Abstract
The Ets family transcription factor PU.1 is crucial for the regulation of hematopoietic development. Pu.1 is activated in hematopoietic stem cells and is expressed in mast cells, B cells, granulocytes, and macrophages but is switched off in T cells. Many of the transcription factors regulating Pu.1 have been identified, but little is known about how they organize Pu.1 chromatin in development. We analyzed the Pu.1 promoter and the upstream regulatory element (URE) using in vivo footprinting and chromatin immunoprecipitation assays. In B cells, Pu.1 was bound by a set of transcription factors different from that in myeloid cells and adopted alternative chromatin architectures. In T cells, Pu.1 chromatin at the URE was open and the same transcription factor binding sites were occupied as in B cells. The transcription factor RUNX1 was bound to the URE in precursor cells, but binding was down-regulated in maturing cells. In PU.1 knockout precursor cells, the Ets factor Fli-1 compensated for the lack of PU.1, and both proteins could occupy a subset of Pu.1 cis elements in PU.1-expressing cells. In addition, we identified novel URE-derived noncoding transcripts subject to tissue-specific regulation. Our results provide important insights into how overlapping, but different, sets of transcription factors program tissue-specific chromatin structures in the hematopoietic system.
Hematopoietic cell differentiation is driven by sequence-specific transcription factors which regulate cell lineage-specific genetic programs. However, transcription factors do not act on their own; rather, they interact with epigenetic regulatory complexes that modify and remodel chromatin structure and thus create a chromatin environment that is permissive for active transcription (23). These general principles have been discovered through in-depth analyses of the regulation of genes activated or repressed in specific blood cell lineages (reviewed in reference 4). In order to understand how the transcription factor and target gene network responds to developmental cues and how the hematopoietic hierarchy is established, it is important to investigate the regulation of genes central to specific developmental pathways. One such gene is the gene encoding the Ets family transcription factor PU.1. The analysis of PU.1 null mice showed that the transcription factor PU.1 is a key factor for the development of both the myeloid and lymphoid lineages (8, 25, 35). The expression of the PU.1 gene is restricted to hematopoietic cells, and the gene is already expressed at high levels in hematopoietic stem cells (HSCs). Thereafter, PU.1 expression is tightly regulated and is maintained during myelopoiesis and B lymphopoiesis but is down-regulated in erythroid cells and T cells (8). PU.1 is not required for HSC formation but is involved in regulating cell fate decisions in the downstream committed precursor compartments (8). The mechanistic explanation for these observations comes from experiments demonstrating that PU.1 regulates the activity and expression of a number of other transcription factors. It was shown previously that PU.1 restricts erythropoiesis by inhibiting the activity of the GATA-1 gene, a crucial regulator of erythropoiesis (26, 29, 43). PU.1 also functions in balance with C/EBPα and GATA-2 to regulate granulocyte and mast cell development, respectively (7, 41). Furthermore, the down-regulation of PU.1 expression is essential for T-cell development (2). The overexpression of PU.1 in transgenic mice leads to erythroleukemia (26), and it was recently found that reduced levels of PU.1 can lead to myeloid leukemia (34). Hence, it is clear that PU.1 expression levels need to be tightly regulated in development for correctly balanced hematopoiesis.
The complete regulatory region of the PU.1 locus as defined by correct expression in transgenic mice is located on a 91-kb fragment of genomic DNA (24). By using DNase I-hypersensitive site (DHS) mapping, several regulatory regions could be identified. Two elements have been extensively characterized: the Pu.1 promoter and a DHS at kb −14 that represents an enhancer element, the upstream regulatory element (URE), that is absolutely required for high-level expression of the endogenous PU.1 locus and the correct regulation of the PU.1 gene in transgenic mice (Fig. 1) (24, 34). The Pu.1 promoter was shown to be active in myeloid cells and B cells but not T cells, and this tissue-specific expression pattern was mediated by binding sites for the transcription factors Sp1 and Oct-1, as well as PU.1 itself. Reporter gene assays also showed that Oct-1, together with its B-cell-specific cofactor OCAB/BOB1/OBF1 (40), is the most important factor mediating promoter activity in B cells, whereas in myeloid cells PU.1 itself is the main factor driving expression (5, 6). The URE consists of two highly conserved regions that are separated by a 500-bp nonconserved DNA sequence harboring the integration site for the spleen focus-forming virus (SFFV), and both regions are important for Pu.1 regulation (31, 33). In the absence of the URE, Pu.1 expression in unsorted bone marrow cells drops by 80%. Interestingly, the down-regulation of Pu.1 during T-cell development is also blocked in the absence of this element, indicating that it is required not only for the activation of Pu.1 expression in HSCs but also for the repression of expression in T-cell precursors. This repressive activity is dependent on a binding site for the transcription factor T-cell factor/lymphoid enhancer factor (TCF/LEF) and on β-catenin, indicating that the Wnt signaling pathway is involved in repression (33, 34). In addition to the TCF/LEF site, the URE contains numerous other elements required for function, including three binding sites for RUNX1 and C/EBP and two binding sites for PU.1 which can also be bound by other Ets factors (31, 42). This finding indicates that in addition to the promoter, the URE is subject to autoregulation by PU.1. However, it is not clear whether other Ets family members can drive the establishment of an active chromatin structure in the absence of PU.1 in vivo and what the precise role of PU.1 is with respect to instructing Pu.1 chromatin during development. We also do not know which chromatin structure supports the expression of Pu.1 in precursor cells and the different PU.1-expressing cell types or whether the binding of the transcription factors driving expression is developmentally regulated. In the study presented here, we addressed these questions. We show by chromatin fine-structure studies and chromatin immunoprecipitation (ChIP) assays that precursor cells, macrophages, and B cells adopt different chromatin architectures and use different sets of transcription factors to drive Pu.1 expression, and we demonstrate that in T cells, Pu.1 chromatin is in a partially active chromatin conformation and is occupied by transcription factors. We also show that the URE has promoter activity and drives the transcription of noncoding RNAs (ncRNAs). Our study provides a coherent picture of how different transcription factors program Pu.1 chromatin structure in vivo in different cell types and at different developmental stages.
FIG. 1.
Map of the Pu.1 locus indicating the positions of introns and exons, DHSs (closed vertical arrows), cis-regulatory elements, and the transcription start site (marked by a horizontal arrowhead). The positions at which previously characterized transcription factors bind to the promoter and the URE are depicted in detail. Factors indicated in bold have been confirmed by ChIP in this and other studies, as cited in the text; others were confirmed by EMSAs. The open vertical arrow indicates the integration site of the SFFV separating the two parts of the URE.
MATERIALS AND METHODS
Cell isolation and culture.
The generation of PU.1−/− and PUER cells has been described previously (41). Cells were cultured in phenol red-free Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum, 50 μM β-mercaptoethanol, 100 U of penicillin/ml, 100 U of streptomycin/ml, and 5 ng of recombinant mouse interleukin-3 (Biosource)/ml. For 4-hydroxy-tamoxifen (OHT) treatment, cells were plated at 0.2 × 106 to 0.3 × 106 cells/ml in complete medium supplemented with 100 nM OHT (Sigma) and were harvested at the time points indicated in the figures. RAW 264 macrophages and NIH 3T3 fibroblasts were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U of penicillin/ml, and 100 U of streptomycin/ml.
The isolation of the lineage-negative (Lin−) Sca+ c-kit+ (LSK) and common myeloid progenitor (CMP) fractions from mouse bone marrow and the in vitro differentiation of CMPs into macrophages were carried out as described previously (37). For experiments with the URE mutant mice, the precursor pool was expanded by culturing total bone marrow cells for 10 days in Iscove's modified Dulbecco's medium supplemented with 10% horse serum, 20 ng of mouse stem cell factor/ml, 10 ng of mouse Flt3 ligand/ml, 25 ng of mouse thrombopoietin/ml, 100 U of penicillin/ml, and 100 U of streptomycin/ml. Primary macrophages were grown as described previously (37). For splenic B cells and thymic T cells, spleens and thymuses were taken from mice and homogenized immediately. Unwanted cell lineages were removed with Low-Tox-M rabbit reverse complement (VH Bio) using anti-major histocompatibility complex class II/anti-B220 for the thymocytes and anti-GR1/anti-CD11b/anti-Ter119/anti-ERMP20/anti-Thy1.2 for the splenocytes. The isolation of the remaining nucleated cells was performed by density separation using Lympholyte-M (VH Bio). Purity was confirmed by flow cytometry using Thy1.2 for T cells (>99%) and CD19 for B cells (>90%).
URE mutant mice.
To generate knock-in mice containing mutations in either the three RUNX1 sites or the PU.1 site in the URE, the targeting vector pPNT-(−14kb URE)-KI was used as described previously (34). In these mice, the URE was replaced with a mutant URE in which either all three RUNX1 sites were mutated from TGTGGT to TGACCT by site-directed mutagenesis (13) or the PU.1 site GGTGACTGGGCGCTTCCTGTTTTCTCAGGC was replaced with the sequence GGTGACTGGGCGCGCGATGTTTTCTCAGGC (data not shown). The inability of these mutant sequences to bind PU.1 and RUNX1 in vitro was reported in reference 31.
ChIP assays and real-time PCR analysis.
The ChIP assays were performed essentially as described previously (22). If not stated otherwise, antibodies were purchased from Santa Cruz. Immunoprecipitation was performed overnight at 4°C on a rotating wheel with 4 μg of anti-PU.1 (sc-352X), anti-C/EBPβ (sc-150X), anti-Fli-1 (sc-356X), anti-RUNX1 (Calbiochem; catalog no. PC284L), anti-RNA polymerase II (anti-RNA Pol II; sc-900X), anti-phosphoserine 5 RNA Pol II (Abcam 5131), anti-phosphoserine 2 RNA Pol II (Abcam 5095), anti-histone H3 (Abcam 1791), anti-trimethyl lysine 4 histone H3 (Abcam 8580), or anti-acetyl lysine 9 histone H3 (Abcam 4441). The amount of precipitated DNA was measured by real-time quantitative PCR with the ABI Prism 7700 or 7900HT sequence detection system (PerkinElmer Life Sciences) using SYBR green as described in reference 21. The amounts of DNA precipitated were calculated using a standard curve obtained from the amplification of serially diluted mouse genomic DNA. For all ChIP assays, values representing the signals observed with the specific antibody were divided by values representing the signals obtained from an input control. For all ChIP assays examining histone modifications, the signal values were additionally normalized against those obtained with a histone H3 antibody (these were also normalized against the input values) to account for histone content. To correct for antibody-dependent backgrounds, the relative PCR signal in transcription factor ChIP assays was additionally normalized against the signal from the control primer set. The primers were originally designed to recognize the transcribed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene but were later found to hybridize to sequences located in a gene desert on chromosome 2 (the mouse chromosome 2 region from position 28600000 to position 28900000) and were therefore used as a negative control. Primers were designed using Primer Express 1.5 software. Primer sets for the PU.1 locus had the following 5′-to-3′ sequences corresponding to the indicated genomic positions relative to the transcription start site: 5′ URE forward primer (−14422 to −14403), GCC CAG GCT AGG GAA GTT TG, and reverse primer (−14342 to −14364), GAG AGC AGA GCA CTT CAT GGC TA; 3′ URE forward primer (−13687 to −13669), GGG AGG CAG AGC ACA CAT G, and reverse primer (−13602 to −13619), GTT TCC ACA TCG GCA GCA G; PU.1 −5-kb control forward primer (−5429 to −5405), GGC ACA TGG TAG AAG AGA ACA AAC T, and reverse primer (−5374 to −5350), TTG TGT TTT CAC TGT GTG TCT GAT G; PU.1 promoter forward primer (−73 to −50), GTA GCG CAA GAG ATT TAT GCA AAC, and reverse primer (+27 to +4), GCA CAA GTT CCT GAT TTT ATC GAA; PU.1 +0.4-kb DHS forward primer (+409 to +430), CCA TTG GCT TCC TTA GAG CAT G, and reverse primer (+522 to +500), CCT GCC ACT GAA CCC ATC TAT AA; PU.1 +6-kb control forward primer (+5670 to +5696), TTG TTT CTT CAT AAC TGT GAT TTT GCT, and reverse primer (+5778 to +5759), TAT CAC CCA GGC CGT GAC TC; PU.1 +17-kb DHS forward primer (+16763 to +16782), TGG TGA GGC ATG GAA CCT TC, and reverse primer (+16843 to +16823), CTG CCG TTG GCT CTG TAG ATC.
In vivo footprinting analysis.
DNase I treatment of cells and naked DNA was performed exactly as described in reference 21. Dimethylsulfate (DMS) and micrococcal nuclease (MNase) treatment and ligation-mediated PCR (LM-PCR) were performed as described previously in reference 36. After DNA purification, 1 μg of DNA was used as the input for the detection of single-strand breaks (with DMS and DNase I) or double-strand breaks (with MNase). The primer sets used had the following 5′-to-3′ sequences corresponding to the indicated genomic positions relative to the transcription start site: PU.1 promoter forward primer 1 (−239 to −222), biotin-TGG ACT ACT TCA GCA AGG, primer 2 (−198 to −175), CCT TCC ATG GTA GTG CTA GCC TTT, and primer 3 (−191 to −165), TGG TAG TGC TAG CCT TTC TCC CTC CCA; PU.1 promoter reverse primer 1 (+115 to +98), biotin-GAC GGT CGT GGG TCA GAC, primer 2 (+86 to +67), GCC TGC CCC CTG AGC TAC AG, and primer 3 (+76 to +53), TGA GCT ACA GGA GCC CTG GGT GAG; PU.1 5′ enhancer forward primer 1 (−14567 to −14550), biotin-CCA GAG ATC AGT GAG CAG, primer 2 (−14541 to −14520), GGA GGC TCT GGG TAG GTG AGG T, and primer 3 (−14536 to −14509), CTC TGG GTA GGT GAG GTG CCT GAG CTT C; and PU.1 3′ enhancer forward primer 1 (−13847 to −13830), biotin-GTT CTT CTA GGT CAC GAT, primer 2 (−13821 to −13800), ACC CTA ACC CCT GCA CAT GAA A, and primer 3 (−13811 to −13786), CTG CAC ATG AAA GCC AGG GTC TGT GT. Primers were designed manually with assistance from Oligo 5.1 software.
RNA expression analysis.
Total RNA was extracted using TRIzol according to the protocol of the manufacturer (Invitrogen). Any potential residual genomic DNA was removed by treatment with DNase I. For PU.1 mRNA expression, 0.75 μg of total RNA was used for first-strand cDNA synthesis with an oligo(dT) 15-mer primer and Moloney murine leukemia virus reverse transcriptase. For the detection of ncRNA, 0.75 to 1.5 μg of total RNA was used for cDNA synthesis with a biotinylated PU.1 region-specific primer as well as a biotinylated GAPDH gene-specific primer and Moloney murine leukemia virus reverse transcriptase. Synthesized cDNA was immobilized on Dynabeads (Dynal; catalog no. M-270). RNA was removed by alkaline denaturation and serial washing, and cDNA was eluted by heating the bead suspension in 0.1× Tris-EDTA for 15 min at 99°C.
Real-time quantitative PCR was performed on the ABI Prism 7700 or 7900HT sequence detection system (PerkinElmer Life Sciences) using SYBR green. Relative expression was calculated as a ratio to the GAPDH signal. The 5′-end-biotinylated primers with the following 5′-to-3′ sequences were used: 5′HF, CCA GAG ATC AGT GAG CAG; 5′HR, CTG TTG CTG TCA GAT CTA T; SFFVF, CTG GCC TCC CCA AAG CAG; SFFVR, CTG GGA GAA ACG CTC TTG; 3′HF, GTT CTT CTA GGT CAC GAT; 3′HR, GCC AAG ACT AGG ACT CAA; and the GAPDH gene primer, GCA GCC CTG GTG ACC AGG CGC GGA ATA CGG. Primers for real-time analysis had the following 5′-to-3′ sequences corresponding to the indicated genomic positions relative to the transcription start site: 5′ URE forward primer (−14422 to −14403), GCC CAG GCT AGG GAA GTT TG, and reverse primer (−14342 to −14364), GAG AGC AGA GCA CTT CAT GGC TA; SFFV forward primer (−14070 to −14049), ATC GAT CTT GGC AAG GCT TAG A, and reverse primer (−14000 to −13978), GAA CCC ATG TCT CAG ATA TGG CT; PU.1 3′ URE forward primer (−13687 to −13669), GGG AGG CAG AGC ACA CAT G, and reverse primer (−13602 to −13619), GTT TCC ACA TCG GCA GCA G; and GAPDH control forward primer, AAA TCC GTT TCA CAC CGA CCT T, and reverse primer, ACA GCC GCA TCT TTC TTG TGC. Primers for real-time analysis were designed using Primer Express 1.5 software, whereas biotinylated primers were designed manually with assistance from Oligo 5.1 software.
RESULTS
The Pu.1 promoter and the URE adopt differential patterns of transcription factor occupancy and chromatin fine structure in primary macrophages, B cells, and T cells.
We examined the chromatin structures and transcription factor occupancy patterns of the Pu.1 promoter and the URE in different cell types by performing in vivo footprinting experiments with DNase I, MNase, and DMS, which each give different types of information (27, 39). DMS methylates the N-7 position of guanines, and this reaction can be either inhibited or enhanced by the binding of transcription factors. In contrast, nucleosome-DNA interactions cannot be detected by DMS footprinting because they occur via the phosphate backbone. Furthermore, DNA-protein interactions need to be very stable to be detected by DMS, because DMS has sufficient time to react at sites of dynamic interactions during the 5-min incubation period. Unstable interactions can yield partial protection from or enhancements of DMS reactivity (38). MNase can gain access to nucleosomal linker regions and is often used to define nucleosome positioning and detect regions of nucleosome remodeling (21). DNase I can create single-strand nicks at the surfaces of nucleosomes and thereby generate a specific cleavage pattern that is defined by the rotational positioning of nucleosomes and by chromatin folding. DNase I is also frequently used to detect sites occupied by transcription factors. These sites often colocalize with nucleosome-free regions and are responsible for the formation of DNA sequences hypersensitive to DNA cleavage (10). Under conditions of limited digestion, DNase I can also be used to assay the general accessibility of chromatin in different cell types.
Figures 2, 3, and 4 show results from the above-described assays examining the chromatin architecture of the Pu.1 promoter and the URE in primary macrophages, purified splenic B cells, and thymocytes. In these experiments, we wanted to address two questions. Firstly, we wanted to see how chromatin structures and transcription factor occupancy patterns differed between macrophages and B cells, which express PU.1 to various degrees. Secondly, we wanted to examine T-lineage cells in which Pu.1 expression had been switched off to determine whether the Pu.1 locus was still occupied by transcription factors and/or whether its chromatin was still reorganized. For control purposes, purified genomic DNA was subjected to the same modifications in vitro, and we employed 3T3 fibroblasts as an example of a lineage in which Pu.1 has never been expressed. Lesions in DNA were made visible using LM-PCR along with different primer sets, including primers amplifying ribosomal DNA as a control for equal degrees of DNase I digestion. Figure 2 shows results from LM-PCR analyses of DNA purified from chromatin treated with DMS and DNase I for the amplification of both strands of promoter sequences. The positions of DMS-reactive guanines and local alterations in DNase I accessibility are plotted along the DNA sequence in the lower panel of Fig. 2. Regions of high DNase I accessibility were found over the transcription start site in macrophages and B cells, probably reflecting the open complex generated by the binding of RNA Pol II. It was also apparent that the promoters in macrophages and B cells were much more accessible to DNase I cleavage than those in T cells and 3T3 cells. Closer inspection of the chromatin architecture in macrophages and B cells shows pronounced differences between these two cell types in the immediate promoter region, indicating differential transcription factor occupancy. This finding is supported by the results of DMS footprinting experiments (Fig. 2) and those of electrophoretic mobility shift assays (EMSAs) (5, 6; also data not shown). Macrophages and B cells both showed occupancy of a sequence specifically binding the Ets family transcription factor Elf-1 in vitro (data not shown). Macrophage-specific DNA-protein contacts and alterations in DNase I cleavage on both strands were seen at the C/EBP site, the PU.1 site downstream of the transcription start site, a cluster of sites binding the transcription factors Sp1 and Sp3, and a downstream site binding the helix-loop-helix factor activation transcription factor 2 (ATF-2) (ChIP and EMSA data not shown). A B-cell-specific difference in DNase I digestion patterns was seen at the Oct-1 site (Fig. 2, leftmost panel), which may indicate that Oct-1, cooperating with the B-cell-specific cofactor OCAB/BOB1/OBF1, is the main factor responsible for Pu.1 promoter activity in B cells as measured by transient transfection assays (6). We also saw a hyperreactive guanine in both B cells and T cells at a site able to bind Sp1 family factors (such as Sp1 and Sp3) in vitro (data not shown). Macrophages generated a different footprint pattern (Fig. 2, panel second from left), including multiple hyporeactive guanines compared to the single hyperreactive guanine in lymphoid cells, suggesting that the Pu.1 promoter is occupied by different factors in lymphocytes and macrophages.
FIG. 2.
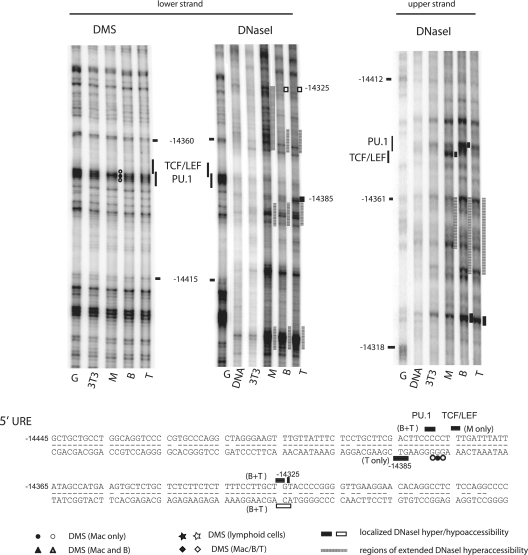
Different transcription factors program differential chromatin structures of the Pu.1 promoter in macrophages (M; Mac), B cells (B), and T cells (T). The upper panels display the results of in vivo footprinting analyses of both strands of the promoter using DMS and DNase I as indicated. 3T3 cells and purified genomic DNA served as controls. G, Maxam-Gilbert G reaction with purified genomic DNA. The far-right panel depicts the analysis of the lower strand of the promoter by an in vivo footprinting reaction after MNase digestion of chromatin. The corresponding regions of nucleosome remodeling compared in macrophages and B cells are indicated by a bracket. Results from an LM-PCR using primers amplifying the ribosomal DNA (rDNA) promoter are shown as the control for equal degrees of DNase I digestion. The positions and the nature of transcription factor binding sites are indicated, and the dashed lines serve to align the different gels. The sequence of the Pu.1 promoter with annotated transcription factor binding sites is shown below, with a schematic indication of the different types of in vivo footprints as explained below the sequence. Base pairs conserved in human and mouse sequences are indicated by horizontal lines. The closed symbols indicate regions of hyperreactivity, and the open symbols indicate regions of hyporeactivity; the cell types in which these regions were observed are indicated. Only reproducible alterations are highlighted.
FIG. 3.
Different transcription factors program differential chromatin structures of the 3′ URE in macrophages (M; Mac), B cells (B), and T cells (T). Results from the analysis of the bottom strand are shown. For more explanation, refer to the legend to Fig. 2. G, Maxam-Gilbert G reaction with purified genomic DNA.
FIG. 4.
Different transcription factors program differential chromatin structures of the 5′ URE in macrophages (M; Mac), B cells (B), and T cells (T). For more explanation, refer to the legend to Fig. 2. G, Maxam-Gilbert G reaction with purified genomic DNA.
In order to obtain information about further aspects of chromatin architecture of the Pu.1 promoter in the different cell types, we digested chromatin with MNase and then subjected the digestion product to an LM-PCR designed to visualize double-strand cuts (Fig. 2, rightmost panel). The analysis of promoter chromatin by MNase indicated differences in chromatin architecture not only between different Pu.1-expressing cell types but also between the two non-Pu.1-expressing cell types (3T3 and T cells). The LM-PCR analysis of DNA digested into mainly mono- and dinucleosomes (data not shown) revealed extensive nucleosome remodeling over the transcription start site in macrophages and B cells. This remodeling was indicated by the loss of signal in the larger-fragment range which occurred over the same sequences that were highly DNase I accessible on both strands. In contrast, 3T3 cells showed prominent cuts across a region of 150 bp, indicating alternatively positioned nucleosomes over the promoter. Although a very similar pattern was seen in T cells, the signal intensity was reduced in the higher-fragment-size range, indicating partial nucleosome destabilization.
Similar tissue-specific differences in chromatin architecture and transcription factor occupancy were seen at the URE (Fig. 3 and 4). However, in contrast to the promoter, both parts of the enhancer exhibited increased general DNase I accessibility in macrophages, B cells, and T cells relative to that in 3T3 cells. In the 3′ enhancer segment (Fig. 3), all three hematopoietic cell types showed occupancy at the single RUNX1 site adjacent to the PU.1 site. Macrophage-specific DMS footprints were seen at a site able to bind Sp1 and Sp3 in vitro (EMSA data not shown), a site binding PU.1 and Fli-1 (see below), and a potential Ets site. B cells and T cells showed similar patterns of transcription factor occupancy, with alterations in DMS reactivity seen at two sites able to bind Sp1 and Sp3 in vitro and the upstream RUNX1 sites. These differences between lymphoid cells and macrophages were also apparent in the chromatin architecture, as assayed by DNase I footprinting. Macrophages showed strong protection from DNase I cleavage compared to 3T3 cells downstream of the bp −13703 position, indicating the occupancy of the C/EBP and Sp1 and Sp3 sites and a hyperreactive region between bp −13633 and −13619, whereas lymphoid cells showed enhanced DNase I accessibility between bp −13669 and −13703.
The 5′ part of the enhancer has been implicated as an important contributor to the repression of Pu.1 in T cells (33). While macrophages showed a clear DMS footprint over the PU.1 binding site, we were unable to see any specific alterations in DMS reactivity on either strand in B cells and T cells (Fig. 4 and data not shown). This is due partly to the fact that the 5′ URE sequence is comparably AT rich, which prevents it from being an ideal substrate for DMS modification. The 5′ URE was highly DNase I accessible in macrophages, B cells, and T cells, and once again there were differences in chromatin architecture between lymphoid cells and macrophages. Macrophages displayed a region of extended DNase I sensitivity downstream of the TCF/LEF site and a strong DHS downstream of the PU.1 binding site. Little difference in chromatin structure was seen between B cells and T cells, with the exception of a relative increase in DNase I accessibility upstream of the PU.1 and TCF/LEF sites at bp −14385.
In summary, our data indicate that the Pu.1 promoter and enhancer bind different sets of transcription factors in the different cell types and adopt differential chromatin architectures. In addition, we can show that, although Pu.1 is not expressed in T cells, chromatin is in an open conformation and is bound by transcription factors.
Pu.1 chromatin is in the active conformation in the absence of PU.1, and myeloid differentiation leads to an alteration in transcription factor binding.
Pu.1 is switched on in HSCs, and expression is retained throughout myelopoiesis. We therefore asked the question of how transcription factor occupancy and chromatin architecture were regulated in these different cell types. In addition, previous experiments suggested that PU.1 autoregulates Pu.1 expression (31), and we therefore examined which aspect of transcription factor occupancy and chromatin remodeling was regulated by PU.1. To this end, we used a myeloid precursor cell line that was derived from the fetal livers of PU.1 knockout mice, as well as a derivative of this cell line (PUER) in which the knockout phenotype could be eliminated with a tamoxifen-inducible version of PU.1 (19, 41). In both cell types, the regulatory region of Pu.1 is completely intact. These cells are unable to differentiate into macrophages unless PU.1 is induced. Figure 5 shows results from experiments in which we employed DMS footprinting and ChIP assays to examine transcription factor occupancy in 3T3 cells, macrophages, PU.1 knockout cells, PUER cells, and PUER cells induced with tamoxifen for 48 h. In parallel, we tested alterations in chromatin architecture by DNase I in vivo footprinting. All experiments with PUER cells used the RAW 264 macrophage cell line as a control instead of primary macrophages.
FIG. 5.
(A and B) Analysis of developmental alterations of transcription factor occupancy and chromatin fine structure at the Pu.1 promoter (A) and the 3′ URE (B) by in vivo DMS and DNase I footprinting (as indicated). For more explanation, refer to the legend to Fig. 2. +OHT and −OHT, PUER cells with and without OHT induction, respectively; M, macrophages; G, Maxam-Gilbert G reaction with purified genomic DNA. (C) Results from a DMS in vivo footprinting experiment analyzing transcription factor occupancy at the 3′ URE in macrophages, 3T3 cells, and two independent cultures of myeloid progenitor cells (prog 1 and prog 2). (D to G) Results from ChIP assays examining the binding of PU.1, RUNX1, C/EBPb, and Fli-1 to the different Pu.1 cis-regulatory elements plus one control region in the absence of PU.1 (PU.1−/−; PUER −OHT) and after 48 h of PU.1 induction (+OHT). The data shown are averages of results from two independent chromatin preparations analyzed in triplicate. RAW, RAW 264 cells; 5′H enh and 3′H enh, 5′ URE and 3′ URE, respectively.
Figure 5A shows that there were few PU.1-dependent alterations in transcription factor occupancy and chromatin architecture at the promoter. After induction, PU.1 associated with the promoter (Fig. 5D), but C/EBPβ binding was independent of PU.1 (Fig. 5F). At the 3′ URE (Fig. 5B), the PU.1 site was occupied in the presence and absence of PU.1, as indicated by DMS hyporeactivity at the PU.1 site, although PU.1 was not present, as indicated by ChIP (Fig. 5D). Instead, we identified the Ets family member Fli-1 binding to the 3′ enhancer element, and this factor continued to contribute to Pu.1 regulation in the presence of PU.1 (Fig. 5G). Interestingly, Fli-1 was also able to bind to the promoter but was unable to associate with the PU.1 site in the 5′ URE. This site was bound exclusively by PU.1. We also noticed differences in the occupancy of the RUNX1 site adjacent to the PU.1 site. Here we saw preferential protection of a guanine in PU.1−/− and uninduced PUER precursor cells, whereas the same base was only partially protected in macrophages and induced PUER cells (Fig. 5B). ChIP assays confirmed that RUNX1 associated preferentially with the 3′ enhancer in precursor cells and only weakly in mature cells (Fig. 5 E). To test whether the presence of PU.1 destabilized RUNX1 binding, we grew primary myeloid precursor cells from the bone marrow of normal mice and prepared differentiated macrophages from these cells (Fig. 5C). Both cell types are known to express high levels of PU.1 (37). Here also we observed full protection only in precursor cells, indicating that the stabilization of RUNX1 binding in these cells is not dependent simply on the absence of PU.1 protein but is a function of the developmental stage. It should be noted that the transcription factor occupancy pattern in precursor cells is identical to that seen in PU.1 knockout cells (Fig. 5C and data not shown).
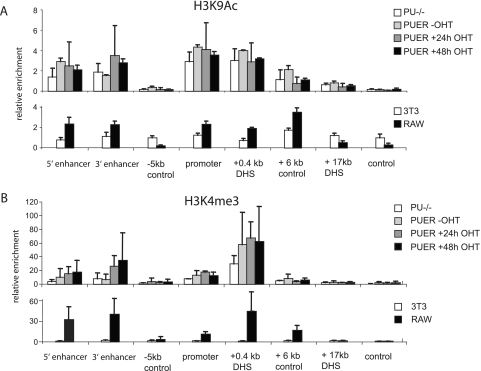
We next wanted to test whether the alterations in transcription factor occupancy after PU.1 induction and macrophage differentiation had consequences with respect to the histone modification status of Pu.1. Fig. 6 shows results from ChIP assays examining the active histone marks histone H3 lysine 9 acetylation (Fig. 6A) and histone 3 lysine 4 trimethylation (Fig. 6B). 3T3 cells and RAW 264 cells served as negative and positive controls, respectively. Histone acetylation was found mainly over the regulatory regions and did not change significantly after PU.1 induction. This result was consistent with the finding that the patterns of binding site occupancy are the same in PU.1-expressing and non-PU.1-expressing cells. However, we observed a high level of histone 3 lysine 4 trimethylation at the 3′ URE, which increased severalfold after the induction of PU.1.
FIG. 6.
Distribution of histone H3 lysine 9 acetylation (H3K9ac) (A) and histone H3 lysine 4 trimethylation (H3K43me3) (B) across the Pu.1 locus in the indicated cell types before and after PU.1 induction. The data shown are averages of results from two independent chromatin preparations analyzed in triplicate. PUER −OHT, PUER cells without OHT induction; RAW, RAW 264 cells.
The URE is bound by RNA Pol II and drives the expression of noncoding transcripts.
The enzymes mediating histone 3 lysine 4 trimethylation are recruited by the serine 5-phosphorylated form of RNA Pol II (30). Our finding that the URE was associated with trimethylated histone 3 lysine 4 indicated that this element could be bound by RNA Pol II. We therefore performed a ChIP experiment testing which forms of RNA Pol II were associated with Pu.1 cis elements (Fig. 7). Using an antibody that recognized all forms of RNA Pol II, we showed that both parts of the URE in all Pu.1-expressing cell types were indeed bound by this enzyme (Fig. 7A). Moreover, the levels of RNA Pol II at the 3′ URE dwarfed those at the promoter. The situation was different when we used an antibody against the serine 5-phosphorylated form of RNA Pol II. Here we observed similar levels at the URE and the promoter. The serine 2-phosphorylated form of RNA Pol II is associated mostly with the 3′ ends of genes, and this pattern was also observed here. In keeping with the increase in histone 3 lysine 4 trimethylation after PU.1 induction, we saw an increase in phosphorylated RNA polymerase at the 3′ URE.
FIG. 7.
Distribution of total RNA Pol II (A), serine 5-phosphorylated (Ser5P) RNA Pol II (B), and serine 2-phosphorylated (Ser2P) RNA Pol II (C) across the Pu.1 locus in the indicated cell types before and after PU.1 induction. RAW, RAW 264 cells; ND, not determined; −5kb, kb −5 control. The insert in panel A shows the RNA Pol II enrichment at the promoter compared to levels in the controls. The data shown are averages of results from two independent chromatin preparations analyzed in triplicate. PUER −OHT, PUER cells without OHT induction.
We addressed the question of whether there were ncRNA transcripts originating from the URE. To avoid any contamination with genomic DNA, we used sequence-specific biotinylated oligonucleotides to prime the cDNA synthesis as described in reference 39 and amplified specific cDNA sequences with a set of real-time-PCR primers. We detected transcripts going in both directions, with overlapping RNA species present within the SFFV integration site (Fig. 8A). ncRNA transcripts were detected only in macrophages, not in 3T3 cells (Fig. 8B to D).
FIG. 8.
The URE gives rise to ncRNA transcripts. (A) Strategy to detect URE transcripts using biotinylated specific primers (arrows) for cDNA synthesis followed by purification of the products on magnetic beads and amplification with different quantitative PCR primers, as indicated in the figure. The figure also shows a representation of the detected RNAs with presumptive transcriptional start regions as determined by the acquisition (solid line) or loss (dashed line) of the PCR signal. The lower panel depicts actual quantitative PCR results with cDNA prepared from 3T3 cells and macrophages as indicated. Note that mRNA and ncRNA levels cannot be compared directly. (B to D) Levels of PU.1 mRNA, the 5′ URE transcript, and the 3′ URE transcript in the HSC-enriched LSK cell population, CMPs in vitro differentiated for 2 and 7 days (CMP+2 and CMP+7, respectively), splenic B cells (spl. B), thymocytes (th.T), macrophages (Mac), and 3T3 cells.
The next experiments examined the tissue specificity of URE transcription compared to that of Pu.1 mRNA transcription in splenic B cells, T cells, mature macrophages, and a population of bone marrow LSK cells expressing high levels of c-kit and enriched with HSCs (Fig. 8B to D). We also measured ncRNA transcript levels in purified CMP cells and cells differentiated from these cells in vitro (37). As expected, Pu.1 mRNA was expressed in all precursor cell types, B cells, and macrophages (Fig. 8B). This pattern of expression was also reflected in the ncRNA transcripts (Fig. 8C and D). However, we observed interesting differences in expression levels in the different cell types. While the level of 5′ URE transcripts was highest in the HSC fraction and higher in B cells than in macrophages (Fig. 8C), the level of 3′ URE transcripts was low in precursor cells and B cells but was up-regulated during macrophage differentiation (Fig. 8D).
URE transcripts are regulated by RUNX1 and PU.1.
The experiments represented in Fig. 8 indicated that the activity of the ncRNAs is under developmental regulation. We therefore performed a series of experiments aimed at elucidating which transcription factors are responsible for this differential activity. To this end, we first exploited the PUER system to test whether URE activity is regulated by PU.1. To distinguish between immediate (direct) and delayed (indirect) PU.1 effects, we measured 5′ and 3′ URE transcripts during a time course of PU.1 induction (Fig. 9A and B). From these experiments, it was obvious that PU.1 expression immediately up-regulated 3′ URE transcripts (Fig. 9B), whereas PU.1 expression and cell differentiation had no or little effect on the expression of the 5′ URE transcript.
FIG. 9.
Transcription from the URE is regulated by PU.1 and RUNX1. (A and B) Levels of expression of the 5′ URE transcript (A) and the 3′ URE transcript (B) during a time course of PU.1 induction. PUER + OHT, PUER cells induced by OHT. (C and D) Levels of expression of the 5′ URE transcript (C) and the 3′ URE (D) transcript in myeloid precursor cells derived from the bone marrow of wild-type (WT) mice, mice carrying a mutation in the 3′ URE PU.1 site (PU.1mut), and mice carrying mutations in all three RUNX1 binding sites in the 3′ URE (Runx1mut). The data shown are averages of results from two independent experiments.
To test in a more direct way how URE transcripts are regulated, we analyzed cells from two novel mouse lines in which 3′ URE sequences at the endogenous Pu.1 locus were altered by point mutagenesis. Point mutations were introduced into all three RUNX1 binding sites and into the PU.1 binding site as described in Materials and Methods. The introduction of these mutations strongly reduced Pu.1 mRNA levels in total bone marrow, with the RUNX1 mutations having a more pronounced effect than the PU.1 binding site mutation (13). Details of the characterization of these mice and the effect on hematopoietic development will be published elsewhere (13; P. Zhang, G. Huang, and D. G. Tenen, unpublished data). We grew precursor cells from the bone marrow of the mutant mice as well as wild-type littermates and measured levels of Pu.1 ncRNA transcripts (Fig. 9C and D). These experiments confirmed that in these cells also, the mutation of the PU.1 binding site reduced Pu.1 mRNA expression to approximately 60% of the wild-type level but that the mutation of the RUNX1 sites had a more pronounced effect and reduced mRNA expression to 25% of the wild-type level (data not shown). Both sets of mutations affected the expression of both URE transcripts, and the RUNX1 mutations completely abolished ncRNA transcription from the 3′ URE.
Taken together, our experiments demonstrate that (i) the URE is associated with RNA Pol II, (ii) the URE gives rise to ncRNA transcripts, (iii) the 3′ URE transcript is regulated by PU.1 and RUNX1, and, most importantly, (iv) URE transcript levels are strictly correlated with the ability of the URE to enhance mRNA synthesis.
DISCUSSION
Pu.1 chromatin structure is differentially programmed by distinct sets of transcription factors in Pu.1-expressing cell types.
The studies of transcription factor occupancy and chromatin architecture at Pu.1 presented here demonstrate how overlapping, but distinct, sets of transcription factors program chromatin structure at the promoter and the URE in macrophages and B cells. In the two cell types, patterns of chromatin folding at the promoter were similar, as demonstrated by DNase I digestion (Fig. 2). However, this folding was achieved by differential sets of transcription factors. The only transcription factors binding to this element shared between macrophages and B cells are Elf-1 (positioned at bp −75) and ATF-2 (positioned at bp −32). In macrophages, these factors cooperated with C/EBP, PU.1, and factors binding to the Sp1 site next to the ATF-2 site, whereas in B cells they cooperated with Oct-1/BOB1/OCAB (6). The GC-rich region binding Sp1 and Sp3 in vitro was also occupied in B cells. However, here we observed altered DMS reactivity at a different guanine compared to that in macrophages, indicating that a different factor was binding to this sequence. At the URE, both chromatin architecture and transcription factor occupancy patterns differed significantly between macrophages and B cells (Fig. 3 and 4). The only transcription factor binding site which was occupied in both cell types was the single RUNX1 site at bp −13660 in the 3′ URE. In macrophages, RUNX1 binding to this site cooperated with a factor binding the juxtaposed Ets consensus sequence (PU.1 or Fli-1) and factors binding to the GC-rich region binding Sp1 and Egr2-like elements. In contrast that in to B cells, the upstream RUNX1 binding site did not appear to be occupied in macrophages. The 5′ URE was bound by PU.1 in macrophages, but this site was not occupied in B cells. These results are consistent with the findings of studies with the granulocyte-macrophage colony-stimulating factor enhancer demonstrating that enhancers functioning in different cell types use different combinations of transcription factors to establish alternative chromatin structures (3). This arrangement indicates that it is the transcription factors that dictate the chromatin architecture at specific sequences.
Pu.1 chromatin in T cells is still bound by transcription factors.
Pu.1 expression is observed in common lymphoid progenitor cells but is then silenced during T-cell development (2). It has been shown previously that the URE adopts a DHS in T cells but that the promoter does not (24). Although our studies similarly showed strongly reduced DNase I and MNase accessibility within the Pu.1 promoter in thymocytes, we nevertheless found that the Sp1-Sp3 site immediately upstream of the transcription start site was still occupied by factors (Fig. 2). At the 3′ URE, the same transcription factor binding sites were occupied in B cells and T cells, leading to the formation of chromatin fine structures that were highly similar in both lymphoid cell types but differed from that in macrophages (Fig. 4). The TCF/LEF site in the 5′ URE is essential for repressing Pu.1 activity in T cells in the absence of Wnt signaling (33). The data presented here suggest that the absence of Pu.1 expression in T cells is a result of both a differential promoter complex and the inhibition of URE activity by the 5′ URE.
Taken together, these data confirm that Pu.1 is not epigenetically silenced in T cells and requires an active repression mechanism involving sequence-specific DNA binding proteins. This active repression can be counteracted by overexpressing myeloid-specific transcription factors, including PU.1 itself (20). This finding is reminiscent of previous results from our laboratory demonstrating that the silencing of the myeloid-specific c-fms locus in B cells requires the continuous presence of the B-cell-specific transcription factor Pax5 and that this repression goes along with a partially active chromatin structure (39). These examples demonstrate that the chromatin of a number of lineage-specific genes is still plastic and can be reprogrammed. Such chromatin plasticity may be one of the driving forces behind the epigenetic reprogramming events observed during the first steps of leukemogenesis, some of which involve aberrant expression of PU.1 (28).
Transcription factor occupancy at the URE is developmentally regulated.
A number of different experiments have shown that many lineage-specific genes are activated in a stepwise fashion following the hierarchical and sequential expression of different transcription factors (15, 16, 19). Pu.1 has the opposite problem, i.e., it has to ensure that expression levels are kept up during a large number of cellular differentiation steps and in the presence of changing transcription factor levels and signaling events. Pu.1 achieves this by assembling a changing set of transcription factors at the promoter and the URE leading to little change in the histone modification state and the chromatin structure of Pu.1 during myelopoiesis. We confirm that PU.1 indeed contributes to the regulation of its own expression, but Fli-1 (and possibly other Ets factors such as Elf-1) can also occupy a subset of PU.1 sites and cooperate with C/EBP and RUNX1 to maintain active chromatin. In this case, it is interesting that Fli-1, although expressed in T cells (1), cannot bind to the 5′ URE site and cannot compensate for the absence of PU.1 in T cells.
Our finding that RUNX1 binds to the Pu.1 URE predominantly in precursor cells may shed light on recent results describing the effects of conditional gene targeting experiments. These studies showed that, although RUNX1 is vital for hematopoietic development, it is dispensable for adult hematopoiesis and myelopoiesis (11, 14). The data presented here raise the possibility that the association of RUNX1 with Pu.1 cis regulatory elements is essential for the initiation of PU.1 expression during the formation of HSCs in the embryo but is dispensable for its maintenance once stem cells have formed. This hypothesis is currently being tested.
The URE gives rise to transcripts that are indicative of URE enhancer activity.
Many cis-regulatory elements of genes have now been shown to recruit RNA Pol II and to direct the expression of ncRNAs (reviewed in references 9 and 32). Such enhancer-driven transcripts have been implicated in gene activation in development but do not seem to be necessary for the establishment of acetylated chromatin (12, 18). This also appears to be the case here. To study the regulation of these transcripts, we investigated the cis-regulatory requirements of URE promoter activity to provide the groundwork for more elaborate experiments determining a possible role of these transcripts in Pu.1 developmental regulation. Our data indicate that URE transcript levels are a true reflection of the impact of the URE on Pu.1 mRNA synthesis. Point mutations within the 3′ URE that affected enhancer activity, as indicated by a drop in Pu.1 mRNA expression, also had a strong impact on URE transcript levels. URE transcripts were seen only in cell types in which the URE was active, such as myeloid precursor cells, macrophages, and B cells but not T cells. The latter example also demonstrates that an open chromatin structure does not necessarily lead to ncRNA synthesis and suggests that active repression mechanisms silencing URE and Pu.1 bona fide promoter activity also repress URE transcription. These data demonstrate that the different activities are interlinked, and we could indeed show that the URE and promoter physically interact (A. K. Ebralidze, F. Guibal, U. Steidl, P. Zhang, S. Lee, F. Rosenbauer, G. Huang, V. Petkova, T. Dayaram, J. Klupp, M. Hoogenkamp, C. Bonifer, and D. G. Tenen, submitted for publication).
We noted a significant discrepancy in the distribution of unmodified RNA Pol II compared to that of the transcribing form of the enzyme phosphorylated at serine 5 and serine 2. Most of the unmodified enzyme was localized at the URE, which is in contrast to the previously observed localization at the human growth hormone gene (12) and which suggests that not all RNA Pol II localized at the URE may be involved in transcription. It has been postulated that the recruitment of RNA Pol II to upstream regulatory sequences prior to the onset of transcription at the bona fide promoter serves as a nucleation event for developmentally regulated enhancer-promoter communication (17). It will be interesting to test whether this is also true for the URE and/or whether URE transcripts themselves play a role in mediating this process.
Acknowledgments
Work in C. Bonifer's laboratory is funded by grants from the Biotechnology and Biological Sciences Research Council, the Leukemia Research Fund, the City of Hope Medical Center, and the Wellcome Trust. Work in P. N. Cockerill's laboratory is supported by Yorkshire Cancer Research and the Association of International Cancer Research. D. G. Tenen's laboratory is funded by NIH grant CA41456. H. Tagoh is a Kay Kendall Fellow.
Footnotes
Published ahead of print on 4 September 2007.
REFERENCES
- 1.Anderson, M. K., G. Hernandez-Hoyos, R. A. Diamond, and E. V. Rothenberg. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126: 3131-3148. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. K., A. H. Weiss, G. Hernandez-Hoyos, C. J. Dionne, and E. V. Rothenberg. 2002. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity 16: 285-296. [DOI] [PubMed] [Google Scholar]
- 3.Bert, A. G., B. V. Johnson, E. W. Baxter, and P. N. Cockerill. 2007. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol. Cell. Biol. 27: 2870-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifer, C. 2005. Epigenetic plasticity of hematopoietic cells. Cell Cycle 4: 211-214. [PubMed] [Google Scholar]
- 5.Chen, H., D. Ray-Gallet, P. Zhang, C. J. Hetherington, D. A. Gonzalez, D. E. Zhang, F. Moreau-Gachelin, and D. G. Tenen. 1995. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene 11: 1549-1560. [PubMed] [Google Scholar]
- 6.Chen, H., P. Zhang, H. S. Radomska, C. J. Hetherington, D. E. Zhang, and D. G. Tenen. 1996. Octamer binding factors and their coactivator can activate the murine PU.1 (spi-1) promoter. J. Biol. Chem. 271: 15743-15752. [DOI] [PubMed] [Google Scholar]
- 7.Dahl, R., J. C. Walsh, D. Lancki, P. Laslo, S. R. Iyer, H. Singh, and M. C. Simon. 2003. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat. Immunol. 4: 1029-1036. [DOI] [PubMed] [Google Scholar]
- 8.Dakic, A., D. Metcalf, L. Di Rago, S. Mifsud, L. Wu, and S. L. Nutt. 2005. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 201: 1487-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, A. 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22: 38-45. [DOI] [PubMed] [Google Scholar]
- 10.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57: 159-197. [DOI] [PubMed] [Google Scholar]
- 11.Growney, J. D., H. Shigematsu, Z. Li, B. H. Lee, J. Adelsperger, R. Rowan, D. P. Curley, J. L. Kutok, K. Akashi, I. R. Williams, N. A. Speck, and D. G. Gilliland. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106: 494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, Y., F. Elefant, S. A. Liebhaber, and N. E. Cooke. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23: 365-375. [DOI] [PubMed] [Google Scholar]
- 13.Huang, G., P. Zhang, H. Hirai, S. Elf, X. Yan, Z. Chen, S. Koschmieder, Y. Okuno, T. Dayaram, J. D. Growney, R. A. Shivdsani, D. G. Gilliland, N. A. Speck, S. D. Nimer, and D. G. Tenen. PU.1 is a major downstream target of AML1/RUNX1 in adult hematopoiesis. Nat. Genet., in press. [DOI] [PubMed]
- 14.Ichikawa, M., T. Asai, T. Saito, S. Seo, I. Yamazaki, T. Yamagata, K. Mitani, S. Chiba, S. Ogawa, M. Kurokawa, and H. Hirai. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10: 299-304. [DOI] [PubMed] [Google Scholar]
- 15.Im, H., J. A. Grass, K. D. Johnson, S. I. Kim, M. E. Boyer, A. N. Imbalzano, J. J. Bieker, and E. H. Bresnick. 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. USA 102: 17065-17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki, H., S. Mizuno, Y. Arinobu, H. Ozawa, Y. Mori, H. Shigematsu, K. Takatsu, D. G. Tenen, and K. Akashi. 2006. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 20: 3010-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8: 465-471. [DOI] [PubMed] [Google Scholar]
- 18.Kim, A., H. Zhao, I. Ifrim, and A. Dean. 2007. Beta-globin intergenic transcription and histone acetylation dependent on an enhancer. Mol. Cell. Biol. 27: 2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krysinska, H., M. Hoogenkamp, R. Ingram, N. Wilson, H. Tagoh, P. Laslo, H. Singh, and C. Bonifer. 2007. A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol. Cell. Biol. 27: 878-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laiosa, C. V., M. Stadtfeld, H. Xie, L. de Andres-Aguayo, and T. Graf. 2006. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity 25: 731-744. [DOI] [PubMed] [Google Scholar]
- 21.Lefevre, P., C. Lacroix, H. Tagoh, M. Hoogenkamp, S. Melnik, R. Ingram, and C. Bonifer. 2005. Differentiation-dependent alterations in histone methylation and chromatin architecture at the inducible chicken lysozyme gene. J. Biol. Chem. 280: 27552-27560. [DOI] [PubMed] [Google Scholar]
- 22.Lefevre, P., S. Melnik, N. Wilson, A. D. Riggs, and C. Bonifer. 2003. Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Mol. Cell. Biol. 23: 4386-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128: 707-719. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., Y. Okuno, P. Zhang, H. S. Radomska, H. Chen, H. Iwasaki, K. Akashi, M. J. Klemsz, S. R. McKercher, R. A. Maki, and D. G. Tenen. 2001. Regulation of the PU.1 gene by distal elements. Blood 98: 2958-2965. [DOI] [PubMed] [Google Scholar]
- 25.McKercher, S. R., B. E. Torbett, K. L. Anderson, G. W. Henkel, D. J. Vestal, H. Baribault, M. Klemsz, A. J. Feeney, G. E. Wu, C. J. Paige, and R. A. Maki. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15: 5647-5658. [PMC free article] [PubMed] [Google Scholar]
- 26.Moreau-Gachelin, F., F. Wendling, T. Molina, N. Denis, M. Titeux, G. Grimber, P. Briand, W. Vainchenker, and A. Tavitian. 1996. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol. Cell. Biol. 16: 2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246: 780-786. [DOI] [PubMed] [Google Scholar]
- 28.Nagel, S., M. Scherr, A. Kel, K. Hornischer, G. E. Crawford, M. Kaufmann, C. Meyer, H. G. Drexler, and R. A. MacLeod. 2007. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 67: 1461-1471. [DOI] [PubMed] [Google Scholar]
- 29.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95: 2543-2551. [PubMed] [Google Scholar]
- 30.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11: 709-719. [DOI] [PubMed] [Google Scholar]
- 31.Okuno, Y., G. Huang, F. Rosenbauer, E. K. Evans, H. S. Radomska, H. Iwasaki, K. Akashi, F. Moreau-Gachelin, Y. Li, P. Zhang, B. Gottgens, and D. G. Tenen. 2005. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol. Cell. Biol. 25: 2832-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasanth, K. V., and D. L. Spector. 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 21: 11-42. [DOI] [PubMed] [Google Scholar]
- 33.Rosenbauer, F., B. M. Owens, L. Yu, J. R. Tumang, U. Steidl, J. L. Kutok, L. K. Clayton, K. Wagner, M. Scheller, H. Iwasaki, C. Liu, B. Hackanson, K. Akashi, A. Leutz, T. L. Rothstein, C. Plass, and D. G. Tenen. 2006. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat. Genet. 38: 27-37. [DOI] [PubMed] [Google Scholar]
- 34.Rosenbauer, F., K. Wagner, J. L. Kutok, H. Iwasaki, M. M. Le Beau, Y. Okuno, K. Akashi, S. Fiering, and D. G. Tenen. 2004. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 36: 624-630. [DOI] [PubMed] [Google Scholar]
- 35.Scott, E. W., M. C. Simon, J. Anastasi, and H. Singh. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573-1577. [DOI] [PubMed] [Google Scholar]
- 36.Tagoh, H., P. N. Cockerill, and C. Bonifer. 2006. In vivo genomic footprinting using LM-PCR methods, p. 285-314. In S. Pells (ed.), Nuclear reprogramming, vol. 325. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 37.Tagoh, H., R. Himes, D. Clarke, P. J. Leenen, A. D. Riggs, D. Hume, and C. Bonifer. 2002. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 16: 1721-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tagoh, H., S. Melnik, P. Lefevre, S. Chong, A. D. Riggs, and C. Bonifer. 2004. Dynamic reorganization of chromatin structure and selective DNA demethylation prior to stable enhancer complex formation during differentiation of primary hematopoietic cells in vitro. Blood 103: 2950-2955. [DOI] [PubMed] [Google Scholar]
- 39.Tagoh, H., A. Schebesta, P. Lefevre, N. Wilson, D. Hume, M. Busslinger, and C. Bonifer. 2004. Epigenetic silencing of the c-fms locus during B-lymphopoiesis occurs in discrete steps and is reversible. EMBO J. 23: 4275-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teitell, M. A. 2003. OCA-B regulation of B-cell development and function. Trends Immunol. 24: 546-553. [DOI] [PubMed] [Google Scholar]
- 41.Walsh, J. C., R. P. DeKoter, H. J. Lee, E. D. Smith, D. W. Lancki, M. F. Gurish, D. S. Friend, R. L. Stevens, J. Anastasi, and H. Singh. 2002. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 17: 665-676. [DOI] [PubMed] [Google Scholar]
- 42.Yeamans, C., D. Wang, I. Paz-Priel, B. E. Torbett, D. G. Tenen, and A. D. Friedman. 1 August 2007, posting date. C/EBPα binds and activates the PU.1 distal enhancer to induce monocyte lineage commitment. Blood. doi: 10.1182/blood-2007-03-080291. [DOI] [PMC free article] [PubMed]
- 43.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96: 2641-2648. [PubMed] [Google Scholar]