Abstract
Nrf2 is the regulator of the oxidative/electrophilic stress response. Its turnover is maintained by Keap1-mediated proteasomal degradation via a two-site substrate recognition mechanism in which two Nrf2-Keap1 binding sites form a hinge and latch. The E3 ligase adaptor Keap1 recognizes Nrf2 through its conserved ETGE and DLG motifs. In this study, we examined how the ETGE and DLG motifs bind to Keap1 in a very similar fashion but with different binding affinities by comparing the crystal complex of a Keap1-DC domain-DLG peptide with that of a Keap1-DC domain-ETGE peptide. We found that these two motifs interact with the same basic surface of either Keap1-DC domain of the Keap1 homodimer. The DLG motif works to correctly position the lysines within the Nrf2 Neh2 domain for efficient ubiquitination. Together with the results from calorimetric and functional studies, we conclude that different electrostatic potentials primarily define the ETGE and DLG motifs as a hinge and latch that senses the oxidative/electrophilic stress.
The timely removal of transcription factors or their regulators is essential for regulated gene expression to react to different environmental stresses, including oxidative/electrophilic attacks, inflammation, and hypoxia (17, 19, 21, 40). The ubiquitous proteasomal degradation pathway determines the concentrations of various proteins in cells via programmed proteolysis to control both cellular quality and activity (4). Although there is only one type of proteasome in humans, there are numerous substrate recognition patterns for labeling target proteins for destruction with multiple ubiquitins, hence discriminating one cellular process from another (34). Nrf2 is the transcription factor for cytoprotective enzymes that counteract oxidative and electrophilic attacks (11, 27). The cellular concentration of Nrf2 remains low in homeostatic or unstressed conditions due to a rapid turnover modulated by Keap1 (Kelch-like ECH-associating protein 1) and proteasomal degradation (13). Unlike IκB and HIF1α, the regulators of inflammatory and hypoxic responses (1, 35, 36), Nrf2 is constitutively recognized by its substrate adaptor Keap1 and recruited to the Cul3 (Cullin 3)-based E3 ligase for subsequent ubiquitination and proteasomal degradation (7, 9, 12, 20, 45). Constitutive substrate recognition is achievable in the case of the Nrf2-Keap1 system, since no posttranslational modification of the substrate protein is required (33, 40). In contrast, in the cases of IκB and HIF1α, stimuli-induced phosphorylation or hydroxylation of the recognition signal of the substrate is needed for ubiquitination and degradation to follow (17, 35).
The target proteins of programmed proteolysis often need to be phosphorylated before they are recognized by their respective proteasomal degradation machineries (5, 17, 43). For instance, β-catenin (10), IκB (17), Gli3 (38), and Sic1 (37) are phosphorylated before being presented for ubiquitination. In some cases, double serine residues are phosphorylated to form the substrate recognition signal (β-catenin and IκB), while in other cases, phosphorylation of a serine or threonine residue at multiple sites within the substrate (Sic1 and Gli3) is required for efficient recruitment to the degradation machinery. Indeed, a minimum of six of the nine Cdc4-phosphodegron (CPD) sites in Sic1 need to be phosphorylated to form multiple suboptimal binding sites (30). This regulatory mechanism helps to set a higher phosphorylation threshold for controlling premature proteolysis of Sic1 and hence a higher precision of entry into S phase (29). It is also believed that β-TrCP (β-transducin repeat-containing protein [25]), a WD40 repeat-containing F-box protein of SCFβ-TrCP, recognizes a doubly phosphorylated destruction sequence or degron in its target proteins (38, 44). However, exceptions were recently identified in that β-TrCP also recognizes nonphosphorylated binding motifs, such as those in the cell cycle regulators Xenopus Cdc25A (15) and Wee1 (42). In both cases, one or two serine residues in the β-TrCP-binding motif are substituted with serine phosphomimetic residues (e.g., aspartate or glutamate), albeit dependency on phosphorylation for substrate recognition still remains. Alanine mutation of any of these single acidic residues within Cdc25A results in attenuation of β-TrCP binding and SCFβ-TrCP-dependent ubiquitination (15). Hence, the composition of either phosphorylation-prone serine/threonine residues or conserved acidic residues within the substrate recognition motif plays a major role in efficient recruitment of the target proteins for proteasomal degradation.
Recently, a two-site substrate recognition hinge-and-latch model was proposed to be the key regulatory mechanism for the oxidative/electrophilic stress response (39, 40). A similar model referred to as a tethering mechanism has also been proposed (26). In this model, a Keap1 homodimer enlists Nrf2 via the evolutionarily conserved DLG and ETGE motifs within the regulatory Neh2 domain of Nrf2 (18, 22). Unlike other examples of stimuli-inducible protein degradation, constitutive substrate recruitment to Keap1 occurs through recognition of the primary sequences of the DLG and ETGE motifs and does not require posttranslational modification of the destruction signals (33, 39, 40). Here, we attempted to validate this model by analyzing a crystal complex of Keap1-DC (double glycine repeat domain [DGR] plus the C-terminal region of Keap1) and a 15-residue-long peptide comprised of the DLG motif of Nrf2 (DLG peptide) (Keap1-DC-DLG peptide complex). We also studied the Keap1-DC-DLG peptide complex by isothermal calorimetry, in vivo ubiquitination assays, protein stability tests, and luciferase reporter assays. The results demonstrated that the binding nature of the Keap1-DC-DLG peptide complex is similar to that of the Keap1-DC-ETGE peptide complex (24, 33), yet there exist critical differences that contribute to the establishment of the hinge and latch system. This study clearly highlights the importance of the DLG motif as the latch to lock and unlock Nrf2 for ubiquitination.
MATERIALS AND METHODS
Plasmid construction.
Gene fragments bearing different alanine substitution mutations in either the DLG or ETGE motif of mouse Neh2 were prepared by a PCR overlap extension method utilizing pET15b-Neh2(ΔETGE) or pET15b-Neh2(Δ1-33) (39) as the template, respectively. The resulting Neh2 mutant PCR products were inserted into pET21a (Novagen) via NdeI and XhoI sites. A gene fragment encoding mouse Keap1-DC (Thr-309 to Cys-624) was inserted into pET21a via NdeI and XhoI sites. Single point substitutions of Gln26Ala, Asp27Ala, and Asp29Ala were introduced into the mammalian expression plasmid pcDNAmNrf2 by a QuikChange site-directed mutagenesis kit (Stratagene). Keap1 in the pcDNA3 vector is as described previously (16).
Protein expression and purification.
The mouse Keap1-DC domain was expressed in Escherichia coli, purified, and crystallized as described (32). Prokaryotically expressed His-tagged Neh2 mutants and Keap1-DC were expressed and purified as described previously (39).
Crystallization and data collection.
The best crystals were obtained in a drop containing 0.8 M lithium sulfate, 0.5 M ammonium sulfate, and 0.1 M sodium citrate, pH 5.2, at 20°C. For cocrystallization of the protein with the DLG peptide (22ILWRQDIDLGVSREV36, mouse Nrf2), purchased from Promega, the peptide and protein solutions were mixed (10:1 molar ratio) and incubated at 4°C for about 1 day before setting up the crystallization experiment. Diffraction data were collected under cryogenic conditions, using a Rigaku RA-Micro7 Cu Kα rotating-anode X-ray generator, operated at 40 kV and 20 mA and equipped with a Rigaku RAXIS IV++ imaging plate area detector and an X-stream low-temperature system. The data were integrated and scaled with HKL2000 (31). The results of the data reduction statistics are summarized in Table 1.
TABLE 1.
Summary of data collection and refinement statistics
| Statistic | Value for the Keap1-DC-Neh2 peptide complexa |
|---|---|
| Data collection statistics | |
| Source | RAXIS IV++ |
| Wavelength (Å) | 1.5418 |
| Space group | P61 |
| Unit cell dimensions (Å) | a = b = 103.13, c = 56.14, γ = 120° |
| Resolution (Å) | 20.0-1.9 |
| Completeness (%) | 98.2 (88.0) |
| Redundancy | 8.6 (3.6) |
| Rmerge (%)b | 8.6 (39.8) |
| Refinement statistics | |
| No. of complex molecules | |
| (asymmetric unit) | 1 |
| Resolution limit (Å) | 20.0-1.9 |
| Sigma cutoff | 0 |
| No. of reflections | 25,143 |
| R factor/Rfree (%)c | 17.2/21.1 |
| No. of protein residues | 295 |
| No. of peptide residues | 6 |
| No. of SO4 ions | 7 |
| No. of water molecules | 307 |
| rms deviationsd | |
| Bond lengths (Å) | 0.016 |
| Bond angles (°) | 1.55 |
Numbers in parentheses are the values for the highest resolution shell.
Rmerge = Σ|Iobs − <I>|/Σ <I> summed over all observations and reflections, where Iobs is observed intensity and <I> is the mean intensity.
The crystallographic R factor (Rcryst) = Σ|Fobs − Fcalc|/ΣFobs, where Fobs is observed structure factors and Fcalc is calculated structure factors. Rfree was calculated with 5% of data omitted from refinement.
rms, root mean square.
Determination and refinement of the structure.
The structure of the Keap1-DC-DLG peptide complex was determined by the molecular replacement method, employing the Keap1 structure (33) as a search model and the program Molrep from the CCP4 suite (6). It gave a distinct peak with an R factor (residual factor or reliability factor) and correlation coefficient of 0.363 and 0.690, respectively, at a resolution between 20 Å and 3.0 Å. The model was refined with CNS (Crystallography & NMR System) (2), and several rounds of manual fitting and refitting were carried out using the program O (14) with careful inspection of the 2Fo-Fc and Fo-Fc difference Fourier maps and omit electron density maps. During the final stage of refinement, Refmac5 (28), which is incorporated into the CCP4 suite, was used for refinement of the structure. The current refined model consists of 301 residues, 7 SO4 ions, and 307 water molecules, with a final Rwork and Rfree of 17.2% and 21.1%, respectively, at a resolution of 1.9 Å. The electron densities of 15 residues at the N terminus and 11 residues at the C terminus were absent. The stereochemistry of the Keap1-DC complex was excellent, as checked with PROCHECK (23). The refinement statistics are summarized in Table 1.
ITC titration experiments.
Isothermal calorimetry (ITC) titration experiments were performed at 25°C on a VP-ITC system (MicroCal Inc.). All proteins were prepared as previously described (39). In a typical experiment, 10-μl aliquots of 0.35 mM Neh2 or Neh2 mutants were injected 28 times at 4-min intervals from a stirring syringe (300 rpm) into the sample cell containing 1.45 ml of 0.04 mM Keap1-DC. The heat changes in binding of the last five injections after saturation were averaged and used to correct the heat of protein dilution. All runs were performed in triplicate. Binding data were analyzed using the computer program Origin version 5.0, supplied by MicroCal Inc. Data processing was carried out as reported previously (39).
In vivo ubiquitination assay.
293T cells were transfected, using Lipofectamine (Invitrogen), with His-tagged ubiquitin (0.5 μg) and 1 μg of wild-type or mutant (Gln26Ala, Asp27Ala, or Asp29Ala) Nrf2 plasmids, either in the absence or presence of Keap1 expression vector (0.5 μg). The extent of Nrf2 ubiquitination was evaluated by Western blotting using a monoclonal anti-Nrf2 antibody (41). Input controls were examined by anti-Nrf2 and monoclonal anti-Keap1 (41) using total cell extracts. The in vivo ubiquitination assay was performed as previously described (20).
Nrf2 in vivo stabilization analysis.
Eukaryotic expression vectors expressing wild-type Nrf2 or Nrf2 mutants (Gln26Ala, Asp27Ala, or Asp29Ala; 1 μg each) were transfected into Cos7 cells, using FuGENE 6 transfection reagent (Roche), either with or without full-length Keap1 expression vector (0.75 μg). A plasmid of enhanced green fluorescent protein (EGFP), pEGFP-N1 (25 ng), was also transfected as the transfection efficiency control. Cells were lysed 36 h after transfection and boiled in Laemmli sample buffer in the presence of 2-mercaptoethanol (final concentration of 2%). Cell extracts were then subjected to immunoblot analysis using anti-Nrf2, anti-Keap1, and anti-EGFP (Molecular Probes) antibodies.
Cycloheximide chase analysis.
293T cells were transfected, using Lipofectamine, with eukaryotic expression vectors of wild-type Nrf2 or Nrf2 mutants (Gln26Ala, Asp27Ala, or Asp29Ala; 1 μg each), Keap1 (0.5 μg), and pEGFP-N1 (25 ng) as the transfection control. Cells were treated with 35 μg/ml of cycloheximide at 24 h following transfection. Cells were then harvested at 0, 15, 30, and 45 min after chemical addition and boiled in Laemmli sample buffer. Protein levels were monitored by immunoblot analysis using anti-Nrf2 and anti-EGFP antibodies. Anti-α-tubulin (Sigma) antibody was used as an input control.
Luciferase reporter assay.
Cos7 cells were transfected with 100 ng of wild-type Nrf2 or Nrf2 mutant (Gln26Ala, Asp27Ala, or Asp29Ala) plasmids either in the absence or presence of Keap1 expression vector (10 ng). After 24 h, cells were treated with either dimethyl sulfoxide or diethyl maleate (DEM) for 12 h. The luciferase reporter assay was performed as previously published (16).
Protein structure accession number.
The atomic coordinates have been deposited in the Protein Data Bank with the accession code 2DYH.
RESULTS
Structure of Keap1-DC complexed with DLG peptide.
The newly proposed hinge-and-latch model for Nrf2 activation/derepression has deciphered the roles of the two Nrf2 motifs, the ETGE and DLG motifs, in the interaction of Nrf2 with Keap1 (39, 40). The crystal structure of the Keap1-DC-ETGE peptide complex conforms to this model (33). To gain further insight into this model, we performed cocrystallization and structural analysis of Keap1-DC in complex with a peptide containing the DLG motif. This peptide contains Ile-22 to Val-36 of mouse Nrf2 (Fig. 1A). The structure of this complex was solved by the molecular replacement method using the apo form of the Keap1-DC structure (33) at 2.5-Å resolution. The difference Fourier map clearly showed the electron density at the peptide-bound region. Although we used a peptide 15 amino acids long (Ile-22 to Val-36), the electron density was visible for only six amino acids, from Trp-24 to Asp-29. The Trp-24 and Ile-28 residues were truncated to Ala, as the electron densities of the side chains were not visible. Since the electron density of the side chain atoms after Cδ was not visible in Arg-25 either, these atoms were not included in refinement of the structure. The final R factor and Rfree values were 17.2% and 21.1%, respectively, for the 301 residues, 7 sulfate ions, and 307 water molecules, refined to a resolution of 1.9 Å (Table 1).
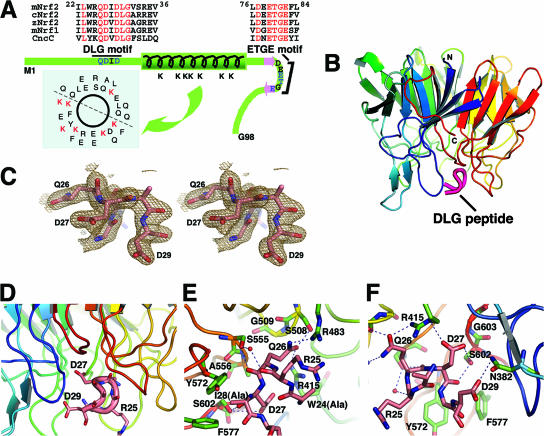
FIG. 1.
Overall tertiary structure of mouse Keap1-DC (mKeap1-DC) complexed with the DLG peptide. (A) Schematic diagram of the Neh2 domain depicting the secondary structures and the two conserved DLG and ETGE motifs. The sequences of mouse Nrf2 (mNrf2), mouse Nrf1 (mNrf1), chicken Nrf2 (cNrf2), zebra fish Nrf2 (zNrf2), and Cap ‘n’ collar isoform C (CncC) are shown. The green bar represents a 33-amino-acid-long α-helix, and the two pink arrows denote β-strands. The ETGE motif is located in the loop region of the antiparallel β-sheet. The DLG motif is N terminal to the α-helix. Within these two motifs, residues that have a direct intermolecular interaction with Keap1 are shown in blue. The distribution of the seven lysines in the α-helical region of Neh2 is shown by the helical wheel. Regions corresponding to the DLG and ETGE motifs of the CNC protein family are aligned, and the residues in red are conserved. (B) The ribbon model of the tertiary structure of the mKeap1-DC β-propeller domain (blue to red) and the DLG peptide (magenta) was generated with PyMOL (http://pymol.sourceforge.net/). (C) Stereo view of a portion of the refined peptide in the protein-bound form showing Gln-26, Asp-27, and Asp-29. The final 2mFo − DFc difference Fourier map is contoured at 1 σ. (D) Close-up view of the peptide-binding region. The backbone and side chains of the bound peptide are shown as a pink tube and sticks, respectively. (E and F) Close-up view of the interface between mKeap1-DC and the DLG peptide, showing interfaces close to Gln26 (E) and Asp27 (F) of the DLG peptide. Residues potentially interacting between Keap1-DC (green) and the DLG peptide (pink) are in ball-and-stick representation. Nitrogen and oxygen atoms are shown in dark blue and red, respectively. The rest of the ribbon model for the backbone of Keap1-DC is colored as described above for panel A) Hydrogen bonds are represented by dashed lines. Potential structural water molecules are depicted as red circles.
As is the case for the ETGE peptide (33), the DLG peptide binds to Keap1-DC at the bottom side of the six-bladed β-propeller (Fig. 1B and C), which is highly basic and mainly occupied by arginine residues (Fig. 1D). The exposed region of the DLG peptide in the protein complex is surrounded by Tyr-334, Asn-382, Arg-483, Tyr-525, Tyr-572, and Phe-577, while in the buried region, the peptide is surrounded by Arg-415, Ser-508, Gly-509, Ser-555, Ala-556, and Gly-603 of Keap1 (Fig. 1E and F). Although the DLG peptide is positioned close to the fourth, fifth, and sixth blades of the Keap1-DC domain, the peptide interacts with residues from almost all of the six blades.
The DLG peptide possesses a tight four-residue β-hairpin conformation comprised of the residues Arg-25, Gln-26, Asp-27, Ile-28, and Asp-29 (Fig. 1E and F). The structure of this peptide is stabilized by two intramolecular hydrogen bonds. The carbonyl group of Arg-25 accepts a hydrogen bond from the amino group of Ile-28, which has a left-handed helical conformation. In addition, the carboxyl group of Trp-24 contributes to an electrostatic interaction with the amino group of Asp-29. The formation of such a β-hairpin conformation in the peptide allows the residues of the DLG motif to interact firmly with the residues of Keap1-DC.
The Keap1-DC-DLG peptide complex generates eight potential intermolecular electrostatic interactions between the residues of the DLG peptide and the Keap1-DC domain, as opposed to the 13 electrostatic interactions found in the Keap1-DC-ETGE peptide complex (33). Gln-26 of Neh2 is a conserved residue within the CNC (cap ‘n’ collar) protein family (Fig. 1A) (18) and has significant intermolecular interactions with Ser-508, Arg-415, Arg-483, and Ser-555 of Keap1-DC (Fig. 1E). For instance, the side chain of Gln-26 is wedged between Arg-415 and Arg-483. Furthermore, the Oɛ1 of Gln-26 makes hydrogen bonds with Ser-508 and Arg-415. The Nɛ2 atom of Gln-26 is firmly positioned through an electrostatic interaction with the side chain atom of Arg-483. In addition to these side chain interactions, the main-chain carbonyl group of Gln-26 interacts electrostatically with the side chain of Ser-555.
Asp-27 of the DLG motif interacts with multiple residues, such as Ser-602, Gly-603, and Arg-415, of Keap1 (Fig. 1F). The Oδ1 atom of Asp-27 supplies hydrogen bonds to the side chain of Ser-602, while the other side chain atom Oδ2 of Asp-27 interacts electrostatically with the guanidinium group of Arg-415. In addition, the carbonyl group of Asp-27 is also hydrogen bonded to the side chain atom of Ser-602. On the other hand, the side chain of the next residue, Ile-28, which adopts a left-handed helical conformation, was absent in the structure. However, this residue is nicely positioned in a hydrophobic pocket produced by Tyr-572 and Phe-577. Moreover, Asp-29, the last residue visible in the electron density map of the DLG peptide, contributes to an electrostatic interaction with Asn-382 of Keap1 (Fig. 1F).
DLG and ETGE peptides bind to Keap1-DC in a similar manner.
Structural comparison of the Keap1-DC-DLG peptide complex and the Keap1-DC-ETGE peptide complex (33) revealed that both DLG and ETGE interact with Keap1 (Fig. 2A and B) in a very similar manner by binding through the bottom region of the β-propeller structure (Fig. 1B and D). Superimposition of the structures of these two complexes over the main chain atoms revealed that the overall structure of Keap1-DC in these two complexes is nearly the same (0.23-Å root mean square deviation; Fig. 2C). However, a small variation was observed in the loop connecting strands β2 and β3 of the second blade of the Keap1-DC domain. At the binding interface, the side chains of the Keap1-DC residues have a very similar conformation within the two complexes, except for Arg-415, Arg-483, and Asn-382 (Fig. 2C).
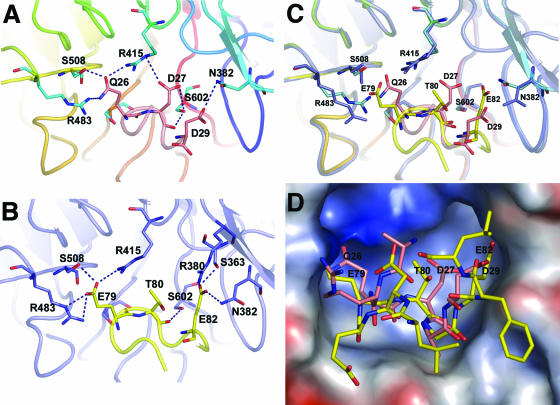
FIG. 2.
Superimposition of the DLG peptide complex with the ETGE peptide complex. (A) Close-up view of the interface between mouse Keap1-DC (mKeap1-DC) and the DLG peptide. The backbone and side chains of the bound peptide are shown as a pink tube and sticks, respectively. The side chains of the interacting residues from Keap1-DC are shown as blue sticks. Dashed lines indicate intermolecular hydrogen bonding. Residues showing intermolecular interaction are labeled. (B) Close-up view of the interface between mKeap1-DC and the ETGE peptide. The backbone and side chains of the bound peptide are shown as a yellow tube and sticks, respectively. The side chains of the interacting residues from Keap1-DC are shown as blue sticks. Dashed lines indicate intermolecular hydrogen bonding. (C) Close-up view of the superimposed protein-peptide interfaces. Some of the residues of Keap1-DC (shown as sticks) potentially interacting with ETGE and DLG are shown in blue and cyan, respectively. The rest of the β-propeller domain is illustrated by a blue ribbon in the case of the ETGE peptide complex and blue to red as described in the legend to Fig. 1 in the case of the DLG peptide complex. Backbone and interacting residues of the ETGE peptide are displayed as yellow ribbons and sticks, respectively, whereas those of the DLG peptide are pink. Nitrogen and oxygen atoms are shown in dark blue and red, respectively. (D) Electrostatic surface potential of mKeap1-DC in the mKeap1-DC peptide complexes. Surface acidic, basic, and neutral residues are in red, blue, and white, respectively. The protein-bound ETGE (yellow) and DLG (pink) peptides are shown as sticks.
Although both peptide structures are quite flexible, they adopt a similar kind of tight β-turn conformation and also orientate in essentially the same manner with respect to the Keap1-DC structure (Fig. 2C). The binding profiles of Gln-26, Asp-27, and Asp29 of the DLG peptide are similar to those of Glu-79, Thr-80, and Glu-82 of the ETGE peptide with the equivalent residues in Keap1-DC. In spite of their conformational similarity, the ETGE peptide possesses a comparatively higher number of electrostatic interactions with Keap1-DC than with the DLG peptide: approximately 13 in the ETGE peptide complex versus 8 in the DLG peptide complex. Furthermore, the ETGE peptide is substantially embedded into the Keap1-DC binding cleft compared to the DLG peptide. For example, the side chain of Glu-79 in the ETGE is deeply buried in the pocket, whereas the equivalent residue in the DLG, Gln-26, is only partially entrenched (Fig. 2D). These results thus demonstrate that the nature of binding of the DLG and ETGE peptides to Keap1-DC is similar, yet critical differences exist. Especially, the differential negative electrostatic potential may play a role in generating the distinct binding affinities of the ETGE and DLG motifs to Keap1-DC.
Acidic residues define the differential binding affinities of the ETGE and DLG motifs.
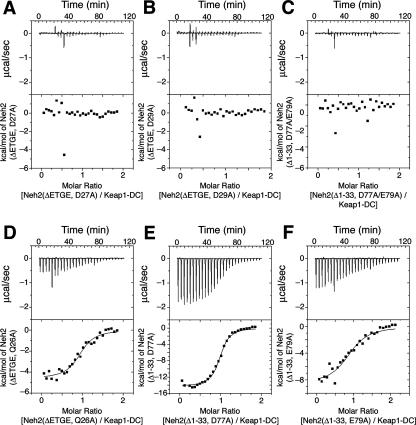
To gain further insight into the differential binding affinities shown by the ETGE and DLG motifs, we carried out systematic point mutations and ITC analysis. The results unveiled a trend in acidic residue utilization as the destruction signal of Nrf2. Previous calorimetric analysis (39) showed that the DLG motif has a binding constant of 1 × 106 M−1 when interacting with Keap1-DC, whereas the ETGE motif has a higher-affinity binding constant of 2 × 108 M−1. However, mutating Asp-27 or Asp-29 to alanine within an ETGE deletion mutant construct [Neh2(ΔETGE,D27A) and Neh2(ΔETGE,D29A), respectively] resulted in no observable affinity to Keap1-DC (Fig. 3A and B). The raw isotherms of the protein titration resembled the heat of buffer dilution (data not shown). Although Gln-26 was found to interact extensively with Keap1-DC, alanine substitution did not have a large effect on the binding affinity (Table 2; Fig. 3D). Similarly, alanine mutations in Ile-28, Arg-25, and Asp-21, which are not conserved within the CNC family, did not attenuate the function of the DLG motif (Table 2). Leu-30 is a conserved residue, and a Leu-30 mutation modestly decreased the affinity (Table 2). On the other hand, although the side chain of Trp-24 was not seen in the crystal structure, when the Trp-24 was mutated to Asp or Glu, no binding affinity to Keap1-DC was observed (Table 3 and raw data not shown). ITC analysis of Leu-30 and Trp-24 suggests that the conservation of these two residues is important for the binding of the DLG motif to Keap1.
FIG. 3.
Different acidic residue concentrations account for the differential binding affinities of the ETGE and DLG motifs to Keap1. Representative ITC titration profiles of the titration of Keap1-DC with Neh2(ΔETGE,D27A) (A), Neh2(ΔETGE,D29A) (B), Neh2(Δ1-33,D77A/E79A) (C), Neh2(ΔETGE,Q26A) (D), Neh2(Δ1-33,D77A) (E), and Neh2(Δ1-33,E79A) (F). The top graphs represent the raw ITC thermograms, and the bottom graphs represent the fitted binding isotherms. The integrated binding isotherms are plotted against the molar ratio of different mutants of Neh2 to Keap1-DC.
TABLE 2.
Thermodynamic parameters for binding of DLG mutants to Keap1-DC at 25°Ca
| Alanine mutation | n | Ka (106 M−1) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| D21A | 0.92 ± 0.03 | 1.1 ± 0.0 | −4.4 ± 0.0 | +3.8 ± 0.1 | −8.2 ± 0.0 |
| R25A | 0.97 ± 0.00 | 4.8 ± 0.1 | −8.4 ± 0.0 | +0.7 ± 0.0 | −9.2 ± 0.0 |
| Q26A | 0.96 ± 0.02 | 0.9 ± 0.1 | −4.4 ± 0.2 | +3.7 ± 0.2 | −8.1 ± 0.0 |
| I28A | 0.84 ± 0.01 | 1.3 ± 0.1 | −5.6 ± 0.2 | +2.7 ± 0.3 | −8.3 ± 0.0 |
| L30A | 0.84 ± 0.01 | 0.4 ± 0.0 | −5.4 ± 0.0 | +2.3 ± 0.1 | −7.6 ± 0.0 |
n is the stoichiometry, and Ka is the binding constant. ΔH, ΔS, and ΔG are the change in binding enthalpy, entropy, and Gibbs energy, respectively. −RT lnKa = ΔG = ΔH − TΔS, where T and R are the absolute temperature and gas constant, respectively. Values are shown as means ± standard deviations from triplicate runs. The ETGE motif was deleted from all mutants.
TABLE 3.
Neh2 mutants that showed no observable Keap1-DC binding affinities in ITC analysis
| Neh2 construct | Modification of DLG | Modification of ETGE |
|---|---|---|
| Neh2(ΔETGE,W24D) | Trp24 to Asp | ETGE deletion |
| Neh2(ΔETGE,W24E) | Trp24 to Glu | ETGE deletion |
| Neh2(ΔETGE,D27A) | Asp27 to Ala | ETGE deletion |
| Neh2(ΔETGE,D29A) | Asp29 to Ala | ETGE deletion |
| Neh2(Δ1-33,D77A/E79A) | N-terminal deletion (amino acids 1 to 33) | Asp77 to Ala and Glu79 to Ala |
A parallel analysis was also performed using a DLG deletion mutant construct lacking the first 33 amino acids (Δ1-33) of Neh2 [Neh2 (Δ1-33)]. Alanine substitution mutation of conserved acidic residues within the ETGE motif, such as Asp-77 and Glu-79, reduced the binding capacity by at least 2 to 3 orders of magnitude; the binding capacity of wild-type ETGE was 2 × 108 M−1, whereas the binding capacities of Neh2(Δ1-33,D77A) and Neh2(Δ1-33,E79A) were 1.7 × 106 and 2 × 105 M−1, respectively (Table 4; Fig. 3E and F). A double point mutation, Asp77Ala and Glu79Ala, totally diminished the affinity of the ETGE motif to Keap1-DC (Fig. 3C). A mutation in Thr-80 also accounted for a decrease in the binding affinity (Table 4), presumably due to its intramolecular bonding with Asp-77 and Glu-82 for stabilizing the conformation of the β-turn and its direct interaction with Keap1-DC at the binding interface (24, 33). Alanine substitution of Glu-82 also attenuated the binding affinity to a magnitude of 106 M−1 (data not shown), while mutating Glu-78, a residue not conserved among the CNC family, did not affect the affinity (Table 4). Conversely, alanine substitution of single or double hydrophobic residues (Leu76Ala or Leu76Ala and Phe83Ala), believed to be involved in stabilizing the antiparallel β-sheet structure of the native or Keap1-unbound Neh2 domain (39), induced a modest reduction in the binding affinity (Table 4). These results suggest that acidic residues define the differential binding affinities of the ETGE and DLG motifs.
TABLE 4.
Thermodynamic parameters for binding of ETGE mutants to Keap1-DC at 25°Ca
| Alanine mutation | n | Ka (106 M−1) | ΔH (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) |
|---|---|---|---|---|---|
| L76A | 0.77 ± 0.00 | 87 ± 9.5 | −26.4 ± 0.1 | −15.6 ± 0.2 | −10.8 ± 0.0 |
| D77A | 0.96 ± 0.00 | 1.7 ± 0.1 | −13.9 ± 0.5 | −5.4 ± 0.6 | −8.5 ± 0.0 |
| E78A | 1.03 ± 0.03 | 113 ± 1.6 | −19.8 ± 0.5 | −8.8 ± 0.6 | −11.0 ± 0.0 |
| E79A | 0.85 ± 0.04 | 0.2 ± 0.0 | −9.2 ± 0.5 | −1.9 ± 0.6 | −7.3 ± 0.1 |
| T80A | 1.00 ± 0.01 | 0.5 ± 0.0 | −3.7 ± 0.1 | +4.0 ± 0.1 | −7.7 ± 0.1 |
| L76A F83A | 1.03 ± 0.01 | 59 ± 4.5 | −21.9 ± 0.0 | −11.3 ± 0.1 | −10.6 ± 0.0 |
n is the stoichiometry, and Ka is the binding constant. ΔH, ΔS, and ΔG are the change in binding enthalpy, entropy, and Gibbs energy, respectively. −RT lnKa = ΔG = ΔH − TΔS, where T and R are the absolute temperature and gas constant, respectively. Values are shown as means ± standard deviations from triplicate runs. The first 33 residues of Neh2, including the DLG motif, were deleted from all mutants.
The DLG motif regulates Nrf2 ubiquitination and protein stability.
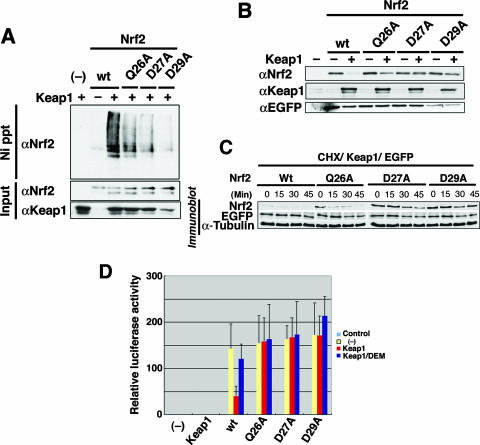
To explore the roles played by the DLG motif in regulating Keap1-mediated ubiquitination of Nrf2, we correlated the binding affinities of DLG mutants with their abilities to direct Keap1-dependent ubiquitination and Nrf2 stability in cells. To this end, in vivo Nrf2 ubiquitination was evaluated in 293T cells. While wild-type Nrf2 was highly ubiquitinated in the presence of Keap1, all three DLG mutants (Gln26Ala, Asp27Ala, and Asp29Ala) showed reduced levels of Nrf2 ubiquitination (Fig. 4A). In particular, mutation of the acidic residue Asp-27 or Asp-29 provoked a strong repression of Nrf2 ubiquitination. While Gln26Ala had only a moderate effect on Keap1-DC association in the calorimetric binding assay, this mutation clearly reduced Nrf2 ubiquitination, although comparatively to a lesser extent. This observation suggests that the effect of this DLG mutation may be augmented in the in vivo context compared to that in the in vitro binding assay. Similarly, when one of these three residues was mutated in the DLG motif, Nrf2 was highly stable compared to wild-type Nrf2 (Fig. 4B).
FIG. 4.
The DLG motif facilitates the Keap1-mediated ubiquitination and protein turnover of Nrf2. (A) Keap1-mediated ubiquitination of wild-type (wt) Nrf2 (lanes 2 and 3) and Gln26Ala (Q26A), Asp27Ala (D27A), and Asp29Ala (D29A) mutants of Nrf2 were examined in the absence (−) or presence (+) of Keap1 by an in vivo ubiquitination assay in 293T cells. Lane 1 is a negative control without Nrf2 input (−). Modified Nrf2 was retrieved by nickel beads (Ni precipitation [Ni ppt]) and monitored by anti-Nrf2 antibody (αNrf2). The protein inputs for Nrf2 and Keap1 were detected by immunoblotting using anti-Nrf2 (αNrf2) and anti-Keap1 (αKeap1) antibodies, respectively. (B) The protein stabilities of wild-type Nrf2 (wt) or mutant Nrf2 (Q26A, D27A and D29A) in steady state were evaluated by immunoblotting in the absence (−) or presence (+) of Keap1 (αNrf2). Keap1 protein input in Cos7 cells and transfection efficiency controls using EGFP plasmid (anti-EGFP antibody [αEGFP]) are shown in the middle and bottom blots, respectively. (C) Protein stability of wild-type (Wt) and mutant Nrf2 after cycloheximide (35 μg/ml) treatment was monitored in 293T cells at 0, 15, 30, and 45 min. Transfection efficiency (EGFP) and protein input (α-Tubulin) controls are as shown. (D) The transactivation activities of wild-type and mutant Nrf2 (as indicated under the bar graph) in Cos7 cells were studied using a luciferase reporter assay in the absence (yellow bars) or presence (red and blue bars) of Keap1. Cells were treated with (blue bars) or without (red bars) the phase II enzyme inducer DEM. The leftmost bar (−) is a negative control with vehicle only.
Cycloheximide chase analysis was also performed to examine more closely how DLG mutations affect the Nrf2 protein stability. Both Asp27Ala and Asp29Ala mutants were stabilized significantly (Fig. 4C, D27A and D29A). Gln26Ala was also more stable than wild-type Nrf2 (Fig. 4C, Q26A), but it was less stable than the other two Asp-Ala mutants. Whereas all three DLG mutations (i.e., Gln26Ala, Asp27Ala, and Asp29Ala) resulted in attenuated Keap1-mediated ubiquitination (Fig. 4A) and a subsequent increase in Nrf2 stability (Fig. 4B and C), the Gln26Ala case is clearly distinguishable from the Asp27Ala and Asp29Ala cases. The latter two mutations almost completely abolished the affinity to Keap1 (Fig. 3A and B), but the Gln26Ala mutant still retained the affinity to Keap1-DC (Fig. 3D), albeit it was less than that of wild-type Nrf2 (Table 2). We surmise that the alanine substitution of Gln-26 gives rise to the reduction of the affinity and alteration of the conformation of the bound DLG motif with Keap1 in such a way that it affects the orientation of the downstream lysines for efficient ubiquitination. These results thus demonstrate the importance of the DLG motif in the efficiency of binding of Nrf2 to Keap1 and consequently in the Keap1-mediated ubiquitination and rapid turnover of Nrf2.
Importantly, the activity of transactivation of these Nrf2-DLG mutants no longer appeared to be suppressed by Keap1 (Fig. 4D). Unlike wild-type Nrf2, these DLG mutants did not respond to DEM, an electrophilic Nrf2 inducer (Fig. 4D). The Gln26Ala mutant showed a severe reduction in the sensitivity to Keap1 in this transactivation-repression assay. These results strongly support our contention that the hinge-and-latch mechanism is regulating Nrf2 activation upon exposure to electrophiles. Since Nrf2 with point mutations in its DLG motif loses its binding affinity to Keap1 (latch unlocked), it cannot be ubiquitinated by Keap1-Cul3 E3 ligase efficiently, even if the Keap1-ETGE binding (hinge) is preserved (Fig. 5A and B). Thus, the mutant Nrf2 is released from the repressive control of Keap1 and is free to activate the expression of cellular defense enzymes.
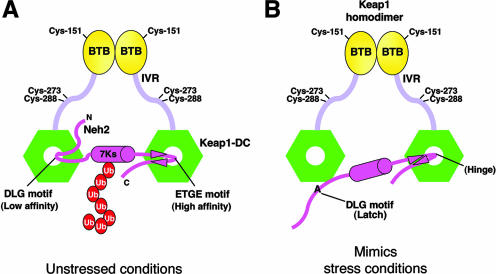
FIG. 5.
Alanine mutation in the DLG motif mimics oxidative stress conditions. (A) Schematic diagram showing the two-site substrate recognition model. Keap1 is composed of BTB (yellow ovals), intervening region (IVR) (light violet bars), and DGR/CTR (Keap1-DC; green hexagons) domains. Some of the important reactive cysteines are labeled. Keap1 homodimer binds with one Neh2 domain (magenta hockey stick) of Nrf2. The middle magenta cylinder represents the α-helix, and the magenta arrowheads represent the β-strands of Neh2. All seven lysines (7Ks) of Neh2 are located within the α-helical region. The multiple ubiquitin (Ub) units are shown as red ovals. Neh2 binds to Keap1-DC via both the ETGE and DLG motifs under unstressed conditions. This complex configuration facilitates efficient ubiquitination of the lysines in the α-helix of Neh2. (B) Alanine mutations in the DLG motif reduce its affinity to Keap1 as well as debilitate Keap1-mediated ubiquitination of Nrf2, which mimics conditions of oxidative stress. The DLG motif, therefore, acts as a latch to lock and unlock the lysines of Neh2 in an appropriate spatial orientation for targeting ubiquitin transfer by the E2 enzyme in different cellular redox states.
DISCUSSION
We recently proposed the hinge-and-latch model for the interaction between Nrf2 and Keap1 and the induction of cellular defense enzymes (39, 40). The Keap1 homodimer recruits its substrate, Nrf2, by binding to the evolutionarily conserved DLG and ETGE motifs within the Neh2 domain of Nrf2 (18, 22). The structural plasticity of its Neh2 domain allows Nrf2 to link two Keap1 molecules in tandem on either side of the central Neh2 α-helix, thereby presenting the lysines of Nrf2 for E2-catalyzed ubiquitination. Our previous nuclear magnetic resonance chemical shift perturbation study suggested that both DLG and ETGE interact with an overlapping binding surface on Keap1-DC composed of conserved arginine residues (39). The present crystal data enable us to visualize the binding interface between the weaker binding DLG motif and Keap1. Superimposition of the ETGE and DLG peptides in complex with Keap1-DC revealed striking similarities in terms of the substrate binding interface and the backbone β-hairpin turn of the two Keap1-bound Neh2 motifs. Nonetheless, critical differences exist in the recognition signal sequences between the Keap1-DC-ETGE complex and the Keap1-DC-DLG complex. Differential negative electrostatic potentials participate in recruitment of the ETGE and DLG motifs. Thus, this study has revealed the importance of the DLG motif in ubiquitination and modulation of cellular Nrf2 activity as the latch that locks and unlocks Nrf2.
Calorimetric binding analysis demonstrated that acidic residues are utilized as the destruction signals. Gln-26 of the DLG motif interacts more extensively with Keap1 residues at the interface than Asp-27 or Asp-29 does. However, a single point mutation in either of the latter two acidic residues induced a marked decrease in the intermolecular interaction of DLG with Keap1-DC, whereas a Gln26Ala mutant showed only a modest reduction in this binding assay. One plausible explanation for this observation is that Keap1 recruits Nrf2 via a basic patch on the surface by recognizing the two acidic aspartates of the flexible DLG motif. Docking of the aspartates on the binding surface of Keap1 allows Gln-26 to form an extensive hydrogen bond network with the Keap1 residues and anchor its side chain between two arginine residues (Arg-415 and Arg-483). By removing either of the two conserved aspartic acids, the substrate recognition signal may become weaker and hence may attenuate the drive for an intermolecular interaction between DLG and Keap1-DC. Therefore, acidic residues seem to serve as the recognition signals for the recruitment of Nrf2 to Keap1 via both the DLG and ETGE motifs. On the other hand, the side chain of Arg-25 was found exposed to solvent in the crystal structure. Indeed, replacement of this nonconserved positive residue with a negative residue did not affect the binding affinity to Keap1 (data not shown).
When correlating reduced binding affinity to physiological functions, we found that all three single DLG mutations (Gln26Ala, Asp27Ala, and Asp29Ala) resulted in attenuated Keap1-mediated ubiquitination and a subsequent increase in Nrf2 stability. This is consistent with the calorimetric binding assay except for Gln26Ala. To our surprise, Gln26Ala still retained an affinity to bind Keap1-DC, although it is enfeebled. However, by cycloheximide analysis, Gln26Ala is much more stable than wild-type Nrf2. This suggests that both proper atomic intermolecular interaction and solid binding affinity between the DLG motif and Keap1 may be essential for the efficient Nrf2 protein turnover. We surmise that the difference seen in Gln26Ala may be due to conformational changes at the DLG region of the Keap1-bound Gln26Ala Nrf2 mutant in the functional assays. Since the region N terminal to the central α-helix of native Neh2 is flexible in nature (39), the tight four-residue β-hairpin structure of the DLG motif in the Keap1-DC-DLG complex is presumed to acquire a conformation that is induced by its binding with Keap1. Removal of a long polar side chain of the glutamine in the Gln26Ala Nrf2 mutant may disable anchorage of the DLG region deep in the binding pocket and may affect the formation of a stable β-hairpin loop in the bound form of the DLG motif. The plausible absence of a stably anchored β-hairpin loop may alter the angle or orientation of downstream residues, including positioning of the lysine residues in one side of the α-helix of Neh2 (39). These multiple lysines in the Neh2 domain have been shown to be the major target for Keap1-mediated Nrf2 ubiquitination (45). Since a defined and correct orientation of these ubiquitin-targeted lysines is crucial for their presentation to the catalytic pocket of the E2 enzyme (30), any deviation may lead to a lower efficiency in ubiquitination. Therefore, the weaker Keap1-binding motif could be highly sensitive to changes in its binding conformation or affinities to the Keap1 homodimer.
In agreement with this notion, previous studies suggest that oxidative/electrophilic stress modifies redox-sensitive reactive cysteines in the intervening region of Keap1 (8). These stimuli-induced modifications seem to trigger conformational change in Keap1 and the modular arrangement of the entire Cul3-based E3 ligase enzyme relative to the substrate Nrf2. Oxidative/electrophilic stress modification of Keap1 is believed to activate Nrf2 by hampering Keap1-mediated Nrf2 ubiquitination (21, 39, 40). Hence, the vulnerability of the DLG motif to changes reflects its important role for derepression/activation of Nrf2 in the oxidative/electrophilic stress response. Interestingly, an Nrf2 mutant containing Gln26Ser, Asp29Ala, Leu30Gly, and Gly31Glu substitutions simultaneously was found stable in Cos1 cells (26). The present study has dissected each of these important residues into specific and distinct roles.
Unlike the DLG motif, the ETGE motif possesses a higher concentration of acidic residues (77DEETGE82). Among the four acidic residues, only Glu-78 is not conserved among the CNC family factors. Except for Glu-78, alanine mutation in any of the other conserved aspartate and glutamate residues showed a reduction of the binding affinity of at least 2 orders of magnitude. In addition, Asp-77 seems to be important for substrate recognition. Asp-77 was suggested to participate in intramolecular bonding with Glu-79, Thr-80, and Gly-81 in the Keap1-bound form of the ETGE peptide (24, 33), and this residue also makes contacts with Keap1 via water molecules (24). On the other hand, analysis using single and double mutations of the hydrophobic residues (Leu76Ala or Leu76Ala and Phe83Ala), which were believed to play a role in stabilizing the antiparallel β-sheet conformation of native or Keap1-unbound Neh2 (39), showed a very modest reduction in binding affinity. Analogously, Arg-25 and Ile-28 in the DLG region also form intramolecular bonding with each other in the protein-peptide complex, but alanine mutation of either one of these two residues did not decrease the binding affinity of DLG to Keap1. These results suggest that multiple conserved acidic residues have a stronger driving force in substrate recognition, although there may still be some contribution by the preformed β-hairpin structure in the native Neh2 (39) to the binding of the ETGE motif with Keap1.
Mutation of any of the three conserved acidic residues (Asp-77, Glu-79, and Glu-82) converts the binding capacity of the ETGE motif close to that of the DLG motif, with an association constant of a similar or lower order, i.e., 105 to 106 M−1. Similarly, concurrent mutation of two acidic residues (Asp-77 and Glu-79) totally abolished the binding. This coincides nicely with the results of mutagenesis studies performed on the DLG motif, where a single aspartate residue (Asp-27 or Asp-29 for DLG and Glu-82 for ETGE) was insufficient for promoting interaction of the DLG with Keap1, albeit the simultaneous presence of Gln-26 in DLG (or Thr-80 in the ETGE motif).
The high-affinity ETGE motif allows efficient recruitment of Nrf2 to the Cul3-based E3 ligase. As summarized in Fig. 5, the ETGE motif remains attached to Keap1 in the event of an electrophilic stress (21, 40), acting as a hinge of the Keap1 repression gate. On the other hand, the DLG motif seemingly functions as a latch to lock and unlock the lysines in space for targeting them to the catalytic pocket of the E2 enzyme (39, 40). By hampering the affinity or altering the binding conformation of the DLG motif to Keap1, the system may switch to mimic stress conditions. Indeed, single point mutation of Asp-27 or Asp-29 almost totally debilitates Keap1-dependent ubiquitination of Nrf2 and elevates Nrf2 stability in cells. Not only were these mutants free from Keap1 repression, they simultaneously failed to respond to phase II enzyme inducers, as they are no longer under the control of the redox sensor. Mutation of Gln26 also enforced similar phenomena. Taken together, these data demonstrate that the DLG motif is truly a factor in the regulation of Keap1-mediated ubiquitination and stability of Nrf2. The hinge-and-latch mechanism provides a sensitive and rapid response to oxidative stress, since any stress signal is transduced directly to the protein adaptor of the E3 ligase.
Recognition of substrates by means of negatively charged moieties seems to be common in other β-propeller-like substrate adaptors, such as β-TrCP for β-catenin (10), human Cdc25A (3), and Gli3 (38) degradation or Cdc4 for Sic1 degradation (37). SCFβ-TrCP can recognize both phosphorylated and nonphosphorylated degrons. Phosphorylation of serine and threonine residues in the recognition sequences or the presence of phosphomimetic aspartic and glutamic acids (DpSGX2-4pS versus DDGX2D or DEGX2E, etc.) are the essential criteria for substrate recruitment (15, 42, 44). Removal of the conserved serines or alanine substitution in either one of the phosphomimetic negatively charged residues within the targeting consensus considerably deflated substrate association with the basic binding pocket in β-TrCP (10, 15). On the other hand, at least six out of the nine CPDs of Sic1 must be phosphorylated for Cdc4 recognition. Of these phosphodegrons, there are some suboptimal binding sequences due to the presence of basic residues flanking the recognition signals. These basic residues may have diluted the negative charge potential of the phosphate moieties of the phosphorylated serines or threonines and led to electrostatic repulsion against the basic region in and around the CPD binding pocket in Cdc4 (30). Therefore, a negative electrostatic potential, produced by acidic residues or phosphorylated serines and threonines, may play an important role in substrate recognition by the conserved basic substrate-binding pocket in β-propeller structures, as demonstrated in the present study.
In contrast to our findings, a 2:2 binding stoichiometry between Keap1 and Nrf2 was recently suggested by means of ectopically expressed Keap1 and Nrf2 or their mutants (24). The same study also proposed that the ETGE motif is self-sufficient for binding to Keap1. As shown clearly in our previous study using a DLG deletion mutant (39), ETGE alone is sufficient for Nrf2 to bind to Keap1 and the binding affinity is similar to the higher affinity of wild-type Neh2. Since ETGE has a much higher binding affinity compared to the binding affinity of DLG, a mixture of an equal molar ratio of Neh2 and Keap1 will result in a 1:1 or 2:2 binding stoichiometry, primarily due to competition of the higher binding affinity ETGE site. This competitive effect, however, will be titrated out as Keap1 increases. At a 1:2 (Neh2:Keap1-DC) ratio, both ETGE and DLG were found to interact with Keap1. Since the two-site binding is very sensitive to the competitive effect of the ETGE motif, these experiments were performed with good control over the concentrations of Keap1 and Neh2 or Neh2 mutants (39). Furthermore, there may be other examples of the two-site binding mechanism (26), and our present data have provided a solid demonstration that the DLG motif is indispensable to Keap1-mediated ubiquitination and Nrf2 stability. As the cellular concentrations of the ectopically expressed Keap1, tagged Neh2, and tagged Nrf2 could not be well defined, the estimated 2:2 stoichiometry might be biased by the competitive effect of ETGE (24). In living cells, however, Keap1 levels should exceed Nrf2 due to the short-lived nature of the transcription factor. It has been shown that endogenous Nrf2 concentration is lower than that of endogenous Keap1 in either mouse liver homogenates or mouse embryonic fibroblast cells under homeostatic conditions (41). Therefore, two-site binding is most probable in real cell conditions.
Taken together, the present study has revealed the determinants for defining the differential affinities of the two Keap1-binding motifs in Nrf2 and their respective roles in the regulation of Keap1-dependent Nrf2 ubiquitination and degradation in the electrophilic stress response. Combining these results with structural information will aid in the rational design of small compounds to manipulate the transactivation activity of Nrf2 for therapeutic treatments of stress-related diseases.
Acknowledgments
We are grateful to M. Kobayashi, H. Motohashi, and K. Itoh for helpful discussions. We are indebted to T. Tanaka for use of the isothermal calorimeter, Y. Nakamura for helping in data collection, and T. O'Connor for critical reading of the manuscript.
This work was supported in part by the RIKEN Structural Genomics/Proteomics Initiative; the National Project on Protein Structural and Functional Analyses; the Japanese Ministry of Education, Culture, Sports, Science and Technology; and ERATO-JST.
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Bonizzi, G., and M. Karin. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25: 280-288. [DOI] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D 54: 905-921. [DOI] [PubMed] [Google Scholar]
- 3.Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N. V. Dorrello, A. Hershko, M. Pagano, and G. F. Draetta. 2003. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426: 87-91. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover, A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79: 13-21. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17: 7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50: 760-763. [DOI] [PubMed] [Google Scholar]
- 7.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24: 8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99: 11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25: 162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart, M., J. P. Concordet, I. Lassot, I. Albert, R. del los Santos, H. Durand, C. Perret, B. Rubinfeld, F. Margottin, R. Benarous, and P. Polakis. 1999. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9: 207-210. [DOI] [PubMed] [Google Scholar]
- 11.Holtzclaw, W. D., A. T. Dinkova-Kostova, and P. Talalay. 2004. Protection against electrophile and oxidative stress by induction of phase 2 genes: the quest for the elusive sensor that responds to inducers. Adv. Enzyme Regul. 44: 335-367. [DOI] [PubMed] [Google Scholar]
- 12.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13: 76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh, K., K. I. Tong, and M. Yamamoto. 2004. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 36: 1208-1213. [DOI] [PubMed] [Google Scholar]
- 14.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. D 47: 110-119. [DOI] [PubMed] [Google Scholar]
- 15.Kanemori, Y., K. Uto, and N. Sagata. 2005. β-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc. Natl. Acad. Sci. USA 102: 6279-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang, M. I., A. Kobayashi, N. Wakabayashi, S. G. Kim, and M. Yamamoto. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 101: 2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18: 621-663. [DOI] [PubMed] [Google Scholar]
- 18.Katoh, Y., K. Iida, M. I. Kang, A. Kobayashi, M. Mizukami, K. I. Tong, M. McMahon, J. D. Hayes, K. Itoh, and M. Yamamoto. 2005. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 433: 342-350. [DOI] [PubMed] [Google Scholar]
- 19.Kim, W., and W. G. Kaelin, Jr. 2003. The von Hippel-Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13: 55-60. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24: 7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, A., M. I. Kang, Y. Watai, K. I. Tong, T. Shibata, K. Uchida, and M. Yamamoto. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26: 221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, M., K. Itoh, T. Suzuki, H. Osanai, K. Nishikawa, Y. Katoh, Y. Takagi, and M. Yamamoto. 2002. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7: 807-820. [DOI] [PubMed] [Google Scholar]
- 23.Lakowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283-291. [Google Scholar]
- 24.Lo, S. C., X. Li, M. T. Henzl, L. J. Beamer, and M. Hannink. 2006. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25: 3605-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis, T. 1999. A ubiquitin ligase complex essential for the NF-κB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 13: 505-510. [DOI] [PubMed] [Google Scholar]
- 26.McMahon, M., N. Thomas, K. Itoh, M. Yamamoto, and J. D. Hayes. 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 281: 24756-24768. [DOI] [PubMed] [Google Scholar]
- 27.Motohashi, H., and M. Yamamoto. 2004. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10: 549-557. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D 53: 240-255. [DOI] [PubMed] [Google Scholar]
- 29.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514-521. [DOI] [PubMed] [Google Scholar]
- 30.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243-256. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307-326. [DOI] [PubMed] [Google Scholar]
- 32.Padmanabhan, B., M. Scharlock, K. I. Tong, Y. Nakamura, M. I. Kang, A. Kobayashi, T. Matsumoto, A. Tanaka, M. Yamamoto, and S. Yokoyama. 2005. Purification, crystallization and preliminary X-ray diffraction analysis of the Kelch-like motif region of mouse Keap1. Acta Crystallogr. Sect. F 61: 153-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanabhan, B., K. I. Tong, T. Ohta, Y. Nakamura, M. Scharlock, M. Ohtsuji, M. I. Kang, A. Kobayashi, S. Yokoyama, and M. Yamamoto. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21: 689-700. [DOI] [PubMed] [Google Scholar]
- 34.Pickart, C. M., and M. J. Eddins. 2004. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695: 55-72. [DOI] [PubMed] [Google Scholar]
- 35.Schofield, C. J., and P. J. Ratcliffe. 2005. Signalling hypoxia by HIF hydroxylases. Biochem. Biophys. Res. Commun. 338: 617-626. [DOI] [PubMed] [Google Scholar]
- 36.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14: 1983-1991. [PubMed] [Google Scholar]
- 37.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209-219. [DOI] [PubMed] [Google Scholar]
- 38.Tempé, D., M. Casas, S. Karaz, M. F. Blanchet-Tournier, and J. P. Concordet. 2006. Multisite protein kinase A and glycogen synthase kinase 3β phosphorylation leads to Gli3 ubiquitination by SCFβTrCP. Mol. Cell. Biol. 26: 4316-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong, K. I., Y. Katoh, H. Kusunoki, K. Itoh, T. Tanaka, and M. Yamamoto. 2006. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 26: 2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tong, K. I., A. Kobayashi, F. Katsuoka, and M. Yamamoto. 2006. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. 387: 1311-1320. [DOI] [PubMed] [Google Scholar]
- 41.Watai, Y., A. Kobayashi, H. Nagase, M. Mizukami, K. Itoh, and M. Yamamoto. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells, in press. [DOI] [PubMed]
- 42.Watanabe, N., H. Arai, Y. Nishihara, M. Taniguchi, N. Watanabe, T. Hunter, and H. Osada. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc. Natl. Acad. Sci. USA 101: 4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willems, A. R., T. Goh, L. Taylor, I. Chernushevich, A. Shevchenko, and M. Tyers. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R Soc. Lond. B 354: 1533-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winston, J. T., P. Strack, P. Beer-Romero, C. Y. Chu, S. J. Elledge, and J. W. Harper. 1999. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13: 270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, D. D., S.-C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24: 10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]