Abstract
Hypoxia is a pervasive microenvironmental factor that affects normal development as well as tumor progression. In most normal cells, hypoxia stabilizes hypoxia-inducible transcription factors (HIFs), particularly HIF-1, which activates genes involved in anaerobic metabolism and angiogenesis. As hypoxia signals a cellular deprivation state, HIF-1 has also been reported to counter the activity of MYC, which encodes a transcription factor that drives cell growth and proliferation. Since many human cancers express dysregulated MYC, we sought to determine whether HIF-1 would in fact collaborate with dysregulated MYC rather countering its function. Here, using the P493-6 Burkitt's lymphoma model with an inducible MYC, we demonstrate that HIF-1 cooperates with dysregulated c-Myc to promote glycolysis by induction of hexokinase 2, which catalyzes the first step of glycolysis, and pyruvate dehydrogenase kinase 1, which inactivates pyruvate dehydrogenase and diminishes mitochondrial respiration. We also found the collaborative induction of vascular endothelial growth factor (VEGF) by HIF-1 and dysregulated c-Myc. This study reports the previously unsuspected collaboration between HIF-1 and dysregulated MYC and thereby provides additional insights into the regulation of VEGF and the Warburg effect, which describes the propensity for cancer cells to convert glucose to lactate.
Animal development relies on mechanisms that coordinate normal cell proliferation, growth, metabolism, and response to the local tissue microenvironment such as hypoxia for organogenesis (21, 50). Tumorigenesis, on the other hand, usurps similar mechanisms through genetic alterations that activate oncogenes, such as the MYC oncogene, or inactivate tumor suppressor genes to promote cell proliferation (28). The proliferation of cancer cells in a three-dimensional multicellular cluster, however, is limited by diffusion of oxygen and nutrients, such that a phase of metabolic adaptation must be endured via the activation of hypoxia-inducible factors (HIFs) and, in particular, HIF-1 (6, 25, 72). HIF-1 is induced in low oxygen tensions and acts as a transcription factor to activate genes that permit metabolic adaptation, such as hexokinase 2 (HK2) and pyruvate dehydrogenase kinase 1 (PDK1) (16, 37, 54, 61, 81). HIF-1 also induces vascular endothelial growth factor (VEGF), which promotes neovascularization (67). Intriguingly, the c-Myc oncogenic transcription factor also induces genes involved in glycolysis and VEGF, but in contrast to HIF-1, c-Myc promotes mitochondrial biogenesis under nonhypoxic conditions (3, 38, 46, 49, 85). We surmise that c-Myc couples glycolytic and oxidative metabolism to cell growth and proliferation in normal cells. With normal development, it stands to reason that oxygen and nutrient deprivation signaled by HIF-1 activity is expected to be associated with cessation of cell proliferation (18, 22, 48). Indeed, multiple mechanisms for HIF-1 to counteract c-Myc activity have been reported. For example, HIF-1 binds c-Myc and inhibits c-Myc's transcriptional activity as well as activating an antagonist of c-Myc, Mxi-1, thereby contributing to cell cycle arrest and genomic instability (9, 24, 42-44, 80). Alternatively, HIF-1, whose consensus binding site (5′-G/ACGTG-3′) overlaps with the c-Myc consensus site (5′-CACGTG-3′), may displace c-Myc from HIF-1/c-Myc response elements (55). In addition, hypoxia-induced proteasomal degradation of c-Myc may be independent of HIF-1 and protects hypoxic cells from c-Myc-mediated apoptosis (9).
These HIF-1-mediated mechanisms are likely to be crucial for normal development where c-Myc is expressed normally and subjected to homeostatic regulation by HIF-1. However, in cancers where MYC is frequently dysregulated, it is not known whether these normal homeostatic mechanisms are bypassed, permitting the cooperation between dysregulated c-Myc and HIF-1. As such, we seek to determine here how dysregulated c-Myc might cooperate with HIF-1 to regulate genes that alter metabolism and recruit new blood vessels. In particular, we focused on HK2, PDK1, and VEGF, all of which have been previously shown to be HIF-1 targets as well as being bound or regulated by c-Myc (3, 16, 37, 38, 47, 54, 61, 77).
Among three isoforms (HIF-1 to -3), HIF-1 has been mainly implicated in tumor progression (20, 25, 69, 72, 74). HIF-1 is a heterodimeric transcription factor composed of two subunits, HIF-1α and HIF-1β. Oxygen regulation of HIF-1 levels depends on the HIF-1α oxygen-dependent domain, which contains two prolyl residues (402 and 564), which are hydroxylated by prolyl hydroxylase in the presence of oxygen (4, 33, 34, 52, 70, 73). Recognition by the von Hippel-Lindau (VHL) protein targets hydroxylated HIF-1α for ubiquitination and subsequent proteasomal degradation (30, 56). Hence, in the tumor microenvironment, the activation of HIF-1 is critical for tumor cell survival and proliferation.
In contrast to HIF-1, which responds to hypoxia or specific signal transduction pathways, the c-Myc transcription factor is expressed in nonhypoxic proliferating normal cells and is frequently dysregulated in human cancers. The transcriptional network of c-Myc, a basic helix-loop-helix transcription factor that heterodimerizes with Max, has been linked to many biological processes including development, cellular proliferation and growth, cell cycle, apoptosis, and energy metabolism (1, 8, 12, 27, 58). Oncogenic activation through dysregulated expression of c-Myc contributes to tumorigenesis of various tissues in transgenic mice and many types of human cancers including lymphomas (14, 32, 57, 65, 66). Little is known, however, about the role of dysregulated c-Myc expression, compared with its physiological expression, in HIF-1-mediated responses to hypoxia.
Here, we report the use of the human P493-6 B cells that conditionally overexpress human MYC upon tetracycline withdrawal (71). The P493-6 model recapitulates human Burkitt's lymphoma as well as responding to hypoxia in vitro and in vivo with hypoxic activation of HIF-1. Furthermore, HIF-2α is not expressed in the P493 system, and hence, this system is not confounded by another hypoxia-responsive factor (17). As such, we utilized the P493-6 system to study transcriptional regulation by HIF-1 and dysregulated c-Myc. In this study, we provide evidence that HIF-1 cooperates with dysregulated c-Myc to promote lymphomagenesis. Our study further demonstrates that c-Myc and HIF-1 enhance transcription of key switches of glucose metabolism such as HK2 and PDK1, which stimulate glycolysis. HK2 catalyzes the first step of glycolysis by phosphorylating glucose to form glucose-6-phosphate, which is further catabolized by glycolysis. PDK1, which is induced by hypoxia, phosphorylates and inactivates pyruvate dehydrogenase, which is required for the conversion of pyruvate to acetyl coenzyme A (acetyl-CoA) (29). PDK1, therefore, inhibits mitochondrial respiration by limiting the availability of acetyl-CoA for mitochondrial oxidative phosphorylation (37, 61). We also demonstrate that induction of VEGF, a proangiogenic factor, is stimulated cooperatively by c-Myc and HIF-1. Our study uncovers the cooperation between dysregulated c-Myc and hypoxia-induced HIF-1 in promoting glucose metabolism and angiogenesis, which confer an adaptive advantage for tumor progression under the hypoxic tumor microenvironment.
MATERIALS AND METHODS
Cell culture and hypoxic exposures.
The human B-cell line P493-6 carrying an inducible MYC repression system (71) was maintained in RPMI 1640 medium with 10% fetal bovine serum (GIBCO/BRL) and 1% streptomycin and penicillin (Invitrogen). P493-6 cells stably expressing the constitutively active form of HIF-1α (CA5-HIF-1α) or empty vector (EV) were generated by retroviral infections and selected with 800 μg/ml G418 as described previously (35). The CA5-HIF-1α mutant derived from adenoviral vector was inserted into retroviral vector pQCXIN (Clontech). P493-HIF-1α short hairpin RNA (shRNA) stably transduced cells were generated by retrovirus infection and G418 selection (800 μg/ml) as described previously (45). Transformed parental rat fibroblasts (TGR), cells null for Myc (HO15.19), and HO15.19 cells overexpressing human MYC (HO15.19-c-Myc) were maintained in high-glucose Dulbecco modified Eagle medium with 1 mM sodium pyruvate, 10% fetal bovine serum, and 1% streptomycin and penicillin. Incubation of P493-6 cells in 0.1 μg/ml tetracycline for 48 h led to significant repression of MYC. Removal of tetracycline induces MYC overexpression as described previously (38, 71). For HIF-1 induction, cells were exposed to hypoxia (0.1% O2 for P493-6 cells or 1% O2 for HO15.19 cells) or 100 μM CoCl2. Nonhypoxic cells (20% O2) were maintained at 37°C in a 5% CO2-95% air incubator. Hypoxic cells were maintained in a control atmosphere chamber (Plas-Labs) at 37°C. Oxygen tension was monitored with a calibrated Series 200 Percent oxygen analyzer (Alpha Omega Instruments).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay and real-time PCR quantification were performed as described previously (38). Briefly, P493-6 cells not treated or treated with tetracycline for 48 h under conditions of hypoxia (0.1% O2) or normoxia (20% O2) were cross-linked by formaldehyde. Fragmented chromatin was immunoprecipitated using rabbit polyclonal MYC (sc-764; Santa Cruz Biotechnology) and polyclonal human HIF-1α antibodies (NB100-134; Novus). Rabbit immunoglobulin G (IgG) antibody (Zymed) was used as a control. The total input was the supernatant from the no-antibody control (see Table S1 in the supplemental material for the primer sequences for the ChIP assay).
RNA interference.
Short interfering RNAs (siRNAs) targeting human HK2 and human PDK1 were purchased from Dharmacon Research Inc. P493-6 cells (3 × 106) were electroporated with 100 nM siRNAs targeting PDK1 (5′-CUACAUGAGUCGCAUUUCAdTdT-3′), HK2 (siGENOME SMART POOL), or scrambled control siRNA (5′-CACGCUCGGUCAAAAGGUUdTdT-3′) using the Amaxa Nucleofection Kit according to the manufacturer's instructions. On the following day, 5 × 105 viable cells were subjected to hypoxic exposure (0.1% O2) or CoCl2 (100 μM) treatment. At indicated times, cells were subjected to glucose uptake and immunoblot assays. Conditioned media were used for lactate measurement.
RNA analysis.
Human HK2 and VEGF mRNA levels were determined by Northern blot analysis or quantitative real-time reverse transcription-PCR (RT-PCR). Total RNA was isolated from P493-6 cells using TRIzol (Invitrogen). Five micrograms of RNA was used in Northern blot analysis. RNA was subjected to 1.2% agarose gel electrophoresis and transferred to a nylon membrane (Nytran). The membrane was probed with human HK2, which was labeled with 32P using a random prime labeling kit (Stratagene). An ethidium bromide-stained agarose gel of 18S rRNA was used as a loading control. Quantitative real-time RT-PCR was performed using the TaqMan One Step RT-PCR Master Mix kit (PE Applied Biosystems) with probes and primers (see Table S2 in the supplemental material for the probe and primer sequences for the real-time RT-PCR). The expression level of human 18S rRNA was determined with a predeveloped mixture of TaqMan probe and primers (PE Applied Biosystems) and used for normalization. All PCRs were performed in triplicate.
Western blotting.
Equal amounts of protein extracted from cells or tumor xenografts were subjected to electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gels. Monoclonal anti-c-Myc (Oncogene Research Products), monoclonal anti-HIF-1α (Affinity Bioreagent), polyclonal anti-HK2 (Santa Cruz), polyclonal anti-PDK1 (Stressgen Bioreagents), anti-HK1 (Santa Cruz), polyclonal anti-enolase 1 (Santa Cruz), polyclonal anti-lactate dehydrogenase (anti-LDHA; Abcam), and monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPD; Advanced ImmunoChemical Inc.) antibodies were used for immunoblotting. Monoclonal antiactin (Sigma), antitubulin (Calbiochem), and polyclonal anti-topoisomerase I (Santa Cruz) antibodies were used as loading controls. CoCl2-treated Cos7 nuclear extract was used for positive control for HIF-1α (active motif). Quantification of Western blotting data was performed using Epi Chemi II Darkroom (UVP Laboratory Products). Optical densities of each band for HK2 or PDK1 were normalized by those of actin bands.
Hexokinase enzyme activity.
Hexokinase enzyme activity was determined by the formation of NADH at 340 nm using a spectrophotometer (Du 800 BeckMan) as described previously (62). NADH production is coupled to the formation of glucose-6-phosphate from glucose by glucose-6-phosphate dehydrogenase. Total cell lysates from P493-6 cells were incubated with the assay medium containing 25 mM triethylamino-Cl (Sigma), 15 mM MgCl2 (Sigma), 5 mM ATP (Amersham), 1 mM NADP+ (Sigma), 1 mM dithiothreitol (Sigma), 0.5 mM glucose (Sigma), 0.45 mM KCN (Sigma), 13 μg/ml oligomycin (Sigma), and 1.2 units/ml glucose-6-phosphate dehydrogenase (Roche). The rate of NADH formation was normalized to total protein concentration.
Lactate production.
Accumulation of lactate in the culture medium was determined using a lactate assay kit (Biomedical Research Service, State University of New York at Buffalo) or the Yellow Springs Instruments 2300 STAT glucose/lactate analyzer (Yellow Springs Instruments) assay. The lactate assay kit is based on the reduction of the tetrazolium salt 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyltetrazolium chloride in an NADH-coupled enzymatic reaction to formazan, which is water soluble and exhibits an absorption maximum at 492 nm. Total viable cell number was used for normalization.
Glucose uptake.
Glucose uptake was determined by intracellular 2-[3H]deoxyglucose level (59). P493-6 cells were incubated in medium containing 0.1 mM 2-deoxyglucose and 10 nM 2-[3H]deoxyglucose for 10 min. Cells were washed with cold phosphate-buffered saline and lysed by the addition of 10 mM NaOH containing 0.1% Triton X-100. Lysates were subjected to a liquid scintillation counting for 3H level. Total protein concentration was used for normalization.
ELISA for VEGF production.
The culture medium of P493-6 cells was collected, and 100-μl aliquots were used to determine VEGF levels. Immunoreactive VEGF was quantified using a sandwich enzyme-linked immunosorbent assay (ELISA; Quantikine VEGF immunoassay kit; R & D Systems) according to the manufacturer's recommended protocol.
In vivo xenograft model.
Cells (3 × 107) were injected subcutaneously into SCID mice. In vivo suppression of MYC expression in the P493-6 xenograft was achieved by treatment with 0.01% doxycycline in drinking water. Tumor volumes were measured every 4 days.
RESULTS
HIF-1 promotes c-Myc-mediated tumorigenesis.
We found that reduced HIF-1α expression by shRNA dramatically inhibited c-Myc-mediated tumorigenesis of P493-6 cells (17). These studies indicate that HIF-1α is necessary for P493-6 cell tumorigenesis in vivo. To determine whether HIF-1α could synergize with c-Myc in P493-6 tumorigenesis, we introduced a constitutively active form of HIF-1α (CA5-HIF-1α) with the oxygen-dependent domain deleted and several prolines mutated as previously described (35) into P493-6 cells and found that tumor growth is significantly enhanced compared with control (Fig. 1A). However, no increase in proliferation or viability was observed in CA5-HIF-1α P493-6 cells in vitro (data not shown). These results indicate that HIF-1α contributes to c-Myc-mediated tumorigenesis in vivo.
FIG. 1.
Effects of constitutively stable HIF-1α (CA5-HIF-1α) on tumorigenesis and expression of HK2 and PDK1. (A) Tumor growth of P493-6 cells expressing CA5-HIF-1α or control EV with or without the tetracycline analog doxycycline (0.01%, wt/vol) treatment through drinking water. Tumor volumes were determined by caliper measurements. Error bars represent standard errors of the means (n = 20 for non-tetracycline-treated tumors, n = 10 for tetracycline-treated tumors). (B) Immunoblot assay (top) and its quantification (bottom) of lysates from xenografts of P493-6 cells expressing CA5-HIF-1α or control EV. The average optical density from two bands for each CA5-HIF-1α and EV control P493-6 xenograft is represented. Error bars represent ranges of two bands. (C) Immunoblot assay (top) and its quantification of induction (n-fold) at 48 h (bottom) of HK2, PDK1, LDHA, HK1, CA5-HIF-1α, and c-Myc in CA5-HIF-1α or control EV P493-6 cells after withdrawal of tetracycline under nonhypoxic conditions. Actin is shown as a loading control. *, P < 0.02 by Student's t test.
We examined the expression of selected proteins involved in metabolic adaptation to hypoxia in control versus CA5-HIF-1α-overexpressing P493-6 cells after the induction of c-Myc by tetracycline withdrawal (Fig. 1B and C). We noted that Myc was slightly increased and HK2 and PDK1 protein levels were both higher in P493-6 cells expressing CA5-HIF-1α than in those expressing control EV, suggesting that both HK2 and PDK1 are induced cooperatively by c-Myc and hypoxia. Despite the high levels of endogenous HIF-1α induced by the hypoxic microenvironment of tumor xenografts, additional ectopic CA5-HIF-1α expression further induced HK2 and PDK1 (1.6- and 1.7-fold higher than EV control, respectively) (Fig. 1B, bottom). In particular, while HK2 is induced by c-Myc in both conditions, the expression of PDK1 appears responsive to c-Myc only in the presence of stabilized HIF-1α (CA5-HIF-1α) activity (Fig. 1C). Intriguingly, CA5-HIF-1α appears to be induced by c-Myc. We surmise that the effect of c-Myc on ectopic CA5-HIF-1 expression may be transcriptional or posttranscriptional, which has not yet been further investigated. Nonetheless, the enhanced inductions of PDK1 and HK2 correlate with ectopic CA5-HIF-1α levels, supporting the idea that ectopic CA5-HIF-1α indeed contributes to HK2 and PDK1 expression. Enhanced expression of these proteins through c-Myc and HIF-1 was also observed in the rat fibroblasts whose Myc is nullified (HO15.19) or reconstituted with human MYC (HO15.19-c-Myc) (46, 53). It is notable that under hypoxia, HO15.19 cells displayed minimal induction of HK2 compared to HO15.19-c-Myc or TGR cells that expressed higher levels of HK2, suggesting that c-Myc is necessary for significant induction of HK2 (see Fig. S1 in the supplemental material). However, PDK1 induction appears to be mainly regulated by HIF-1 or hypoxia in rat fibroblasts. These studies further support the cooperative effects of c-Myc and HIF-1 in another system (see Fig. S1 in the supplemental material).
c-Myc and HIF-1 cooperate to enhance glucose metabolism by inducing HK2.
Since both gain-of-function and loss-of-function studies demonstrate the importance of HIF-1α in c-Myc-mediated tumorigenesis of P493-6 cells and the expression of HK2 protein is increased cooperatively by c-Myc and HIF-1, we sought to determine whether HIF-1 and dysregulated c-Myc could cooperatively alter glucose catabolism and transcriptionally regulate HK2. P493-6 cells were treated with tetracycline and/or hypoxia to modulate the levels of c-Myc and HIF-1α. As illustrated in Fig. 2A, in the presence of tetracycline (0.1 μg/ml), MYC expression was virtually completely repressed. MYC is rapidly induced by removal of tetracycline from P493 cells, which were previously treated with tetracycline for 48 h. To induce HIF-1α expression, P493-6 cells were exposed to hypoxia (0.1% O2) or 100 μM of CoCl2. HIF-1α expression peaked by 29 h of hypoxia or CoCl2 treatment. In these conditions, P493-6 cells were viable up to 72 h and displayed no sign of apoptosis. Cells stopped proliferating by hypoxia or c-Myc suppression (tetracycline treatment) or both. Only nonhypoxic P493-6 cells with high c-Myc expression were proliferating (see Fig. S2 in the supplemental material).
FIG. 2.
Enhanced glycolysis by c-Myc and HIF-1 in P493-6 cells. (A) Immunoblot assays of c-Myc and HIF-1α expression in P493-6 cells. P493-6 cells were pretreated with tetracycline (0.1 μg/ml) for 48 h. After washing, cells were cultured with or without tetracycline in hypoxia (0.1% O2) or normoxia as indicated in the bottom. To induce HIF-1α in nonhypoxic conditions, P493-6 cells were incubated in medium containing 100 μM CoCl2. Equal amounts of nuclear protein were subjected to immunoblot assay to determine the expressions of c-Myc and HIF-1α. Topoisomerase 1 is shown as a loading control for nuclear extracts. (B) Cellular glucose uptake in indicated conditions at 48 h was determined by measuring intracellular 2-[3H]deoxyglucose. Error bars represent standard errors of the means from two independent experiments. (C) Lactate accumulation in the medium was measured using a lactate assay kit. Lactate concentrations were normalized to viable cell number. Error bars represent standard errors of the means from two independent experiments.
We determined the glycolytic activity by measuring glucose uptake and lactate production. We found that P493-6 cells expressing both c-Myc and HIF-1 display additive increases in both glucose uptake and lactate production, indicating elevated glycolytic metabolism (Fig. 2B and C). Since HK2, among the four isoforms of HK, is expressed highly in human cancers and known to be a direct target of both c-Myc and HIF-1, we sought to determine whether increased glucose uptake and glycolysis are associated with increased HK2 expression. We found that HK2 was significantly enhanced at the mRNA (Fig. 3A; see also Fig. S3A in the supplemental material) and protein (Fig. 3B) levels in P493-6 cells expressing both c-Myc and HIF-1α compared with P493-6 cells expressing either c-Myc or HIF-1α alone. Consistent with mRNA and protein levels, hexokinase enzymatic activity was also enhanced by c-Myc and HIF-1 (Fig. 3C). We also examined the levels of other glycolytic enzymes including HK1, ENO1, GAPD, and LDHA, known to be regulated by either c-Myc, HIF-1, or both (38, 75). No increase of other glycolytic enzyme genes including the ENO1, GAPD, HK1, and LDHA genes by c-Myc and HIF-1 was observed in P493 cells, supporting the major roles of HK2 in enhanced glycolytic metabolism by c-Myc and HIF-1 (see Fig. S3B in the supplemental material). HK2 expression, in contrast to that of the other enzymes, was also increased in P493-6 xenografts as well as P493-6 cells bearing constitutively active HIF-1α (CA5-HIF-1α) in nonhypoxic conditions (Fig. 1B and C).
FIG. 3.
Enhanced HK2 expression by c-Myc and HIF-1. (A) Northern blot analysis of HK2 expression in P493-6 cells. Pretreatment with tetracycline for 48 h to suppress MYC expression is followed by incubating cells in different conditions as illustrated. Ethidium bromide staining of 18S is shown as a loading control. (B) Immunoblot assays of HK2 expression in P493-6 cells. Actin is shown as a loading control. (C) HK enzymatic activities. Total cell lysates from P493-6 cells cultured with or without tetracycline in hypoxia (left) or CoCl2 treatment (right) for 29 h were subjected to HK activity assay. HK activity is determined by NADH production at 340 nm and normalized by total protein concentration. Error bars represent standard deviations from three independent experiments.
In addition to CoCl2 treatment, which stabilizes HIF-1α under nonhypoxic conditions, we employed P493-6 cells expressing shRNA targeting HIF-1α to further confirm that cooperative induction of HK2 under hypoxia is mediated by HIF-1α and not by non-HIF-1 pathways (Fig. 4A). As illustrated in Fig. 4B, induction of HK2 was significantly attenuated in HIF-1α-knockdown P493-6 cells under high c-Myc and hypoxic conditions. It should be noted that c-Myc levels were not elevated by reduced HIF-1α expression (Fig. 4A).
FIG. 4.
Effect of decreased HIF-1α expression on induction of HK2 and PDK1. (A) Immunoblot of HIF-1α and c-Myc expression in P493-6 cells stably expressing shRNA targeting HIF-1α or control vector. HIF-1α shRNA or control P493-6 cells were pretreated with tetracycline (0.1 μg/ml) for 48 h. After washing, cells were cultured in hypoxia (0.1% O2). Actin is shown as a loading control. (B) Immunoblot of HK2, PDK1, and p21 expression. HIF-1α shRNA or control P493-6 cells were pretreated with tetracycline (0.1 μg/ml) for 48 h. After washing, cells were cultured with or without tetracycline in hypoxia (0.1% O2) or normoxia as indicated in the bottom. Actin is shown as a loading control.
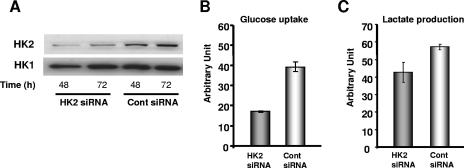
Next, we sought to determine the role of HK2 in increased glucose uptake and lactate production in hypoxia. P493-6 cells were transfected with siRNA targeting HK2 (Fig. 5A). Increased glucose uptake in hypoxic P493-6 cells was abrogated by suppression of HK2 expression (Fig. 5B). Lactate production was also reduced, although to a lesser extent, by HK2 siRNA treatment under hypoxia (Fig. 5C). Taken together, these results demonstrate the cooperative induction of HK2 expression by HIF-1 and dysregulated c-Myc activity, which together promote increased glucose uptake and lactate production.
FIG. 5.
Effect of decreased HK2 expression on glycolysis in P493-6 cells. (A) Immunoblot assays of HK2 expression in P493-6 cells electroporated with pools of four different siRNAs targeting HK2 (siGENOME SMART POOL) or control scrambled siRNA in hypoxia (0.1% O2). HK1 is shown as a loading control. (B and C) Glucose uptake (B) and lactate accumulation (C) of P493-6 cells. Cells in indicated conditions at 48 h were subjected to measurement of intracellular 2-[3H]deoxyglucose or lactate accumulation in the culture medium. Protein concentration from viable cells or total number of viable cells is used for normalization of glucose uptake or lactate accumulation, respectively. Error bars represent standard errors of the means from two or three independent experiments.
DNA binding of c-Myc and HIF-1 in the regulatory regions of HK2 gene.
Since earlier studies reported that HK2 is a direct target of either HIF-1 in hypoxia or c-Myc in nonhypoxic conditions, we sought to determine the binding of both HIF-1 and Myc to the HK2 promoter area under hypoxia. By DNA sequence analysis, we identified five potential c-Myc binding sites (E-boxes, 5′-CACGTG-3′) and about 50 potential HIF-1 binding sites (hypoxia response elements, 5′-G/ACGTG-3′) from 5 kb upstream to the HK2 first intron. We performed a scanning ChIP with 22 primer pairs, which were designed to scan at approximately 1-kb intervals the human HK2 genomic region spanning 25 kb from 5 kb upstream of the transcription start site through intron 1. As reported in our previous study, scanning ChIP confirmed a c-Myc binding region illustrated as a binding signal in highly conserved E boxes located in intron 1 (Fig. 6, upper panel, regions 6) (38). We found HIF-1 binding in the promoter region (Fig. 6, middle panel, region 3 or 4), which also contains highly conserved HIF response elements. Samples from no-antibody control and control IgG antibody samples display background signals, indicating the specificity of c-Myc or HIF-1 binding (see Fig. S4 in the supplemental material). It is notable that the HIF-1 binding region is located at least 1 kb away from the intronic c-Myc binding region, suggesting that c-Myc and HIF-1 could independently and cooperatively transactivate the HK2 gene.
FIG. 6.
Scanning ChIP assay of the HK2 gene. The human HK2 gene was scanned by ChIP to determine the c-Myc (top panel) or HIF-1 (middle panel) DNA binding regions. Labeled regions (amplicons 1 to 22) of the HK2 gene were quantitatively measured by real-time PCR. PCR was performed on the fragmented chromatin precipitated from non-tetracycline-treated hypoxic (72-h) P493-6 cells with anti-c-Myc or anti-HIF-1α, without antibody, or control IgG antibody (see Fig. S2 in the supplemental material for the background signals of no-antibody and IgG antibody controls). Binding activity is represented as a percentage of total input of the chromatin DNA. The bottom panel indicates the locations of conserved canonical E boxes (large arrows), conserved HIF-1 binding sites (small arrows), and exon 1 in the human HK2 genomic sequence. The amplicons for scanning ChIP are indicated by the lines above the HK2 gene and labeled. Error bars represent standard deviations from two or three independent duplicate reactions.
c-Myc and HIF-1 cooperate to induce a metabolic switch toward glycolysis by PDK1.
PDK1 was found to be directly induced by HIF-1 under hypoxic conditions, which mediate a metabolic switch toward glycolysis by suppressing mitochondrial respiration (37, 61). In addition, c-Myc has been reported to bind the promoter region of PDK1 in promoter arrays by ChIP assay (47). As reported earlier, c-Myc alone in nonhypoxic conditions has minimal effect on PDK1 expression (37). In the presence of HIF-1α induced by either hypoxia or CoCl2, c-Myc further enhances PDK1 expression (Fig. 7A and B). Similarly, ectopic expression of constitutively active CA5-HIF-1α permits c-Myc induction of PDK1 expression in P493-6 cells and tumor xenografts (Fig. 1B and C). Furthermore, HIF-1α knockdown by shRNA reduced PDK1 induction under hypoxic conditions (Fig. 4B). However, we note a variation in the PDK1 expression in the control cells upon Myc induction under normoxic conditions compared with results shown in Fig. 1C. We surmise that this results from fluctuation of the relatively low expression of PDK1 in nonhypoxic conditions. These observations nonetheless suggest that the increased conversion of pyruvate into lactate observed in hypoxic tumor cells may result from the cooperative induction of PDK1 by c-Myc and HIF-1.
FIG. 7.
Enhanced PDK1 expression by c-Myc and HIF-1 in P493-6 cells. (A) mRNA levels of PDK1 are determined by real-time RT-PCR. Error bars represent standard deviations from two independent duplicate reactions. (B) Immunoblot assays of PDK1 expression in hypoxic (or CoCl2 treated) or nonhypoxic P493-6 cells that were treated with or without tetracycline. Actin is shown as a loading control. (C) Immunoblot assay of decreased PDK1 expression by PDK1 siRNA or control siRNA in P493-6 cells. CoCl2 treatment was used to induce HIF-1-dependent PDK1 expression. Actin is shown as a loading control. (D and E) Lactate accumulation in the culture medium of P493-6 cells with or without CoCl2 treatment to induce PDK1 expression. PDK1 is inhibited by treatment with either siRNA (D) or a PDK inhibitor, dichloroacetate (DCA) (E). Lactate concentrations are normalized to total viable cells. Error bars represent standard errors of the means from two to three independent experiments.
These observations may explain the intriguing findings illustrated in Fig. 2C that in the presence of HIF-1α, c-Myc synergistically increased lactate production while lactate production was not increased by c-Myc alone. Inhibition of PDK1 by siRNA transfection (Fig. 7C) or treatment with the PDK inhibitor dichloroacetate results in significantly reduced lactate production (Fig. 7D and E), confirming a role for PDK1 in the generation of lactate mediated by c-Myc and HIF-1. However, CoCl2 treatment is known to stabilize HIF-1α, which in turn activates target glycolytic enzyme genes such as LDHA. In this regard, CoCl2 treatment presumably increases lactate production not only by PDK1 induction but also through elevated LDHA. Since LDHA elevated by CoCl2 is likely to increase basal lactate production, which is blunted by the inhibition of PDK1, complete reduction of lactate production by PDK1 inhibition would not be expected. (Fig. 7D).
DNA binding of c-Myc and HIF-1 in the regulatory regions of the PDK1 gene.
To determine the nature of c-Myc and HIF-1 collaboration in stimulating PDK1 expression, we performed a scanning ChIP to pinpoint the binding regions of these two factors. We previously reported that HIF-1-associated chromatin fragments enriched by immunoprecipitation cover the boundary of exon 1 and intron 1, which contains a cluster of consensus HIF-1 binding sites (37). To further pinpoint HIF-1 binding sites, phylogenetic footprinting was employed to identify conserved consensus HIF-1 binding sites. As illustrated in Fig. 8, highly conserved consensus sites are located in exon 1 (5′-G/ACGTG-3′) that are significantly bound by HIF-1, while those in intron 1 are canonical c-Myc binding sites (E boxes, 5′-CACGTG-3′) and mainly bound by c-Myc. The specificity of c-Myc and HIF-1 binding was verified by control experiments with no antibody and control IgG samples (see Fig. S5 in the supplemental material). As similarly seen in the HK2 gene, ChIP suggests that c-Myc and HIF-1 bind nonoverlapping genomic regions.
FIG. 8.
Scanning ChIP assay of the PDK1 gene. Labeled regions (amplicons 1 to 9) of the PDK1 gene were quantitatively amplified by real-time PCR. Error bars represent standard deviations from two or three independent duplicate reactions. Symbols are identical to those in Fig. 6.
c-Myc and HIF-1 cooperate to induce VEGF.
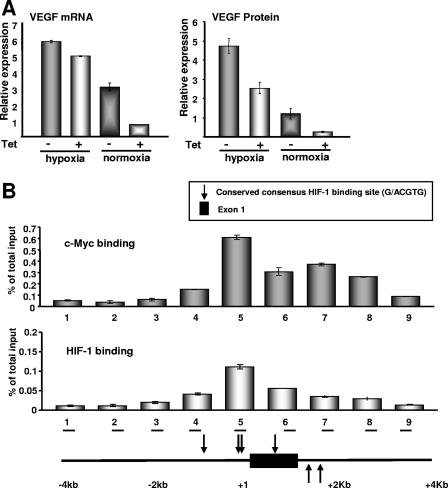
In addition to metabolic adaptation through the regulation of genes such as HK2 and PDK1, the progression of tumors is highly dependent on angiogenesis, which is one of the hallmarks of cancers. A key mechanism for the promotion of angiogenesis during tumor development is HIF-1-mediated transcriptional activation of VEGF, which mitogenically stimulates endothelial cells for their recruitment into new blood vessels (67). Earlier studies using transgenic animal models have reported that c-Myc-deficient mice display serious defects in the development of blood vessels, angiogenesis, and erythropoiesis, suggesting that c-Myc participates in regulating VEGF expression and angiogenesis (67, 77). Using the P493-6 system, we sought to determine whether VEGF expression, similar to that of HK2 and PDK1, is also regulated by c-Myc and HIF-1. VEGF expression at the mRNA (measured by real-time RT-PCR) and protein (measured by ELISA) levels is induced by c-Myc. Hypoxia further enhances VEGF expression (Fig. 9A and B). A scanning ChIP assay revealed the association of c-Myc and HIF-1 with chromatin fragments flanking the promoter region of the VEGF gene (Fig. 9B, region 5; see also Fig. S6 in the supplemental material). Although no conserved canonical c-Myc binding site (E box) exists, DNA binding of c-Myc appears to localize in the same genomic region where HIF-1 binds. c-Myc binding appears to trail into intron 1, which is characteristic of c-Myc binding in other genes (84). These data suggest that c-Myc and HIF-1 could bind overlapping and nonoverlapping binding sites in VEGF.
FIG. 9.
Enhanced VEGF expression by c-Myc and HIF-1. (A) mRNA and protein levels of VEGF were determined by real-time RT-PCR and ELISA, respectively. Error bars represent standard deviations from two independent duplicate reactions. (B) DNA binding of c-Myc and HIF-1 to the VEGF gene was determined by scanning ChIP assay as described for Fig. 6.
DISCUSSION
In this study, we provide molecular clues underlying our recent observations indicating that HIF-1α is essential for c-Myc-mediated tumorigenesis (17). Moreover, our in vivo xenograft assay indicates that gain-of-function via a stabilized HIF-1α mutant enhances the c-Myc-mediated tumorigenic potential of P493-6 cells (Fig. 1). Proliferation of P493-6 cells, however, slowed considerably in hypoxia in vitro (see Fig. S2 in the supplemental material and see also below), while in vivo tumor growth was accelerated by HIF-1 and c-Myc. It is notable that a large number of P493-6 cells are needed for in vivo tumorigenesis, implying that there are additional factors involved in tumorigenesis (17). However, the in vitro model that we use in this study allows for an experimental system to study HIF-Myc interactions, which could be modulated further in vivo. Notwithstanding this caveat, we demonstrate here that hypoxic adaptation, which includes increased glucose uptake, enhanced glycolysis, and elevated VEGF levels, is stimulated by the cooperation of HIF-1 with dysregulated c-Myc expression.
Various recent studies suggest that HIF-1α negatively regulates the transcriptional activity of c-Myc and suppresses c-Myc's physiological roles (9, 24, 42-44, 55, 80). It stands to reason that in a normal cell, which expresses c-Myc when cell proliferative cues are received to augment MYC expression, such as in wound healing, hypoxia should signal diminished blood flow and limitation of nutrients to a tissue. Hence, hypoxia via the induction of HIF-1 would be expected to trigger cell growth arrest, except in endothelial cells, and altered cellular metabolism. We have, in fact, found that hypoxia causes cell growth arrest and a decrease in endogenous MYC expression in vitro (Fig. 2A; see also Fig. S2 in the supplemental material) (18). However, no p27 expression was observed in P493-6 cells even under hypoxic conditions, which is consistent with a previously reported study (60). Very low levels of p21 were detected; however, we did not observe an increase in p21 under hypoxic conditions (Fig. 4B). Rather, p21 appears to be decreased in hypoxia by an unknown mechanism.
The observation in Fig. 1 that constitutive HIF-1 expression augments Myc-mediated P493-6 tumorigenesis in vivo appears to contradict the inability of P493-6 cells in vitro to proliferate in hypoxia as shown in Fig. 2A and in Fig. S2 in the supplemental material. How then could HIF-1 increase Myc-mediated tumorigenesis? We surmise that among the large number of P493-6 cells needed for in vivo tumorigenesis, a subpopulation acquires additional factors that are permissive for establishing a tumor mass that is dependent on both Myc and HIF. Although our attempt to study Myc-HIF interactions in vitro is nuanced by these differences, we believe that insights gleaned from our studies are necessary to further understand the interplay between Myc and HIF in vivo (17).
These earlier studies do not, however, address the interaction of HIF-1 with c-Myc when MYC expression is dysregulated, as one would expect to find in a tumor that may result from the activation of the MYC oncogene. Studies with various model systems revealed that c-Myc function is an essential contributor to cellular proliferation and normal embryonic development through modulating its target genes involved in cellular adhesion, proliferation, growth, and metabolism (12, 27, 84). Previously, we have also reported that c-Myc promotes mitochondrial biogenesis and respiration, in addition to its induction of glycolytic enzyme genes (38, 46, 59, 78). In the current study, we found that P493-6 cells bearing dysregulated c-Myc consume more glucose in nonhypoxic conditions but that lactate production is not increased, since c-Myc promotes mitochondrial biogenesis and respiration as reported previously (Fig. 2B and C). These activities of c-Myc suggest that it could couple the cell growth and cell cycle machinery with the cellular energy production system in normal cells. Because many cancer cells display the Warburg effect or have the propensity to take up glucose and convert glucose to lactate, rather than metabolizing glucose through oxidative phosphorylation (2, 7, 15, 19, 36, 76, 82, 83), we sought to understand how dysregulated expression of c-Myc might interact with HIF-1 in hypoxia to mediate the Warburg effect and adaptation to the tumor microenvironment.
Using the P493-6 system, we determined whether enforced overexpression of c-Myc could liberate c-Myc from homeostatic regulation by HIF-1α. HIF-1 is known to activate transcription of genes encoding glycolytic enzymes and PDK1, which appears to be a key hypoxic adaptation through a metabolic switch from oxidative phosphorylation into glycolysis under a hypoxic microenvironment (37, 61). Previously, we comprehensively dissected the transcriptional network of c-Myc and its target glycolytic enzyme genes, demonstrating that c-Myc transactivates virtually all glycolytic enzyme genes, many of which are also activated by HIF-1α in hypoxia (38, 75). In contrast to previous studies, HIF-1α induced by hypoxia or CoCl2 further accelerates glucose uptake and lactate production in P493-6 cells. This indicates that HIF-1-mediated shutdown of mitochondrial function by the HIF-1 target PDK1 is required for converting pyruvate into lactate (Fig. 7). Scanning ChIP assays for DNA binding of c-Myc and HIF-1 in HK2 and PDK1 genes demonstrate that c-Myc and HIF-1 independently bind different genomic regions (Fig. 6 and 8). These observations suggest that in the context of dysregulated c-Myc, HIF-1α no longer inhibits c-Myc activity but rather the two cooperate to transactivate certain common target genes, such as HK2, PDK1, and VEGF.
It is intriguing that c-Myc and HIF-1 could cooperatively activate HK2 and PDK1, which are the critical regulators of glycolytic metabolism and mitochondrial function. Particularly, in addition to its rate-limiting role in glucose metabolism, HK2 is overexpressed in most human cancers and suppresses proapoptotic molecules including Bad and Bax at the mitochondrial outer membrane, by which HK2 functions as a key coordinator by integrating glucose metabolism with apoptosis (13, 23, 26, 49, 63, 64, 68). The PDK1 gene, recently identified as a HIF-1 direct target gene, inactivates mitochondrial tricarboxylic acid cycle and respiration through inhibiting the mitochondrial pyruvate dehydrogenase complex that converts pyruvate into acetyl-CoA (37, 61). Inhibition of PDK by dichloroacetate was recently shown to be antitumorigenic, suggesting that PDK activity is critically important for in vivo tumorigenesis (5). c-Myc binding in PDK promoters has been reported in a ChIP study (47), and our scanning ChIP assay verified that c-Myc binds the intron 1 region of the PDK1 gene even under nonhypoxic conditions (data not shown) and even when PDK1 expression is not induced. These observations are compatible with our finding in the P493-6 system that c-Myc is frequently bound to genes which are not induced at the steady-state mRNA level (84). Other signals, such as hypoxia, would provide additional transcription factors, such as HIF-1, to induce genes bound by c-Myc. Our study suggests that hypoxia via HIF-1 is necessary for the full induction of PDK1 that presumably counteracts c-Myc-mediated activation of mitochondrial functions in nonhypoxic conditions. Conversely, HIF-1 minimally induces PDK1 with low c-Myc expression (Fig. 7A and B). These observations suggest that the Warburg effect could well result from the cooperation between HIF-1 and dysregulated c-Myc.
In addition to the efficient metabolic switch to glycolysis, development of new blood vessels or angiogenesis is a required adaptive process to improve delivery of oxygen and nutrients into hypoxic tumor regions. According to the data from various in vivo model systems, c-Myc has been suggested to be a potent angiogenesis inducer during embryonic development and tumorigenesis (3, 77). A recent study demonstrated that hypoxia further increases c-Myc-mediated VEGF expression and angiogenesis in an in vivo transgenic model, suggesting that HIF-1 cooperates with c-Myc to induce VEGF expression and angiogenic phenotypes (40). These studies, however, did not examine the mechanism by which c-Myc and HIF-1 could cooperate to activate VEGF expression. Similarly to HK2 and PDK1, VEGF is induced by the cooperation between HIF-1 and dysregulated c-Myc (Fig. 9A). Further, we identified strong DNA binding of c-Myc and HIF-1 in a genomic region that contains a cluster of conserved putative binding sites for HIF-1 (Fig. 9B). Our study supports the molecular mechanism underlying enhanced induction of VEGF by c-Myc and HIF-1 and suggests that cooperative roles of c-Myc and HIF-1 may contribute to a broader range of oncogenic processes.
The effects of HIF-2α can be excluded since HIF-2α is not detectable at both mRNA and protein levels in P493-6 cells. However, there is emerging evidence that HIF-2α may play more prominent roles in tumorigenesis (10, 11, 39, 41, 51, 79). Although recent studies reported that adaptive responses to hypoxia such as induction of glycolytic enzyme genes are predominantly mediated by HIF-1α (25, 31), additional studies are necessary to address the effects of HIF-2α on transcriptional and oncogenic activities of c-Myc.
It is intriguing that no significant enhanced expression of other glycolytic enzyme proteins including HK1, LDHA, ENO, and GAPD was observed in the P493-6 cells under conditions with both HIF-1 and c-Myc expressed (see Fig. S3B in the supplemental material). The differences between these enzymes and HK2, PDK1, and VEGF will require further studies. It is conceivable, however, that expression of glycolytic enzymes is also regulated at the posttranscriptional level, since c-Myc binds to and induces mRNA levels of LDHA, ENO, and GAPD (38). Further studies will be needed to address how c-Myc and HIF-1 work together at the transcriptional level. It will be of great interest to ask whether unique or additional coactivators are recruited to the regulatory regions when the factors are both bound.
In summary, we report here that the tumorigenic potential of P493 cells is increased by the cooperation of dysregulated c-Myc with HIF-1, and we demonstrate that this cooperation accelerates glycolytic metabolism and angiogenesis through the induction of key proteins, HK2, PDK1, and VEGF. Our ChIP studies pinpoint the binding of c-Myc and HIF-1 to these critical genes, whose levels are significantly elevated by both transcription factors in a cooperative manner. In particular, our study underscores the importance of distinguishing the effects of HIF-1 on the normal roles of proto-oncogenes from those of dysregulated oncogenes that escape from normal homeostatic mechanisms.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grants CA52497, CA57341, NHLBI NO1-HV-28180, and the Johns Hopkins Institute for Cell Engineering. C. V. Dang is Johns Hopkins Family Professor in Oncology Research.
Footnotes
Published ahead of print on 4 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adhikary, S., and M. Eilers. 2005. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6: 635-645. [DOI] [PubMed] [Google Scholar]
- 2.Ashrafian, H. 2006. Cancer's sweet tooth: the Janus effect of glucose metabolism in tumorigenesis. Lancet 367: 618-621. [DOI] [PubMed] [Google Scholar]
- 3.Baudino, T. A., C. McKay, H. Pendeville-Samain, J. A. Nilsson, K. H. Maclean, E. L. White, A. C. Davis, J. N. Ihle, and J. L. Cleveland. 2002. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 16: 2530-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22: 4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, S., S. L. Archer, J. Allalunis-Turner, A. Haromy, C. Beaulieu, R. Thompson, C. T. Lee, G. D. Lopaschuk, L. Puttagunta, G. Harry, K. Hashimoto, C. J. Porter, M. A. Andrade, B. Thebaud, and E. D. Michelakis. 2007. A mitochondria-k(+) channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 11: 37-51. [DOI] [PubMed] [Google Scholar]
- 6.Brahimi-Horn, M. C., J. Chiche, and J. Pouyssegur. 2007. Hypoxia signalling controls metabolic demand. Curr. Opin. Cell Biol. 19: 223-229. [DOI] [PubMed] [Google Scholar]
- 7.Bui, T., and C. B. Thompson. 2006. Cancer's sweet tooth. Cancer Cell 9: 419-420. [DOI] [PubMed] [Google Scholar]
- 8.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18: 2916-2924. [DOI] [PubMed] [Google Scholar]
- 9.Corn, P. G., M. S. Ricci, K. A. Scata, A. M. Arsham, M. C. Simon, D. T. Dicker, and W. S. El-Deiry. 2005. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol. Ther. 4: 1285-1294. [DOI] [PubMed] [Google Scholar]
- 10.Covello, K. L., J. Kehler, H. Yu, J. D. Gordan, A. M. Arsham, C. J. Hu, P. A. Labosky, M. C. Simon, and B. Keith. 2006. HIF-2α regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 20: 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covello, K. L., M. C. Simon, and B. Keith. 2005. Targeted replacement of hypoxia-inducible factor-1α by a hypoxia-inducible factor-2α knock-in allele promotes tumor growth. Cancer Res. 65: 2277-2286. [DOI] [PubMed] [Google Scholar]
- 12.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danial, N. N., C. F. Gramm, L. Scorrano, C. Y. Zhang, S. Krauss, A. M. Ranger, S. R. Datta, M. E. Greenberg, L. J. Licklider, B. B. Lowell, S. P. Gygi, and S. J. Korsmeyer. 2003. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424: 952-956. [DOI] [PubMed] [Google Scholar]
- 14.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15: 2023-2030. [DOI] [PubMed] [Google Scholar]
- 15.Fantin, V. R., J. St.-Pierre, and P. Leder. 2006. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9: 425-434. [DOI] [PubMed] [Google Scholar]
- 16.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16: 4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, P., H. Zhang, R. Dinavahi, L. Feng, Y. Xiang, V. Raman, Z. Bhujwalla, D. W. Felsher, L. Cheng, J. Pevsner, L. A. Lee, G. L. Semenza, and C. V. Dang. 2007. HIF-dependent anti-tumorigenic effect of anti-oxidants in vivo. Cancer Cell 12: 230-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, L. B., Q. Li, M. S. Park, W. M. Flanagan, G. L. Semenza, and C. V. Dang. 2001. Hypoxia inhibits G1/S transition through regulation of p27 expression. J. Biol. Chem. 276: 7919-7926. [DOI] [PubMed] [Google Scholar]
- 19.Gatenby, R. A., and R. J. Gillies. 2004. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4: 891-899. [DOI] [PubMed] [Google Scholar]
- 20.Giaccia, A., B. G. Siim, and R. S. Johnson. 2003. HIF-1 as a target for drug development. Nat. Rev. Drug Discov. 2: 803-811. [DOI] [PubMed] [Google Scholar]
- 21.Giaccia, A. J., M. C. Simon, and R. Johnson. 2004. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 18: 2183-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goda, N., H. E. Ryan, B. Khadivi, W. McNulty, R. C. Rickert, and R. S. Johnson. 2003. Hypoxia-inducible factor 1α is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 23: 359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golshani-Hebroni, S. G., and S. P. Bessman. 1997. Hexokinase binding to mitochondria: a basis for proliferative energy metabolism. J. Bioenerg. Biomembr. 29: 331-338. [DOI] [PubMed] [Google Scholar]
- 24.Gordan, J. D., J. A. Bertout, C. J. Hu, J. A. Diehl, and M. C. Simon. 2007. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11: 335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordan, J. D., and M. C. Simon. 2007. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 17: 71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlob, K., N. Majewski, S. Kennedy, E. Kandel, R. B. Robey, and N. Hay. 2001. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 15: 1406-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandori, C., and R. N. Eisenman. 1997. Myc target genes. Trends Biochem. Sci. 22: 177-181. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100: 57-70. [DOI] [PubMed] [Google Scholar]
- 29.Holness, M. J., and M. C. Sugden. 2003. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 31: 1143-1151. [DOI] [PubMed] [Google Scholar]
- 30.Hon, W. C., M. I. Wilson, K. Harlos, T. D. Claridge, C. J. Schofield, C. W. Pugh, P. H. Maxwell, P. J. Ratcliffe, D. I. Stuart, and E. Y. Jones. 2002. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417: 975-978. [DOI] [PubMed] [Google Scholar]
- 31.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23: 9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hueber, A. O., and G. I. Evan. 1998. Traps to catch unwary oncogenes. Trends Genet. 14: 364-367. [DOI] [PubMed] [Google Scholar]
- 33.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464-468. [DOI] [PubMed] [Google Scholar]
- 34.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468-472. [DOI] [PubMed] [Google Scholar]
- 35.Kelly, B. D., S. F. Hackett, K. Hirota, Y. Oshima, Z. Cai, S. Berg-Dixon, A. Rowan, Z. Yan, P. A. Campochiaro, and G. L. Semenza. 2003. Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ. Res. 93: 1074-1081. [DOI] [PubMed] [Google Scholar]
- 36.Kim, J. W., and C. V. Dang. 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66: 8927-8930. [DOI] [PubMed] [Google Scholar]
- 37.Kim, J. W., I. Tchernyshyov, G. L. Semenza, and C. V. Dang. 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3: 177-185. [DOI] [PubMed] [Google Scholar]
- 38.Kim, J. W., K. I. Zeller, Y. Wang, A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 2004. Evaluation of Myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 24: 5923-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, W. Y., M. Safran, M. R. Buckley, B. L. Ebert, J. Glickman, M. Bosenberg, M. Regan, and W. G. Kaelin, Jr. 2006. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. EMBO J. 25: 4650-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knies-Bamforth, U. E., S. B. Fox, R. Poulsom, G. I. Evan, and A. L. Harris. 2004. c-Myc interacts with hypoxia to induce angiogenesis in vivo by a vascular endothelial growth factor-dependent mechanism. Cancer Res. 64: 6563-6570. [DOI] [PubMed] [Google Scholar]
- 41.Kondo, K., J. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin, Jr. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1: 237-246. [DOI] [PubMed] [Google Scholar]
- 42.Koshiji, M., and L. E. Huang. 2004. Dynamic balancing of the dual nature of HIF-1α for cell survival. Cell Cycle 3: 853-854. [DOI] [PubMed] [Google Scholar]
- 43.Koshiji, M., Y. Kageyama, E. A. Pete, I. Horikawa, J. C. Barrett, and L. E. Huang. 2004. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 23: 1949-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koshiji, M., K. K. To, S. Hammer, K. Kumamoto, A. L. Harris, P. Modrich, and L. E. Huang. 2005. HIF-1α induces genetic instability by transcriptionally downregulating MutSα expression. Mol. Cell 17: 793-803. [DOI] [PubMed] [Google Scholar]
- 45.Krishnamachary, B., D. Zagzag, H. Nagasawa, K. Rainey, H. Okuyama, J. H. Baek, and G. L. Semenza. 2006. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 66: 2725-2731. [DOI] [PubMed] [Google Scholar]
- 46.Li, F., Y. Wang, K. I. Zeller, J. J. Potter, D. R. Wonsey, K. A. O'Donnell, J. W. Kim, J. T. Yustein, L. A. Lee, and C. V. Dang. 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 25: 6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 100: 8164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, L., T. P. Cash, R. G. Jones, B. Keith, C. B. Thompson, and M. C. Simon. 2006. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell 21: 521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majewski, N., V. Nogueira, R. B. Robey, and N. Hay. 2004. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol. Cell. Biol. 24: 730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maltepe, E., and M. C. Simon. 1998. Oxygen, genes, and development: an analysis of the role of hypoxic gene regulation during murine vascular development. J. Mol. Med. 76: 391-401. [DOI] [PubMed] [Google Scholar]
- 51.Maranchie, J. K., J. R. Vasselli, J. Riss, J. S. Bonifacino, W. M. Linehan, and R. D. Klausner. 2002. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 1: 247-255. [DOI] [PubMed] [Google Scholar]
- 52.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20: 5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8: 1039-1048. [PubMed] [Google Scholar]
- 54.Mathupala, S. P., A. Rempel, and P. L. Pedersen. 2001. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J. Biol. Chem. 276: 43407-43412. [DOI] [PubMed] [Google Scholar]
- 55.Mazure, N. M., C. Chauvet, B. Bois-Joyeux, M. A. Bernard, H. Nacer-Cherif, and J. L. Danan. 2002. Repression of alpha-fetoprotein gene expression under hypoxic conditions in human hepatoma cells: characterization of a negative hypoxia response element that mediates opposite effects of hypoxia inducible factor-1 and c-Myc. Cancer Res. 62: 1158-1165. [PubMed] [Google Scholar]
- 56.Min, J. H., H. Yang, M. Ivan, F. Gertler, W. G. Kaelin, Jr., and N. P. Pavletich. 2002. Structure of an HIF-1α-pVHL complex: hydroxyproline recognition in signaling. Science 296: 1886-1889. [DOI] [PubMed] [Google Scholar]
- 57.Nesbit, C. E., J. M. Tersak, and E. V. Prochownik. 1999. MYC oncogenes and human neoplastic disease. Oncogene 18: 3004-3016. [DOI] [PubMed] [Google Scholar]
- 58.Orian, A., S. S. Grewal, P. S. Knoepfler, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2005. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harbor Symp. Quant. Biol. 70: 299-307. [DOI] [PubMed] [Google Scholar]
- 59.Osthus, R. C., H. Shim, S. Kim, Q. Li, R. Reddy, M. Mukherjee, Y. Xu, D. Wonsey, L. A. Lee, and C. V. Dang. 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 275: 21797-21800. [DOI] [PubMed] [Google Scholar]
- 60.Pajic, A., D. Spitkovsky, B. Christoph, B. Kempkes, M. Schuhmacher, M. S. Staege, M. Brielmeier, J. Ellwart, F. Kohlhuber, G. W. Bornkamm, A. Polack, and D. Eick. 2000. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int. J. Cancer 87: 787-793. [DOI] [PubMed] [Google Scholar]
- 61.Papandreou, I., R. A. Cairns, L. Fontana, A. L. Lim, and N. C. Denko. 2006. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 3: 187-197. [DOI] [PubMed] [Google Scholar]
- 62.Parry, D. M., and P. L. Pedersen. 1983. Intracellular localization and properties of particulate hexokinase in the Novikoff ascites tumor. Evidence for an outer mitochondrial membrane location. J. Biol. Chem. 258: 10904-10912. [PubMed] [Google Scholar]
- 63.Pastorino, J. G., and J. B. Hoek. 2003. Hexokinase II: the integration of energy metabolism and control of apoptosis. Curr. Med. Chem. 10: 1535-1551. [DOI] [PubMed] [Google Scholar]
- 64.Pastorino, J. G., N. Shulga, and J. B. Hoek. 2002. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 277: 7610-7618. [DOI] [PubMed] [Google Scholar]
- 65.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2: 764-776. [DOI] [PubMed] [Google Scholar]
- 66.Ponzielli, R., S. Katz, D. Barsyte-Lovejoy, and L. Z. Penn. 2005. Cancer therapeutics: targeting the dark side of Myc. Eur. J. Cancer 41: 2485-2501. [DOI] [PubMed] [Google Scholar]
- 67.Pugh, C. W., and P. J. Ratcliffe. 2003. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9: 677-684. [DOI] [PubMed] [Google Scholar]
- 68.Rathmell, J. C., C. J. Fox, D. R. Plas, P. S. Hammerman, R. M. Cinalli, and C. B. Thompson. 2003. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol. Cell. Biol. 23: 7315-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 17: 3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5: 343-354. [DOI] [PubMed] [Google Scholar]
- 71.Schuhmacher, M., M. S. Staege, A. Pajic, A. Polack, U. H. Weidle, G. W. Bornkamm, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9: 1255-1258. [DOI] [PubMed] [Google Scholar]
- 72.Semenza, G. L. 2002. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8: S62-S67. [DOI] [PubMed] [Google Scholar]
- 73.Semenza, G. L. 2001. HIF-1, O2, and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 107: 1-3. [DOI] [PubMed] [Google Scholar]
- 74.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3: 721-732. [DOI] [PubMed] [Google Scholar]
- 75.Semenza, G. L., P. H. Roth, H. M. Fang, and G. L. Wang. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269: 23757-23763. [PubMed] [Google Scholar]
- 76.Shaw, R. J. 2006. Glucose metabolism and cancer. Curr. Opin. Cell Biol. 18: 598-608. [DOI] [PubMed] [Google Scholar]
- 77.Shchors, K., E. Shchors, F. Rostker, E. R. Lawlor, L. Brown-Swigart, and G. I. Evan. 2006. The Myc-dependent angiogenic switch in tumors is mediated by interleukin 1β. Genes Dev. 20: 2527-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shim, H., C. Dolde, B. C. Lewis, C. S. Wu, G. Dang, R. A. Jungmann, R. Dalla-Favera, and C. V. Dang. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 94: 6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11: 72-82. [DOI] [PubMed] [Google Scholar]
- 80.To, K. K., M. Koshiji, S. Hammer, and L. E. Huang. 2005. Genetic instability: the dark side of the hypoxic response. Cell Cycle 4: 881-882. [DOI] [PubMed] [Google Scholar]
- 81.Wang, G. L., and G. L. Semenza. 1993. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl. Acad. Sci. USA 90: 4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Warburg, O. 1956. On respiratory impairment in cancer cells. Science 124: 269-270. [PubMed] [Google Scholar]
- 83.Warburg, O. 1956. On the origin of cancer cells. Science 123: 309-314. [DOI] [PubMed] [Google Scholar]
- 84.Zeller, K. I., X. Zhao, C. W. Lee, K. P. Chiu, F. Yao, J. T. Yustein, H. S. Ooi, Y. L. Orlov, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, V. A. Kuznetsov, W. K. Sung, Y. Ruan, C. V. Dang, and C. L. Wei. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA 103: 17834-17839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang, H., P. Gao, R. Fukuda, G. Kumar, B. Krishnamachary, K. I. Zeller, C. V. Dang, and G. L. Semenza. 2007. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11: 407-420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.