Abstract
Compartmentalization of Src tyrosine kinases (SFK) plays an important role in signal transduction induced by a number of extracellular stimuli. For example, Src mitogenic signaling induced by platelet-derived growth factor (PDGF) is initiated in cholesterol-enriched microdomain caveolae. How this Src subcellular localization is regulated is largely unknown. Here we show that the Tom1L1-clathrin heavy chain (CHC) complex negatively regulates the level of SFK in caveolae needed for the induction of DNA synthesis. Tom1L1 is both an interactor and a substrate of SFK. Intriguingly, it stimulates Src activity without promoting mitogenic signaling. We found that, upon association with CHC, Tom1L1 reduced the level of SFK in caveolae, thereby preventing its association with the PDGF receptor, which is required for the induction of mitogenesis. Similarly, the Tom1L1-CHC complex reduced also the level of oncogenic Src in cholesterol-enriched microdomains, thus affecting both its capacity to induce DNA synthesis and cell transformation. Conversely, Tom1L1, when not associated with CHC, accumulated in caveolae and promoted Src-driven DNA synthesis. We concluded that the Tom1L1-CHC complex defines a novel mechanism involved in negative regulation of mitogenic and transforming signals, by modulating SFK partitioning at the plasma membrane.
The cytoplasmic tyrosine kinases of the Src family (SFK) play important roles in signal transduction induced by growth factors leading to DNA synthesis, cytoskeletal rearrangement, and receptor endocytosis (5). How growth factors use SFK for transmitting these signals is largely unknown. Signal specificity may be dictated by phosphorylation of appropriate substrates. Additionally, it may be achieved spatially through recruitment of a specific pool of SFK within the cell. Indeed, platelet-derived growth factor (PDGF)-induced DNA synthesis requires SFK activation in the cholesterol-enriched domains, the caveolae, while cytoskeleton rearrangement requires SFK association with F-actin assembly for dorsal ruffle formation (27). Accordingly, these pools are regulated by distinct mechanisms: mitogenic activity involves direct association of SFK with the receptor in caveolae, while SFK-induced F-actin assembly is mediated by the lipid second messenger sphingosine 1 phosphate. This lipid is likely to promote kinase activation via binding to a heterotrimeric Gi protein-coupled receptor. Therefore, regulation of SFK subcellular localization may be an important feature of signaling specificity. The molecular mechanism that governs such a compartmentalization is an important issue that remains unexplained.
Cholesterol-enriched microdomains are membrane organelles with specific physical features that are distinct from the contiguous membrane (26). While subjected to intense debates, they are thought to function as lipid scaffolds to regulate signal transduction induced by a number of extracellular stimuli, including T-cell receptor complexes (6, 18). Caveolae define a subclass of these membrane structures in nonlymphoid cells with a diameter of 50 to 100 nm and represent the major cholesterol-enriched microdomains present in fibroblasts. They are composed of caveolins, the main structural proteins, cholesterol, and sphingolipids, and a number of signaling molecules, including growth factor receptors and SFK. Compelling pieces of evidence indicate that they regulate signal transduction induced by growth factors and integrins in nontransformed cells (19).
Src is subjected to strict control in nontransformed cells, and constitutive kinase activation leads to oncogenic properties (17). Catalytic regulation involves intramolecular interactions (e.g., SH2 with the phosphorylated Tyr527 tail and the SH3 with a linker between the SH2 and the catalytic core) that stabilize the kinase in a close and inactive conformation. Opening the conformation by various means is predicted to stimulate the catalytic activity. Moreover, most SrcSH2 and/or SH3 binders increase Src activity in vivo and exhibit mitogenic and/or transforming activity (2). Nevertheless, we and others have recently identified Tom1L1 as a novel substrate and Src binder that does not induce mitogenic activity while promoting kinase activity in vitro (8, 25). This adapter belongs to the Tom1 family of proteins and presents a VHS (Vps27, Hrs, and STAM) and a GAT (GGA and Tom1) homology domain implicated in the regulation of vesicular trafficking (3, 16), a linker region, and a unique C terminus for phosphorylation and interaction with Src. Here we show that Tom1L1 interacts with clathrin heavy chain (CHC) in vivo, a structural component of clathrin-coated vesicles. Tom1L1, when bound to CHC, negatively regulates Src mitogenic and transforming activities by reducing its level in cholesterol-enriched microdomains including caveolae. Conversely, Tom1L1, when not associated with CHC, relocates in the caveolae and promotes Src-driven DNA synthesis. Therefore, the Tom1L1-CHC complex defines a novel mechanism for regulation of Src mitogenic and transforming activities, by influencing the kinase's membrane partitioning.
MATERIALS AND METHODS
Reagents.
pBABE and pSGT constructs encoding murine Tom1L1, YFPP (Tom1L1 R419D/P421A/P424A/Y457F), ΔL (deletion of amino acids 292 to 386), ΔL/YFPP, SrcY527F, PDGF receptor β (PDGFRβ), and Cav-3DGV were described in references 8, 23, and 27. ΔC (deletion at amino acid 386 of murine Tom1L1) and the ΔL/L401A (L401A/L402A/L407A) construct were obtained by PCR using the Quick Change site-directed mutagenesis system (Stratagene). Green fluorescent protein (GFP)-Tom1L1 constructs were obtained by subcloning Tom1L1 into pEGFP. Constructs encoding CHC Discosoma sp. red (DsRed) (30) and the glutathione S-transferase (GST)-CHC terminal domain (Gst-CHC-TD) (7) were obtained from P. Coopman and D. Drubin, respectively. Control (scramble) and small interfering RNAs (siRNA) specific to murine Tom1L1 (8), murine CHC (AACCGCATGGAGACATAATAT), and human CHC (10) were purchased from QIAGEN. Mock or short hairpin RNA (shRNA) specific to human Tom1L1 was obtained from the pSiren retroviral vector containing shRNA specific to luciferase (mock) or shRNA that targets the GACAAGAGACTGCTAAAT sequence of human Tom1L1. Polyclonal Tom1L1.1 to -3 antibodies were raised against GST fusion proteins containing the full-length (anti-Tom1L1.1), amino acids 291 to 474 (anti-Tom1L1.2), and amino acids 1 to 291 (anti-Tom1L1.3) of the murine Tom1L1 and were described in reference 8. Antibodies specific to Src, Fyn, and Yes (cst1), PDGFRβ (PRC), mT (762) myc tag (9E10), tubulin, and 4G10 have been described in references 4, 8, and 27. Anti-CHC antibodies used for immunoprecipitation (X22) and for Western blotting (TD.1) were from Alexis Biochemicals and Sigma, respectively; anti-pan-caveolin was from Transduction Laboratory, EC10 (anti-avian Src) was from UBI, antibodies coupled to fluorescent probes were from Molecular Probes, bromodeoxyuridine (BrdU) was from Sigma, anti-BrdU was from Pharmingen, PDGF-BB was from AbCys, and SU6656 was from Calbiochem. Purified GST fusion proteins and SFK were described in reference 8.
Cells culture, transfection, retroviral infection, immunofluorescence, DNA synthesis, and cell transformation.
NIH 3T3, SrcY527F-NIH 3T3 (Src 527), HEK 293, and HeLa cells were cultured as described in references 1 and 8. HEK 293 cells stably expressing PDGFRβ were obtained followed by infection of retroviruses expressing the human receptor (gift of A. Kazlauskas, Harvard Medical School, Boston, MA). Transfection and retroviral infection procedures were described in references 4 and 13. Cell transformation assays were performed using NIH 3T3 cells infected with indicated retroviruses, transfected or not with the indicated siRNA using Lipofectamine reagent (Invitrogen), and maintained in 10% fetal calf serum for 10 to 12 days. After staining with crystal violet (1%), the number of foci was visually scored. For BrdU incorporation assays, NIH 3T3 cells were seeded onto coverslips and made quiescent by serum starvation for 30 h. Cells were next stimulated or not with PDGF (5 to 20 ng/ml) in the presence of BrdU (0.1 mM) for 18 h. When indicated, cells were treated with SU6656 (2 μM) 30 min before stimulation and/or BrdU addition. Cells were then fixed and processed for immunofluorescence as described in reference 9. The percentage of transfected cells that incorporated BrdU for each coverslip was calculated using the following formula: % BrdU-positive cells = [(number of BrdU-positive transfected cells)/(number of transfected cells)] × 100. For siRNA experiments, cells were transfected and serum starvation was started 48 h afterwards. Caveola immunostaining was performed using anti-pan caveolin on cells fixed with ice-cold methanol, which allows detection of caveolin present in mature caveolae (20). Src (or Tom1L1)-caveolin colocalization at the cell periphery was detected by confocal analysis of 20 to 30 cells for each experiment and calculated as follows: % = [(number of transfected cells that exhibit colocalization at the cell periphery)/(number of transfected cells)] × 100. Cells were observed with a Carl Zeiss LSM510 META confocal microscope and a 100× PL APO (NA = 1.4) oil immersion objective. Confocal images were acquired using the single-track mode and Ar 488-nm and HeNe 543-nm excitation. Fluorescein isothiocyanate (FITC) and rhodamine channels were acquired using a BP 505/530 filter and a custom 550- 603 filter (ChS), respectively. For CHC colocalization, cells were fixed with 3.7% formaldehyde before immunostaining. Fixed cells were observed with a DMRB oil immersion microscope APO 63X (Leica). Acquisition was performed with a cooled charge-coupled device Micromax camera (Princeton Instruments) driven by MetaMorph 6.2 (Molecular Device). Stacks of images were restored using Huygens 2.3 (Scientific Volume Imaging) and an MLE algorithm.
Biochemistry.
Cell lysates, pull-down assays, immunoprecipitation, Western blotting, kinase assays, and reimmunoprecipitations were performed as described in references 8 and 27. For biochemical analysis, cells were stimulated for 1 h on ice with PDGF (25 ng/ml) as described in reference 22. Fractionation experiments were performed essentially as described in reference 27. Briefly, scrapped cells were centrifuged and pellets were suspended in 2× lysis buffer containing 1% Triton X-100, 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 75 U/ml aprotinin, and 1 mM vanadate for 20 min. Cell suspensions were homogenized with a Dounce homogenizer and centrifuged to remove nuclei. Supernatants were subjected to (5 to 42.5% [wt/vol]) sucrose gradient centrifugation, and nine fractions were collected from the top to the bottom of each gradient. Caveola-enriched fractions (CEF) corresponded to collected fractions 2 to 4 and were treated as described in reference 27 before biochemical analysis. Purification of GST fusion proteins and GST cleavage were performed as described in reference 8. Tyrosine-phosphorylated Tom1L1 proteins were generated as follows: after excision of the GST sequence, proteins were phosphorylated in vitro by incubation with Gst-FynSH1 bound to glutathione beads with 0.1 mM ATP for 30 min at 30°C. ATP was then removed from the supernatant using G-50 minicolumns (Amersham).
RESULTS
CHC-Tom1L1 complex formation.
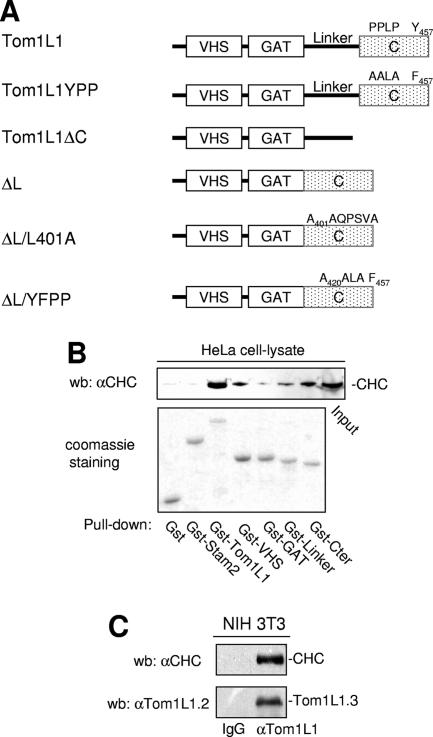
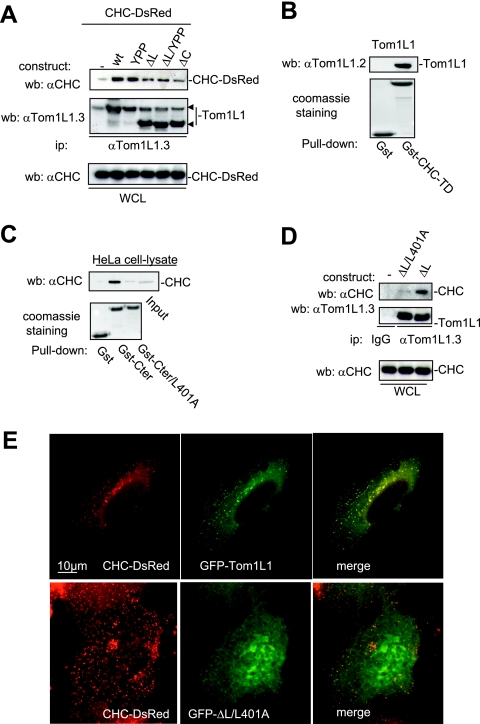
The negative mitogenic regulation of Tom1L1 requires its C terminus and the linker regions (8). We then searched for interactors involved in this activity through an affinity purification strategy coupled to mass spectrometry using HeLa cell lysates. CHC was found to be the main interactor of Tom1L1 (V. Simon and S. Roche, unpublished observations), and it was also recently reported by Katoh et al. (12). The CHC association with Gst-Tom1L1 was confirmed by Western blotting using CHC-specific antibody (Fig. 1B). The formation of endogenous Tom1L1-CHC complex was suggested by coimmunoprecipitation of CHC with Tom1L1 in NIH 3T3 cells (Fig. 1C). The molecular mechanism by which CHC associates with Tom1L1 was next addressed. First, CHC exhibited in vitro affinity for the VHS, the linker, and the C-terminus regions of Tom1L1 (Fig. 1B). The involvement of both the linker and the C-terminus regions in CHC association was further investigated in vivo. Deletion of either domain reduced the association of Tom1L1 with CHC-DsRed, which were coexpressed in HEK 293 cells (Fig. 2A). Mutations in Tom1L1 binding sites for the Src SH2 (Y457) and SH3 (P420PLP) domains, however, had no effect, indicating that SFK are not involved in this process (mutants YPP and ΔL/YPP in Fig. 2A). Most clathrin partners bind to its N terminus via Leu motifs called “clathrin boxes” (14). We found that a similar mechanism regulates CHC-Tom1L1 complex formation: the N-terminal domain of CHC fused to GST (Gst-CHC-TD) associated with Tom1L1 in vitro (Fig. 2B). Moreover, three potential “clathrin boxes” were found in the C terminus, and mutation of the first Leu-rich motif, L401LQPVSL, into AAQPVSA strongly affected the interaction of Gst-Tom1L1 C terminus (Gst-Cter) with CHC in vitro (Fig. 2C; mutant Gst-Cter/L401A). Finally we investigated the importance of the linker and the L401LQPVSL sequences in Tom1L1-CHC interaction in vivo. The Tom1L1 ΔL/L401 mutant that was deleted from the linker sequence and in which Leu401, Leu402 and Leu407 were replaced with Ala barely associated with endogenous CHC when expressed in HEK 293 cells (Fig. 2D). Moreover, while GFP-Tom1L1 and CHC-DsRed exhibited a strong colocalization when coexpressed in NIH 3T3 cells (see Fig. 2E, upper panels), the GFP-Tom1L1 ΔL/L401 mutant did not colocalize with CHC-DsRed (Fig. 2E, lower panels). We thus concluded that the linker and the Leu-rich motif L401LQPVSL present in the C terminus are required for association of Tom1L1 with CHC.
FIG. 1.
Association of CHC with Tom1L1. (A) Modular structure of Tom1L1 wild type and of the mutants used in this study. The VHS and GAT homology domains, the linker region, and the C terminus (C) are indicated. Src binding sites (i.e., PP421LP and Tyr457), the mutations of these Src binding sites (i.e., A420ALA and Phe457 in Tom1L1YPP), and the mutation of the CHC binding site A401AQPSVA (in ΔL/L401A) are indicated. (B) Association of CHC with Tom1L1 in vitro. HeLa cell lysates were incubated with the indicated GST fusion proteins or control GST beads, and the presence of CHC in the pull-down assays was revealed by Western blotting (wb) with a specific antibody. Input (15% of the cell lysate) was loaded as a positive control. αCHC, anti-CHC. (C) Association of CHC with Tom1L1 in NIH 3T3 cells. The CHC level associated with Tom1L1 in NIH 3T3 cells was assessed by Western blotting with anti-CHC antibody after immunoprecipitation of Tom1L1 with control (immunoglobulin G [IgG]) or anti-Tom1L1.3 (αTom1L1.3) antibody as shown. The level of immunoprecipitated Tom1L1 is also shown.
FIG. 2.
CHC-Tom1L1 complex formation. (A) Association of CHC with Tom1L1 involves both the linker and the C-terminus sequences. HEK 293 cells were transfected with CHC-DsRed and the indicated Tom1L1 constructs. Tom1L1 proteins were immunoprecipitated (ip) with the anti-Tom1L1.3 (αTom1L1.3) antibodies, and the presence of associated CHC-DsRed was revealed by Western blotting (wb) with anti-CHC (αCHC) antibodies. The levels of associated CHC-Ds-Red, immunoprecipitated Tom1L1, and expressed CHC-DsRed are shown. wt, wild type; WCL, whole-cell lysate. (B) In vitro association of Tom1L1 with Gst-CHC-TD fusion protein. The indicated fusion protein bound to glutathione beads was incubated with the purified Tom1L1. The presence of Tom1L1 was revealed by Western blotting with the indicated antibody. (C) Association of Tom1L1 C terminus with CHC involves a Leu-rich motif at the C terminus. HeLa cell lysates were incubated with indicated GST fusion proteins or control GST beads, and the interaction with CHC was revealed by Western blotting with a specific antibody. Input (5% of the cell lysates) was included as a positive control. (D) Regulation of CHC-Tom1L1 complex formation by the linker and the Leu-rich motif L401LQPSVL. Tom1L1 was immunoprecipitated from lysates of HEK 293 cells transiently expressing the indicated constructs with the anti-Tom1L1.3 antibodies or a control immunoglobulin G (IgG), and the presence of associated CHC was revealed by Western blotting using anti-CHC antibodies. The levels of expressed and associated CHC and immunoprecipitated Tom1L1 are shown. (E) Colocalization of CHC with Tom1L1. Shown is the representative fluorescence of CHC-DsRed, GFP-Tom1L1 (top panels), or a GFP-Tom1L1 mutant that does not associate with CHC (ΔL/L401A) (bottom panel). Also shown is the merged image of a cotranfected NIH 3T3 cell as obtained after deconvolution (Huygens software).
CHC-Tom1L1-Src complex formation.
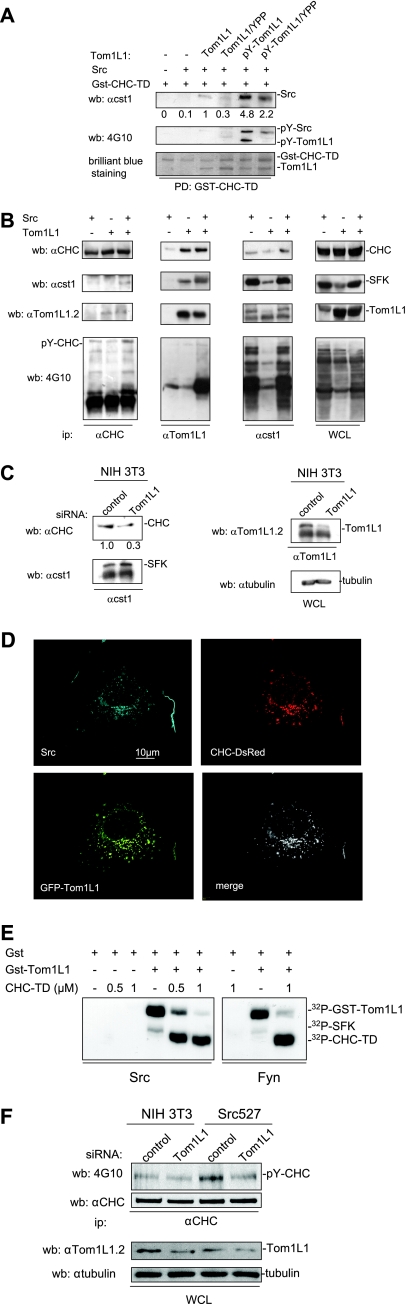
Since Tom1L1 binds also to Src, we searched for CHC-Tom1L1-Src ternary complex formation in vitro. Purified Tom1L1 associated with bound Gst-CHC-TD at a ratio of about 1:1 (Fig. 3A, Coomassie brilliant blue staining), confirming the strong interaction between Tom1L1 and CHC. While no interaction was detected when only Gst-CHC-TD and Src were used in vitro, an association was clearly observed in the presence of Tom1L1 (Fig. 3A). This suggests that Tom1L1 can bridge Src to CHC. Indeed, the association between Src and CHC was barely detected in the presence of Tom1L1/YPP, which harbors reduced affinity for Src (8) but retains its strong interaction with Gst-CHC-TD (Fig. 3A). We then addressed the impact of Tom1L1 phosphorylation on the ternary complex formation. pY-Tom1L1, which has been phosphorylated by the catalytic domain of Fyn, was purified in a free form (8) and incubated with bound Gst-CHC-TD for complex formation. As shown in Fig. 3A (bottom panel), Fyn phosphorylation did not affect the capacity of Tom1L1 to interact with Gst-CHC-TD but induced a 4.8-fold increase in Src association with the heterodimer. Src binding was, however, reduced by twofold when we used the Tom1L1/YPP mutant that exhibited lower tyrosine phosphorylation content (i.e., pY-Tom1L1/YPP) and lower affinity to Src. This piece of data indicates that tyrosine phosphorylation of Tom1L1 regulates association of Src with the Tom1L1-CHC complex.
FIG. 3.
CHC-Tom1L1-Src ternary complex formation. (A) Src-CHC-Tom1L1 ternary complex formation in vitro. Association of indicated purified proteins with Gst-CHC-TD bound to beads. The presence of Src, phosphorylated Src, and phosphorylated Tom1L1 was shown by Western blotting (wb) with the indicated antibodies (αcst1). Association of Gst-CHC-TD with Tom1L1 was revealed by Coomassie brilliant blue staining of the complex separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. (B) Src-CHC-Tom1L1 complex formation in HEK 293 cells that coexpress Src and Tom1L1. Each member of the complex was immunoprecipitated with the indicated antibodies (anti-CHC [α-CHC], anti-Tom1L1 [αTom1L1], and [αcst1]), and the presence of coassociated protein was detected by Western blotting with the antibodies shown. The tyrosine phosphorylation content of each immunoprecipitate is shown. Levels of tyrosine-phosphorylated proteins, CHC, SFK, and Tom1L1 from a whole-cell lysate (WCL) are also shown. (C) Endogenous Tom1L1 bridges SFK to CHC in NIH 3T3 cells. The CHC level associated with SFK was assessed by Western blotting with the indicated antibody after immunoprecipitation of SFK from NIH 3T3 cells that were transfected with the indicated siRNA. The levels of SFK, Tom1L1 and tubulin are also shown. Quantification of associated CHC is indicated. αtubulin, antitubulin antibody. (D) Colocalization of Src, Tom1L1, and CHC. Representative fluorescences of CHC-DsRed, GFP-Tom1L1, and immunostained avian Src are shown with the merged image of an NIH 3T3 cell coexpressing all three components after deconvolution, as described in Materials and Methods. (E) SFK phosphorylates CHC-TD in the presence of Tom1L1. Shown are the results of an in vitro kinase assay using purified Src or Fyn and in the presence of the indicated concentrations of CHC-TD and 1 μM of GST or Gst-Tom1L1, as indicated. Labeled SFK, Gst-Tom1L1, and CHC-TD are shown. (F) Tom1L1 regulates Src-induced CHC phosphorylation in Src-transformed cells. CHC was immunoprecipitated from NIH 3T3 cells or NIH3 3T3 cells stably expressing SrcY527F (Src 527) as shown and that were transfected with control or siRNA Tom1L1 as indicated. The levels of CHC and tyrosine-phosphorylated CHC (pY-CHC) are shown and were assessed by Western blotting with the indicated antibodies. The levels of Tom1L1 and tubulin from indicated cell lysates (WCL) are also shown.
The interaction of SFK with CHC-Tom1L1 was next evaluated in vivo using HEK 293 cells coexpressing Src and/or Tom1L1 (Fig. 3B). CHC was detected in SFK immunoprecipitates from cells expressing endogenous level of Tom1L1. Nevertheless, coimmunoprecipitation was significantly enhanced by Tom1L1 overexpression. In contrast, CHC-Tom1L1 complex formation was not affected by Src coexpression. It should be mentioned that similar lower levels of SFK and Tom1L1 were detected in CHC immunoprecipitates—the lower levels obtained may be due to low efficacy of antibodies to precipitate native clathrin (M. Franco and S. Roche, unpublished observations). Accordingly, a role for endogenous Tom1L1 in CHC-SFK interaction was also suggested in NIH 3T3 cells: CHC was detected in SFK immunoprecipitates, and its level was reduced by Tom1L1 depletion (Fig. 3C). Finally, immunofluorescence analysis using cells coexpressing Src, CHC-DsRed, and GFP-Tom1L1 showed that all three components colocalized in fibroblasts (Fig. 3D). Triple colocalization was observed especially in clathrin-coated vesicles and at the plasma membrane. We concluded that Tom1L1 participates in Src-CHC association.
CHC has been reported as a Src substrate (29); therefore, we also investigated the role of Tom1L1 in CHC tyrosine phosphorylation. CHC-TD alone was not phosphorylated by Src in vitro, even in the presence of higher concentrations of the protein (Fig. 3E, left panel). Nevertheless, in vitro phosphorylation was readily detected in the presence of Gst-Tom1L1. This effect was not restricted to Src as similar results were obtained with the tyrosine kinase Fyn (Fig. 3E, right panel). It should be mentioned that under these conditions, Src and Fyn preferentially phosphorylated CHC-TD, suggesting that the association with Tom1L1 allows unmasking of CHC phosphorylation sites. The role of Tom1L1 in CHC tyrosine phosphorylation was next investigated in vivo. Coexpression experiments in HEK 293 cells suggested that Src-induced CHC phosphorylation was favored by the presence of overexpressed Tom1L1 (Fig. 3B, left panels). Indeed, we found that endogenous CHC tyrosine phosphorylation was enhanced in NIH 3T3 cells stably expressing oncogenic SrcY527F and this was abrogated by Tom1L1 depletion (Fig. 3F). We concluded that Tom1L1 additionally regulates Src-induced CHC tyrosine phosphorylation.
The CHC-Tom1L1 complex affects the SFK level in caveolae.
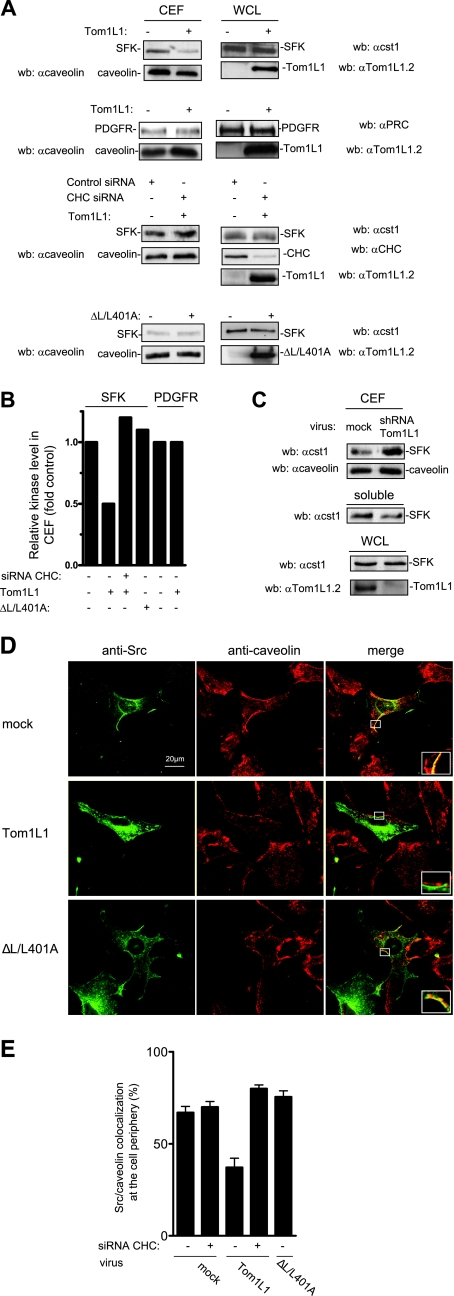
We next investigated whether TomL1-CHC affects SFK subcellular localization in caveolae. This was first addressed biochemically using CEF purified from HEK 293 cells expressing Src together or not with Tom1L1. Triton X-100 cell lysates were homogenized to increase protein solubility and fractionated through a sucrose gradient. CEF were isolated in the light fractions (2-4), as shown by the bulk of caveolin (Fig. 4A) (see also reference 27). We found that Tom1L1 overexpression strongly reduced the level of SFK in CEF without affecting caveolin accumulation. This inhibition was due to kinase delocalization as Tom1L1 did not affect the whole SFK. Quantification of these experiments indicated that about 50% of SFK was excluded from the CEF (Fig. 4B). In contrast, Tom1L1 did not influence the PDGFR level in these fractions (Fig. 4A and B). This finding indicates that Tom1L1 is not a general regulator of tyrosine kinases partitioning at the plasma membrane. We next investigated the role of CHC on Tom1L1 regulation of SFK level at the CEF. As shown in Fig. 4A and B, down-regulation of CHC by a specific siRNA strongly reduced the capacity of Tom1L1 to deplete SFK from CEF. Similarly, the reduction of SFK level was not observed with the ΔL/L401A Tom1L1 mutant that cannot associate with CHC (Fig. 4A and B). This mutant still retains some capacity to bind Src (8), indicating that the absence of effects on the SFK level was not due to its inability to associate with SFK. Finally, we investigated whether a similar mechanism occurs with endogenous Tom1L1 (Fig. 4C). Accordingly, down-regulation of Tom1L1 level by 80% induced a twofold accumulation of endogenous SFK at the CEF, concomitant with a significant reduction of SFK level in soluble fractions (fractions 7 to 9). Thus, we concluded that the reduction of SFK level in caveolae is dependent on the association of Tom1L1 with CHC.
FIG. 4.
Tom1L1 reduces SFK localization in caveolae through CHC association. (A) Tom1L1 reduces SFK but not PDGFR level in CEF. Shown are the levels of SFK, PDGFR, and caveolin in CEF purified from HEK 293 cells that were transfected with the indicated constructs and control or CHC siRNAs when indicated. The levels of expressed SFK, PDGFR, and Tom1L1 from the whole-cell lysate (WCL) are also shown. wb, Western blotting. αcaveolin, anticaveolin; αcst1, anti-cst1; αTom1L1.2, anti-Tom1L1.2; αPRC, anti-PRC; αCHC, anti-CHC. (B) Quantitative analysis of SFK and PDGFR levels in CEF obtained from two independent experiments. (C) Depletion of Tom1L1 enhances SFK accumulation in CEF. SFK and caveolin levels in CEF and soluble fractions (fractions 7 to 9 [“soluble”]) purified from HEK 293 cells that were mock infected (luciferase shRNA) or infected with Tom1L1 shRNA as indicated. The level of tubulin and Tom1L1 is also shown. A representative example (D) and its statistical analysis (E) (mean ± standard deviation [n = 3]) of Src-caveolin colocalization at the periphery of NIH 3T3 cells are shown. Cells infected with control (mock) or indicated Tom1L1 retroviruses were seeded on coverslips and transfected with avian Src together with siRNA (when indicated) for 48 h. Cells were then fixed and processed for immunofluorescence as described in Materials and Methods. Shown is a representative example of avian Src immunostaining, caveolin immunostaining, and the merged image from indicated infected NIH 3T3 cells obtained by confocal analysis as described in Materials and Methods. A threefold magnification of the merged image at the cell periphery is also included. The percentage of transfected cells that exhibited Src-caveolin colocalization at the cell periphery was calculated as described in Materials and Methods. Results are expressed as the mean ± standard deviation of three independent experiments. The levels of Tom1L1 and CHC are shown in panel A.
The role of Tom1L1-CHC complex on SFK membrane partitioning was next confirmed by an immunofluorescence approach. Src, together or not with Tom1L1, was expressed in fibroblasts by retroviral infection in order to get moderate level of ectopic protein and to prevent aberrant subcellular localization. We immunostained caveolae with an antibody that recognized the caveolin present in these membrane domains and detected its expression in restricted area of the plasma membrane (Fig. 4D), as previously reported (20). An anti-Src antibody was used to analyze the impact of Tom1L1 expression on Src distribution. An example of such experiments is shown in Fig. 4D, and the statistical analysis is shown in Fig. 4E. About 70% of infected cells exhibited a Src-caveolin colocalization at the cell periphery. While Tom1L1 overexpression did not affect membrane caveolin distribution, it induced a twofold reduction in the Src-caveolin localization (Fig. 4D). Again this effect was dependent on clathrin association with Tom1L1 as CHC down-regulation restored Src caveolar distribution. Similarly, mutation of the CHC binding sites in Tom1L1 (ΔL/L401A) abrogated this effect. Collectively, these data support the idea that the CHC-Tom1L1 complex acts as a negative regulator of SFK caveolar localization.
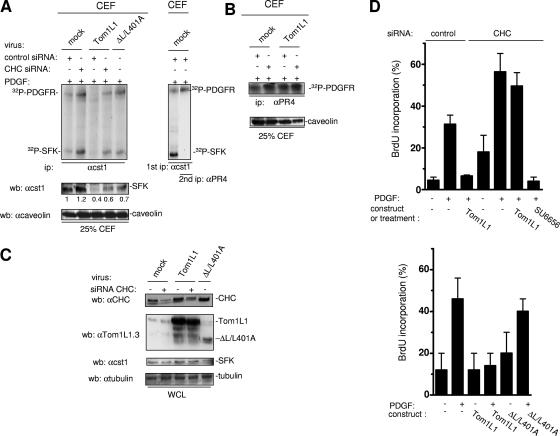
Tom1L1-CHC inhibits SFK-PDGFR association in caveolae and mitogenesis.
The biological meaning of SFK membrane partitioning was next investigated in PDGF-stimulated NIH 3T3 cells, as we have previously reported that SFK directly associate with activated PDGFR in caveolae to regulate mitogenic signaling (27, 28). Tom1L1 overexpression reduced SFK levels in CEF of PDGF-stimulated NIH 3T3 cells (Fig. 5A, left panels). No changes were detected on the whole level of SFK, excluding a protein degradation mechanism (Fig. 5C). In contrast, PDGFR activity was not affected in these fractions (Fig. 5B), confirming that Tom1L1 does not regulate partitioning of this receptor at the plasma membrane. Tom1L1-induced SFK caveolar depletion was reversed by CHC down-regulation or by mutation of CHC binding sites in Tom1L1 (ΔL/L401A) (Fig. 5B). These observations are consistent with a Tom1L1-CHC inhibitory mechanism in PDGF-stimulated fibroblasts. Therefore, we hypothesized that, due to SFK delocalization, Tom1L1 should reduce SFK-PDGFR complex formation in caveolae. Association of PDGFR with SFK was revealed by in vitro kinase assay of immunoprecipitated SFK from CEF (Fig. 5A, left panel). As previously reported (27), phosphorylated SFK was detected in association with a 180-kDa phosphoprotein, which was further identified as the PDGFR by reimunoprecipitation with a specific antibody (Fig. 5A, right panel). Moreover, Tom1L1 overexpression reduced the level of SFK-PDGFR complexes in CEF. As expected, this effect was abolished by down-regulation of CHC or mutation of the CHC binding sites in Tom1L1. We concluded that the effect of Tom1L1 on SFK-PDGFR complex formation is primarily due to a reduction of SFK in caveolae. Higher overexpression of this adapter may additionally compete with the receptor for association with SFK (8).
FIG. 5.
Tom1L1-CHC inhibits SFK-PDGFR coupling in caveolae and PDGF-induced DNA synthesis. (A) The Tom1L1-CHC complex inhibits SFK-PDGFR complex formation in caveolae. In vitro kinase assays of immunoprecipitated (ip) SFK (left panel) are shown from CEF purified from PDGF-stimulated NIH 3T3 cells that were infected with control (mock) or indicated viruses and transfected with control or CHC siRNA as indicated. The level of caveolin and quantified level of SFK in CEF are shown. (Right panel) Reimmunoprecipitation of the labeled 180-kDa protein observed in SFK immunoprecipitate with the PDGFRβ-specific PR4 antibody (αPR4). Labeled PDGFR and SFK are shown. wb, Western blotting; αcst1, anti-cst1; αcaveolin, anticaveolin. (B) The Tom1L1-CHC does not affect PDGFR activity in caveolae. The results of in vitro kinase assays of immunoprecipitated PDGFRβ with the indicated antibody are shown from CEF purified from PDGF-stimulated NIH 3T3 cells that were infected with control (mock) or indicated viruses and transfected with control or CHC siRNA when indicated. The level of caveolin in CEF is shown. (C) Shown are the levels of CHC, Tom1L1, SFK, and tubulin from whole-cell lysates (WCL) of NIH 3T3 cells infected with control (mock) or indicated viruses and transfected with control or CHC siRNAs when indicated. αtubulin, antitubulin. (D) PDGF mitogenic inhibition induced by Tom1L1 requires association with CHC. NIH 3T3 cells seeded onto coverslips and transfected or not with indicated constructs or siRNA were made quiescent by serum starvation for 30 h, treated or not with the SFK inhibitor SU6656 (2 μM), and stimulated or not with PDGF (5 ng/ml), as indicated, in the presence of BrdU for 18 h. Cells were then fixed and processed for immunofluorescence. The percentage of transfected cells that incorporated BrdU was calculated as described in Materials and Methods. Results are expressed as the mean ± standard deviation of three to five independent experiments.
The role of CHC on Tom1L1 biological activity was next assessed on mitogenic response induced by a low concentration of PDGF (Fig. 5D and E). DNA synthesis was monitored by adding BrdU to the medium. We found that PDGF induced a 30% BrdU incorporation and that retroviral expression of Tom1L1 abolished this cellular response (Fig. 5D). Like previously reported with Tom1L1 (8), CHC down-regulation enhanced S-phase entry in both quiescent and stimulated cells. This was consistent with a better SFK-PDGFR coupling observed in caveolae (Fig. 5A) and an increase in Src mitogenic signaling (M. Franco and S. Roche, unpublished data). Most importantly, Tom1L1 mitogenic inhibition was not observed any longer in cells with a reduced level of CHC. Nonetheless, these cells still required SFK activities for mitogenic signaling as the SFK inhibitor SU6656 still inhibited PDGF-induced DNA synthesis (Fig. 5D). This suggested that Tom1L1 inhibits mitogenesis by a CHC-dependent mechanism. This hypothesis was further confirmed by the absence of inhibitory effect observed with the ΔL/L401A mutant, which still associates with Src but not CHC (Fig. 5E). Therefore, the negative effect on the mitogenic response of Tom1L1 is dependent upon its association with CHC.
Tom1L1-CHC affects oncogenic Src membrane partitioning and Src mitogenic and transforming activities.
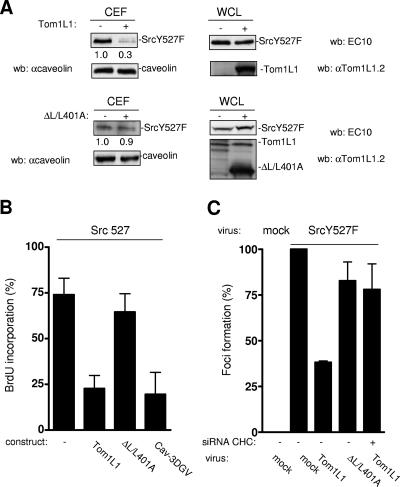
We then asked whether a similar regulation occurs in the presence of oncogenic Src. We first looked at the level of avian SrcY527F in CEF from HEK 293 cells expressing a low level of this kinase. As shown in Fig. 6A, SrcY527F was readily detected in caveolar fractions. Again, Tom1L1 overexpression strongly reduced the Src level without affecting caveolin accumulation. Inhibition was due to SrcY527F delocalization as Tom1L1 did not have an effect on the whole level of the protein. Quantification of these experiments indicated that up to 70% of the expressed Src was excluded from caveolar fractions. Interestingly, this exclusion was largely reduced when the Tom1L1 mutant ΔL/L401A was used, indicating that this activity is dependent on its association with CHC. The impact of membrane compartmentation was next investigated on SrcY527F mitogenic signaling in the absence of extracellular stimuli. NIH 3T3 cells stably expressing a low level of SrcY527F (Src 527-NIH 3T3) were serum starved for 30 h, and then BrdU was added for an extra 18 h in order to record de novo DNA synthesis. Under these conditions, 75% of the cells incorporated BrdU (Fig. 5B). We then addressed the role of cholesterol-enriched microdomains on this cellular response. The amino-terminally truncated mutant of caveolin 3, Cav-3DGV, has been described to reduce the levels of both caveolae and cholesterol from the plasma membrane (24, 27). We have also observed that this mutant blocks Src mitogenic signaling induced by PDGF (27). Interestingly, Cav-3DGV inhibited SrcY527F-driven DNA synthesis (Fig. 6B), suggesting that membrane cholesterol-enriched domains regulate this Src biological activity. The role of the Tom1L1-CHC complex in this cellular response was next investigated. Tom1L1 reduced SrcY527F-induced BrdU incorporation by 70%. Inhibition was dependent on the association with CHC, as no significant effect was observed with ΔL/L401A. In the presence of higher levels of SrcY527F, this inhibitory effect can be completely abolished (8). This may be explained by the inability of the Tom1L1-CHC complex to deplete enough Src from the cholesterol-enriched microdomains to prevent mitogenic signaling (G. Collins and S. Roche, unpublished data). The influence of the Tom1L1-CHC complex was also tested on Src transforming activity. SrcY527F was transduced by retroviral infection in NIH 3T3 cells for efficient induction of foci. Cytomegalovirus-driven SrcY527F expression exhibited lower biological activity in this assay, probably due to active protein degradation (G. Collin and S. Roche, unpublished data). Coinfection of Tom1L1 viruses reduced focus induction by 60% (Fig. 6C). This inhibition was largely overcome by CHC down-regulation (CHC siRNA) or by using the Tom1L1 mutant, which cannot associate with CHC (ΔL/L401A), suggesting that the Tom1L1 inhibitory effect is dependent upon its association with CHC. Therefore Tom1L1-CHC also inhibits Src transforming activity, in addition to Src-driven DNA synthesis.
FIG. 6.
Tom1L1-CHC affects SrcY527F membrane partitioning, SrcY527F-induced DNA synthesis, and focus formation. (A) Tom1L1-CHC reduces SrcY527F level in CEF. Shown are the levels of avian Src and caveolin from CEF purified from HEK 293 cells that were transfected with SrcY527F together or not with the indicated Tom1L1 constructs. The levels of expressed Src, Tom1L1, and tubulin expressed from the whole-cell lysate (WCL) are shown. wb, Western blotting; αcaveolin, anticaveolin; αTom1L1.2, anti-Tom1L1.2. (B) Tom1L1-CHC inhibits Src-driven DNA synthesis. NIH 3T3 cells stably transformed by SrcY527F (Src 527), seeded onto coverslips, and transfected or not with the indicated constructs were incubated in 0.5% serum for 30 h and further incubated in the presence of BrdU for 18 h. Cells were then fixed and processed for immunofluorescence. The percentage of transfected cells that incorporated BrdU was calculated as described in Materials and Methods. Results are expressed as the mean ± standard deviation of three independent experiments. (C) Tom1L1-CHC inhibits SrcY527F transforming activity. The statistical analysis of inhibition of SrcY527F-induced foci by Tom1L1-CHC is shown. NIH 3T3 cells were transfected or not with siRNA CHC as shown and then infected with control (mock) or indicated retroviruses. After 12 days of growth, foci were stained and scored as described in Materials and Methods. Focus formation (percentage of foci obtained relative to foci induced by SrcY527F) is represented as the mean ± standard deviation of three independent experiments.
Tom1L1 that does not associate with CHC accumulates in caveolae and promotes Src-driven DNA synthesis.
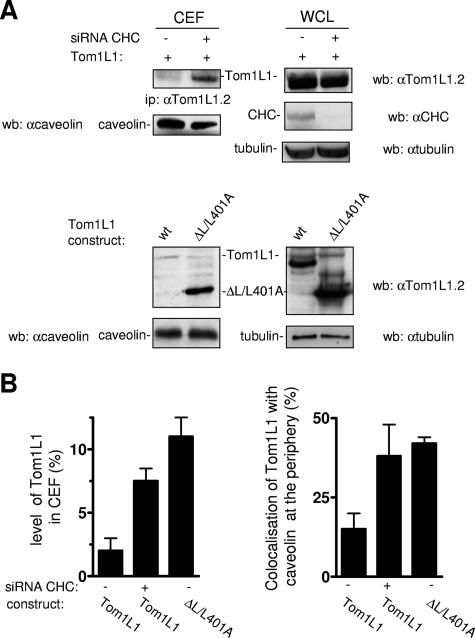
We next investigated the impact of CHC on Tom1L1 membrane distribution. Tom1L1 was barely detected in CEF when expressed in HEK 293 cells (Fig. 7A and B). However, down-regulation of CHC increased the level of Tom1L1 in CEF by 3.5-fold while not affecting that of caveolin. Accordingly, a fourfold-higher level of ΔL/L401A was observed in caveolar fractions compared to the wild-type protein. These results were next confirmed by an immunofluorescence approach in fibroblasts: while less than 25% of cells showed colocalization of Tom1L1 with caveolin at the cell periphery, CHC knock-down increased this percentage by twofold, and a similar scenario was observed when CHC binding sites in Tom1L1 were deleted (Fig. 7B, right panel). We thus concluded that Tom1L1 is mostly excluded from caveolae when in association with CHC.
FIG. 7.
Tom1L1 that does not associate with CHC accumulates in caveolae. (A and B) Tom1L1 that does not associate with CHC accumulates in caveolae. (A) Tom1L1 and caveolin levels in CEF of HEK 293 cells transfected with Tom1L1 constructs and control or CHC siRNAs, when indicated. The levels of CHC, Tom1L1, and tubulin from the whole-cell lysate (WCL) are also shown. wb, Western blotting; ip, immunoprecipitation; wt, wild type; αcaveolin, anticaveolin; αTom1L1.2, anti-Tom1L1.2; αCHC, anti-CHC; αtubulin, antitubulin. (B, left panel) Statistical analysis (mean ± standard deviation [n = 3]) of the percentage of the Tom1L1 level in CEF shown in panel A. (B, right panel) Statistical analysis of Tom1L1-caveolin colocalization at the periphery of NIH 3T3 cells. Cells seeded on coverslips were transfected with the indicated Tom1L1 construct together or not with the indicated siRNA for 48 h. Cells were then fixed and processed for immunofluorescence as described in Materials and Methods. The percentage of transfected cells that exhibited Tom1L1-caveolin colocalization at the cell periphery was calculated as described in Materials and Methods. The results are expressed as the mean ± standard deviation of three independent experiments.
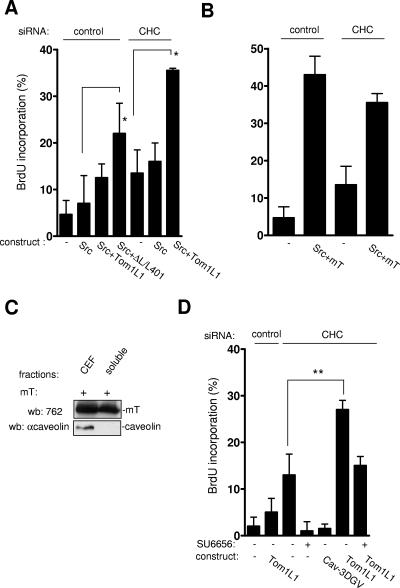
The biological significance of Tom1L1 accumulation in caveolae was next investigated with Src-driven DNA synthesis (Fig. 8A). As previously reported, wild-type Src did not induce BrdU incorporation even in the presence of Tom1L1 (8). In contrast, the Tom1L1 mutant ΔL/L401A, which does not associate with CHC, increased BrdU incorporation by 20%. This suggested that the inability of Tom1L1 to promote Src mitogenic signaling was due to its association with clathrin. The moderate effect obtained with ΔL/L401A could be due to the absence of the linker sequence that has been also implicated in the interaction and activation of Src (8). The negative role of the association with CHC on DNA synthesis was confirmed by a siRNA approach. While Src per se still had no effect in CHC-depleted cells, Tom1L1 promoted Src-driven DNA synthesis for 20% of expressing cells. We next addressed the specificity of CHC inhibition. The middle T (mT) antigen of the polyomavirus is another interactor and activator of Src catalytic activity. In vivo, mT promotes Src mitogenic and transforming activity (11). Accordingly, mT triggered Src-driven DNA synthesis in 40% of expressing cells; however, this effect was not increased by CHC depletion (Fig. 8B). We hypothesized that the capacity of mT to induce Src mitogenic activity was due to its localization in cholesterol-enriched microdomains. Indeed, mT was preferentially found in CEF (Fig. 8C)—unlikely from Tom1L1, whose membrane partitioning is regulated by CHC. We concluded that the inability of Tom1L1 to induce Src mitogenic signaling was related to its exclusion from caveolae due to its association with CHC. Finally, we wished to confirm the role of endogenous Src in this cellular process. We noticed that CHC depletion alone enhanced DNA synthesis from 10 to 15% (Fig. 8D). This cellular effect was abrogated by treatment of the SFK inhibitor SU6656 and expression of the Cav-3DGV mutant, implicating a Src mitogenic signaling regulated by caveolae and/or membrane cholesterol. Similarly, we found that Tom1L1 alone also enhanced DNA synthesis in cells with reduced CHC that was inhibited by the SFK inhibitor SU6656 (Fig. 8D). We concluded that Tom1L1 interacts with endogenous Src when present in caveolae for induction of DNA synthesis.
FIG. 8.
Tom1L1 that does not associate with CHC increases Src-driven DNA synthesis. (A) Tom1L1 that does not associate with CHC increases wild-type Src-driven DNA synthesis. NIH 3T3 cells seeded onto coverslips and transfected or not with the indicated Tom1L1 constructs and CHC siRNA, as indicated, were incubated in 0.5% serum for 30 h and further incubated in the presence of BrdU for 18 h. Cells were then fixed and processed for immunofluorescence. (B) CHC does not regulate the capacity of mT antigen to enhance Src-driven DNA synthesis. Shown is BrdU incorporation (mean ± standard deviation [n = 4]) of serum-starved NIH 3T3 cells that were transfected with indicated constructs and the indicated siRNAs as shown. (C) mT antigen is preferentially localized in CEF. Shown are the level of mT antigen in CEF and soluble fractions (fractions 7 to 9 [“soluble”]) from HEK 293 cells expressing mT antigen. The level of caveolin is also shown. wb, Western blotting; αcaveolin, anticaveolin. (D) Tom1L1 enhances DNA synthesis in CHC-depleted NIH 3T3 cells in a Src-dependent manner. NIH 3T3 cells seeded onto coverslips and transfected or not with the indicated constructs and CHC siRNA, as indicated, were incubated in 0.5% serum for 30 h, treated or not with SU6656 (2 μM) as shown, and further incubated in the presence of BrdU for 18 h. Cells were then fixed and processed for immunofluorescence. The percentage of transfected cells that incorporated BrdU was calculated as described in Materials and Methods. The percentage of transfected cells that incorporated BrdU was calculated as described in Materials and Methods. Results are expressed as the mean ± standard deviation of three to four independent experiments. *, P < 0.05, and **, P < 0.01, by Student's t test.
DISCUSSION
SFK play important roles in signal transduction induced by growth factors, and they are subjected to complex regulation for specific signaling, including catalytic activation, substrate specificity, and subcellular compartmentalization (5, 28). Our results point to membrane cholesterol-enriched domains as important regulators of Src proliferative function both upon a physiological activation (PDGF) and oncogenic activation (SrcY527F, mT of polyomavirus). We have identified a novel mechanism of negative regulation of Src proliferative and transforming activities through depletion of SFK from cholesterol-enriched domains. By associating with the complex Tom1L1-CHC, Src is delocalized from these organelles, thus preventing association with growth factor receptors to induce mitogenic signals. Alternatively exclusion of oncogenic Src from cholesterol-enriched membrane below a threshold may prevent the induction of DNA synthesis and focus formation. This mechanism may have important implications for Src signaling specificity (i.e., proliferation versus differentiation) and during tumorigenesis (8, 15). This may also explain why Tom1L1 does not promote Src mitogenic and/or oncogenic function while strongly stimulating catalytic activity in vitro. One can hypothesize that other Src activators, which do not localize in cholesterol-enriched microdomains, could also not promote Src mitogenic and transforming activities.
Finally, this report raises important issues regarding the mechanism and the function of SFK delocalization by Tom1L1-CHC. Our data suggest that CHC is primarily responsible for Src exclusion from caveolae. Tom1L1-Tyr457 phosphorylation by Src may increase ternary complex formation for efficient delocalization of the kinase. Finally, the nature of Src vesicular relocalization has not been investigated in this study. Nevertheless, the association with CHC strongly suggests that it could accumulate in clathrin-coated vesicle for endocytosis of membrane receptors to be identified. While probably not involved in PDGFR internalization (G. Collins and C. Bénistant, unpublished data), CHC and Tom1L1 have been implicated in EGFR endocytosis (14, 21) and our data suggest that Tom1L1 allows CHC phosphorylation by Src for enhanced receptor internalization (29). Therefore, the balance between SFK localization in cholesterol-enriched microdomains and that in clathrin-coated vesicles may play a crucial role for normal and tumor cell growth.
Acknowledgments
We thank P. Coopman, D. Drubin, A. Kazlauskas and J. Keen for various reagents; P. Jouin and J. Poncet for mass spectrometric analysis; the RIO platform for imaging analysis; W. J. Hong for sharing unpublished data; and our colleagues for critical reading of the manuscript.
This work was supported by grants from the CNRS, University of Montpellier II, INCa and ARC. M.F. was supported by the ARC. G.C. is supported by the Ligue Nationale Contre le Cancer. C.B. and S.R. are INSERM investigators.
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Blake, R. A., M. A. Broome, X. Liu, J. Wu, M. Gishizky, L. Sun, and S. A. Courtneidge. 2000. SU6656, a selective Src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 20: 9018-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boggon, T. J., and M. J. Eck. 2004. Structure and regulation of Src family kinases. Oncogene 23: 7918-7927. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacino, J. S. 2004. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5: 23-32. [DOI] [PubMed] [Google Scholar]
- 4.Boureux, A., O. Furstoss, V. Simon, and S. Roche. 2005. c-Abl tyrosine kinase regulates a Rac/JNK and a Rac/Nox pathway for DNA synthesis and c-myc expression induced by growth factors. J. Cell Sci. 118: 3717-3726. [DOI] [PubMed] [Google Scholar]
- 5.Bromann, P. A., H. Korkaya, and S. A. Courtneidge. 2004. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene 23: 7957-7968. [DOI] [PubMed] [Google Scholar]
- 6.Douglass, A. D., and R. D. Vale. 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 121: 937-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engqvist-Goldstein, A. E., R. A. Warren, M. M. Kessels, J. H. Keen, J. Heuser, and D. G. Drubin. 2001. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J. Cell Biol. 154: 1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franco, M., O. Furstoss, V. Simon, C. Benistant, W. J. Hong, and S. Roche. 2006. The adaptor protein Tom1L1 is a negative regulator of Src mitogenic signaling induced by growth factors. Mol. Cell. Biol. 26: 1932-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furstoss, O., K. Dorey, V. Simon, D. Barilla, G. Superti-Furga, and S. Roche. 2002. c-Abl is an effector of Src for growth factor-induced c-myc expression and DNA synthesis. EMBO J. 27: 514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, F., A. Khvorova, W. Marshall, and A. Sorkin. 2004. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 279: 16657-16661. [DOI] [PubMed] [Google Scholar]
- 11.Ichaso, N., and S. M. Dilworth. 2001. Cell transformation by the middle T-antigen of polyoma virus. Oncogene 20: 7908-7916. [DOI] [PubMed] [Google Scholar]
- 12.Katoh, Y., H. Imakagura, M. Futatsumori, and K. Nakayama. 2006. Recruitment of clathrin onto endosomes by the Tom1-Tollip complex. Biochem. Biophys. Res. Commun. 341: 143-149. [DOI] [PubMed] [Google Scholar]
- 13.Lassaux, A., M. Sitbon, and J. L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 79: 6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Roy, C., and J. L. Wrana. 2005. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 6: 112-126. [DOI] [PubMed] [Google Scholar]
- 15.Li, W., C. Marshall, L. Mei, L. Dzubow, C. Schmults, M. Dans, and J. Seykora. 2005. Srcasm modulates EGF and Src-kinase signaling in keratinocytes. J. Biol. Chem. 280: 6036-6046. [DOI] [PubMed] [Google Scholar]
- 16.Lohi, O., A. Poussu, Y. Mao, F. Quiocho, and V. P. Lehto. 2002. VHS domain—a longshoreman of vesicle lines. FEBS Lett. 513: 19-23. [DOI] [PubMed] [Google Scholar]
- 17.Martin, G. S. 2001. The hunting of the Src. Nat. Rev. Mol. Cell Biol. 2: 467-475. [DOI] [PubMed] [Google Scholar]
- 18.Munro, S. 2003. Lipid rafts: elusive or illusive? Cell 115: 377-388. [DOI] [PubMed] [Google Scholar]
- 19.Pike, L. J. 2005. Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochim. Biophys. Acta 9: 9. [DOI] [PubMed] [Google Scholar]
- 20.Pol, A., S. Martin, M. A. Fernandez, M. Ingelmo-Torres, C. Ferguson, C. Enrich, and R. G. Parton. 2005. Cholesterol and fatty acids regulate dynamic caveolin trafficking through the Golgi complex and between the cell surface and lipid bodies. Mol. Biol. Cell 16: 2091-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puertollano, R. 2005. Interactions of TOM1L1 with the multivesicular body sorting machinery. J. Biol. Chem. 280: 9258-9264. [DOI] [PubMed] [Google Scholar]
- 22.Puri, C., D. Tosoni, R. Comai, A. Rabellino, D. Segat, F. Caneva, P. Luzzi, P. P. Di Fiore, and C. Tacchetti. 2005. Relationships between EGFR signaling-competent and endocytosis-competent membrane microdomains. Mol. Biol. Cell 16: 2704-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roche, S., G. Alonso, A. Kazlauskas, V. M. Dixit, S. A. Courtneidge, and A. Pandey. 1998. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr. Biol. 8: 975-978. [DOI] [PubMed] [Google Scholar]
- 24.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1: 98-105. [DOI] [PubMed] [Google Scholar]
- 25.Seykora, J. T., L. Mei, G. P. Dotto, and P. L. Stein. 2002. 'Srcasm: a novel Src activating and signaling molecule. J. Biol. Chem. 277: 2812-2822. [DOI] [PubMed] [Google Scholar]
- 26.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31-39. [DOI] [PubMed] [Google Scholar]
- 27.Veracini, L., M. Franco, A. Boureux, V. Simon, S. Roche, and C. Benistant. 2006. Two distinct pools of Src family tyrosine kinases regulate PDGF-induced DNA synthesis and actin dorsal ruffles. J. Cell Sci. 20: 2921-2934. [DOI] [PubMed] [Google Scholar]
- 28.Veracini, L., M. Franco, A. Boureux, V. Simon, S. Roche, and C. Benistant. 2005. Two functionally distinct pools of Src kinases for PDGF receptor signalling. Biochem. Soc. Trans. 33: 1313-1315. [DOI] [PubMed] [Google Scholar]
- 29.Wilde, A., E. C. Beattie, L. Lem, D. A. Riethof, S. H. Liu, W. C. Mobley, P. Soriano, and F. M. Brodsky. 1999. EGF receptor signaling stimulates SRC kinase phosphorylation of clathrin, influencing clathrin redistribution and EGF uptake. Cell 96: 677-687. [DOI] [PubMed] [Google Scholar]
- 30.Zyss, D., P. Montcourrier, B. Vidal, C. Anguille, F. Merezegue, A. Sahuquet, P. H. Mangeat, and P. J. Coopman. 2005. The Syk tyrosine kinase localizes to the centrosomes and negatively affects mitotic progression. Cancer Res. 65: 10872-10880. [DOI] [PubMed] [Google Scholar]