Abstract
We explored the mechanisms of chromatin compaction and transcriptional regulation by poly(ADP-ribose) polymerase 1 (PARP-1), a nucleosome-binding protein with an NAD+-dependent enzymatic activity. By using atomic force microscopy and a complementary set of biochemical assays with reconstituted chromatin, we showed that PARP-1 promotes the localized compaction of chromatin into supranucleosomal structures in a manner independent of the amino-terminal tails of core histones. In addition, we defined the domains of PARP-1 required for nucleosome binding, chromatin compaction, and transcriptional repression. Our results indicate that the DNA binding domain (DBD) of PARP-1 is necessary and sufficient for binding to nucleosomes, yet the DBD alone is unable to promote chromatin compaction and only partially represses RNA polymerase II-dependent transcription in an in vitro assay with chromatin templates (∼50% of the repression observed with wild-type PARP-1). Furthermore, our results show that the catalytic domain of PARP-1, which does not bind nucleosomes on its own, cooperates with the DBD to promote chromatin compaction and efficient transcriptional repression in a manner independent of its enzymatic activity. Collectively, our results have revealed a novel function for the catalytic domain in chromatin compaction. In addition, they show that the DBD and catalytic domain cooperate to regulate chromatin structure and chromatin-dependent transcription, providing mechanistic insights into how these domains contribute to the chromatin-dependent functions of PARP-1.
Chromatin is the physiological template for nuclear processes involving genomic DNA, including transcription, replication, recombination, and repair. Nucleosomes, the fundamental repeating units of chromatin, are protein-DNA complexes containing 146 bp of DNA wrapped around a protein core containing two copies each of four core histone proteins (H2A, H2B, H3, and H4) (22). Chromatin exists in various conformations, including open conformations where individual nucleosomes are separated, freely accessible, and mobile, as well as compact conformations where nucleosomes are juxtaposed and partially occluded in higher-order structures (6, 11, 37). The extent of nucleosome mobility and chromatin compaction are two determinants of the activity of chromatin-dependent processes, such as transcription by RNA polymerase II (Pol II) (11, 25, 33).
A variety of chromatin “architectural” proteins are known to bind to nucleosomes, promote chromatin compaction, and modulate transcriptional responses (23, 28). For example, the linker histone H1 binds at or near the dyad axis of nucleosomes and promotes the formation of a higher-order structure known as the 30-nm fiber (28, 38). In the 30-nm fiber, nucleosomes are tightly packed, and the nucleosomal DNA is less accessible and more refractory to transcription (28). We have recently shown that poly(ADP-ribose) polymerase 1 (PARP-1) is a nucleosome binding protein with linker histone-like properties. Like H1, PARP-1 binds at or near the dyad axis of the nucleosome and promotes the compaction of chromatin into higher-order structures (15). In biochemical assays, these PARP-1-dependent structures are refractory to transcription (15). The underlying mechanisms of PARP-1-dependent chromatin compaction and transcriptional repression are not clear.
PARP-1, the founding member of a superfamily of PARP proteins, is an abundant nuclear protein (∼1.5 to 2 million molecules per cell) involved in a variety of nuclear processes, including the regulation of chromatin structure and transcription (1, 5, 16, 19). PARP-1 contains an amino-terminal zinc finger DNA binding domain (DBD) and a carboxyl-terminal allosterically regulated catalytic domain (CAT) with low basal activity (5, 29). This enzymatic activity allows PARP-1 to polymerize ADP-ribose monomers from donor NAD+ molecules into poly(ADP-ribose) chains on target proteins (5, 16). In vivo, PARP-1 is the major target for poly(ADP-ribosyl)ation (PARylation) through an automodification reaction (5). AutoPARylation of PARP-1 in the presence of NAD+ inhibits PARP-1's DNA binding, nucleosome binding, chromatin compaction, and transcriptional repression activities (15). In addition to the DBD and CAT, PARP-1 contains a number of other less well-characterized structural and functional domains that likely play important roles in its activity. These include a central automodification domain containing a BRCA1 C terminus (BRCT) motif and an NAD+-binding domain (NBD) comprising the carboxyl-terminal-most module of the CAT (29, 30). The roles played by the individual PARP-1 domains in the regulation of chromatin structure and transcription by PARP-1 have not been determined.
In the current study, we explored the mechanisms of chromatin compaction and transcriptional regulation by PARP-1 by using a complementary set of biochemical and single-molecule biophysical techniques, including in vitro chromatin assembly, binding, and transcription assays, as well as atomic force microscopy (AFM). Using these approaches, we have defined the domains of PARP-1 required for nucleosome binding, chromatin compaction, and transcriptional repression. Our results suggest key roles for the DBD and CAT of PARP-1 in the regulation of chromatin structure and chromatin-dependent transcription. In addition, they provide mechanistic insights into how these domains contribute to the chromatin-dependent functions of PARP-1.
MATERIALS AND METHODS
Purification of recombinant and native proteins.
A set of bacterial constructs for expressing His6-tagged wild-type and deletion mutant human PARP-1 proteins were made by PCR and assembled in pET19b. The PARP-1 proteins were expressed in Escherichia coli and purified using standard nickel-nitrilotriacetic acid (NTA) affinity chromatography with a 1 M NaCl wash to remove contaminating low-molecular-weight DNA fragments, as described previously (15). The proteins comprising the recombinant ACF chromatin assembly system (i.e., Drosophila NAP-1, Acf1, and ISWI) were expressed in Sf9 insect cells using recombinant baculoviruses and purified as described previously (13). Recombinant estrogen receptor alpha (ERα) was expressed in Sf9 insect cells and purified as described previously (17). Native Drosophila core histones were purified as described previously (2). Native calf thymus histone H1 was purchased from Calbiochem. The purities and concentrations of the purified proteins were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie brilliant blue R-250. All experiments were run with a minimum of two independent preparations of protein.
Assembly and purification of chromatin templates.
In vitro chromatin assembly was carried out with two different plasmid DNAs, an ∼10.5-kb plasmid (a pFastbac1 derivative containing a large cDNA insert) (27), which can accommodate ∼64 physiologically spaced nucleosomes, and an ∼3.2- kb plasmid (pERE, containing four estrogen response elements (EREs) upstream of the adenovirus E4 core promoter) (18), which can accommodate ∼19 physiologically spaced nucleosomes.
Chromatin assembly was performed using two different methods: (i) Drosophila embryo extract (S190) (2) and (ii) recombinant ACF (13). The S190-based chromatin assembly reactions were set up as described previously (2, 18). Briefly, S190 extract was incubated with purified core histones, DNA plasmid template, and an ATP-regenerating system for 4 to 5 h at 27°C. RNase (Roche) was added during the last half hour of the incubation to degrade the RNA in the S190 extract. Assembled chromatin was purified from the extract by two sequential chromatography steps: fractionation through a 4-ml, 15% to 40% sucrose gradient (10 mM HEPES, pH 7.6, 200 mM NaCl, 1 mM EDTA, 0.01% NP-40, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol), followed by size exclusion using a 4-ml Sepharose CL-4B column with elution in a buffered solution containing 5 mM triethanolamine-HCl (pH 7.0) and 0.2 mM EDTA (14, 34). Fractions were analyzed for DNA content by phenol-chloroform extraction, followed by agarose gel electrophoresis with ethidium bromide staining, and for protein content by SDS-PAGE with silver staining.
The recombinant ACF-based chromatin assembly reactions were set up using recombinant dACF-1, dISWI, and dNAP-1 as described previously (8, 13, 15). To generate “tailless” chromatin (or mock-trypsinized control chromatin), ACF-assembled chromatin was incubated with (or without) trypsin (Worthington Biochemicals) under conditions determined empirically to efficiently cleave the amino-terminal tails of the core histones (i.e., 2.8 μg trypsin/μg DNA; 27°C for 25 min). Digestion with 50% more trypsin generated histone fragments of similar sizes, indicating formation of a stably protected octamer core (see Fig. 3C). Trypsinization (or mock-trypsinization) reactions were stopped by the addition of a 10-fold molar excess of soybean trypsin inhibitor (Sigma), followed by purification of the chromatin by size exclusion chromatography, as described above. Any potential residual trypsin was removed by affinity chromatography with immobilized trypsin inhibitor (Pierce).
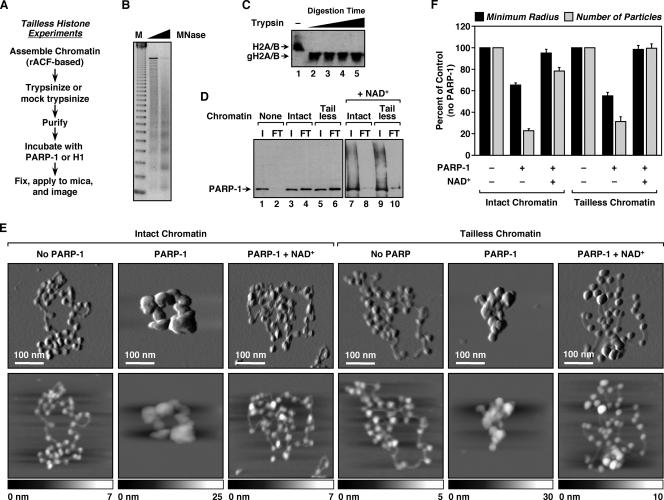
FIG. 3.
Histone tails are dispensable for PARP-1-mediated chromatin compaction. (A) Overview of the preparation of ACF-assembled tailless chromatin samples for AFM imaging (see Materials and Methods). (B) Agarose gel electrophoresis of MNase-digested chromatin assembled using recombinant ACF and pERE (see Materials and Methods). M, 123-base pair ladder. (C) Western blot for H2A and H2B illustrating the effective removal of histone tails by trypsin treatment. The digestion condition shown in lane 2 (i.e., complete, not excessive, digestion) was used for all studies with tailless chromatin. gH2A/B, globular (i.e., tailless) H2A and H2B. (D) PARP-1 chromatin binding assays. (Lanes 1 to 6) PARP-1 was subjected to small-scale size exclusion chromatography in the absence or presence of intact or tailless chromatin to assess its chromatin binding ability (see Materials and Methods). Two percent of the input (I) and 10% of the FT were analyzed by Western blotting for PARP-1. In this assay, unbound PARP-1 has a high residence time on the resin and thus is not evident in the FT. In contrast, chromatin-bound PARP-1 passes through the column with the FT. (Lanes 7 to 10) The effect of NAD+ on PARP-1 binding to chromatin was assayed in a similar manner. (E) AFM images of intact and tailless pFASTbac1-cDNA chromatin molecules alone, with a saturating amount of PARP-1, or with a saturating amount of PARP-1 plus NAD+, as indicated. Scan probe oscillation amplitude images (top) and topographical images (bottom) are as described for Fig. 1C. (F) Quantification of AFM images like those shown in panel E, as described in the legend to Fig. 1D. All determinations are based on at least 25 molecules derived from two independent preparations of chromatin. Each bar is the mean plus standard error of the mean.
Limited digestion with micrococcal nuclease (MNase) was used to assess the assembly of chromatin using either S190 or ACF. Aliquots of the assembled chromatin were digested with increasing amounts of MNase (Sigma) as described previously (2, 13). The samples were deproteinized by digestion with proteinase K and phenol-chloroform extraction and then analyzed by agarose gel electrophoresis with ethidium bromide staining.
AFM.
Purified chromatin was diluted to a DNA concentration of ∼0.5 ng/μl and then incubated alone, with PARP-1, or with histone H1 for 30 min at 27°C. Typical PARP-1/nucleosome and H1/nucleosome ratios used for saturating binding were ∼2:1. When used, NAD+ was added at a concentration of 300 μM after the binding of PARP-1, with continued incubation at 27°C for an additional 30 min. Samples were fixed for 2 to 4 h at 4°C with 0.5% glutaraldehyde. Magnesium acetate was added to each sample at a concentration of 2 mM, and the samples were applied to freshly cleaved mica, incubated at room temperature for 2 min, rinsed with double-distilled H2O, and dried under nitrogen (3). Imaging was performed in AC mode (tapping mode) on a PicoPlus atomic force microscope (Molecular Imaging) using silicon AC160TS cantilevers (Asylum Research) at Cornell University's Nanobiotechnology Center. The typical driving frequency was 300 kHz, and the scan speeds were ∼2 μm/s. The digitized images were used to extract the following quantitative information about the chromatin molecules using Scion Image (Scion Corp., Frederick, MD): (i) the average minimum radius of a circle needed to enclose an entire molecule of chromatin, (ii) the average number of discernible particles per molecule of chromatin, and (iii) the average height of the chromatin molecules. For each condition tested and quantified, at least 15 molecules of chromatin (but typically 20 to 30 molecules) derived from two independent preparations were analyzed.
Chromatin binding assays.
Chromatin was assembled using recombinant ACF (and trypsinized, in some cases) as described above, but purification by Sepharose CL-4B size exclusion was omitted. The chromatin was centrifuged for 1 min at 3,000 rpm in a microcentrifuge to remove any aggregated material prior to the binding assay. Wild-type or mutant PARP-1 protein was added to aliquots of chromatin at a PARP-1/nucleosome ratio of 1:2, and the samples were incubated for 30 min at 27°C. When used, NAD+ was added at a concentration of 300 μM after the binding of PARP-1, with continued incubation at 27°C for an additional 30 min. The binding reaction mixtures were fractionated using spin columns with a 0.75-ml bed volume of Sepharose CL-4B resin. The input and flowthrough (FT) fractions were analyzed for DNA content by phenol-chloroform extraction, followed by agarose gel electrophoresis with ethidium bromide staining, and for PARP-1 content by Western blotting with antibodies recognizing the DBD or NBD of PARP-1.
In vitro transcription assays.
ERα-dependent transcription with ACF-assembled pERE chromatin templates and HeLa cell nuclear extract was performed as described previously (15, 17). Briefly, pERE was assembled into chromatin using recombinant ACF. After assembly, aliquots of chromatin were incubated with or without 10 nM ERα, 100 nM 17β-estradiol (E2), and 50 nM of PARP-1 (wild type or mutant) at 27°C for an additional 30 min. Fifteen-microliter aliquots of chromatin containing 75 ng of DNA were then transcribed using a HeLa cell nuclear extract as a source of the Pol II transcriptional machinery. The RNA products from the transcription reactions were analyzed by primer extension (17), and the data were quantified using a PhosphorImager (Molecular Dynamics). All reactions were performed in duplicate, and each experiment was run more than four times to ensure reproducibility.
PARP-1 auto(ADP-ribosyl)ation assays.
PARP-1 automodification assays were performed as described previously (15). Briefly, 2.5 pmol of purified PARP-1 protein (wild type or mutant) was incubated with 0.67 μM [32P]NAD+ in a 15-μl reaction mixture containing 30 mM HEPES, pH 7.4, and 12.5 mM MgCl2. Sheared salmon sperm DNA was added as an allosteric activator of PARP-1 enzymatic activity. The reaction mixtures were incubated at room temperature for 20 min and then analyzed on 6% polyacrylamide-SDS gels with subsequent autoradiography.
RESULTS
Imaging PARP-1-dependent chromatin compaction by AFM.
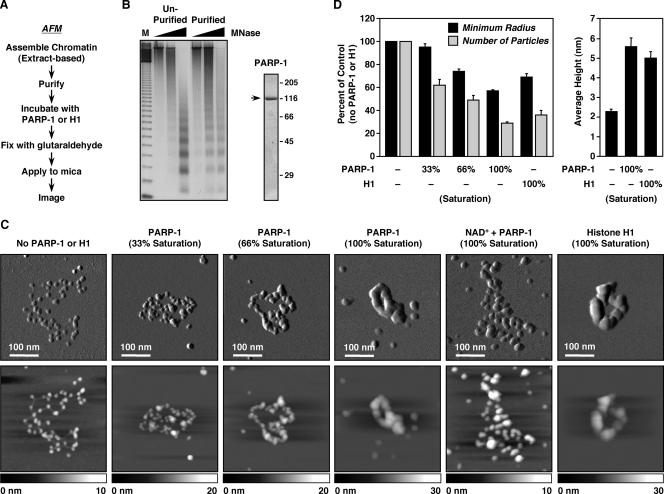
To explore the mechanisms of PARP-1-dependent chromatin compaction, we used AFM to image single molecules of chromatin in the absence or presence of PARP-1. In our initial studies, chromatin assembled on an ∼10.5-kb plasmid by using an extract-based in vitro chromatin assembly system (i.e., S190) (2) was purified, incubated with various amounts of purified recombinant PARP-1, and prepared for AFM imaging (Fig. 1A and B). Purified chromatin in the absence of PARP-1 presented an open beads-on-a-string conformation with clearly visible nucleosomes and linker DNA (Fig. 1C), similar to the conformation observed for histone H1-depleted native chromatin (21). The addition of increasing amounts of PARP-1, however, promoted a dose-dependent compaction of the chromatin, resulting in a well-condensed, torus-shaped structure with multiple subdomains but no resolvable individual nucleosomes (Fig. 1C; see also Fig. S1 and S2 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). (Note that for the studies described here, chromatin “compaction” refers to intrafiber chromatin folding. See below for a description of localized compaction by PARP-1.) This structure is similar to the structure observed upon addition of histone H1 to chromatin assembled in vitro using either S190 extract (as in Fig. 1C) or salt dialysis (12). Similar effects of PARP-1 were observed with linearized chromatin molecules, although the torus-shaped structures seen with circular chromatin molecules were not observed, as expected (see Fig. S3 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm).
FIG. 1.
PARP-1 compacts in vitro-assembled chromatin. (A) Overview of the preparation of S190-assembled chromatin samples for AFM imaging (see Materials and Methods). (B) (Left) Agarose gel electrophoresis of MNase-digested chromatin assembled using S190 and pFASTbac1-cDNA before and after purification by glycerol gradient and size exclusion chromatography (see Materials and Methods). M, 123-base pair ladder. (Right) SDS-PAGE analysis of purified baculovirus-expressed FLAG-tagged PARP-1 visualized by staining with Coomassie brilliant blue R-250. (C) AFM images of S190-assembled pFASTbac1-cDNA chromatin with increasing amounts of PARP-1, a saturating amount of PARP-1 plus 300 μM NAD+, or a saturating amount of H1. The amount of PARP-1 or H1 required to saturate compaction was determined empirically (see Materials and Methods). (Top) Scan probe oscillation amplitude images. The length scale is indicated by the white bars. (Bottom) Topographical images. The height scale is shown along the bottom of each image. (D) Quantification of AFM images like those shown in panel C. (Left) Average minimum radius of the circle needed to enclose an entire molecule of chromatin (black bars) and average number of discernible particles per molecule of chromatin (gray bars) under the conditions indicated. The values are plotted as percentages of chromatin with no PARP-1 or H1 added. (Right) Average height of nucleosomal arrays with or without saturating amounts of PARP-1 or H1. All determinations for PARP-1 are averages of at least 20 individual molecules of chromatin derived from three independently prepared batches of chromatin. All determinations for H1 are averages of at least 15 molecules of chromatin derived from two independent preparations of chromatin. Each bar is the mean plus standard error of the mean.
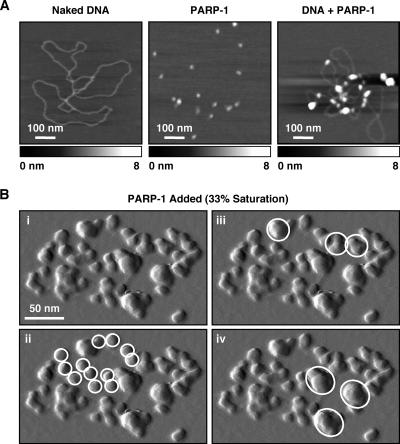
The compacting activity of PARP-1 was specific to chromatin and did not promote the formation of similarly compacted structures on naked DNA templates (Fig. 2A). PARP-1, however, did show dispersed binding to naked plasmid DNA, especially at crossovers (Fig. 2A), as previously described (29). The addition of NAD+ to PARP-1-bound chromatin reversed the compaction induced by PARP-1, resulting again in an open, beads-on-a-string structure (Fig. 1C), as previously noted (15). NAD+-mediated decondensation also resulted in an increase in the apparent particle size, possibly due to incomplete release of PARP-1 from nucleosomes, as well as littering of the sample surface with globular structures, possibly representing automodified PARP-1 molecules.
FIG. 2.
PARP-1 promotes the localized compaction of chromatin, but not naked DNA. (A) AFM topographical images, as described for Fig. 1C, of naked pFastbac1-cDNA plasmid DNA (left) purified recombinant PARP-1 protein (center), and PARP-1 plus naked pFastbac1-cDNA plasmid DNA (right). (B) (i) Enlargement of a scan probe oscillation amplitude AFM image from Fig. 1C showing an ACF-assembled chromatin array with a subsaturating amount (33%) of PARP-1 added. In ii through iv, the same image is marked with white circles to highlight unbound nucleosomes (ii) and supranucleosomal structures of increasing size, representing localized compaction (iii and iv).
The AFM images were used to extract quantitative information about the chromatin molecules, including (i) the average minimum radius of a circle required to encompass an entire molecule of chromatin, (ii) the average number of discernible particles per molecule of chromatin, and (iii) the average height of the chromatin molecules (Fig. 1D). The average minimum radius of a circle required to enclose an entire chromatin molecule decreased in a dose-dependent manner as more PARP-1 was added, as did the average number of discrete particles per molecule. Furthermore, the average height of the chromatin molecules increased by nearly threefold in the presence of saturating amounts of PARP-1, indicating the formation of supranucleosomal structures. The addition of H1 induced similar changes in these parameters (Fig. 1D); comparable results have been observed for the heterochromatin-associated Polycomb group protein complex (7). Our direct observations by AFM imaging of chromatin compaction by PARP-1 provide strong support for our conclusions drawn from previous biochemical assays (15).
PARP-1 promotes the localized compaction of chromatin.
To explore the underlying mechanisms of PARP-1-dependent chromatin compaction in more detail, we examined more closely AFM images of chromatin molecules containing subsaturating amounts of PARP-1. At 33% saturation, the chromatin was organized into supranucleosomal particles interspersed with free nucleosomes (Fig. 1C). At 66% saturation, the chromatin was organized into a chain of adjacent supranucleosomal particles (Fig. 1C). Under both of these conditions, the chromatin molecules were well spread and the individual particles were well dispersed (Fig. 1C). Higher magnification of chromatin with subsaturating amounts of PARP-1 revealed more clearly the “neighborhoods” of supranucleosomal particles interspersed with free nucleosomes at 33% saturation (Fig. 2B). Taken together, these images suggest that (i) PARP-1 brings together groups of local, possibly adjacent, nucleosomes to form supranucleosomal structures (as opposed to bridging between nucleosomes located at a distance on the same molecule of chromatin [7] or on adjacent molecules of chromatin, i.e., interfiber interactions), and (ii) these PARP-1-bound supranucleosomal structures organize along the linear path of the DNA into higher-order structures. In this regard, note that at 100% saturation, the chromatin molecules contain a series of subdomains organized in a torus-shaped structure, which is expected from the topological constraints imposed by using circular plasmid DNA to assemble the chromatin (Fig. 1C; see also Fig. S1 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm).
Histone tails are dispensable for PARP-1-dependent chromatin compaction.
Having developed a system for directly visualizing chromatin compaction by PARP-1, we proceeded to investigate the structural aspects of nucleosomes and PARP-1 required for this activity. We began by assessing the requirement for the amino-terminal tails of the core histones. To do so, we generated “tailless” chromatin by digesting ACF-assembled chromatin with trypsin until a minimal, but stable, nucleosomal protein core remained (Fig. 3A through C). The chromatin was then purified to fully remove tryptic activity (data not shown).
To determine if PARP-1 binds tailless chromatin as effectively as intact chromatin, we devised a chromatin binding assay based on size exclusion chromatography using Sepharose CL-4B resin (see Materials and Methods). In this assay, free PARP-1 remained with the resin in the column and thus was not evident in the FT (Fig. 3D, lanes 1 and 2). In contrast, chromatin-bound PARP-1 passed through the column and appeared in the FT (Fig. 3D, lanes 3 and 4). Interestingly, PARP-1 bound to intact (i.e., mock-trypsinized) and tailless chromatin equally well (Fig. 3D, compare lanes 4 and 6), suggesting that PARP-1's major contacts with nucleosomes occur via the DNA (15) or the globular domains of the core histones, not the amino-terminal tails. The addition of NAD+ promoted the autoPARylation of PARP-1 and its release from both intact and tailless chromatin, as indicated by the higher-molecular-weight smearing of the PARP-1 signal in the input and a reduced level of PARP-1 in the FT (Fig. 3D, lanes 7 through 10).
In AFM imaging experiments, tailless chromatin exhibited a beads-on-a-string conformation in the absence of PARP-1 and a compact conformation in the presence of PARP-1, similar to what was observed with intact control chromatin (Fig. 3E). The addition of NAD+ promoted the reversal of compaction with the tailless chromatin, again similar to what was observed with intact control chromatin (Fig. 3E). The effects of PARP-1 and NAD+ on tailless chromatin were readily apparent upon quantification of the results (Fig. 3F). Taken together, the results from the binding assays and AFM imaging experiments indicate that NAD+-dependent modulation of chromatin binding and compaction by PARP-1 do not require the amino-terminal tails of the core histones.
The DBD and CAT of PARP-1 cooperate to promote chromatin compaction.
Next, we sought to determine the structural and functional domains of PARP-1 required for chromatin compaction. We showed previously, using point mutants, that PARP-1's DNA binding activity is required for binding to nucleosomes and, hence, for the compaction of chromatin by PARP-1 (15). Furthermore, we showed that PARP-1's catalytic activity is required for the NAD+-dependent release of PARP-1 from chromatin and subsequent decondensation, although the catalytic activity itself is not required for the binding and compaction of chromatin by PARP-1 (15). To evaluate the roles of these and other PARP-1 domains in more detail, we generated a panel of PARP-1 deletion mutants, including mutants lacking the BRCT motif (ΔBRCT) and the CAT (ΔCAT) (Fig. 4A and 5A and B). BRCT motifs are protein-protein interaction modules that can function as phosphoprotein binding domains (10, 29). The region of PARP-1 containing the BRCT motif has been shown previously to mediate homodimerization of PARP-1 molecules (31). The CAT, which forms a structured region at the carboxyl terminus of PARP-1, contains a discrete NBD (29, 30). It mediates the NAD+-dependent activities of PARP-1 and provides additional protein-protein interactions that contribute to the homodimerization of PARP-1 molecules (24).
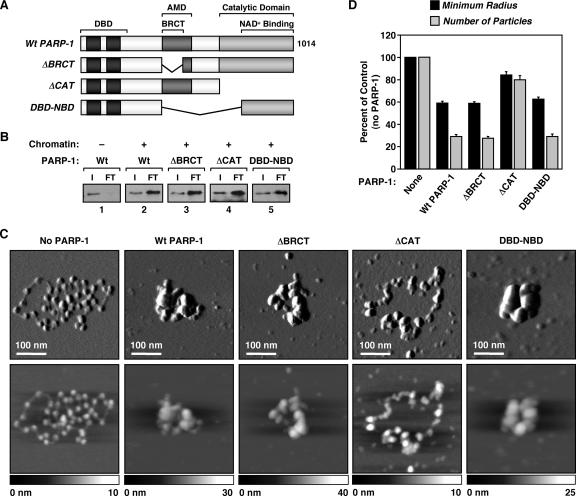
FIG. 4.
The CAT is required for chromatin compaction by PARP-1. (A) Schematic diagram of the PARP-1 deletion mutants used in these studies. (B) PARP-1 chromatin binding assays as described in the legend to Fig. 3D using the PARP-1 mutants shown in panel A. (C) AFM images of ACF-assembled pFASTbac1-cDNA chromatin alone or with the addition of wild-type or mutant PARP-1, as indicated. Scan probe oscillation amplitude images (top) and topographical images (bottom) are as described for Fig. 1C. (D) Quantification of AFM images like those shown in panel C, as described in the legend to Fig. 1D. All determinations are based on at least 30 molecules derived from two independent preparations of chromatin. Each bar is the mean plus standard error of the mean.
FIG. 5.
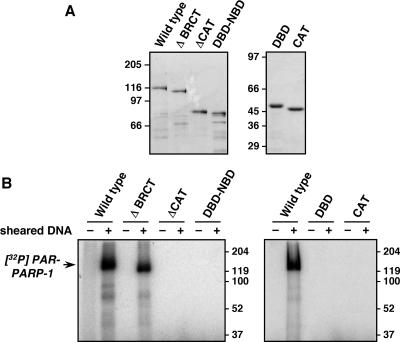
Purification and enzymatic activities of PARP-1 mutants. The PARP-1 mutants shown in Fig. 4A and 6A were expressed in E. coli and purified by nickel-NTA affinity chromatography (see Materials and Methods). (A) Purified wild-type and mutant PARP-1 proteins were analyzed by SDS-PAGE with staining by Coomassie brilliant blue R-250. Molecular mass markers are shown. (B) Automodification activities of wild-type and mutant PARP-1 proteins. The automodification activities of the indicated PARP-1 proteins were determined in the presence of [32P]NAD+, as described in Materials and Methods. Sheared DNA was added as an allosteric activator of PARP-1. The samples were analyzed by SDS-PAGE with detection of the 32P signal by phosphorimager analysis.
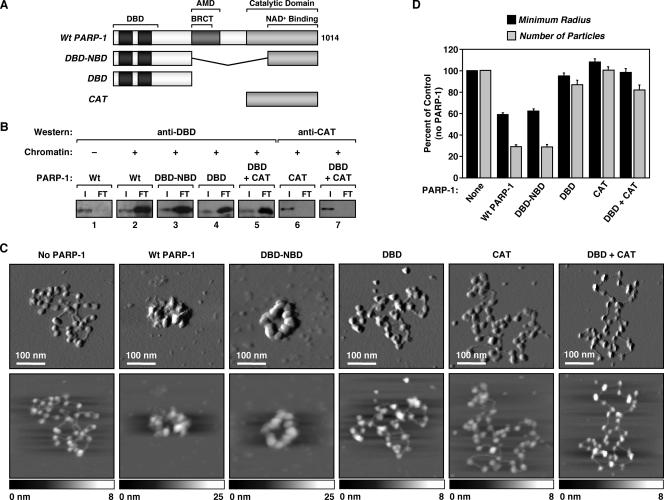
In chromatin binding assays, both the PARP-1 protein with the BRCT motif deleted and that with the CAT deleted (ΔBRCT and ΔCAT, respectively) bound to chromatin like wild-type PARP-1 (Fig. 4B, lanes 2 through 4, compare input/FT ratios; see also Fig. S4A at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). In AFM imaging experiments, ΔBRCT promoted the compaction of chromatin in a manner similar to that of wild-type PARP-1 (Fig. 4C and D). In contrast, ΔCAT failed to promote the compaction of chromatin, as is apparent from both the imaging (Fig. 4C; see also Fig. S5 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm) and quantification (Fig. 4D). ΔCAT, however, increased the apparent particle size, consistent with its binding to nucleosomes (Fig. 4C; see also Fig. S5 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Interestingly, a PARP-1 protein containing only the extended DBD plus the NBD (DBD-NBD) exhibited activities similar to those of wild-type PARP-1 in both chromatin binding and compaction assays (Fig. 4B through D). Together, our results indicate that the DBD and CAT/NBD of PARP-1 are required for chromatin compaction, while the BRCT motif is dispensable.
To determine the role of the DBD and CAT/NBD in PARP-1-dependent chromatin compaction in more detail, we performed a series of experiments with the two domains separated, or linked as in the DBD-NBD protein (Fig. 5A and B and 6A). In chromatin binding assays, the DBD alone and DBD-NBD bound to chromatin in a manner similar to that of wild-type PARP-1 (Fig. 6B; see also Fig. S4A at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). In contrast, the CAT alone or CAT in the presence of, but physically separated from, DBD did not bind to chromatin (Fig. 6B). These results indicate that direct binding to nucleosomes is mediated primarily by the DBD of PARP-1. In AFM imaging experiments, DBD-NBD promoted the compaction of chromatin in a manner similar to that of wild-type PARP-1, while DBD alone, CAT alone, or DBD and CAT added simultaneously as separate polypeptides did not promote the compaction of chromatin (Fig. 6C and D; see also Fig. S5 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Collectively, our results from the chromatin binding and compaction assays indicate that the binding of PARP-1 to chromatin requires the DBD, while compaction of chromatin by PARP-1 requires both the DBD and the CAT/NBD.
FIG. 6.
The DBD and CAT are necessary and sufficient for chromatin compaction by PARP-1 but do not function in trans. (A) Schematic diagram of the PARP-1 deletion mutants used in these studies. (B) PARP-1 chromatin binding assays as described in the legend to Fig. 3D using the PARP-1 mutants shown in panel A. DBD-NBD indicates a single fusion peptide containing both domains, whereas DBD + CAT indicates separate polypeptides added in trans. Wt, wild type. (C) AFM images of ACF-assembled pFASTbac1-cDNA chromatin alone or with the addition of wild-type or mutant PARP-1, as indicated. Scan probe oscillation amplitude images (top) and topographical images (bottom) are as described for Fig. 1C. (D) Quantification of AFM images like those shown in panel C, as described in the legend to Fig. 1D. All determinations are based on at least 25 molecules derived from two independent preparations of chromatin. Each bar is the mean plus standard error of the mean.
The DBD and CAT of PARP-1 cooperate to promote efficient transcriptional repression.
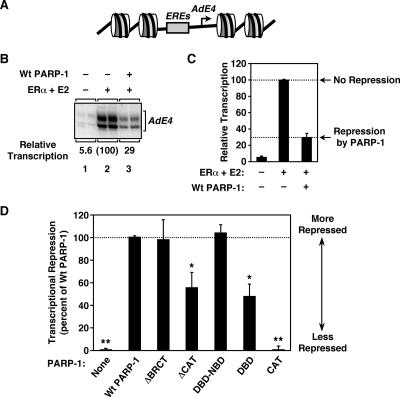
To explore the relationship between chromatin compaction and transcriptional repression by PARP-1, we examined the abilities of the PARP-1 mutants to repress activator-dependent in vitro transcription by Pol II. In this case, we used ERα, a ligand-regulated, DNA binding transcription factor, as the activator. We have shown previously that both PARP-1 and H1 repress ERα-dependent transcription from a minimal enhancer-promoter construct assembled into chromatin (4, 15). We assembled a plasmid DNA template containing four ERα binding sites (EREs) upstream of the adenovirus E4 promoter (Fig. 7A) into chromatin by using ACF and transcribed it by using a HeLa cell nuclear extract as a source of Pol II transcription machinery. The addition of ERα and its ligand, E2, stimulated transcription ∼20-fold over the basal level in the absence of PARP-1 (Fig. 7B, compare lanes 1 and 2). The addition of wild-type PARP-1 reduced ERα/E2-dependent transcription by about threefold (Fig. 7B, compare lanes 2 and 3, and C), as we observed previously for H1 (4). Similar effects of PARP-1 were observed with linearized chromatin templates (see Fig. S3 at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Of the PARP-1 mutants tested, only ΔBRCT and DBD-NBD exhibited abilities to repress transcription similar to that of wild-type PARP-1 (Fig. 7D). In contrast, CAT showed no repression, whereas ΔCAT and DBD showed about a 50% reduction in repression relative to wild-type PARP-1 (Fig. 7D; see also Fig. S4B at http://www.mbg.cornell.edu/cals/mbg/research/kraus-lab/sm.cfm). Together, these results indicate that some level of transcriptional repression is conferred by the DBD alone but the DBD plus the CAT is required for maximal transcriptional repression. The implications of these results are discussed below.
FIG. 7.
Both the DBD and the CAT are required for efficient transcriptional repression by PARP-1. (A) Schematic diagram of chromatinized pERE, a plasmid template containing four EREs upstream of the adenovirus E4 core promoter. (B) Effect of wild-type (Wt) PARP-1 on ERα-dependent transcription with chromatin templates. pERE was assembled into chromatin using ACF and transcribed using a HeLa cell nuclear extract with or without ERα plus E2 and in the presence or absence of wild-type PARP-1, as indicated. The RNA products from the in vitro transcription reactions were analyzed by primer extension, and the data were quantified by phosphorimager analysis. All reactions were performed in duplicate, and the experiment was run at least four times to ensure reproducibility. (C) Quantification of in vitro transcription experiments like the one shown in panel B. Each bar is the mean plus standard error of the mean (SEM) from more than four independent experiments. (D) Effects of PARP-1 mutants on ERα-dependent transcription with chromatin templates. pERE was assembled into chromatin, transcribed in the presence or absence of wild-type or mutant PARP-1, and analyzed as described in the legend to panel B. The PARP-1 mutants are the same as those shown in Fig. 4, 5, and 6. The data are expressed as percentages of the transcriptional repression observed with wild-type PARP-1 (i.e., a percentage of “no repression” minus “repression by PARP-1”; see panel C). Each bar is the mean plus SEM from more than four independent experiments. Bars that are marked similarly with asterisks are statistically the same, while bars that are marked differently with asterisks are statistically different, as determined by analysis of variance; P < 0.05.
DISCUSSION
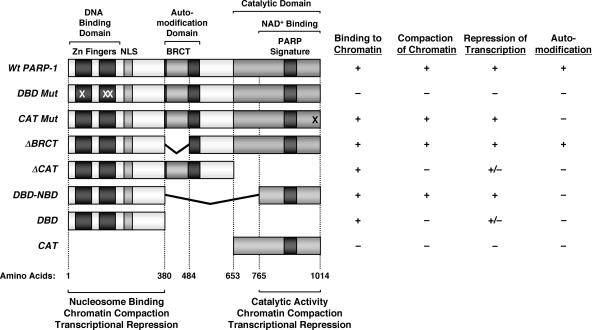
A growing body of evidence has highlighted the importance of PARP-1 as a key regulator of chromatin structure and transcription in a number of cellular and physiological contexts (16, 19). In the studies described here, we examined the molecular mechanisms underlying PARP-1's activities with chromatin. Specifically, we (i) analyzed the compaction of individual chromatin molecules by PARP-1 using AFM, (ii) showed that the amino-terminal tails of the core histones are not required for nucleosome binding or chromatin compaction by PARP-1, and (iii) defined the domains of PARP-1 required for nucleosome binding, chromatin compaction, and transcriptional repression (Fig. 8). Taken together, our results indicate that both the DBD and CAT of PARP-1 are required for efficient chromatin binding, compaction, and transcriptional modulation. These results shed new light on the roles of the DBD and CAT in the chromatin-dependent functions of PARP-1, as well as the underlying mechanisms by which PARP-1 regulates chromatin structure and transcription.
FIG. 8.
Summary of binding, chromatin compaction, transcriptional repression, and automodification characteristics of PARP-1 mutants. (Left) Schematic diagrams showing the structural and functional domains of PARP-1, as well as a set of PARP-1 deletion and point mutants. The locations of the point mutations in DBD mut (C21G, C125G, and L139P) and CAT mut (E988K) are marked with Xs. The amino acid start points and end points for the mutants are shown. NLS, nuclear localization signal. (Right) The PARP-1 deletion mutants shown were expressed in E. coli and purified by nickel-NTA affinity chromatography (see Materials and Methods). The purified proteins were screened for four different activities, as indicated: (i) binding to chromatin, as determined by in vitro chromatin binding assays (Fig. 4B and 6B); (ii) compaction of chromatin, as determined by AFM imaging (Fig. 4C and D and 6C and D); (iii) repression of transcription, as determined by in vitro transcription assays (Fig. 7D); and (iv) automodification, as determined by an in vitro PARylation assay in the presence of [32P]NAD+ (Fig. 5B). The data for DBD mut and CAT mut are from Kim et al. (15). Activity key: + indicates at least 75% of wild-type PARP-1 activity, − indicates less than 20% of wild-type PARP-1 activity, and +/− indicates ∼40 to 60% of wild-type PARP-1 activity.
Role of the DBD in the chromatin-dependent activities of PARP-1.
PARP-1's DBD is its primary point of contact with nucleosomal DNA (15, 29). As shown here, the DBD is necessary and sufficient for binding to nucleosomes, yet the DBD alone is unable to promote chromatin compaction (Fig. 8). Furthermore, the DBD alone can partially repress transcription by Pol II in an in vitro transcription assay with chromatin templates, but only about half as effectively as wild-type PARP-1. These results revealed a previously uncharacterized intrinsic transcriptional repression activity for the PARP-1 DBD that is mediated by nucleosome binding but functions independently of chromatin compaction. As suggested by previous studies with linker histones, this may result from the restriction of nucleosome mobility.
The binding of linker histone to a nucleosome stabilizes the interaction of an additional ∼20 bp of DNA with the periphery of the nucleosome (32), which acts to reduce nucleosome mobility (26, 35, 36) and repress transcription (4, 20, 36). Like H1, PARP-1 also binds nucleosomal DNA at or near the dyad axis of the nucleosome, as well as to linker DNA where it enters and exits the nucleosome, stabilizing an additional ∼10 to 15 bp of DNA with the periphery of the nucleosome (15). Central to this activity of PARP-1 is its ability to bind simultaneously to two DNA helices (i.e., the entering and exiting linker DNAs) (15, 29). The pattern of nucleosome binding by PARP-1 (15), as well as the activity of the isolated PARP-1 DBD in the transcription assays, is consistent with restricted nucleosome mobility and increased nucleosome stability as a means for transcriptional repression. Additional studies will be required, however, to show this directly.
Role of the CAT in the chromatin-dependent activities of PARP-1.
Our studies have revealed a novel function for the CAT, which we have shown to play a critical role in promoting chromatin compaction. Furthermore, chromatin compaction mediated by the CAT is required for maximal transcriptional repression by PARP-1 (Fig. 8). The CAT, which forms a structured region at the carboxyl terminus of PARP-1 (30), contributes to PARP-1 activity in two ways. First, it provides the intrinsic enzymatic activity of PARP-1 and thus mediates the NAD+-dependent activities of the protein (29). Second, it mediates homodimerization of PARP-1 molecules (24). Self-association of PARP-1 through the carboxyl-terminal region could promote the localized compaction of chromatin by bringing together groups of adjacent nucleosomes, as suggested in Fig. 1C and 2B. In this regard, note that the abilities of many chromatin architectural proteins to promote the compaction of chromatin correlate with their abilities to bind to two or more sites in chromatin at the same time, either as self-associating dimers/oligomers (e.g., HP1, Polycomb, Sir3p, and MENT) or as multivalent monomers (e.g., MeCP2) (23). In this model, the role of PARP-1's DBD is to tether the CAT to the nucleosome. In fact, as we show in Fig. 6, a PARP-1 molecule consisting only of the DBD and the NBD (i.e., DBD-NBD) exhibits wild-type activity in chromatin binding and compaction assays, yet the DBD and CAT added in trans result in no binding of the CAT and, consequently, no compaction. These results fit well with the fact that the amino-terminal tails of core histones are dispensable for chromatin compaction by PARP-1 (Fig. 3E), since the tails are not required by PARP-1 for DNA binding (Fig. 3) or homodimerization (24). One caveat of our analysis is that we have not shown a role for the CAT in chromatin compaction independent of the DBD. This, however, is likely due to the fact that the DBD is required to tether the CAT to a nucleosome (see below).
Distinct regulatory mechanisms involving the DBD and CAT of PARP-1.
One model that is consistent with our data suggests that PARP-1 promotes the NAD+-dependent regulation of chromatin structure and transcription through at least two mechanisms. First, PARP-1 acts on individual nucleosomes to repress transcription, an activity that is mediated by the DBD but is inefficient in the absence of the carboxyl-terminal region of PARP-1. Second, PARP-1 acts to promote the localized compaction of groups of adjacent nucleosomes, an activity that (i) requires both the CAT and the DBD and (ii) contributes an additional level of transcriptional repression. In this scenario, the DBD acts to tether the CAT to nucleosomes, while the CAT (perhaps more specifically the NBD [see the results with DBD-NBD]) drives the compaction of adjacent nucleosomes, possibly through self-association. A detailed test of this hypothesis will require more information about the specific residues in PARP-1 mediating dimerization, as well as mutants with alterations in these residues. NAD+ has the potential to regulate both activities of PARP-1. AutoPARylation of PARP-1 in the presence of NAD+ reverses the binding of PARP-1 to nucleosomes, which in turn leads to the reversal of both transcriptional repression and chromatin compaction (Fig. 3) (15). In addition, allosteric effects of NAD+ on the structure of the CAT, as suggested by previous structural studies (30), might also block PARP-1 self-association through the CAT.
Collectively, our results indicate that the independent activities provided by the DBD and CAT cooperate to regulate chromatin structure and transcription. Interestingly, the dual mechanism for PARP-1 action is similar to the dual effects on chromatin noted for linker histones, which also act at the levels of both individual nucleosomes and higher-order chromatin structures (9). These dual actions for PARP-1 and linker histones should provide multiple opportunities for the regulation of chromatin structure and function in vivo, as well as a means by which gene-specific transcriptional regulation by these factors can be achieved (9).
Acknowledgments
We acknowledge and thank Michael Hottiger for PARP-1 constructs; Jim Kadonaga and Takashi Ito for the recombinant ACF chromatin assembly system; Mi Young Kim for reagents and advice; Kit Umbach, Magnus Bergkvist, and the Cornell University Nanobiotechnology Center for assistance with AFM; Matthew Gamble for assistance with the statistical analyses; and members of the Kraus and Lis laboratories for critical reading of the manuscript.
This work was supported by a grant from the NIH/NIDDK (DK069710 to W.L.K.), postdoctoral fellowships from the Susan G. Komen Breast Cancer Foundation (to D.D.R.) and the New York State Health Research Science Board (to T.Z.), and Cornell University's Nanobiotechnology Center, an STC Program of the NSF (Agreement no. ECS-9876771).
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Amé, J. C., C. Spenlehauer, and G. de Murcia. 2004. The PARP superfamily. Bioessays 26: 882-893. [DOI] [PubMed] [Google Scholar]
- 2.Bulger, M., and J. T. Kadonaga. 1994. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet. 5: 241-262. [Google Scholar]
- 3.Bustamante, C., G. Zuccheri, S. H. Leuba, G. Yang, and B. Samori. 1997. Visualization and analysis of chromatin by scanning force microscopy. Methods 12: 73-83. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, E., A. S. Zarifyan, and W. L. Kraus. 2002. Histone H1 represses estrogen receptor alpha transcriptional activity by selectively inhibiting receptor-mediated transcription initiation. Mol. Cell. Biol. 22: 2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Amours, D., S. Desnoyers, I. D'Silva, and G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342: 249-268. [PMC free article] [PubMed] [Google Scholar]
- 6.Dillon, N. 2006. Gene regulation and large-scale chromatin organization in the nucleus. Chromosome Res. 14: 117-126. [DOI] [PubMed] [Google Scholar]
- 7.Francis, N. J., R. E. Kingston, and C. L. Woodcock. 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574-1577. [DOI] [PubMed] [Google Scholar]
- 8.Fyodorov, D. V., and J. T. Kadonaga. 2003. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371: 499-515. [DOI] [PubMed] [Google Scholar]
- 9.Georgel, P. T., and J. C. Hansen. 2001. Linker histone function in chromatin: dual mechanisms of action. Biochem. Cell Biol. 79: 313-316. [PubMed] [Google Scholar]
- 10.Glover, J. N., R. S. Williams, and M. S. Lee. 2004. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29: 579-585. [DOI] [PubMed] [Google Scholar]
- 11.Horn, P. J., and C. L. Peterson. 2002. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science 297: 1824-1827. [DOI] [PubMed] [Google Scholar]
- 12.Huynh, V. A., P. J. Robinson, and D. Rhodes. 2005. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. Mol. Biol. 345: 957-968. [DOI] [PubMed] [Google Scholar]
- 13.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13: 1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamakaka, R. T., M. Bulger, and J. T. Kadonaga. 1993. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 7: 1779-1795. [DOI] [PubMed] [Google Scholar]
- 15.Kim, M. Y., S. Mauro, N. Gevry, J. Lis, and W. L. Kraus. 2004. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119: 803-814. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M. Y., T. Zhang, and W. L. Kraus. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19: 1951-1967. [DOI] [PubMed] [Google Scholar]
- 17.Kraus, W. L., and J. T. Kadonaga. 1999. Ligand- and cofactor-regulated transcription with chromatin templates, p. 167-189. In D. Picard (ed.), Steroid/nuclear receptor superfamily: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 18.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12: 331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, W. L., and J. Lis. 2003. PARP goes transcription. Cell 113: 677-683. [DOI] [PubMed] [Google Scholar]
- 20.Laybourn, P. J., and J. T. Kadonaga. 1991. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science 254: 238-245. [DOI] [PubMed] [Google Scholar]
- 21.Leuba, S. H., G. Yang, C. Robert, B. Samori, K. van Holde, J. Zlatanova, and C. Bustamante. 1994. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. USA 91: 11621-11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251-260. [DOI] [PubMed] [Google Scholar]
- 23.McBryant, S. J., V. H. Adams, and J. C. Hansen. 2006. Chromatin architectural proteins. Chromosome Res. 14: 39-51. [DOI] [PubMed] [Google Scholar]
- 24.Mendoza-Alvarez, H., and R. Alvarez-Gonzalez. 2004. The 40 kDa carboxy-terminal domain of poly(ADP-ribose) polymerase-1 forms catalytically competent homo- and heterodimers in the absence of DNA. J. Mol. Biol. 336: 105-114. [DOI] [PubMed] [Google Scholar]
- 25.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. [DOI] [PubMed] [Google Scholar]
- 26.Pennings, S., G. Meersseman, and E. M. Bradbury. 1994. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA 91: 10275-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3: 247-253. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, P. J., and D. Rhodes. 2006. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 16: 336-343. [DOI] [PubMed] [Google Scholar]
- 29.Rolli, V., A. Ruf, A. Augustin, G. E. Schulz, J. Ménissier-de Murcia, and G. de Murcia. 2000. Poly(ADP-ribose) polymerase: structure and function, p. 35-79. In G. de Murcia and S. Shall (ed.), From DNA damage and stress signalling to cell death: poly ADP-ribosylation reactions. Oxford University Press, New York, NY.
- 30.Ruf, A., J. Mennissier de Murcia, G. de Murcia, and G. E. Schulz. 1996. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc. Natl. Acad. Sci. USA 93: 7481-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277: 23028-23036. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, R. T. 1978. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17: 5524-5531. [DOI] [PubMed] [Google Scholar]
- 33.Svejstrup, J. Q. 2004. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim. Biophys. Acta 1677: 64-73. [DOI] [PubMed] [Google Scholar]
- 34.Thoma, F., T. Koller, and A. Klug. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83: 403-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varga-Weisz, P. D., T. A. Blank, and P. B. Becker. 1995. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 14: 2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vignali, M., and J. L. Workman. 1998. Location and function of linker histones. Nat. Struct. Biol. 5: 1025-1028. [DOI] [PubMed] [Google Scholar]
- 37.Woodcock, C. L. 2006. Chromatin architecture. Curr. Opin. Struct. Biol. 16: 213-220. [DOI] [PubMed] [Google Scholar]
- 38.Woodcock, C. L., A. I. Skoultchi, and Y. Fan. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14: 17-25. [DOI] [PubMed] [Google Scholar]