Abstract
During embryonic development, the anterior-posterior body axis is specified in part by the combinatorial activities of Hox genes. Given the poor DNA binding specificity of Hox proteins, their interaction with cofactors to regulate target genes is critical. However, few regulatory partners or downstream target genes have been identified. Herein, we demonstrate that Hox11 paralogous proteins form a complex with Pax2 and Eya1 to directly activate expression of Six2 and Gdnf in the metanephric mesenchyme. We have identified the binding site within the Six2 enhancer necessary for Hox11-Eya1-Pax2-mediated activation and demonstrate that this site is essential for Six2 expression in vivo. Furthermore, genetic interactions between Hox11 and Eya1 are consistent with their participation in the same pathway. Thus, anterior-posterior-patterning Hox proteins interact with Pax2 and Eya1, factors important for nephrogenic mesoderm specification, to directly regulate the activation of downstream target genes during early kidney development.
The Hox genes are conserved among all metazoans and specify positional information along the body axes. In mammals, 39 Hox genes are arranged into four chromosomal clusters, which are organized into 13 paralogous groups along the chromosome in a 3′-to-5′ manner. This arrangement of genes leads to spatiotemporal colinearity of Hox gene expression, with 3′ genes expressed more anteriorly and earlier in development than 5′ genes (16, 28). The unique, combinatorial expression of Hox proteins is important for the regulation of anterior-posterior patterning, skeletal morphogenesis, mesodermal organ development, neural patterning, and multiple other cellular and developmental processes (reviewed in reference 64). Despite genetic analyses of a variety of organisms, the molecular details regarding which cofactors interact with Hox proteins to regulate transcription are poorly understood, and very few downstream Hox targets have been identified.
The mammalian kidney serves as an ideal model organ to study Hox protein function. The embryonic kidney is derived from the intermediate mesoderm and exhibits anterior-posterior patterning as it develops (reviewed in reference 14). The most anterior region of the intermediate mesoderm, the pronephros, is a rudimentary structure in mammals. Caudal to the pronephros are the mesonephric tubules, a linear array of nephron-like structures that are transient filtering units. The most posterior intermediate mesoderm generates the metanephric, or adult, kidney, which forms adjacent to the hind-limb buds. Adult kidney development begins when the metanephric mesenchyme induces an outgrowth, the ureteric bud, from the adjacent nephric duct epithelia. The ureteric bud invades the metanephric mesenchyme, provides inductive Wnt signals, and subsequently undergoes branching morphogenesis to generate the radial pattern of the kidney (reviewed in reference 75). In mice, Hox11 paralogous genes are essential for early patterning of the metanephric mesenchyme, as the loss of Hoxa11 and Hoxd11 results in misrouted ureters and hypoplastic kidneys, while the loss of all three Hox11 paralogs results in a complete failure of the metanephric mesenchyme to induce ureteric bud induction (11, 46, 70).
Hox proteins recognize a degenerate ATTA or TTAT sequence, which confers very little locus specificity (17; reviewed in reference 64). Thus, it is likely that cofactors interact with Hox proteins in order to promote high-affinity binding and locus-specific regulatory activities. The three-amino-acid-loop extension proteins Pbx and Meis/Prep are known Hox cofactors that regulate target gene expression (reviewed in reference 39). Pbx1, Pbx3, and Meis1 are expressed in the kidney; however, the loss of Pbx1 and Meis function results in kidney phenotypes that are less severe and affect later stages than those of the Hox11 paralogous gene mutants (12, 24, 60, 61). Thus, it is likely that Hox11 proteins are interacting with other cofactors to specify the early intermediate mesoderm along the anterior-posterior axis.
Prior to ureteric bud induction, the condensing metanephric mesenchyme expresses a unique combination of markers, including the Hox11 paralogs (Hoxa11, Hoxc11, and Hoxd11), Osr1, Pax2, Eya1, Wt1, Six1, Six2, and Gdnf (14). In Hox11 triple mutants, the expression of many early kidney patterning markers in the uninduced metanephric mesenchyme are unperturbed; however, both Six2 expression and Gdnf expression are absent (70). Six2 regulates metanephric progenitor cell renewal (63). Gdnf ligand activates coreceptors c-ret and Gdnfrα in the Wolffian duct to promote ureteric bud outgrowth and invasion and is a key regulator of continued branching morphogenesis (6, 23, 40, 50, 58, 59, 62, 66, 67). Pax2 mutant mice have reduced levels of Six2 expression and do not express Gdnf in the mesenchyme (65). Even though Pax2 can directly regulate Gdnf expression in cell culture, it is not sufficient in vivo, as Gdnf expression is absent in Hox11 and Eya1 mutants, while Pax2 expression is normal in these mutants (5, 70, 71). Eya1 mutants, like Hox11 mutants, exhibit no ureteric bud induction and also lack Six2 and Gdnf expression (71).
The Pax, Eya, and Six gene families encompass members of a common regulatory network that is conserved from Drosophila melanogaster to mammals (reviewed in reference 3). Eya proteins have no intrinsic DNA binding domain but can localize to the nucleus and function as coactivators of transcription (43, 51). The Eya1 protein also has a protein phosphatase activity which is essential for the regulation of some target genes (reviewed in reference 55).
Given the similarities in molecular phenotypes observed in the Hox11, Pax2, and Eya1 mouse mutants in the developing kidney, we propose that Hox11 proteins interact with Pax2 and Eya1 to regulate essential patterning genes within the posterior intermediate mesoderm. All three proteins physically interact and synergistically up-regulate Gdnf and Six2 promoter activities. We identified a binding site critical for this activation within the Six2 promoter region and demonstrated that this site is essential for driving expression within the renal mesenchyme. Furthermore, renal development is sensitized to a partial loss of Hox11 paralogous gene function in an Eya1 heterozygous genetic background, suggesting that these proteins work in the same regulatory network. These data point to a novel Hox11-Eya1-Pax2 network that translates anterior-posterior positional information within the developing mammalian kidney. Further, our data demonstrate that Six2 and Gdnf are direct downstream targets of the group of Hox11 paralogous genes.
MATERIALS AND METHODS
Six2- and Gdnf-luciferase constructs.
The 3.03-kb Six2 promoter region, at base pairs −266 to −3296 upstream of the ATG start site, was PCR amplified from a Six2 bacterial artificial chromosome clone and inserted into a pGL3-Basic vector (Promega). A previously reported Gdnf promoter (5) was subcloned into pGL3-Basic. The QuikChange II XL site-directed mutagenesis kit (Stratagene) was used to mutate the Pax2 binding site or delete the Hox binding site in the 3.03-kb Six2 promoter-luciferase vector. Mutations and deletions were confirmed by sequence analysis (see materials and methods in the supplemental material).
Protein expression vectors.
The Hoxa11 protein coding sequence (NCBI IMAGE clone 8734051) was cloned into a p3XFlag-CMV10 expression vector (Sigma), which places three FLAG tags at the N terminus of Hoxa11. The Eya1 protein coding sequence (NCBI IMAGE clone 6848408) was cloned into a pCS2+MT expression vector, which places six Myc tags at the N terminus of Eya1 (57, 68). The Pax2 protein expression vector was previously described (34).
Luciferase assays.
MDCK cells were plated at 50,000 cells per well in a 24-well plate, cultured in minimum essential medium with 10% fetal bovine serum, 50 IU of penicillin per ml, and 50 μg of streptomycin per ml at 37°C, and transfected the second day with FuGENE6 (Roche). For each well, 0.64 ml of FuGENE was used per 0.32 mg of DNA, which contained 75 ng of reporter plasmid and 20 ng pRL-CMV renilla luciferase for standardization. Twenty-four hours posttransfection, cells were lysed in Passive lysis buffer (Promega). Luciferase activity was measured using a Vector3 PerkinElmer luminometer. Counts were standardized using pGL3 empty vector or cotransfected renilla luciferase (Dual-Luciferase assay system; Promega). Each transfection was performed in triplicate.
Northern blot analysis.
RNA from MDCK cells was prepared using TRIzol (Invitrogen) and resuspended in RNA storage solution (Ambion). Twenty micrograms of RNA per sample was electrophoresed for 2 h in 1% agarose denature gel (Ambion), blotted onto a Hybond-N+ membrane (Amersham Biosciences), and prehybridized with 8 ml ULTRAhyb ultrasensitive hybridization buffer (Ambion). The membrane was then hybridized at 68°C overnight with 8 × 106 cpm of a Six2 RNA probe (HindIII-SfoI fragment from NCBI IMAGE clone 6394139 [ATG to bp 346 of exon 1]) labeled with [32P]dUTP via in vitro RNA probe reverse transcription. The blot was washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate at 37°C and then twice with 0.1× SSC plus 0.1% sodium dodecyl sulfate at 68°C and exposed to film or analyzed on a Typhoon 9400 variable-mode imager (Amersham). The RNA probes for the Hox11 paralogs, Eya1, and Pax2 have been previously published (15, 26-28, 72).
Coimmunoprecipitation assays.
HEK-293 cells were transfected with Hoxa11-FLAG, Pax2-hemagglutinin (HA), and Eya1-Myc expression vectors, and cell lysates were prepared (Complete Mini; Roche). Mouse anti-HA monoclonal antibody (catalog no. H9658; Sigma), mouse anti-FLAG monoclonal antibody (M2) (catalog no. F3165; Sigma), or mouse anti-Myc monoclonal antibody (catalog no. SC-40; Santa Cruz) was added to 0.5 μg lysate and incubated 4°C overnight. Mouse or rabbit immunoglobulin G (Jackson ImmunoResearch) was used as a negative control. Protein was precipitated using protein G agarose beads (Invitrogen), washed, and resuspended. Western blots were probed with 1:5,000 anti-FLAG antibody for Hoxa11, 1:3,000 anti-Pax2 antibody (catalog no. PRB-276P; Covance) for Pax2, or 1:500 anti-Myc antibody for Eya1.
Pax2-PD pull-down and electromobility shift assays.
The 3.03-kb Six2 upstream region was digested by HpaII and used in the Pax2-paired domain (Pax2-PD) pull-down assay following methods described previously (4).
Gel electromobility shift assays were performed as previously described with modifications (5). Nuclear extracts from untransfected HEK-293 cells and cells transfected with the Hoxa11-FLAG, Pax2-HA, and Eya1-Myc protein expression vectors were isolated as previously described (49). Polyacrylamide gel electrophoresis-purified oligonucleotides (Invitrogen) for the wild-type and mutant probes (see materials and methods in the supplemental material) were annealed and end labeled with P-32 (T4 polynucleotide kinase; Promega). Supershifts were performed using anti-Hoxa11 antibody (7) and anti-HA antibody (see above).
Six2-LacZ transgenic mice.
The 3.03-kb wild-type and mutant upstream Six2 sequences from the luciferase vectors were subcloned upstream from the β-galactosidase gene in the pNASSβ expression vectors (BDB Clontech) and were digested with NheI and KpnI to create 980-bp Six2-LacZ wild-type and mutant pNASSβ reporter vectors. DNA for injection was isolated using NheI and AseI. Purified DNA was microinjected into fertilized eggs obtained by mating (C57BL/6 × SJL)F1 or C57BL/6 female mice with (C57BL/6 × SJL)F1 male mice. Pronuclear microinjection was performed as described previously (41). Embryos were collected at embryonic day 11.5 (E11.5), genotyped for the β-galactosidase gene (2), and stained for β-galactosidase activity by following standard protocols (41).
In situ hybridization and histology.
In situ hybridization and histology were performed as previously described (70).
RESULTS
As Six2 expression is affected in Hox11, Eya1, and Pax2 mutant metanephric mesenchymes, we tested whether the proteins encoded by these genes can activate Six2 expression directly. A 3-kb sequence upstream of the Six2 ATG start site was used to determine whether Six2 expression is regulated in cell culture by Hox11, Pax2, or Eya1 (Fig. 1A). In transfected cells, Hoxa11, Eya1, or Pax2 alone was unable to substantially increase expression from the Six2 reporter construct. However, when the proteins were coexpressed in combination, activation of the reporter was observed. Coexpression of all three proteins resulted in 50-fold activation of the Six2 luciferase reporter (Fig. 1B). Expression of Hoxc11 and Hoxd11 vectors in place of Hoxa11 in these experiments showed similar results, consistent with their redundant genetic function at this early stage of kidney development (see Fig. S1 in the supplemental material) (70). In MDCK cells transiently transfected with Hoxa11, Pax2, and Eya1, a fivefold up-regulation of endogenous Six2 expression was also demonstrated (Fig. 1C). Untransfected MDCK cells had measurable levels of endogenous Pax2 mRNA expression but low to undetectable levels of Hoxa11, Hoxc11, Hoxd11, and Eya1 expression (see Fig. S2 in the supplemental material).
FIG. 1.
Regulation of Six2 expression by a Hox11-Eya1-Pax2 complex. (A) Schematic of the Six2-luciferase vector. A fragment from base pair 3296 to base pair 266 upstream of the Six2 ATG start site was subcloned into a luciferase expression vector. Ex1, exon 1; Ex2, exon 2. (B) Activity of the Six2 luciferase reporter plasmid in transfected MDCK cells with different combinations of Hoxa11, Pax2, and Eya1 protein expression vectors. (C) Northern blot analysis of endogenous Six2 mRNA in MDCK cells after transfection with Hoxa11, Eya1, and Pax2, normalized to β-actin. (D) Whole-cell extracts of HEK-293 cells transfected with Hoxa11-Flag, Pax2-HA, and Eya1-Myc protein expression constructs were subjected to reciprocal coimmunoprecipitations. Immunoblotting (IB) for Hoxa11 (αFlag) demonstrated coimmunoprecipitation with Pax2 (αHA) and Eya1 (αMyc) (lanes 2 and 3). Immunoblotting with Pax2 (αPax2) demonstrated coimmunoprecipitation of Hoxa11 (αFlag) and Eya1 (αMyc) (lanes 5 and 6). Immunoblotting with Eya1 (αMyc) demonstrates coimmunoprecipitation with Hoxa11 (αFlag) and Pax2 (αHA) (lanes 8 and 9). Immunopreciptations (IP) using mouse or rabbit immunoglobulin G (lanes 1, 4, and 7) were negative controls (Ctl).
The cooperative activation of Six2 could be mediated by the formation of a Hox11-Eya1-Pax2 complex binding directly to an upstream cis regulatory sequence. To test for physical interactions, we performed coimmunoprecipitation experiments using Hoxa11, Pax2, and Eya1. In cell lysates expressing Hoxa11, Eya1, and Pax2, immunoprecipitation using antibodies specific for one of the three proteins resulted in the coprecipitation of the other two proteins (Fig. 1D). Thus, Hoxa11, Pax2, and Eya1 can form a complex and physically associate either directly or through interactions with as yet unidentified adaptor proteins within the complex.
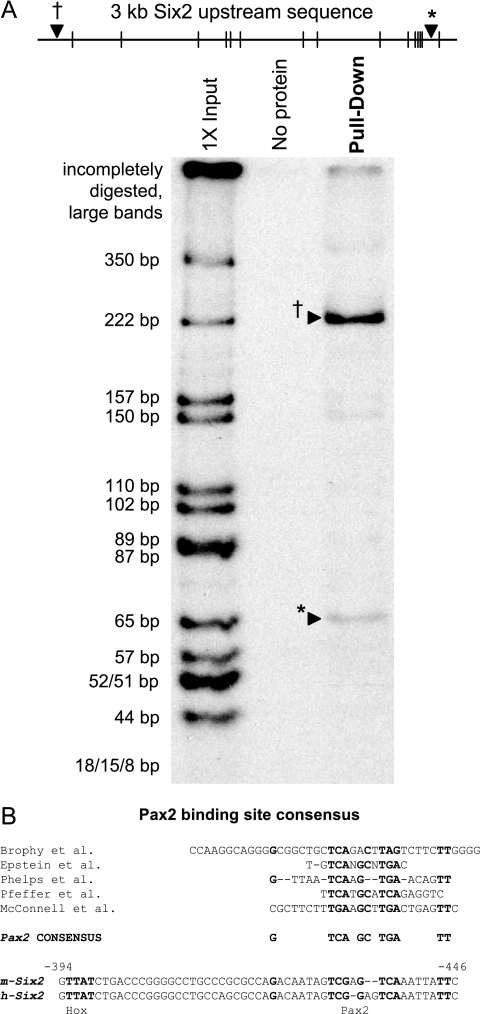
To examine the possibility that the Hox11-Eya1-Pax2 complex could bind directly to sequences upstream of the Six2 coding sequence, we first tested for Pax2 binding, since Pax2 is expected to have the most specificity in terms of a DNA recognition sequence, and the Pax2-PD has been previously shown to bind Pax2 target sequences with high affinity (5, 34). After digestion of the 3.0-kb Six2 reporter sequence with HpaII, two fragments showed strong binding by the Pax2-PD in vitro: one at the 5′ end of the reporter construct and another sequence 450 base pairs upstream of the Six2 coding sequence (Fig. 2A). Subsequent deletion analyses showed that the 5′ putative binding site was not required for Hox11-Eya1-Pax2-mediated activity in cell culture (data not shown). Sequence analysis of the bp −450 region binding site revealed a conserved Pax2 consensus sequence (5, 18, 38, 48, 49) and an adjacent putative Hox binding site (Fig. 2B).
FIG. 2.
Pax2 binds regions upstream of the Six2 protein coding sequence. (A) The Pax2-PD binds two regions of the Six2 promoter in vitro. The black hatch marks indicate the HpaII sites in the 3.0-kb Six2 upstream sequence. A 5′ 222-bp region (†) and a 3′ 65-bp region (*) are pulled down only when the Pax2-PD is present. (B) Sequence analysis of the 65-bp region at bp −450 identified a putative Pax2 binding site and a putative Hox binding site based on sequence conservation to consensus sites.
We next tested the necessity of these binding sites for reporter gene activation in our reporter assay. In addition to testing the wild-type 3-kb Six2 luciferase constructs tested previously, we generated and tested three analogous 3-kb constructs with a mutation of the putative Pax2 binding site at the bp −450 region, a deletion of a nearby putative Hox binding site, or both mutations together. Mutation of the Pax2 or the Hox binding site alone caused a decrease in Hox11-Eya1-Pax2-mediated luciferase activity compared to the activity of the wild-type Six2 construct. However, Hox11-Eya1-Pax2-mediated expression from the Six2 reporter was nearly abolished when both the putative Pax2 and Hox sites were mutated (Fig. 3A).
FIG. 3.
The Hox11-Eya1-Pax2 complex binds at the bp −450 region site and is necessary for Six2 expression. (A) Luciferase activities from the 3.0-kb wild-type Six2 expression construct (Six2) and constructs with the putative Pax2 binding site mutated (Six2/Pax2mut), with the Hox binding site mutated (Six2/HoxΔ), or with both the putative Pax2 and Hox sites mutated (Six2/Pax2mut-HoxΔ) were compared. All plates were cotransfected with or without Hoxa11, Pax2, and Eya1 protein expression vectors in MDCK cells. (B) An 89-bp probe (wt) containing the putative Pax2 and Hox binding sites of the Six2 promoter, incubated with nuclear extracts from HEK-293 cells transfected with Hoxa11, Eya1, and Pax2 (HEP), demonstrated retention on a nondenaturing acrylamide gel (arrow in lane 3). Probe retention was not seen in untransfected extracts (Unt; lane 2). This interaction was competed with excess (50×) unlabeled competitor (lane 4), and supershifts were observed using antibodies to Hoxa11 (αa11) or to an HA tag (αHA) on the Pax2 protein (arrows in lanes 5 and 6, respectively). The transfected extract does not show retention using a probe (mut) with the Pax2 and Hox binding sites mutated (lane 7).
Binding at the bp −450 region site by the Hox11-Eya1-Pax2 complex was further examined by using nuclear extracts from HEK-293 cells transfected with Hoxa11, Eya1, and Pax2 in electrophoretic mobility shift assays (Fig. 3B). An 89-base-pair fragment containing the putative binding site exhibited a slower-migrating complex upon incubation with nuclear lysate expressing Hoxa11, Eya1, and Pax2 (Fig. 3B, lane 3). This slower-migrating species was supershifted upon incubation with antibodies against Hoxa11 or Pax2, indicating that these proteins are part of the DNA/protein complex (Fig. 3B, lanes 5 and 6). The specificity of binding was demonstrated by competition with a molar excess of unlabeled wild-type competitor probe and by loss of retention by using a labeled probe containing both the putative Pax2 and Hox binding sites mutated (Fig. 3B, lanes 4 and 7).
To confirm the importance of this Hox11-Eya1-Pax2 regulatory binding site in vivo, we generated LacZ reporter constructs by using wild-type and mutant Six2 upstream sequences to test expression within the renal mesenchyme of transgenic mice. A 980-bp Six2 construct was used to drive LacZ expression, and this was compared to expression from a construct containing mutations of the putative Pax2 and Hox binding sites. The transgenic constructs were injected into fertilized mouse embryos and assayed for LacZ expression at E11.5. The wild-type Six2 reporter demonstrated LacZ expression in a pattern similar to that of endogenous Six2 expression (44) in 5 of 12 independent transgenic lines. LacZ staining was prominent in the branchial arches, the otic region, and the developing urogenital mesenchyme (Fig. 4A). Of 26 independent transgenic embryos generated with the mutant Six2-LacZ reporter, 19 embryos demonstrated LacZ staining in some region of the developing embryo, but none of the 26 mutant embryos showed any staining in the nephrogenic mesenchyme (Fig. 4A). This confirms that the Hox11-Eya1-Pax2 binding site is necessary for Six2 expression in the nephrogenic mesenchyme in vivo.
FIG. 4.
The Hox11-Eya1-Pax2 binding site is critical for kidney expression in vivo, and the Hox-Eya-Pax network synergistically activates Gdnf expression. (A) Six2-LacZ transient transgenic mice. The top panel shows a schematic of the 980-bp Six2-LacZ reporters (Hox site in red and Pax2 site in blue). (Lower left panel) E11.5 transgenic embryos carrying the wild-type Six2-LacZ constructs exhibit staining in the nephrogenic mesenchyme (arrow) and the branchial arches (asterisk). (Lower right panel) Transgenic mice carrying a construct with the Pax2 and Hox sites mutated retain staining in the branchial arches (asterisk; 19 of 26 embryos) but have no nephrogenic staining (arrow; 26 of 26 embryos). (B through E) No differences in the expression of Hoxd11 in the posterior intermediate mesoderm are seen in Eya1 heterozygous (B) or homozygous (C) embryos at E10.5 or in the metanephric mesenchyme of Pax2 heterozygous (D) or homozygous (E) embryos at E10.5. (F through H) Frontal hematoxylin-eosin-stained histological sections from an E14.5 control embryo (F) and embryos with three mutant Hox11 alleles plus one mutant allele of Eya1 (G and H). The brackets in panels F through H indicate relative kidney sizes. (I) Gdnf upstream sequence-driven luciferase activity in the presence of Hoxa11, Eya1, and/or Pax2 in MDCK cells.
Both Eya1 expression and Pax2 expression are unaffected in the metanephric mesenchyme at preinduction stages in Hox11 triple mutants (70). Further, Eya1 expression is unaffected in Pax2 mutant mesenchymes, and Pax2 expression is initially unaffected in Eya1 mutant mesenchymes (71, 73). We examined the expression of Hoxd11 in the posterior nephrogenic mesenchymes of Eya1 and Pax2 mutant mice, and no changes in expression were seen (Fig. 4B to E). Therefore, while the loss of these genes individually leads to the loss of Gdnf expression and ureteric bud induction, these genes do not affect the expression of one another at early metanephric stages, consistent with their operating in parallel as transcriptional coregulators in this system.
If the Hox11-Eya1-Pax2 complex cooperatively contributes to the expression of early kidney mesenchyme-specific genes, then a reduction in gene dosage may provide genetic evidence for this interaction. Eya1 heterozygotes or three-allele Hox11 mutants have no histological renal phenotype at E14.5 (70, 71; data not shown). However, the addition of a single Eya1 null allele to three mutant Hox11 alleles results in hypoplastic kidneys at E14.5 (Fig. 4F to H). This phenotype is observed regardless of which three Hox11 alleles are missing. Thus, reduced Eya1 gene dosage uncovers a phenotype in the Hox11 three-allele mutant kidneys similar to the one reported in mutants carrying four or more mutant Hox11 alleles (11, 46, 70). These data provide compelling genetic evidence that Hox11 group proteins interact with Eya1.
Hox11 paralogous gene mutants and Eya1 and Pax2 mutant mice show similar kidney phenotypes with a failure to induce ureteric bud formation and loss of Gdnf (5, 70, 71). Thus, we examined Gdnf expression as a second potential candidate for regulation by the Hox11-Eya1-Pax2 complex. The Gdnf reporter construct was activated by Pax2 alone approximately fivefold, in agreement with previously published reports (5). However, coexpression of Hoxa11 (or Hoxc11 or Hoxd11 [data not shown]) with Eya1 and Pax2 increased activation of the Gdnf luciferase reporter more than 40-fold, whereas Hox11 or Eya1 alone had no effect on activation (Fig. 4I). These data demonstrate that the Hox11-Eya1-Pax2 complex can strongly activate multiple target genes in the early renal mesenchyme.
DISCUSSION
During the specification of the body plan, cells and tissues are organized along three axes: anterior-posterior, dorsal-ventral, and mediolateral. The intermediate mesoderm, from which the kidney derives, is first specified along the mediolateral axis and marks a region between the paraxial mesoderm and lateral-plate mesoderm shortly after gastrulation. One model proposes that Bmp signals from the lateral plate and as yet unidentified signals from the paraxial mesoderm provide positional cues to activate genes such as Pax2, Osr1, and Lim1, which mark the intermediate mesoderm (30, 31). The intermediate mesoderm also has anterior-posterior patterning that is clearly represented by the pro-, meso-, and metanephric kidneys. Thus, any position within the developing mesoderm can be specified by a unique combination of anterior-posterior factors and mediolateral factors. How these factors function cooperatively is unclear. Our results suggest a model whereby Hox11 paralogous proteins cooperate with Eya1 and Pax2 to activate Six2 and Gdnf, two genes specific to the posterior intermediate mesoderm that generates the metanephric kidney (Fig. 5). Hox11, Eya1, and Pax2 proteins physically associate, bind to a metanephric-mesenchyme-specific enhancer region within the Six2 promoter, and synergistically activate reporter gene expression. Thus, the direct interactions between anterior-posterior determinants and mediolateral determinants may be essential for determining the positional address of the metanephric mesenchyme.
FIG. 5.
Diagram of a proposed mechanism of Hox11 molecular function. Taken together, this work supports a model wherein Hox11 proteins form a transcriptional complex with Pax2 and Eya1 and directly activate the expression of Six2 and Gdnf during early mammalian metanephric development. Hox11P, a given Hox11 paralogous protein.
Despite the fact that targeted deletions of Hox genes were among the first generated (8, 9, 35) and that mutations in Hox genes affect numerous developmental processes (reviewed in reference 47), few downstream target genes have been identified. Probably the most studied mammalian Hox target is Hoxb1, which is autoregulated by its own expression (13, 54). Working as a complex with Pbx and Meis/Prep, Hoxb1 has been shown to regulate several other anterior Hox genes (19, 20, 29, 36). Here, we demonstrate that Six2 and Gdnf are novel molecular targets of the Hox11 genes and identify a novel set of Hox regulatory partners, Eya1 and Pax2, for this activity.
The Pax-Eya-Six pathway is a conserved regulatory network in organogenesis (3). Initially found in Drosophila, ey (Pax) activates eya (Eya) and so (Six) expression during eye formation (22). Eya can also ectopically activate so/Six, and so/Six and eya both in turn regulate ey/Pax as well as their own expression (51). These genes are also expressed and play important roles in mammalian development. In addition to affecting kidney development, mutations in these genes affect development of other organ systems, such as ear, thymus, and thyroid formation, as well as muscle and eye formation (reviewed in references 3, 25, and 32). How this regulatory cassette contributes to the differentiation of so many unique structures is unclear but presumably relies on interactions with region-specific patterning factors.
More recently, it has been shown that Hox/HomC proteins can bind ey/Pax and affect developmental processes in Drosophila (1, 52). Our current work suggests that this interaction is conserved during mammalian kidney development as well. Further, it is possible that interaction between Hox genes and the Pax-Eya-Six pathway may be more generally conserved. For example, Hoxa3 is necessary for thymus and thyroid formation, as are Eya1 and Six1 (37, 74, 76). Pax1 and Pax9 are also expressed in the thymus, and Pax8 is expressed in the thyroid (42, 53, 69). Similarly, during otic development, Hoxa2 is expressed in the second branchial arch, and Hoxa2 mutant mice have middle ear defects, as do Eya1, Six1, and Pax8 mutants (10, 21, 45, 56, 71). Six2 is expressed in the first and second branchial arches and periotic mesenchyme, and a recent study has shown that Six2 expression increases in these regions in Hoxa2 mutants (33, 44). Because of the striking similarities in the phenotypes of these mutants, it is plausible that a Hox-Eya-Pax complex is also necessary for the development of other organ systems and that Six genes are general developmental targets of Hox genes and possibly this Hox-Eya-Pax complex.
In conclusion, our results demonstrate that a Hox11-Eya1-Pax2 regulatory network is necessary for Six2 and Gdnf expression to promote mammalian kidney development. This study identifies new downstream targets of the Hox11 paralogous genes as well as a novel regulatory complex in which these Hox proteins operate. It will be important to determine if this pathway is conserved in other organ systems during mammalian development.
Supplementary Material
Acknowledgments
We acknowledge Galina Gavrilina and Maggie Van Keuren for the preparation of transgenic mice and also the Transgenic Animal Model Core of the University of Michigan's Biomedical Research Core Facilities. The Eya1 mutant mice were a gift from Richard Maas, and the Hoxa11 antibody was a gift from Honami Naora.
Core support was provided by the University of Michigan Cancer Center, NIH grant number CA46592, and the University of Michigan Center for Organogenesis. We gratefully acknowledge the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for their support of this research program (grant no. 085P1000815). This work was supported in part by National Institutes of Health grant R01-DK071929 and by a University of Michigan Organogenesis predoctoral fellowship award (T32-HD007505) to A.R.Y.
Footnotes
Published ahead of print on 4 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Benassayag, C., S. Plaza, P. Callaerts, J. Clements, Y. Romeo, W. J. Gehring, and D. L. Cribbs. 2003. Evidence for a direct functional antagonism of the selector genes proboscipedia and eyeless in Drosophila head development. Development 130: 575-586. [DOI] [PubMed] [Google Scholar]
- 2.Brodbeck, S., B. Besenbeck, and C. Englert. 2004. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech. Dev. 121: 1211-1222. [DOI] [PubMed] [Google Scholar]
- 3.Brodbeck, S., and C. Englert. 2004. Genetic determination of nephrogenesis: the Pax/Eya/Six gene network. Pediatr. Nephrol. 19: 249-255. [DOI] [PubMed] [Google Scholar]
- 4.Brophy, P. D., K. M. Lang, and G. R. Dressler. 2003. The secreted frizzled related protein 2 (SFRP2) gene is a target of the Pax2 transcription factor. J. Biol. Chem. 278: 52401-52405. [DOI] [PubMed] [Google Scholar]
- 5.Brophy, P. D., L. Ostrom, K. M. Lang, and G. R. Dressler. 2001. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development 128: 4747-4756. [DOI] [PubMed] [Google Scholar]
- 6.Cacalano, G., I. Farinas, L. C. Wang, K. Hagler, A. Forgie, M. Moore, M. Armanini, H. Phillips, A. M. Ryan, L. F. Reichardt, M. Hynes, A. Davies, and A. Rosenthal. 1998. GFRalpha1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21: 53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, W., J. Liu, H. Yoshida, D. Rosen, and H. Naora. 2005. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat. Med. 11: 531-537. [DOI] [PubMed] [Google Scholar]
- 8.Chisaka, O., and M. R. Capecchi. 1991. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 350: 473-479. [DOI] [PubMed] [Google Scholar]
- 9.Chisaka, O., T. S. Musci, and M. R. Capecchi. 1992. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature 355: 516-520. [DOI] [PubMed] [Google Scholar]
- 10.Christ, S., U. W. Biebel, S. Hoidis, S. Friedrichsen, K. Bauer, and J. W. Smolders. 2004. Hearing loss in athyroid pax8 knockout mice and effects of thyroxine substitution. Audiol. Neurootol. 9: 88-106. [DOI] [PubMed] [Google Scholar]
- 11.Davis, A. P., D. P. Witte, H. M. Hsieh-Li, S. S. Potter, and M. R. Capecchi. 1995. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375: 791-795. [DOI] [PubMed] [Google Scholar]
- 12.Di Giacomo, G., M. Koss, T. D. Capellini, A. Brendolan, H. Popperl, and L. Selleri. 2006. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr. Patterns 6: 747-757. [DOI] [PubMed] [Google Scholar]
- 13.Di Rocco, G., A. Gavalas, H. Popperl, R. Krumlauf, F. Mavilio, and V. Zappavigna. 2001. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 276: 20506-20515. [DOI] [PubMed] [Google Scholar]
- 14.Dressler, G. R. 2006. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22: 509-529. [DOI] [PubMed] [Google Scholar]
- 15.Dressler, G. R., U. Deutsch, K. Chowdhury, H. O. Nornes, and P. Gruss. 1990. Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development 109: 787-795. [DOI] [PubMed] [Google Scholar]
- 16.Duboule, D., and P. Dolle. 1989. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8: 1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekker, S. C., K. E. Young, D. P. von Kessler, and P. A. Beachy. 1991. Optimal DNA sequence recognition by the Ultrabithorax homeodomain of Drosophila. EMBO J. 10: 1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein, J., J. Cai, T. Glaser, L. Jepeal, and R. Maas. 1994. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 269: 8355-8361. [PubMed] [Google Scholar]
- 19.Ferretti, E., F. Cambronero, S. Tumpel, E. Longobardi, L. M. Wiedemann, F. Blasi, and R. Krumlauf. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 25: 8541-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferretti, E., H. Marshall, H. Popperl, M. Maconochie, R. Krumlauf, and F. Blasi. 2000. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127: 155-166. [DOI] [PubMed] [Google Scholar]
- 21.Gendron-Maguire, M., M. Mallo, M. Zhang, and T. Gridley. 1993. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell 75: 1317-1331. [DOI] [PubMed] [Google Scholar]
- 22.Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter, and W. J. Gehring. 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125: 2181-2191. [DOI] [PubMed] [Google Scholar]
- 23.Hellmich, H. L., L. Kos, E. S. Cho, K. A. Mahon, and A. Zimmer. 1996. Embryonic expression of glial cell-line derived neurotrophic factor (GDNF) suggests multiple developmental roles in neural differentiation and epithelial-mesenchymal interactions. Mech. Dev. 54: 95-105. [DOI] [PubMed] [Google Scholar]
- 24.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23: 450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holländer, G., J. Gill, S. Zuklys, N. Iwanami, C. Liu, and Y. Takahama. 2006. Cellular and molecular events during early thymus development. Immunol. Rev. 209: 28-46. [DOI] [PubMed] [Google Scholar]
- 26.Hostikka, S. L., and M. R. Capecchi. 1998. The mouse Hoxc11 gene: genomic structure and expression pattern. Mech. Dev. 70: 133-145. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh-Li, H. M., D. P. Witte, M. Weinstein, W. Branford, H. Li, K. Small, and S. S. Potter. 1995. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development 121: 1373-1385. [DOI] [PubMed] [Google Scholar]
- 28.Izpisúa-Belmonte, J. C., H. Falkenstein, P. Dolle, A. Renucci, and D. Duboule. 1991. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10: 2279-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs, Y., C. A. Schnabel, and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19: 5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James, R. G., C. N. Kamei, Q. Wang, R. Jiang, and T. M. Schultheiss. 2006. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development 133: 2995-3004. [DOI] [PubMed] [Google Scholar]
- 31.James, R. G., and T. M. Schultheiss. 2005. Bmp signaling promotes intermediate mesoderm gene expression in a dose-dependent, cell-autonomous and translation-dependent manner. Dev. Biol. 288: 113-125. [DOI] [PubMed] [Google Scholar]
- 32.Kawakami, K., S. Sato, H. Ozaki, and K. Ikeda. 2000. Six family genes—structure and function as transcription factors and their roles in development. Bioessays 22: 616-626. [DOI] [PubMed] [Google Scholar]
- 33.Kutejova, E., B. Engist, M. Mallo, B. Kanzler, and N. Bobola. 2005. Hoxa2 downregulates Six2 in the neural crest-derived mesenchyme. Development 132: 469-478. [DOI] [PubMed] [Google Scholar]
- 34.Lechner, M. S., and G. R. Dressler. 1996. Mapping of Pax-2 transcription activation domains. J. Biol. Chem. 271: 21088-21093. [DOI] [PubMed] [Google Scholar]
- 35.Lufkin, T., A. Dierich, M. LeMeur, M. Mark, and P. Chambon. 1991. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66: 1105-1119. [DOI] [PubMed] [Google Scholar]
- 36.Maconochie, M. K., S. Nonchev, M. Studer, S. K. Chan, H. Popperl, M. H. Sham, R. S. Mann, and R. Krumlauf. 1997. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 11: 1885-1895. [DOI] [PubMed] [Google Scholar]
- 37.Manley, N. R., and M. R. Capecchi. 1995. The role of Hoxa-3 in mouse thymus and thyroid development. Development 121: 1989-2003. [DOI] [PubMed] [Google Scholar]
- 38.McConnell, M. J., H. E. Cunliffe, L. J. Chua, T. A. Ward, and M. R. Eccles. 1997. Differential regulation of the human Wilms tumour suppressor gene (WT1) promoter by two isoforms of PAX2. Oncogene 14: 2689-2700. [DOI] [PubMed] [Google Scholar]
- 39.Moens, C. B., and L. Selleri. 2006. Hox cofactors in vertebrate development. Dev. Biol. 291: 193-206. [DOI] [PubMed] [Google Scholar]
- 40.Moore, M. W., R. D. Klein, I. Farinas, H. Sauer, M. Armanini, H. Phillips, L. F. Reichardt, A. M. Ryan, K. Carver-Moore, and A. Rosenthal. 1996. Renal and neuronal abnormalities in mice lacking GDNF. Nature 382: 76-79. [DOI] [PubMed] [Google Scholar]
- 41.Nagy, A., M. Gerstenstein, K. Vintersten, and R. Behringer (ed.). 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Neubüser, A., H. Koseki, and R. Balling. 1995. Characterization and developmental expression of Pax9, a paired-box-containing gene related to Pax1. Dev. Biol. 170: 701-716. [DOI] [PubMed] [Google Scholar]
- 43.Ohto, H., S. Kamada, K. Tago, S. I. Tominaga, H. Ozaki, S. Sato, and K. Kawakami. 1999. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19: 6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver, G., R. Wehr, N. A. Jenkins, N. G. Copeland, B. N. Cheyette, V. Hartenstein, S. L. Zipursky, and P. Gruss. 1995. Homeobox genes and connective tissue patterning. Development 121: 693-705. [DOI] [PubMed] [Google Scholar]
- 45.Ozaki, H., K. Nakamura, J. Funahashi, K. Ikeda, G. Yamada, H. Tokano, H. O. Okamura, K. Kitamura, S. Muto, H. Kotaki, K. Sudo, R. Horai, Y. Iwakura, and K. Kawakami. 2004. Six1 controls patterning of the mouse otic vesicle. Development 131: 551-562. [DOI] [PubMed] [Google Scholar]
- 46.Patterson, L. T., M. Pembaur, and S. S. Potter. 2001. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development 128: 2153-2161. [DOI] [PubMed] [Google Scholar]
- 47.Pearson, J. C., D. Lemons, and W. McGinnis. 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6: 893-904. [DOI] [PubMed] [Google Scholar]
- 48.Pfeffer, P. L., M. Bouchard, and M. Busslinger. 2000. Pax2 and homeodomain proteins cooperatively regulate a 435 bp enhancer of the mouse Pax5 gene at the midbrain-hindbrain boundary. Development 127: 1017-1028. [DOI] [PubMed] [Google Scholar]
- 49.Phelps, D. E., and G. R. Dressler. 1996. Identification of novel Pax-2 binding sites by chromatin precipitation. J. Biol. Chem. 271: 7978-7985. [DOI] [PubMed] [Google Scholar]
- 50.Pichel, J. G., L. Shen, H. Z. Sheng, A. C. Granholm, J. Drago, A. Grinberg, E. J. Lee, S. P. Huang, M. Saarma, B. J. Hoffer, H. Sariola, and H. Westphal. 1996. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382: 73-76. [DOI] [PubMed] [Google Scholar]
- 51.Pignoni, F., B. Hu, K. H. Zavitz, J. Xiao, P. A. Garrity, and S. L. Zipursky. 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91: 881-891. [DOI] [PubMed] [Google Scholar]
- 52.Plaza, S., F. Prince, J. Jaeger, U. Kloter, S. Flister, C. Benassayag, D. Cribbs, and W. J. Gehring. 2001. Molecular basis for the inhibition of Drosophila eye development by Antennapedia. EMBO J. 20: 802-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poleev, A., H. Fickenscher, S. Mundlos, A. Winterpacht, B. Zabel, A. Fidler, P. Gruss, and D. Plachov. 1992. PAX8, a human paired box gene: isolation and expression in developing thyroid, kidney and Wilms' tumors. Development 116: 611-623. [DOI] [PubMed] [Google Scholar]
- 54.Pöpperl, H., M. Bienz, M. Studer, S. K. Chan, S. Aparicio, S. Brenner, R. S. Mann, and R. Krumlauf. 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81: 1031-1042. [DOI] [PubMed] [Google Scholar]
- 55.Rebay, I., S. J. Silver, and T. L. Tootle. 2005. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 21: 163-171. [DOI] [PubMed] [Google Scholar]
- 56.Rijli, F. M., M. Mark, S. Lakkaraju, A. Dierich, P. Dolle, and P. Chambon. 1993. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 75: 1333-1349. [DOI] [PubMed] [Google Scholar]
- 57.Rupp, R. A., L. Snider, and H. Weintraub. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8: 1311-1323. [DOI] [PubMed] [Google Scholar]
- 58.Sainio, K., P. Suvanto, J. Davies, J. Wartiovaara, K. Wartiovaara, M. Saarma, U. Arumae, X. Meng, M. Lindahl, V. Pachnis, and H. Sariola. 1997. Glial-cell-line-derived neurotrophic factor is required for bud initiation from ureteric epithelium. Development 124: 4077-4087. [DOI] [PubMed] [Google Scholar]
- 59.Sánchez, M. P., I. Silos-Santiago, J. Frisen, B. He, S. A. Lira, and M. Barbacid. 1996. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382: 70-73. [DOI] [PubMed] [Google Scholar]
- 60.Schnabel, C. A., R. E. Godin, and M. L. Cleary. 2003. Pbx1 regulates nephrogenesis and ureteric branching in the developing kidney. Dev. Biol. 254: 262-276. [DOI] [PubMed] [Google Scholar]
- 61.Schnabel, C. A., L. Selleri, Y. Jacobs, R. Warnke, and M. L. Cleary. 2001. Expression of Pbx1b during mammalian organogenesis. Mech. Dev. 100: 131-135. [DOI] [PubMed] [Google Scholar]
- 62.Schuchardt, A., V. D'Agati, V. Pachnis, and F. Costantini. 1996. Renal agenesis and hypodysplasia in ret-k− mutant mice result from defects in ureteric bud development. Development 122: 1919-1929. [DOI] [PubMed] [Google Scholar]
- 63.Self, M., O. V. Lagutin, B. Bowling, J. Hendrix, Y. Cai, G. R. Dressler, and G. Oliver. 2006. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 25: 5214-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svingen, T., and K. F. Tonissen. 2006. Hox transcription factors and their elusive mammalian gene targets. Heredity 97: 88-96. [DOI] [PubMed] [Google Scholar]
- 65.Torres, M., E. Gomez-Pardo, G. R. Dressler, and P. Gruss. 1995. Pax-2 controls multiple steps of urogenital development. Development 121: 4057-4065. [DOI] [PubMed] [Google Scholar]
- 66.Treanor, J. J., L. Goodman, F. de Sauvage, D. M. Stone, K. T. Poulsen, C. D. Beck, C. Gray, M. P. Armanini, R. A. Pollock, F. Hefti, H. S. Phillips, A. Goddard, M. W. Moore, A. Buj-Bello, A. M. Davies, N. Asai, M. Takahashi, R. Vandlen, C. E. Henderson, and A. Rosenthal. 1996. Characterization of a multicomponent receptor for GDNF. Nature 382: 80-83. [DOI] [PubMed] [Google Scholar]
- 67.Trupp, M., E. Arenas, M. Fainzilber, A. S. Nilsson, B. A. Sieber, M. Grigoriou, C. Kilkenny, E. Salazar-Grueso, V. Pachnis, and U. Arumae. 1996. Functional receptor for GDNF encoded by the c-ret proto-oncogene. Nature 381: 785-789. [DOI] [PubMed] [Google Scholar]
- 68.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8: 1434-1447. [DOI] [PubMed] [Google Scholar]
- 69.Wallin, J., H. Eibel, A. Neubuser, J. Wilting, H. Koseki, and R. Balling. 1996. Pax1 is expressed during development of the thymus epithelium and is required for normal T-cell maturation. Development 122: 23-30. [DOI] [PubMed] [Google Scholar]
- 70.Wellik, D. M., P. J. Hawkes, and M. R. Capecchi. 2002. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 16: 1423-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu, P. X., J. Adams, H. Peters, M. C. Brown, S. Heaney, and R. Maas. 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23: 113-117. [DOI] [PubMed] [Google Scholar]
- 72.Xu, P. X., I. Woo, H. Her, D. R. Beier, and R. L. Maas. 1997. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development 124: 219-231. [DOI] [PubMed] [Google Scholar]
- 73.Xu, P. X., W. Zheng, L. Huang, P. Maire, C. Laclef, and D. Silvius. 2003. Six1 is required for the early organogenesis of mammalian kidney. Development 130: 3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu, P. X., W. Zheng, C. Laclef, P. Maire, R. L. Maas, H. Peters, and X. Xu. 2002. Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development 129: 3033-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, J., A. P. McMahon, and M. T. Valerius. 2004. Recent genetic studies of mouse kidney development. Curr. Opin. Genet. Dev. 14: 550-557. [DOI] [PubMed] [Google Scholar]
- 76.Zou, D., D. Silvius, J. Davenport, R. Grifone, P. Maire, and P. X. Xu. 2006. Patterning of the third pharyngeal pouch into thymus/parathyroid by Six and Eya1. Dev. Biol. 293: 499-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.