31). Upon activation of these receptors, cells of the innate immune system activate divergent signal transduction pathways that ultimately converge to activate the IκB kinase (IKK) complex (11, 30). This complex consists, in its simplest form, of the kinases IKKα and IKKβ bound together by NEMO (IKKγ). Upon activation, this kinase complex phosphorylates IκBα, the inhibitor of NF-κB. This phosphorylation event leads to IκBα's ubiquitination and proteasomal degradation, allowing NF-κB transcription factors to stimulate the transcription of inflammatory genes (11). While these initial mechanisms of NF-κB activation have been well studied, the mechanisms by which NF-κB is downregulated are less well understood.
The numerous signaling pathways causing NF-κB activation must be tightly regulated. Failure to mount a sufficient immune response results in susceptibility to infectious disease, and failure to properly downregulate NF-κB results in autoimmune or inflammatory disorders (11). Numerous studies have demonstrated that diverse inflammatory pathways are regulated in large part by coordinated phosphorylation and ubiquitination events (6, 16, 24, 28). The phosphorylation- and ubiquitin-mediated degradation of IκBα is the most well-known example of this coordination. In addition, TLR and RIP2/NOD2 activation results in the K63-linked ubiquitination of TRAF6 and NEMO (1, 2, 7, 30). Unlike K48-linked polyubiquitin chains, which target a protein for proteasomal degradation, the nondegradative K63-linked polyubiquitinated chains are thought to nucleate a signaling complex that activates the kinases TAK1 and IKKα/β (6, 16). In a similar way, K63-linked ubiquitination of TRAF2 and RIP1 is responsible for activating the IKK complex following activation of the tumor necrosis factor (TNF) receptor by TNF-α (6).
Ubiquitination is a rapidly reversible modification, as deubiquitinating enzymes such as A20 (TNFAIP3) decrease the amplitude of ubiquitin-mediated NF-κB signaling (9, 29, 35). A20 contains a deubiquitinase domain at its N terminus and a series of seven zinc fingers at its C terminus (5, 9, 35). These C-terminal zinc fingers mediate K48-linked polyubiquitination. A20 deubiquitinates K63-polyubiquitinated proteins and then helps to conjugate K48-linked polyubiquitin chains such that the protein is targeted for degradation (5, 35). K63-linked polyubiquitination of many NF-κB regulators, such as RIP, TRAF6, TRAF2, and NEMO, positively regulates NF-κB activity (6, 16), and A20 has been shown to interact with and/or deubiquitinate the same proteins to ultimately inhibit the NF-κB signaling pathway (12, 29, 35, 37). The importance of A20 in the downregulation of inflammatory signaling pathways is further illustrated by A20−/− mice, which show uncontrolled whole-body inflammation and hypersensitivity to TNF-α and lipopolysaccharide (LPS) (5, 18). A20 is known to be transcriptionally upregulated following activation of NF-κB, but mechanisms regulating the intrinsic enzymatic activity of A20 have not been reported (17, 23).
IKKβ is a serine/threonine kinase that is essential for the activation of the canonical NF-κB signaling pathway. Its activation of NF-κB is important for cell survival pathways, and a role in insulin signaling pathways was suggested following a report that IKKβ can phosphorylate IRS-1 (10, 25). IKKβ has also been shown to have a role in the development of a number of inflammation-related cancers (reviewed in references 3 and 15). While several IKKβ substrates have been recognized, both the importance of IKKβ and the breadth of signaling pathways it affects suggest that the complete repertoire of intracellular substrates has yet to be identified.
Given this possibility, we developed and utilized an unbiased method to identify novel substrates of IKKβ. The optimal phosphorylation motif for IKKβ was identified using a positional scanning peptide library technique. A combination of several bioinformatic approaches was then used to identify the most likely candidate IKKβ substrates, and the deubiquitinating enzyme A20 was identified as a putative substrate of IKKβ. Serine 381 (S381) on A20 was identified as a target phosphorylation site for IKKβ and was verified as an in vitro and in vivo IKKβ phosphorylation site. We further found that phosphorylation of this site increases the ability of A20 to downregulate NF-κB in response to multiple stimuli. These findings show that phosphorylation of A20 by IKKβ modulates NF-κB signaling and represents a novel mechanism of regulation within innate immune signaling pathways.
MATERIALS AND METHODS
Cell culture, transfection, immunoprecipitation, and Western blotting.
HEK-293T cells were obtained from ATCC and were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). THP-1 cells were obtained from ATCC and were grown in RPMI containing 10% heat-inactivated FBS. mIMCD3 cells were obtained from ATCC and were grown in DMEM-F-12 medium (50%/50%) containing 10% FBS. A20−/− murine embryonic fibroblasts (MEFs) were grown in DMEM containing 10% FBS. NEMO-null MEFs were generous gifts from Tak Mak (Toronto, Ontario, Canada) and Michael Karin (UCSD) and were used as described previously (2). mIMCD3 cells were transduced with retrovirus produced from pBabe-Flag-A20, pBabe-Flag-A20 S381A, or empty pBABE vector. Stable pools were selected with 2 μg/ml puromycin. A20−/− MEFs were transduced with retrovirus produced from pBabe-Flag-A20, pBabe-Flag-A20 S381A, or empty pBABE vector control, and stable pools were selected with 2 μg/ml puromycin. Transfection was performed by calcium phosphate precipitation. For preparation of recombinant glutathione S-transferase-IKKβ (GST-IKKβ) and GST-IKKβ K44A and for preparation of all immunoprecipitates used in kinase assays, cells were lysed in 50 mM Tris (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 1 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium orthovanadate, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 10 nM calyculin A. For immunoprecipitation-ubiquitination assays, cells were lysed in high-salt modified radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 1 M NaCl, 1% NP-40, 0.25% deoxycholic acid, 1 mM EDTA, 1 mM EGTA, 1 mM β-glycerophosphate, 5 mM iodoacetamide, 1 mM PMSF, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 10 nM calyculin A). THP-1 cells, reconstituted A20−/− MEFs, or mIMCD-3 cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.25% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA, 1 mM EGTA, 1 mM β-glycerophosphate, 1 mM PMSF, 1 mM sodium orthovanadate, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and10 nM calyculin A).
Antibodies, plasmids, and reagents.
Anti-Myc (9E10), anti-RIP2, and anti-OMNI antibodies were obtained from Santa Cruz Technology. Anti-A20 (mouse monoclonal antibody) was obtained from Imgenex. Anti-Myc (rabbit), anti-GST (mouse), anti-phospho-IKKα/β, anti-phospho-IκBα, anti-IKKα, anti-IKKβ, anti-IκBα, and anti-NEMO antibodies were obtained from Cell Signaling Technology. Anti-Flag (M2) and antihemagglutinin (anti-HA; HA-11) were obtained from Covance. Antiactin was obtained from Abcam. Ultrapure Escherichia coli LPS was obtained from Invivogen. Recombinant murine TNF-α was obtained from R&D Systems, Inc. BMS-345541 was obtained from EMD Biosciences. OMNI-RIP2, HA-ubiquitin, Myc-NEMO, Myc-NEMO K399R, HA-NOD2, HA-RIP2, and XP-TRAF6 were used as previously described (1). Flag-A20 was a generous gift from Alex Toker (Harvard). GST-IKKβ was generated by PCR cloning into the BamHI and NotI sites of pEBG. A20 S381A and GST-IKKβ K44A were generated using a modification of the QuikChange protocol (Stratagene). pBabe-Flag-A20 was generated by subcloning into the EcoRI site of pBabe.
Positional scanning peptide library assay.
Recombinant GST-IKKβ and GST-IKKβ K44A were generated as described above, and a positional scanning peptide library assay was carried out as described previously (14, 32). The kinase buffer contained 50 mM Tris (pH 7.5), 12 mM MgCl2, 1 mM β-glycerophosphate, 0.1% Tween, 100 μM ATP, and 5 μCi [γ-32P]ATP/reaction mix. Kinase reaction mixes were incubated at 30°C for 2 h.
In vitro kinase assays.
Recombinant GST-IKKβ and immunoprecipitated Flag-A20 were generated as described above. The kinase buffer contained 50 mM Tris (pH 7.5), 12 mM MgCl2, 1 mM β-glycerophosphate, 100 μM ATP, and 10 μCi [γ-32P]ATP/reaction mix. Reaction mixes were incubated at 30°C for 1 h.
MS.
Proteins from gel bands were reduced with 10 mM dithiothreitol (Sigma, St. Louis, MO) and cysteine residues were alkylated with 55 mM iodoacetamide (Sigma, St. Louis, MO), and the proteins were digested with sequencing-grade modified trypsin from Promega (Madison, WI) overnight at 37°C. Extracted tryptic peptides were analyzed by data-dependent reversed-phase microcapillary liquid chromatography-tandem mass spectrometry (LC/MS/MS), using an LTQ 2D linear-ion-trap mass spectrometer (ThermoScientific, San Jose, CA) operated in positive-ion mode at a flow rate of 250 nl/min. A 75-μm (inner diameter) by 15-μm (inner diameter of the tip) PicoFrit microcapillary column (New Objective, Woburn, MA) was self-packed with Magic C18 resin (Michrom Bioresources, Auburn, CA) to 10 cm (length). The column was equilibrated, and peptides were loaded using buffer A (0.1% acetic acid-0.9% acetonitrile-99% water) and then eluted with a gradient from 5% buffer B (acetonitrile) to 38% buffer B, followed by 95% buffer B for washing. The Sequest algorithm in Proteomics Browser software (ThermoScientific, San Jose, CA) was used for database searching of all MS/MS spectra against a reversed NCBInr protein database with the differential modifications oxidation of Met and phosphorylation of Ser, Thr, and Tyr. Spectra that matched phosphorylated residues in the A20 sequence were first identified by the Sequest correlation score and further validated by manual inspection of the fragment ions. The interpretation of the exact site of phosphorylation was aided by the GraphMod algorithm in Proteomics Browser software, which individually scores the modification on each residue in the peptide.
Reporter assays.
Six-well dishes of HEK293T cells were transfected using polyethylenimine with an NF-κB luciferase reporter (Clontech), Renilla luciferase, and the indicated constructs. Transfection efficiency was standardized using Renilla luciferase, and assays were performed according to the manufacturer's instructions (Promega).
Generation of anti-phospho-A20 S381 antibody.
Two rabbits were immunized with the following peptide: Ac-EPSVPQL(phospho-S)LMDVK-Ahx-C-amide (Ahx = aminohexanoic acid, a C6 spacer). The terminal bleeds were depleted over a nonphosphopeptide column and then subjected to three successive purifications over a phosphopeptide column. Enzyme-linked immunosorbent assay analysis showed a >1,000-fold preference for phosphorylated peptide over nonphosphopeptide. This antibody was produced under contract by 21st Century Biochemicals.
RESULTS
Within innate immune signaling pathways, divergent stimuli converge onto the IKK signalosome. Upon activation, IKKβ activates NF-κB transcription factors via the phosphorylation and proteasomal degradation of IκBα (reviewed in reference 11). Little is known about the preferred substrate specificity of IKKβ, and few in vivo substrates have been identified. To address this limitation, we undertook a combined proteomic and bioinformatic approach to perform an unbiased search for novel IKKβ substrates. First, a positional scanning peptide library technique that we recently developed (14, 32) was used to identify the optimal phosphorylation motif for IKKβ. This technique utilizes a set of biotinylated peptide libraries, each of which contains a 50-50 mixture of serine and threonine fixed at the central position. In each library, one position relative to the central Ser/Thr is fixed with one of the 20 amino acids or with phosphothreonine (included to facilitate the identification of kinases which require priming by prior phosphorylation events). All other positions in each library contain an equimolar mixture of natural amino acids (excluding serine, threonine, and cysteine). Recombinant GST-IKKβ was purified from transfected HEK293T cells, and kinase assays were carried out on all 189 libraries simultaneously in solution, using [γ-32P]ATP. Biotinylated peptides were captured using a streptavidin-coated membrane. For each position relative to the phosphorylation site, the relative amino acid preference was determined by the relative level of 32P radiolabel incorporation into the corresponding peptide.
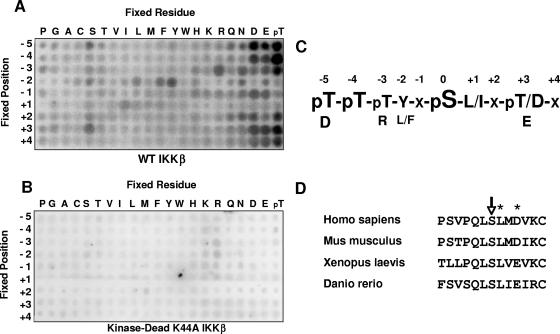
Using this method, we found that IKKβ exhibited significant sequence specificity at multiple positions relative to the phosphorylation site. The kinase strongly prefers peptides that contain phosphothreonine (and, presumably, phosphoserine) at the −4 and −5 positions relative to the phosphorylation site (Fig. 1A). Strong selectivity was also observed for aromatic residues at the −2 position, for hydrophobic residues at the +1 position, and for acidic residues at the +3 position (Fig. 1A and C). A kinase-deficient mutant preparation of IKKβ K44A did not phosphorylate any of the peptides, indicating that these results were not due to a contaminating kinase present in the IKKβ kinase preparation (Fig. 1B). This motif correlates well with several known IKKβ phosphorylation sites, including Ser32 and Ser36 of IκBα (DDRHDS32GLDS and DpSGLDS36MKDE, respectively) as well as Ser536 of the NF-κB transcription factor p65 (DEDFSS536IADM) (8, 20, 27).
FIG. 1.
Determination of preferred phosphorylation motif of IKKβ. (A and B) Recombinant IKKβ or, as a control, kinase-dead IKKβ K44A was used to phosphorylate 189 peptide libraries in individual kinase assays. The general sequence for these libraries was Y-A-X-X-X-Z-X-S/T-X-X-X-X-A-G-K-K-biotin (Z, fixed amino acid; X, equimolar mixture of amino acids). After binding of the libraries to a streptavidin-coated membrane, phosphorylation was visualized by the incorporation of 32P. While IKKβ (A) gave a strong, consistent motif, no motif could be identified using IKKβ K44A (B). (C) Primary and secondary selections determined from IKKβ phosphorylation motif. (D) Evolutionary conservation of A20 S381 and its surrounding sequence.
Bioinformatic approaches were then used to identify candidate IKKβ substrates. First, spot intensities from the peptide library screen were quantified and converted into a matrix that could be used within the signaling search engine Scansite (data not shown). Scansite (http://scansite.mit.edu) allows proteome-wide searches as well as searches within individual proteins for residues which best match the data provided by the matrix (22, 36). Proteome-wide searches frequently yield more candidates than can reasonably be tested, and results include numerous false-positive data (J. E. Hutti et al., unpublished observations). Because of this, we also used the IKKβ matrix to individually search proteins known to be involved in innate immune signaling for sequences that match the IKKβ phosphorylation motif. Once a list of candidate phosphorylation sites was identified, NetPhos (http://www.cbs.dtu.dk/services/NetPhos/) was used to determine whether these sites are likely to be surface exposed and thus potentially phosphorylated in vivo (4). Finally, the evolutionary conservation of candidate sites, in the context of conserved surrounding residues consistent with the optimal motif of IKKβ, was considered in determining the most likely candidate substrates. For instance, potential phosphorylation sites had to be conserved among all sequenced mammalian genomes and, preferably, among those of lower organisms.
Among the many candidate phosphorylation sites analyzed, Ser381 of A20 (TNFAIP3) emerged as a likely IKKβ phosphorylation site. A20 is a known negative regulator of NF-κB activity and has both K63 deubiquitinase and K48 E3 ligase activities (35). Ser381 of this enzyme received a Scansite score comparable to those of known IKKβ phosphorylation sites, and NetPhos predicts that in vivo phosphorylation of Ser381 is very likely (Table 1). In addition, Ser381, as well as the +1 leucine and +3 acidic residues, is evolutionarily conserved in mammals, in Xenopus laevis, and in Danio rerio, the lowest organism in which A20 is found (Fig. 1D).
TABLE 1.
Scansite and NetPhos scores for IKKβ substratesa
| IKKβ substrate | Site | Scansite score | NetPhos score |
|---|---|---|---|
| IκBα | S32 | 0.552 | 0.995 |
| S36 | 0.575 | 0.998 | |
| RelA (p65) | S536 | 0.513 | 0.962 |
| NEMO | S376 | 1.047 | 0.008 |
| FOXO3a | S644 | 0.670 | 0.017 |
| IRS-1 | S312 | 1.113 | 0.919 |
| BCL-10 | S134 | 0.950 | 0.926 |
| S136 | 0.719 | 0.987 | |
| S138 | 0.662 | 0.995 | |
| S141 | 0.757 | 0.922 | |
| S144 | 0.711 | 0.915 | |
| Dok1 | S439 | 1.217 | 0.179 |
| S443 | 0.938 | 0.304 | |
| S446 | 1.127 | 0.030 | |
| S450 | 0.889 | 0.013 | |
| A20 | S381 | 1.043 | 0.973 |
Scansite scores for each protein were determined using a matrix created from IKKβ specificity values (data not shown). A lower score represents a better match to the optimal motif. NetPhos provides scores ranging from 0 to 1. Residues receiving NetPhos scores above 0.5 are considered potential phosphorylation sites, with higher scores representing the most likely sites of phosphorylation. A20 S381 received Scansite and NetPhos scores comparable to those of known IKKβ phosphorylation sites (10, 13, 15, 19, 20, 26, 27, 33).
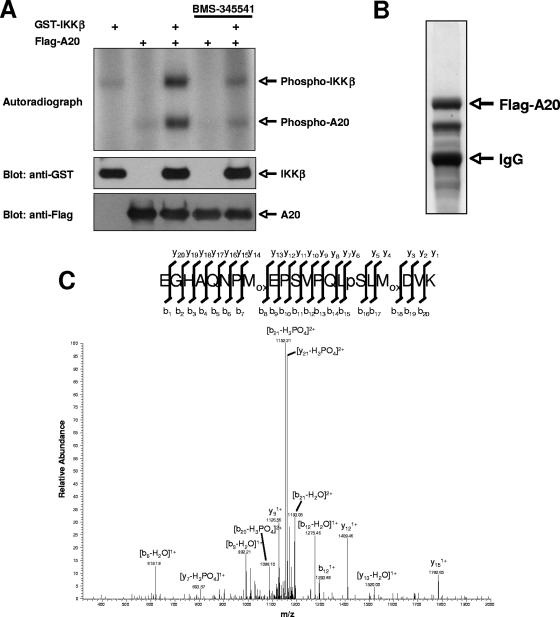
In order to verify that A20 could be phosphorylated directly by IKKβ, Flag-tagged A20 immunoprecipitated from HEK293T cells was incubated with purified GST-IKKβ and [γ-32P]ATP. We found that IKKβ could phosphorylate A20 in vitro and that phosphorylation was blocked by pretreatment with the IKKβ-specific inhibitor BMS-345541 (Fig. 2A). To identify the residue(s) on A20 that is phosphorylated by IKKβ, GST-IKKβ was cotransfected with Flag-A20 into HEK293T cells. Flag-A20 immunoprecipitates were subjected to stringent washing (1% NP-40, 0.25% deoxycholic acid, 1 M NaCl), followed by SDS-polyacrylamide gel electrophoresis and Coomassie staining (Fig. 2B). The stained band corresponding to A20 was subjected to tryptic digestion followed by reversed-phase microcapillary LC/MS/MS. This analysis identified phosphorylation of A20 at Ser381 (Fig. 2C), the same site that was predicted by our bioinformatic analysis.
FIG. 2.
IKKβ phosphorylates A20 on serine 381 in vitro. (A) Flag-A20 was transfected into HEK293T cells and immunoprecipitated using an anti-Flag antibody. Immunoprecipitates were incubated with purified GST-IKKβ in the presence of BMS-345541 (3 μM) or vehicle control for 20 min at room temperature. [γ-32P]ATP was then added to the kinase reaction mix, and an in vitro kinase assay was performed. (B) IKKβ was cotransfected into HEK293T cells with Flag-A20. Flag-A20 was stringently immunopurified and subjected to SDS-polyacrylamide gel electrophoresis. Coomassie staining identified the purified Flag-A20, which was then excised from the gel and submitted for analysis by LC/MS/MS. (C) LC/MS/MS identified phosphorylation of A20 at serine 381.
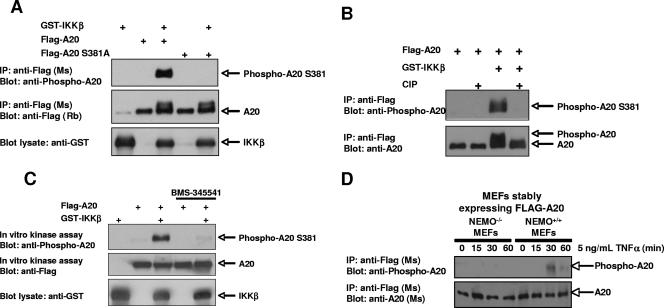
To determine whether this phosphorylation event occurs in vivo, we generated a phospho-specific antibody against this site. The specificity of the antibody was determined by cotransfecting GST-IKKβ with Flag-A20 or the point mutant Flag-A20 S381A. Flag immunoprecipitates were subjected to immunoblotting with the phospho-specific antibody. The antibody recognized wild-type (WT) A20 coexpressed with IKKβ but did not recognize the A20 S381A mutant (Fig. 3A). In addition, the phospho-A20 S381 antibody did not recognize A20 that had been treated with calf intestinal phosphatase, confirming that the antibody is specific for phosphorylated A20 (Fig. 3B).
FIG. 3.
Anti-phospho-A20 S381 antibody recognizes A20 which is phosphorylated by IKKβ. (A) A phospho-specific antibody directed against A20 phospho-S381 was generated. To show specificity, Flag-A20 was cotransfected into HEK293T cells with GST-IKKβ and immunoprecipitated using an anti-Flag antibody. Immunoprecipitates were immunoblotted using the phospho-specific antibody for A20 S381. (B) Flag-A20 was cotransfected with GST-IKKβ into HEK293T cells. Lysates were subjected to immunoprecipitation using an anti-Flag antibody. Immunoprecipitates were treated with calf intestinal phosphatase (CIP) for 45 min at 37°C and then immunoblotted using the anti-phospho-A20 S381 antibody. The phospho-A20 S381 antibody no longer recognized calf intestinal phosphatase-treated A20. (C) Flag-A20 was cotransfected into HEK293T cells with GST-IKKβ. Cells were pretreated with 10 μM BMS-345541 or vehicle control for 3 h prior to lysis in order to facilitate dephosphorylation of A20 in the absence of IKKβ activity. Anti-Flag immunoprecipitates were washed four times with lysis buffer to remove BMS-345541 and three times with kinase buffer. Immunoprecipitates were then incubated a second time with 3 μM BMS-345541 or vehicle control for 20 min at room temperature in order to inhibit coprecipitating IKKβ activity, and an in vitro kinase assay was performed. Immunoblot analysis using the phospho-A20 S381 antibody showed in vitro phosphorylation of A20 S381 by coprecipitating IKKβ. (D) NEMO−/− or isogenic NEMO+/+ cells were transduced with pBABE-A20 retrovirus. Cells were selected in puromycin, and clones (>1,000) were pooled. Cells were treated with TNF-α (5 ng/ml) for the indicated times. Cells were then lysed, and A20 was immunoprecipitated by virtue of its Flag tag. Western blotting showed that in NEMO+/+ cells, A20 was phosphorylated in a TNF-α-dependent manner, while in NEMO−/− cells, A20 showed very little TNF-α-dependent phosphorylation.
Although these data strongly suggested that IKKβ phosphorylates A20 at S381, it was still possible that phosphorylation of A20 S381 is not direct. To address this possibility, a second in vitro kinase assay was performed. In this experiment, GST-IKKβ and Flag-A20 were cotransfected into HEK293T cells. Three hours prior to lysis, cells were treated with BMS-345541 to allow dephosphorylation of A20 in the absence of IKKβ activity. Following lysis, Flag-A20 was immunoprecipitated, and the immunoprecipitates were washed twice with lysis buffer and twice with kinase buffer. Immunoprecipitates were then resuspended in kinase buffer and preincubated for 20 min with BMS-345541 or vehicle control. ATP was then added to each reaction mix. Because no exogenous IKKβ was added following immunoprecipitation, all A20 phosphorylation occurring during this assay must be due to coprecipitating IKKβ. In accordance with this, coimmunoprecipitation experiments indicated that kinase-dead IKKβ can interact with A20 (data not shown). Following completion of the kinase reaction, the reaction mixes were immunoblotted using the phospho-A20 S381 antibody. Under these conditions, A20 was phosphorylated at position S381 and this phosphorylation was blocked by the IKKβ inhibitor BMS-345541, indicating that the kinase that phosphorylates this site coimmunoprecipitates with A20 and is inhibited by BMS-345541 (Fig. 3C). As a last test to determine the IKK dependence of this phosphorylation, NEMO+/+ and NEMO−/− MEFs were transduced with a retrovirus designed to express WT A20. After puromycin selection, clones (>1,000) were pooled and exposed to TNF-α. Lysates were generated, and A20 was immunoprecipitated through its Flag tag. Western blotting showed that in NEMO+/+ MEFs, S381 of A20 was phosphorylated, while in NEMO−/− MEFs (with no TNF-induced IKK activity [reviewed in references 11 and 28]), the S381A position of A20 was not phosphorylated (Fig. 3D). The results shown in Fig. 3C and D, coupled with the results from Fig. 2A, argue that IKKβ can directly phosphorylate A20 at S381.
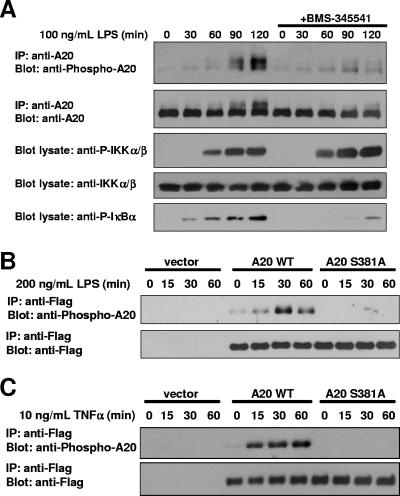
Because A20 is a negative regulator of the NF-κB pathway (5, 9, 35) and because A20−/− mice are characterized by extreme hypersensitivity to LPS and TNF-α (18), we hypothesized that A20 phosphorylation might be involved in the immune response to these stimuli. To test this possibility, partially differentiated THP-1 monocytes were pretreated for 3 h with BMS-345541 or vehicle control and then stimulated with LPS for 0 to 120 min. Endogenous A20 was immunoprecipitated, and the immunoprecipitates were immunoblotted using the phospho-A20 S381 antibody (Fig. 4A). While a strong signal was observed with the phospho-A20 S381 antibody at 90 and 120 min, inhibiting endogenous IKKβ activity with BMS-345541 pretreatment prevented a significant portion of this phosphorylation. To verify these results in a separate LPS-responsive cell line, mIMCD3 cells were transduced with a retrovirus expressing Flag-A20 or Flag-A20 S381A or with a vector control, and stable pools of A20-expressing cells were selected. Cells were stimulated with LPS for 0 to 60 min. Following LPS stimulation, Flag-A20 was immunoprecipitated, and immunoprecipitates were again immunoblotted using the phospho-A20 S381 antibody. In cells transduced with WT A20, a strong signal was observed with the phospho-specific antibody. However, this signal was not observed in cells expressing A20 S381A or the vector control (Fig. 4B). A similar result was obtained following stimulation of the same A20-expressing mIMCD3 cells with TNF-α (Fig. 4C). Together, these data show that A20 is phosphorylated at S381 in multiple cell types (IMCD cells are epithelial, THP-1 cells are hematopoietic, and MEFs are mesenchymal) following stimulation with IKKβ agonists.
FIG. 4.
IKKβ phosphorylates A20 S381 in a stimulus- and time-dependent manner. (A) To induce expression of endogenous A20, THP-1 cells were differentiated for 16 h with 20 nM phorbol myristate acetate. These partially differentiated THP-1 cells were pretreated for 3 h with 10 μM BMS-345541 or vehicle control. Ultrapure E. coli LPS (100 ng/ml) was added for the indicated times. Endogenous A20 was immunoprecipitated, and immunoprecipitates were subjected to immunoblot analysis using a phospho-A20 S381 antibody. Lysates were immunoblotted for phospho-IκBα (S32/S36) to verify IKKβ inhibition by BMS-345541. (B and C) An LPS-responsive kidney epithelial cell line, mIMCD3, was stably transduced with Flag-A20, Flag-A20 S381A, or empty vector control. Cells were stimulated with 200 ng/ml LPS (B) or 10 ng/ml TNF-α (C) for the indicated times. Flag-A20 was immunoprecipitated using an anti-Flag antibody and immunoblotted using a phospho-A20 S381 antibody.
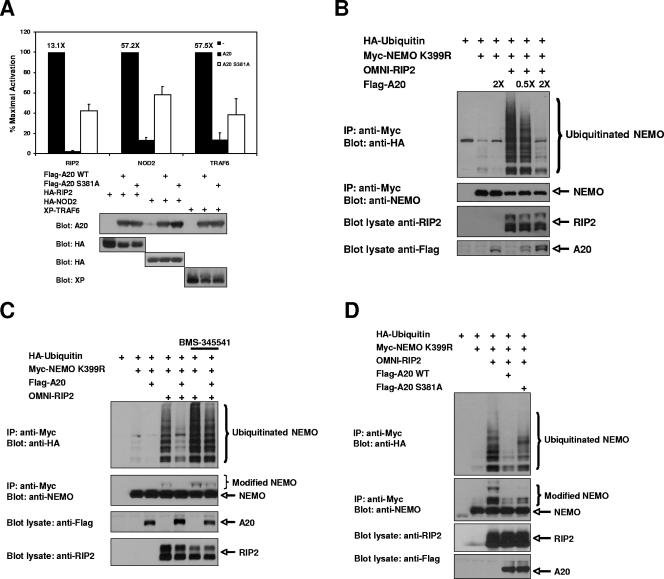
The phosphorylation of A20 by IKKβ could either enhance or inhibit the ability of A20 to downregulate NF-κB. To address this question, we first performed a luciferase reporter assay using an NF-κB-driven promoter transfected with proteins known to strongly induce K63-linked polyubiquitination of components of the NF-κB pathway (RIP2, NOD2, and TRAF6) (1, 2, 7, 30). While overexpression of these proteins gave various degrees of NF-κB activation, the NF-κB activity driven by either RIP2, NOD2, or TRAF6 was almost completely abrogated by coexpression of A20 (Fig. 5A). However, cotransfection of A20 S381A led to dramatically increased NF-κB reporter activation compared to that by WT A20 (Fig. 5A). This observation suggested that IKKβ phosphorylation of A20 at S381 results in increased A20 activity, which might provide a feedback mechanism to prevent uncontrolled inflammation.
FIG. 5.
Phosphorylation of A20 S381 increases the ability of A20 to inhibit the NF-κB response. (A) HEK293T cells were transfected with an NF-κB-luciferase reporter gene, cytomegalovirus-driven Renilla luciferase (to standardize transfection efficiency), and either HA-RIP2, HA-NOD2, or XP-TRAF6 alone or in combination with WT A20 or A20 S381A. An NF-κB-luciferase reporter assay was performed at 24 h posttransfection. Each experiment was performed four times with similar results. Levels of activation are shown over the bars. Error bars represent standard deviations. WT A20 strongly inhibited the NF-κB response induced by RIP2, NOD2, or TRAF6, while A20 S381A showed a significant reduction in the ability to inhibit the NF-κB response. Equivalent protein expression of each of the transfected genes is also shown (bottom panels). (B) HEK293T cells were transfected with Myc-NEMO K399R, HA-ubiquitin, OMNI-RIP2, and increasing amounts of A20. Lysates were subjected to immunoprecipitation using an anti-Myc antibody and were immunoblotted with an anti-HA antibody to identify RIP2-induced NEMO ubiquitination at K285. RIP2-induced NEMO ubiquitination decreased with increasing amounts of A20. (C) HEK293T cells were transfected with Myc-NEMO K399R and HA-ubiquitin in combination with OMNI-RIP2 and Flag-A20. Cells were pretreated with 10 μM BMS-345541 or vehicle control for 6 h prior to lysis. Lysates were subjected to immunoprecipitation using an anti-Myc antibody, washed stringently (1% NP-40 plus 0.25% deoxycholate plus 1 M NaCl), and immunoblotted with an anti-HA antibody to identify RIP2-induced NEMO ubiquitination of K285. A20-expressing cells pretreated with the IKKβ inhibitor showed significantly larger amounts of NEMO ubiquitination. (D) HEK293T cells were transfected with Myc-NEMO K399R, HA-ubiquitin, OMNI-RIP2, and either Flag-A20 WT or Flag-A20 S381A. Lysates were subjected to immunoprecipitation with an anti-Myc antibody, washed stringently (1% NP-40 plus 0.25% deoxycholate plus 1 M NaCl), and immunoblotted with an anti-HA antibody to identify RIP2-induced NEMO ubiquitination of K285. A20 S381A showed increased NEMO ubiquitination compared to WT A20.
Activation of the Crohn's disease susceptibility complex, NOD2/RIP2, or the Toll-like receptor-activated E3 ubiquitin ligase TRAF6 leads to the K63-linked ubiquitination of NEMO at K285 (1, 2, 30), and these K63-linked ubiquitin chains play an important role in coordinating innate immune signaling pathways (6, 16). Consistent with this, K63-linked ubiquitination of NEMO K285 is known to be essential for full IKKβ activity (1, 2). Since A20 is a K63 deubiquitinase, we decided to test whether A20 could suppress ubiquitination of NEMO. HA-ubiquitin, Myc-NEMO K399R, and OMNI-RIP2 were cotransfected into HEK293T cells, and the resulting NEMO ubiquitination produced was consistent with previously published data (1). However, the addition of increasing amounts of A20 resulted in decreased RIP2-induced NEMO ubiquitination (Fig. 5B). NEMO K399R was used because elimination of the background ubiquitination at K399 facilitates the observation of NEMO K285 ubiquitination (1), and these immunoprecipitations were all performed in high-stringency wash buffer (1 M NaCl). In addition, denaturing the lysate prior to immunoprecipitation to eliminate NEMO-binding proteins from the immunoprecipitates gave similar results (data not shown), indicating that the ubiquitination witnessed was due to NEMO ubiquitination and not to a coprecipitating protein. If phosphorylation of A20 by IKKβ increases A20 activity, then inhibition of IKKβ would lead to decreased A20 activity. This hypothesis was tested by observing decreases in the level of RIP2-induced NEMO ubiquitination as a readout for A20 activity. HA-ubiquitin, OMNI-RIP2, and Flag-A20 were cotransfected into 293T cells with Myc-NEMO K399R. Cotransfection of Myc-NEMO K399R with OMNI-RIP2 induced the expected increase in ubiquitination of NEMO at K285, and the addition of Flag-A20 eliminated a large portion of this ubiquitination (Fig. 5C). However, pretreating cells with BMS-345541 prior to lysis caused a large increase in the amount of NEMO ubiquitination, even in the presence of A20. Under these conditions, IKKβ activity was strongly inhibited by BMS-345541, as measured by IκBα degradation (data not shown). Consistent with the findings shown in Fig. 5C, when Flag-A20 S381A replaced WT Flag-A20, the amount of NEMO ubiquitination also increased dramatically (Fig. 5D). Collectively, these findings support the hypothesis that IKKβ activity and phosphorylation of A20 at Ser 381 are required for full A20 activity.
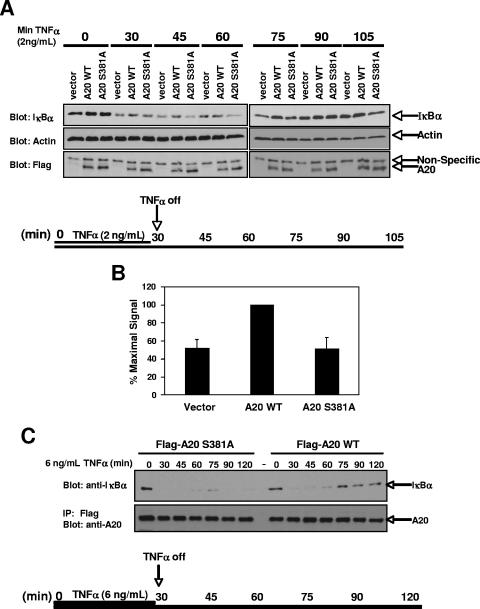
In order to determine whether phosphorylation of A20 at S381 results in altered signaling following inflammatory stimuli in a more endogenous setting, A20−/− MEFs were reconstituted with Flag-A20 WT, Flag-A20 S381A, or empty vector control. As described by Werner et al. (34), reconstituted MEFs were pulsed with a nonsaturating level (2 ng/ml) of TNF-α for 30 min, and then the TNF-α-containing growth medium was replaced with serum-free medium after extensive phosphate-buffered saline (PBS) washing of the cells (Fig. 6A). By then assaying for recovery of IκBα via NF-κB-induced transcriptional activation, these conditions have been shown to assay the NF-κB-dependent negative regulation of TNF-induced signaling (34). Cells were lysed every 15 min following removal of the TNF-α, and the recovery of IκBα levels was followed by immunoblotting. Between 30 and 75 min, we observed consistently higher levels of IκBα in cells reconstituted with WT A20 than in those with vector control (Fig. 6A). Previous studies comparing WT and A20−/− MEFs have shown that this increase is due to the inhibition of NF-κB-activating pathways by A20 and therefore to a more rapid recovery of IκBα levels following TNF-α removal (18, 34). In our studies, the differences in IκBα levels observed between A20−/− MEFs and A20-reconstituted MEFs are comparable to those observed previously using a similar time course (34). Importantly, cells reconstituted with A20 S381A showed a time course of IκBα degradation and resynthesis matching that of vector-transduced cells, supporting the model that phosphorylation of A20 S381 is required for full A20 activity. To quantify this activity, the TNF-α stimulation was repeated for the 45-min time point in four separate experiments (Fig. 6B). At 45 min, the amount of IκBα present in the A20 S381A-transduced A20−/− MEFs matched the amount in the vector-transduced cells and was significantly decreased relative to that in the WT A20-transduced MEFs. At a higher dose of TNF-α (6 ng/ml), this effect was even more pronounced, with very little IκBα produced in the A20-null MEFs reconstituted with A20 S381A (Fig. 6C). The findings presented in Fig. 5 and 6 strongly suggest that IKKβ phosphorylates A20 to increase its activity, thereby helping to limit the duration of NF-κB activity.
FIG. 6.
Phosphorylation of A20 S381 leads to rapid inhibition of NF-κB following LPS stimulation. (A) A20−/− MEFs were stably reconstituted with Flag-A20 WT, Flag-A20 S381A, or vector control and pulsed with 2 ng/ml TNF-α for 30 min. After extensive PBS washes, the TNF-α-containing medium was replaced with serum-free medium and the recovery of IκBα levels was observed over time via immunoblotting of lysates. (B) Reconstituted A20−/− MEFs were pulsed with 2 ng/ml TNF-α for 30 min, the medium was replaced as described above, and cells were allowed to recover for 15 min. Lysates were immunoblotted using an anti-IκBα antibody, and differences were calculated between IκBα levels in WT A20-reconstituted versus A20 S381A- or vector-reconstituted MEFs. The experiment was repeated four times with similar results. Error bars show standard deviations. (C) Stably reconstituted A20−/− MEFs were pulsed with a higher dose (6 ng/ml) of TNF for 30 minutes. After extensive PBS washes, the TNF-containing medium was replaced with serum-free medium and the recovery of IκBα levels was assayed via Western blotting.
DISCUSSION
The innate immune system is responsible for both the initiation and termination of cytokine release associated with the recognition of pathogens, and proper coordination of these opposing functions is required to prevent autoimmune or inflammatory disease states. IKKβ is essential for the initiation of proper innate immune signaling through its activation of NF-κB, and this activation results in the transcriptional upregulation of cytokines and other proinflammatory molecules. However, in some signaling pathways, IKKβ is also responsible for NF-κB downregulation. For example, phosphorylation of BCL10 by IKKβ negatively regulates T-cell receptor signaling (33), and many anti-inflammatory molecules, such as IκBα and A20, are transcriptionally upregulated by NF-κB, emphasizing the multiple roles of IKKβ in the maintenance of proper inflammatory responses (17, 23). The strict requirement of IKKβ for proper regulation of innate immune signaling, coupled with its importance in a large number of other signaling pathways, led to the hypothesis that many IKKβ substrates remain unidentified.
To this end, we developed and utilized an unbiased method to identify novel substrates of IKKβ. The optimal phosphorylation motif for IKKβ was identified using a positional scanning peptide library technique. A combination of several bioinformatic approaches was then used to identify the most likely candidate IKKβ substrates. This method led us to the identification of A20 as a putative novel IKKβ substrate (5, 12, 18, 35, 37). A20 is known to be essential for the downregulation of NF-κB signaling, which immediately suggested that its phosphorylation by IKKβ might have an important role in regulating inflammatory responses.
Direct phosphorylation of A20 by IKKβ was confirmed using in vitro kinase assays, and the phosphorylation site (S381) was mapped using MS. A phospho-specific antibody was then used to verify that A20 S381 is in fact phosphorylated following LPS or TNF-α stimulation in three divergent cell lines, namely, the THP-1 monocytic cell line, the mIMCD3 kidney epithelial cell line, and MEFs. Phosphorylation by IKKβ increases A20 activity, which in turn activates a negative feedback pathway to downregulate NF-κB. Since A20 is known to have both K63 deubiquitinase and K48 E3 ligase activities, we investigated the role of A20 phosphorylation in the ubiquitination of NEMO K285, which is known to be important for attaining full IKKβ activity following TLR2, TLR4, and NOD2/RIP2 activation (1, 2). Indeed, experiments showed that a site-directed mutant, A20 S381A, resulted in higher levels of RIP2-induced NEMO ubiquitination than did WT A20. While we have not conclusively determined whether this increase in NEMO ubiquitination is caused by decreased deubiquitinase activity of A20 or decreased A20 E3 ligase activity, either of these mechanisms would be expected to result in the increased NF-κB activity that was observed.
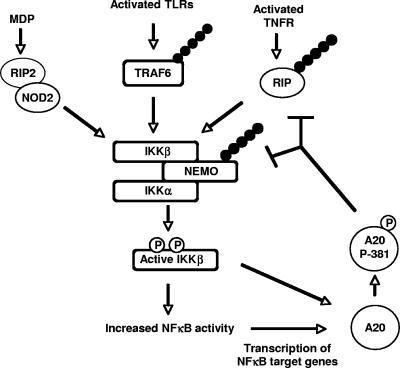
These studies provide evidence that the techniques used here, including determination of a kinase phosphorylation motif followed by bioinformatic identification of candidate substrates, can provide an efficient and unbiased method for identifying novel kinase substrates. In addition, this study has identified a novel mechanism of NF-κB negative feedback. In inactive cells, A20 is very weakly expressed. Upon stimulation with NF-κB agonists, IKKβ is activated, ultimately leading to the NF-κB-dependent transcription of target genes, including A20 (17, 23). If IKKβ activity continues, the newly translated A20 is phosphorylated by IKKβ, causing it to be fully activated. This fully active A20 then helps to more completely inhibit NF-κB signaling (Fig. 7). This sort of dual regulation of A20 activity (via gene transcription and subsequent protein phosphorylation) allows the downregulation of NF-κB activity to be tailored more precisely to the needs of the cell, depending on the strength and duration of the inflammatory signal. Given the importance of K63-linked ubiquitination in innate immune signaling and the dramatic phenotype of the A20−/− mouse (5, 18), it would not be surprising if misregulation of A20 phosphorylation has a role in the development of inflammatory disease states.
FIG. 7.
Model for the role of A20 phosphorylation at Ser381 in the inhibition of NF-κB responses. Recognition of pathogens or proinflammatory cytokines by cells of the innate immune system leads to activation of IKKβ. Active IKKβ leads to the transcription of NF-κB target genes, including A20. If IKKβ activity remains high following A20 translation, IKKβ phosphorylates this newly translated A20 at serine 381. This phosphorylation increases the activity of A20, allowing it to more forcefully downregulate the NF-κB pathway.
Acknowledgments
We thank Alex Toker for providing the Flag-A20 construct, and we thank Michael Karin (UCSD) and Tak Mak (Toronto) for supplying NEMO-null MEFs. We thank members of the Cantley lab for technical assistance and critical comments.
This work was supported by grant 1K08 AI53819-01A1 (D.W.A.), by a Burroughs-Wellcome Career Award for Biomedical Scientists (D.W.A.), by the Cleveland Foundation (D.W.A.), and by NIH grants R01 GM56203 (L.C.C.) and R01 DK071939 (A.M.).
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Abbott, D. W., A. Wilkins, J. M. Asara, and L. C. Cantley. 2004. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14: 2217-2227. [DOI] [PubMed] [Google Scholar]
- 2.Abbott, D. W., Y. Yang, J. E. Hutti, S. Madhavarapu, M. A. Kelliher, and L. C. Cantley. 2007. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol. Cell. Biol. 27: 6012-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basseres, D. S., and A. S. Baldwin. 2006. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene 25: 6817-6830. [DOI] [PubMed] [Google Scholar]
- 4.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294: 1351-1362. [DOI] [PubMed] [Google Scholar]
- 5.Boone, D. L., E. E. Turer, E. G. Lee, R. Ahmad, M. T. Wheeler, C. Tsui, P. Hurley, M. Chien, S. Chai, O. Hitotsumatsu, E. McNally, C. Pickart, and A. Ma. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5: 1052-1060. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z. J. 2005. Ubiquitin signaling in the NFkB pathway. Nat. Cell Biol. 7: 758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351-361. [DOI] [PubMed] [Google Scholar]
- 8.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IkB kinase that activates the transcription factor NFkB. Nature 388: 548-554. [DOI] [PubMed] [Google Scholar]
- 9.Evans, P. C., H. Ovaa, M. Hamon, P. J. Kilshaw, S. Hamm, S. Bauer, H. L. Ploegh, and T. S. Smith. 2004. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 378: 727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, Z., D. Hwang, F. Bataille, M. Lefevre, D. York, M. J. Quon, and J. Ye. 2002. Serine phosphorylation of insulin receptor substrate 1 by inhibitor kB kinase complex. J. Biol. Chem. 277: 48115-48121. [DOI] [PubMed] [Google Scholar]
- 11.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-κB. Genes Dev. 18: 2195-2224. [DOI] [PubMed] [Google Scholar]
- 12.Heyninck, K., and R. Beyaert. 1999. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 442: 147-150. [DOI] [PubMed] [Google Scholar]
- 13.Hu, M. C., D. F. Lee, W. Xia, L. S. Golfman, F. Ou-Yang, J. Y. Yang, Y. Zou, S. Bao, N. Hanada, H. Saso, R. Kobayashi, and M. C. Hung. 2004. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117: 225-237. [DOI] [PubMed] [Google Scholar]
- 14.Hutti, J. E., E. T. Jarrell, J. D. Chang, D. W. Abbott, P. Storz, A. Toker, L. C. Cantley, and B. E. Turk. 2004. A rapid method for determining protein kinase specificity. Nat. Methods 1: 27-29. [DOI] [PubMed] [Google Scholar]
- 15.Karin, M., and F. R. Greten. 2005. NFκB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5: 749-759. [DOI] [PubMed] [Google Scholar]
- 16.Krappmann, D., and C. Schedereit. 2005. A pervasive role of ubiquitin conjugation in activation and termination of IkappaB kinase pathways. EMBO Rep. 6: 321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krikos, A., C. D. Laherty, and V. M. Dixit. 1992. Transcriptional activation of the tumor necrosis factor α-inducible zinc-finger protein, A20, is mediated by kB elements. J. Biol. Chem. 267: 17971-17976. [PubMed] [Google Scholar]
- 18.Lee, E. G., D. L. Boone, S. Chai, S. L. Libby, M. Chien, J. P. Lodolce, and A. Ma. 2000. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289: 2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S., C. Andrieu, F. Saltel, O. Destaing, J. Auclair, V. Pouchkine, J. Michelon, B. Salaun, R. Kobayashi, P. Jurdic, E. D. Kieff, and B. S. Sylla. 2004. IkappaB kinase beta phosphorylates Dok1 serines in response to TNF, IL-1, or gamma radiation. Proc. Natl. Acad. Sci. USA 101: 17416-17421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IkB kinases essential for NF-kB activation. Science 278: 860-866. [DOI] [PubMed] [Google Scholar]
- 21.Meylan, E., and J. Tschopp. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell 22: 561-569. [DOI] [PubMed] [Google Scholar]
- 22.Obenauer, J. C., L. C. Cantley, and M. B. Yaffe. 2003. Scansite 2.0: proteome-wide prediction of cell signalling interactions using short sequence motifs. Nucleic Acids Res. 31: 3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opipari, A. W., Jr., M. S. Bokuski, and V. M. Dixit. 1990. The A20 cDNA induced by tumor necrosis factor α encodes a novel type of zinc-finger protein. J. Biol. Chem. 265: 14705-14708. [PubMed] [Google Scholar]
- 24.Perkins, N. D. 2006. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene 25: 6717-6730. [DOI] [PubMed] [Google Scholar]
- 25.Perkins, N. D. 2007. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell. Biol. 8: 49-62. [DOI] [PubMed] [Google Scholar]
- 26.Prajapati, S., and R. B. Gaynor. 2002. Regulation of Ikappa B kinase (IKK) gamma/NEMO function by IKKbeta-mediated phosphorylation. J. Biol. Chem. 277: 24331-24339. [DOI] [PubMed] [Google Scholar]
- 27.Sakurai, H., H. Chiba, H. Miyoshi, T. Sugita, and W. Toriumi. 1999. IκB kinases phosphorylate NFκB P65 subunit on serine 536 in the transactivation domain. J. Biol. Chem. 274: 30353-30356. [DOI] [PubMed] [Google Scholar]
- 28.Scheidereit, C. 2006. IkB kinase complexes: gateways to NF-kB activation and transcription. Oncogene 25: 6685-6705. [DOI] [PubMed] [Google Scholar]
- 29.Song, H. Y., M. Rothe, and D. V. Goeddel. 1996. The tumor-necrosis factor-inducible zinc-finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc. Natl. Acad. Sci. USA 93: 6721-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strober, W., P. J. Murray, A. Kitani, and T. Watanabe. 2005. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 6: 9-20. [DOI] [PubMed] [Google Scholar]
- 31.Takeda, K., and S. Akira. 2004. TLR signaling pathways. Semin. Immunol. 16: 3-9. [DOI] [PubMed] [Google Scholar]
- 32.Turk, B. E., J. E. Hutti, and L. C. Cantley. 2006. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide libraries. Nat. Protoc. 1: 375-379. [DOI] [PubMed] [Google Scholar]
- 33.Wegener, E., A. Oeckinghaus, N. Papadopoulou, L. Lavitas, M. Schmidt-Supprian, U. Ferch, T. W. Mak, J. Ruland, V. Heissmeyer, and D. Krappmann. 2006. Essential role for IκB kinase β in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol. Cell 23: 13-23. [DOI] [PubMed] [Google Scholar]
- 34.Werner, S. L., D. Barke, and A. Hoffmann. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309: 1857-1861. [DOI] [PubMed] [Google Scholar]
- 35.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2000. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430: 694-699. [DOI] [PubMed] [Google Scholar]
- 36.Yaffe, M. B., G. C. Leparc, J. Lai, T. Obata, S. Volinia, and L. C. Cantley. 2001. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 19: 348-353. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, S. Q., A. Kovalenko, G. Cantarella, and D. Wallach. 2000. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity 12: 301-311. [DOI] [PubMed] [Google Scholar]