Abstract
Vaccinia virus (VACV) encodes enzymes that cap the 5′ end of viral mRNAs, which enhances their stability and translation. Nevertheless, recent studies demonstrated that the VACV D10 protein (VACV-WR_115) decaps mRNA, an enzymatic activity not previously shown to be encoded by a virus. The decapping activity of D10 is dependent on a Nudix hydrolase motif that is also present in the VACV D9 protein (VACV-WR_114), which shares 25% sequence identity with D10. Here, we showed that a purified recombinant VACV D9 fusion protein also decaps mRNA and that this activity was abolished by point mutations in the Nudix hydrolase motif. Decapping was specific for a methylated cap attached to RNA and resulted in the liberation of m7GDP. D9 differed from D10 in requiring a longer capped RNA substrate for optimal activity, having greater sensitivity to inhibition by uncapped RNA, and having lower sensitivity to inhibition by nucleotide cap analogs unattached to RNA. Since D9 is expressed early in infection and D10 late, we suggest that the two proteins enhance mRNA turnover and manipulate gene expression in a complementary and overlapping manner.
The regulation of mRNA turnover allows a cell to rapidly adjust gene expression in an adaptive manner. Vaccinia virus (VACV), the prototype for the laboratory study of poxviruses, modulates both viral and host gene expression during the course of infection (12). The double-stranded DNA genome of VACV encodes approximately 200 proteins that are expressed in sequential stages delineated as early, intermediate, and late (13, 16). Although a transcriptional cascade drives entry into each phase of gene expression, rapid turnover eliminates viral mRNAs soon after their synthesis ceases, thereby augmenting progression through the virus life cycle (1). VACV infection also triggers the degradation of cellular mRNAs (2, 5), which contributes to the shutdown of host protein synthesis.
A clue to the mechanism of accelerated cellular and viral mRNA degradation was obtained during studies designed to identify positive transcriptional regulators. Shors and coworkers (18) reported that overexpression of gene VACV-WR_115 (D10R) and to a lesser extent VACV-WR_114 (D9R) decreased expression of viral proteins and the quantity of transcripts possessing a 5′ cap, a structural feature of both cellular and viral messages. In contrast, a mutant virus with a deletion of the D10R gene displays persistence of cellular and viral transcripts, a delay in shutoff of host protein synthesis, and replication defects (14). Taken together, these data suggested that the D10 protein (the product of the D10R gene) regulates mRNA stability and consequently the amount and duration of host and viral proteins synthesized. Although a D9R deletion mutant virus did not exhibit replication defects in tissue culture cells, a mutant virus with deletions of both the D9R and D10R genes could not be isolated, suggesting that these two proteins have compensating functions (14). Moreover, the individual importance of D9 and D10 is reflected in the conservation of the genes for both proteins in most chordopoxviruses. D10 is the more highly conserved of the two, however, as it is one of approximately 50 proteins whose sequence can be recognized in both poxvirus subfamilies: chordopoxviruses and entomopoxviruses. Maintenance of the two genes in chordopoxviruses may be due to different functions or patterns of expression. In this regard, D9 is synthesized early in infection and D10 at late times (8). Thus, continuing synthesis of D9 or D10 may be necessary for efficient replication in vivo.
The existence of related functions for D9 and D10 is suggested by their 25% sequence identity to each other and their possession of a Nudix hydrolase or MutT motif, a signature sequence characteristic of a diverse group of phosphohydrolases that cleave nucleoside diphosphates linked to another moiety, X (10). The presence of the Nudix hydrolase motif, in conjunction with the observation that overexpression of D9 and D10 reduced the quantity of capped transcripts, led to the hypothesis that D9 and D10 may cleave the m7GpppN cap structure, marking the message for subsequent degradation by cellular exonucleases. Adding support for this hypothesis, cellular Dcp2 Nudix hydrolases have been shown to modulate mRNA turnover through cleavage of the mRNA cap (19, 20).
Recent biochemical investigations demonstrated that purified recombinant VACV D10 has intrinsic mRNA-decapping activity, liberating m7GDP as a product (15). Mutations in the Nudix motif abolished D10 decapping activity, indicating that the site is essential for catalysis and confirming that D10 is the protein responsible for cap cleavage (15). Here we show that D9 also has mRNA-decapping activity, although the two poxvirus Nudix hydrolases appear to have some differences in substrate recognition.
MATERIALS AND METHODS
Sequence alignments.
Sequences were obtained from the Poxvirus Bioinformatics Resource Center (www.poxvirus.org) and aligned with muscle 3.6 (3) using standard settings. Alignments were formatted with ESPript (4).
Plasmid construction.
To create pMAL-c2x-malE-D9-his, a plasmid encoding a maltose binding protein (MBP)-D9 fusion protein appended with a C-terminal 10-histidine tag (MBP-D9-HIS), the D9R open reading frame was amplified by PCR using VACV strain WR genomic DNA as the template and oligonucleotide primers 5′-ATG GGA ATT ACA ATG GAT GAG GAA GTG ATA TTT G and 5′ GGC CGG AAG CTT TCA ATG ATG ATG ATG ATG ATG ATG ATG ATG ATG TTT ACT ATT AAC TAG CAT ATT ATA AAT ATA AGA CAG ATA TTC G. The gel-purified PCR product was cleaved with the restriction endonuclease HindIII and ligated into pMAL-c2x (New England Biolabs, Ipswich, MA), directly downstream of the gene encoding MBP. Specific mutations in the Nudix motif were introduced through use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Synthesis and purification of recombinant D9 protein.
The pMAL-c2x-malE-D9-his plasmid or mutated derivatives were transformed into Escherichia coli strain BL21 (EMD Biosciences, San Diego, CA). The bacteria were propagated in LB broth supplemented with 50 μg/ml carbenicillin and 0.2% (wt/vol) glucose, and protein expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Following 4 h of growth at 25°C, the bacteria were lysed in B-per detergent (Pierce, Rockford, IL), and the clarified lysate was purified over an amylose column (New England Biolabs) followed by a nickel-nitrilotriacetic acid (NTA) column (QIAGEN, Valencia, CA), according to the manufacturer's instructions. The purified protein was dialyzed against a solution containing 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol (DTT), and 2 mM Mg acetate (17). MBP-D10 was expressed and purified as described by Parrish et al. (15).
RNA synthesis.
The 309-nucleotide (nt) RNA substrate was synthesized from the pTRI-β-actin-human template by in vitro transcription using the MEGAshortscript (Ambion, Austin, TX) kit. To generate the 12-, 24-, 36-, and 48-nt transcripts, annealed oligonucleotide primers containing the T7 promoter and the corresponding portion of the 309-nt actin transcript were utilized as templates in an in vitro transcription reaction (11). The purified RNA was cap labeled using recombinant VACV guanylyltransferase/guanine-7-methyltransferase (9), obtained from Ambion or Epicenter Biotechnologies (Madison, WI), in the presence of 3.75 μM [α-32P]GTP (800 Ci/mmol), capping buffer (50 mM Tris-HCl [pH 8.0], 6 mM KCl, 1.25 mM DTT, 1.25 MgCl2, 0.05 mg/ml bovine serum albumin), and 1 mM S-adenosylmethionine. The labeled RNA was separated from unincorporated nucleotides by use of ProbeQuant G-50 gel filtration columns (Amersham Pharmacia Biotech) or G-25 gel filtration columns (Roche Applied Science, Indianapolis, IN).
RNA-decapping assays.
Approximately 1.0 pmol (70 ng) purified recombinant MBP-D9-HIS protein was incubated with 0.02 pmol of cap-labeled RNA in decapping buffer (100 mM K acetate, 10 mM Tris-HCl [pH 7.5], 2 mM MgCl2, 0.5 mM MnCl2, and 2 mM DTT) in a total volume of 15 μl for 30 min at 37°C (17). The reaction products were resolved on polyethyleneimine (PEI)-cellulose thin-layer chromatography (TLC) plates (Alltech Biotechnology, Columbia, MD) that were developed in 0.75 M LiCl. The radioactive signals were detected by autoradiography, and the unlabeled standards were visualized by UV shadowing. In some experiments the reaction product was quantified using a PhosphorImager (GE Healthcare, Piscataway, NJ).
RESULTS
Sequence comparisons of D9 and D10 proteins of chordopoxviruses.
The chordopoxviruses are comprised of eight genera: Orthopoxvirus, Parapoxvirus, Avipoxvirus, Capripoxvirus, Leporipoxvirus, Suipoxvirus, Molluscipoxvirus, and Yatapoxvirus. The genomes of representatives of each genus have been completely sequenced, and most contain two adjacent open reading frames with Nudix hydrolase motifs. The first set of open reading frames in Fig. 1 correspond to D9R (VACV-WR_114) and the second set to D10R (VACV-WR_115). A D10R ortholog is present in all sequenced chordopox species, whereas a D9R ortholog is absent from bovine papular stomatitis virus and orf virus, members of the parapoxvirus genus, and from one attenuated vaccine strain of fowlpox virus derived by tissue culture passage (7). The D9R orthologs of orthopoxvirus strains are 97 to 100% identical in sequence, whereas the identity drops to 50 to 60% or less when D9R is compared to orthologs of other chordopoxvirus genera. A similar degree of identity is found for orthologs of D10R. The Nudix hydrolase motif consists of the conserved 23-amino-acid sequence GX5EX5[UA]XREX2EEXGU. where U represents an aliphatic, hydrophobic residue and X represents any amino acid. The glycine, glutamates, and arginine are perfectly conserved in all D9R and D10R orthologs (Fig. 1). Aside from the highly conserved Nudix hydrolase motif, the D9R family is quite divergent from the D10R family, the members of which also have longer C-terminal segments (Fig. 1).
FIG. 1.
Sequence alignment of chordopoxvirus proteins with Nudix hydrolase motifs. Orthologs of VACV-WR_114 (D9) and VACV-WR_115 (D10) were aligned separately. FWPV, fowlpox virus; MOCV, molluscum contagiosum virus; SWPV, swinepox virus; MYXV, myxoma virus; GTPV, goatpox virus; YMTV; Yaba monkey tumor virus. Completely conserved amino acids are indicated by white on a black background; highly conserved amino acids are boxed. Triangles are below residues defining the Nudix motif.
Recombinant VACV D9 possesses intrinsic mRNA-decapping activity.
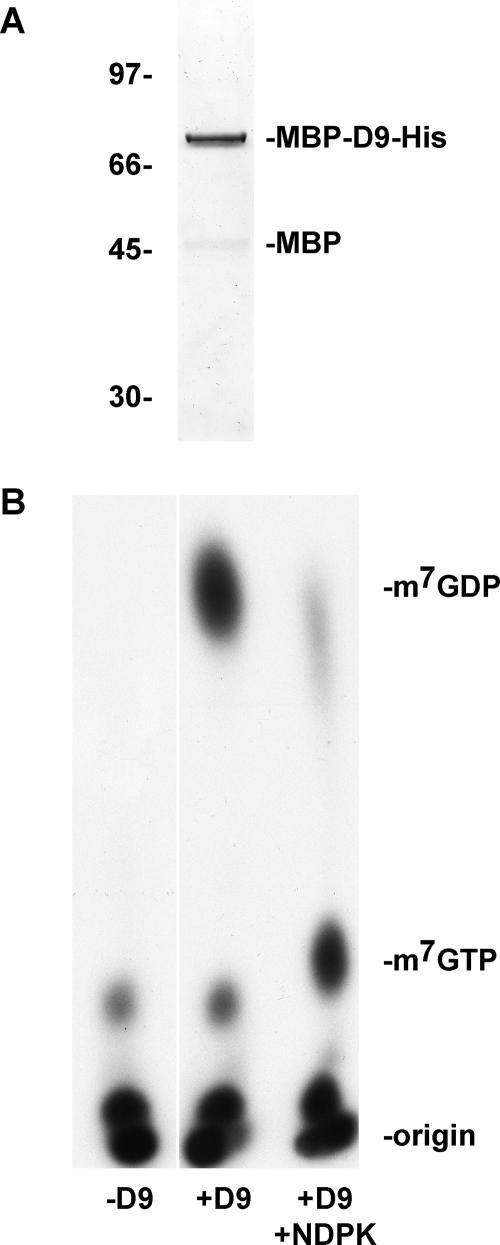
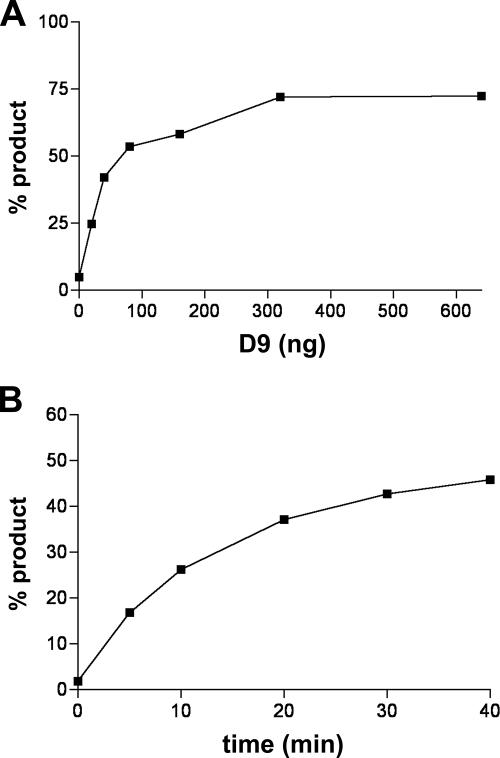
The common Nudix hydrolase motif and similar phenotype resulting from D9 and D10 overexpression raised the possibility that D9 may also cleave the mRNA cap to accelerate mRNA turnover. To determine if VACV D9 possesses mRNA-decapping activity, an MBP-D9 fusion protein appended with a C-terminal His10 tag was expressed in E. coli and MBP-D9-HIS was sequentially purified over amylose and nickel-NTA columns. Resolution of the purified recombinant protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a predominant band corresponding to full-length MBP-D9-HIS and a minor band corresponding to MBP (Fig. 2A), similar to that previously found after purification of the corresponding D10 fusion protein (15). The free MBP most likely represents the remnants of degradation of the full-length fusion protein. To generate a substrate for decapping assays, a 309-nt RNA was synthesized in vitro and subsequently capped and methylated with [α-32P]GTP and unlabeled S-adenosylmethionine using recombinant VACV RNA guanylyltransferase/guanine-7-methyltransferase. The recombinant D9 protein was incubated with the capped RNA substrate, and the product of the reaction was resolved by TLC and visualized by autoradiography. Unlabeled standards were resolved on the same TLC plate and detected by UV shadowing. When D9 was omitted from the reaction, the labeled capped RNA remained largely at the origin; the slightly faster-migrating material may represent residual GTP substrate remaining after purification of the RNA (Fig. 2B). In contrast, addition of recombinant D9 resulted in the release of a radioactive product that comigrated with the unlabeled m7GDP standard (Fig. 2B). To confirm that the product of the reaction was m7GDP, the decapping reaction product was treated with nucleoside diphosphate kinase, an enzyme that specifically converts nucleoside diphosphates into nucleoside triphosphates. As predicted, treatment with kinase converted the putative m7GDP product into a species that comigrated with the m7GTP standard (Fig. 2B). The amount of m7GDP released by D9 increased with enzyme concentration and time (Fig. 3). However, even at a high enzyme excess, only about 75% of the cap was released. This is likely due to the presence of RNA with unmethylated cap, which, as will be shown below, is not a substrate for D9. In addition, the substrates contain variable amounts of uncapped RNA, which inhibits decapping activity (shown below). For these reasons and because of the need for high enzyme/substrate ratios, kinetic constants were not determined.
FIG. 2.
Recombinant VACV D9 possesses intrinsic mRNA-decapping activity. (A) Recombinant VACV D9 was synthesized in E. coli as an MBP-D9 fusion protein with a C-terminal His10 epitope tag. The protein was purified by affinity column chromatography using amylose and nickel-NTA resins, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by silver staining. The sizes (kDa) of protein standards are denoted at the left of the gel and the bands corresponding to MBP-D9-HIS and MPB on the right. (B) One picomole (70 ng) of MBP-D9-HIS (D9) and 0.02 pmol of 32P-cap-labeled actin RNA (309 nt) were incubated in decapping buffer for 30 min at 37°C. A portion of the reaction mixture was incubated with 2 units of nucleotide diphosphate kinase (NDPK) in the presence of 1 mM ATP for 30 min at 37°C. In the presence of ATP, NDPK converts nucleoside diphosphates into nucleoside triphosphates. The reaction products were resolved on PEI-cellulose TLC plates. Radioactive signals were visualized by autoradiography, and unlabeled migration standards were detected by UV shadowing as shown on the right.
FIG. 3.
Effects of enzyme concentration and time on decapping. (A) Effect of D9 concentration on decapping. The reaction was carried out as described in the legend to Fig. 2 except for the use of different amounts of D9. The percent product released as m7GDP in 30 min was determined with a PhosphorImager. (B) Decapping was carried out as for panel A except that 30 ng of D9 was used per reaction, and the amount of m7GDP was quantified at the indicated times.
The Nudix hydrolase motif of VACV D9 is essential for mRNA decapping.
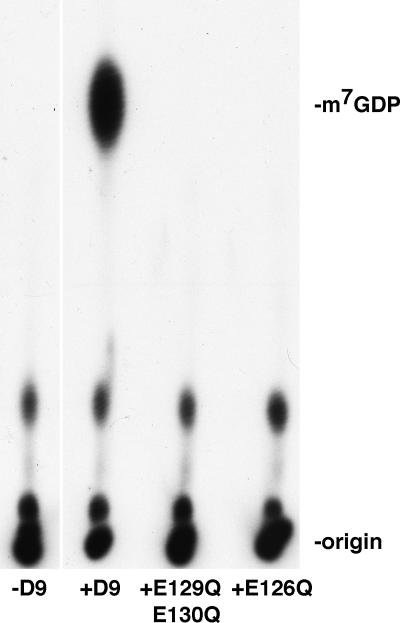
The essential residues for phosphohydrolase activity have been determined for a subset of Nudix hydrolases. In particular, the glutamic acid residues of the EX2EE sequence of the Nudix motif (Fig. 1) are critical for divalent cation binding and therefore essential for enzyme activity. To determine whether the Nudix hydrolase motif of D9 is required for mRNA decapping, mutations were created in the essential divalent cation-binding domain and the mutant proteins were synthesized and purified in parallel to wild-type D9 recombinant protein. Two mutants were purified, one in which the two glutamic acid residues at positions 129 and 130 were converted to glutamine (E129Q/E130Q) and a second in which the glutamic acid residue at position 126 was converted to glutamine (E126Q). Whereas wild-type recombinant D9 decapped the RNA substrate and released m7GDP, equivalent amounts of the mutated proteins did not (Fig. 4). The absence of RNA decapping when the mutated D9 proteins were added to the reaction mixture confirmed that D9 itself and not a copurifying contaminant was responsible for the observed activity and further demonstrated that the Nudix motif of D9 is essential for hydrolysis.
FIG. 4.
The Nudix motif of VACV D9 is required for mRNA cap cleavage. Mutations were generated in the conserved divalent cation domain of the D9 Nudix motif. The mutated proteins were synthesized in E. coli and purified in parallel to wild-type MBP-D9-HIS over an amylose column followed by a nickel-NTA column. E129Q/E130Q possesses two point mutations in which glutamic acid residues at position 129 and 130 were transformed to glutamine. E126Q contains a point mutation at position 126 resulting in the conversion of glutamic acid to glutamine. Equivalent amounts (100 ng) of MBP-D9-HIS, E129Q/E130Q, and E126Q were added to the decapping assay mixture, and products of the reaction were resolved by PEI-cellulose TLC as for Fig. 2B.
D9 cleavage activity is specific for the methylated cap structure.
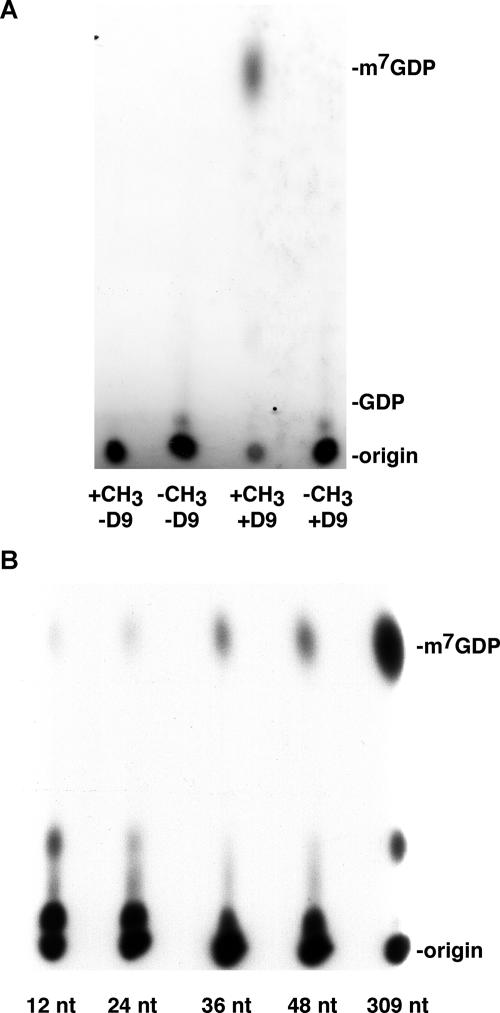
Nudix hydrolases can potentially cleave a wide variety of substrates, so it was important to determine the specificity of D9. A structural hallmark of the mRNA cap is the presence of a methyl group at position 7 of the guanosine residue, which serves as a recognition signal for proteins that selectively bind the 5′ cap. To determine the specificity of D9 for capped mRNA, an unmethylated capped RNA substrate was synthesized by omitting S-adenosylmethionine and incubated with recombinant D9. D9 was incapable of cleaving the unmethylated capped RNA and liberating GDP, demonstrating the specificity of D9 for the biologically relevant mRNA cap structure (Fig. 5A).
FIG. 5.
D9 decapping activity is dependent on a methylated cap and enhanced by increased RNA length. (A) The 309-nt RNA was capped in the absence (−CH3) or presence (+CH3) of S-adenosylmethionine and tested as a substrate for D9 as for Fig. 2B. (B) Equivalent amounts of 12-, 24-, 36-, 48-, and 309-nt 32P-cap-labeled RNAs were incubated with 100 ng MBP-D9-HIS in the presence of decapping buffer, and the products of the reaction were resolved by PEI-cellulose TLC as for Fig. 2B.
D9 decapping activity is dependent on the length of the RNA.
To determine whether D9 decapping activity was influenced by RNA length, 12-, 24-, 36-, 48-, or 309-nt RNAs of overlapping sequence were synthesized by in vitro transcription and 32P cap labeled and methylated using VACV RNA guanylyltransferase/guanine-7-methyltransferase. The decapping activity of D9 increased with RNA length; only trace amounts of the 12- and 24-nt RNA substrates were cleaved (Fig. 5B). In contrast, VACV D10 was shown to cleave substrates of 24 nt or more with similar efficiency (15).
D9 decapping activity is potently reduced by uncapped RNA.
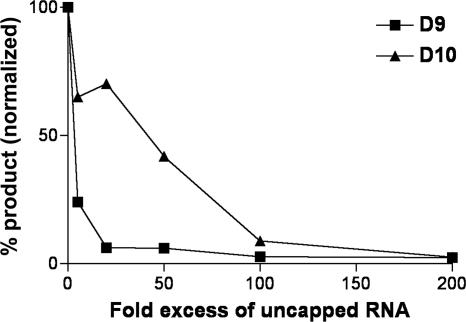
Previous studies demonstrated that uncapped RNA inhibited the decapping activity of VACV D10, suggesting that the protein can interact with RNA independent of the cap. To determine the effects of RNA on D9 activity, increasing molar amounts of uncapped RNA were added to the D9 decapping reaction mixture and the percentage of m7GDP product generated was calculated. Both substrate and competitor RNAs were identical except for the absence of the cap. Decapping by D9 was inhibited 75% by a 5-fold molar excess of unlabeled RNA and 94% by 20-fold excess (Fig. 6). In contrast, a much higher molar excess of RNA was needed to strongly inhibit decapping by D10 (Fig. 6), suggesting that D9 has a greater affinity for RNA.
FIG. 6.
D9 decapping activity is inhibited by uncapped RNA. Uncapped, unlabeled 309-nt actin RNA was added in increasing amounts to the decapping reaction mixture containing 70 ng of either MBP-D9-HIS or MBP-D10, and products were resolved by PEI-cellulose TLC. The percentage of product generated was quantified using a PhosphorImager.
Methylated nucleotides weakly inhibit D9 decapping activity.
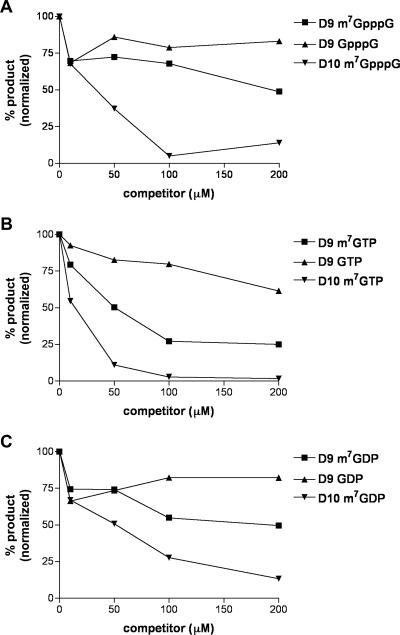
Previous competition studies showed that VACV D10 decapping activity was inhibited by methylated nucleotide derivatives unattached to RNA, with the greatest inhibition observed for m7GTP, followed by m7GpppG and m7GDP (15). We found that higher concentrations of the same methylated nucleotides were needed to inhibit the decapping activity of D9 compared to D10 (Fig. 7A, B, and C). Neither D9 nor D10 was significantly inhibited by unmethylated nucleotide derivatives (Fig. 7A, B, and C). Thus, D9 is more sensitive to RNA and D10 is more sensitive to methylated nucleotides, suggesting that the two enzymes bind differently to their substrates.
FIG. 7.
D9 decapping activity is weakly inhibited by a cap analog and methylated nucleotides. (A) One picomole (70 ng) of MBP-D9-HIS (D9) or MBP-D10 (D10) was incubated with 0.02 pmol of capped and methylated substrate RNA in the presence or absence of methylated (m7GpppG) or unmethylated (GpppG) cap analog, and products of the reaction were resolved by PEI-cellulose TLC and quantified as for Fig. 5. (B) m7GTP or GTP was added to the decapping reaction mixture and analyzed as for panel A. (C) m7GDP or GDP was added to the decapping reaction mixture as for panel A.
DISCUSSION
Nudix hydrolases are present in bacteria, archaea, and eukarya, where they hydrolyze a wide variety of organic pyrophosphates with various degrees of substrate specificity (10). Mammalian cells have about 30 different genes with Nudix hydrolase motifs, including Dcp2, which cleaves mRNA caps (20). Amino acid side chains and sequences outside of the Nudix motif determine substrate specificity. The selectivity of D9 for a methylated cap attached to RNA suggests that it has a specific role as an mRNA-decapping enzyme, similar to VACV D10 (15) and Dcp2 (20). The RNA requirement, as well as the m7GDP product, distinguishes D9 from the cellular scavenger enzyme DcpS, which hydrolyzes free cap structures to release m7GMP (21). Moreover, overexpression of D9 during a VACV infection resulted in decreased levels of viral mRNA and proteins (18). Thus, VACV and by inference most other sequenced chordopoxviruses have two decapping enzymes. The similarity in size and 25% sequence identity of D9 and D10 suggest that they have a common origin, perhaps resulting from gene duplication after the divergence of chordo- and entomopoxviruses. Alternatively, the two genes may be even older, with D9 having been lost in parapoxviruses and entomopoxviruses. Going back further in time, sequence analyses suggest that Nudix hydrolases are part of the ancestral nucleocytoplasmic large DNA virus gene set (6).
Proof that D9 has intrinsic decapping activity came from the purification of a recombinant fusion protein in E. coli and the loss of activity upon mutation of the Nudix hydrolase motif. A comparison of D9 with D10 showed similarities and differences in enzyme activity. Although both enzymes required a minimal-length RNA for decapping, the length was greater for D9. Furthermore, uncapped RNA was a more potent inhibitor of decapping by D9 than of that by D10. Conversely, the decapping activity of D10 was more sensitive to inhibition by methylated cap analogs and methylated nucleotides unattached to RNA. Together, these data suggest that binding of D9 to RNA precedes cap recognition and hydrolysis. In this respect, D9 is similar to the cellular decapping enzyme Dcp2 (17). One caveat is that our experiments were all carried out with MBP fusions to increase solubility and facilitate purification of the enzyme. A region of Dcp2 called Box B, just C terminal to the Nudix hydrolase motif, is important but not sufficient for RNA binding (17). The corresponding region of D9 and D10 has some similarity to Box B, but it is weak. Detailed mutational and structural analyses will be important to map the functional domains of the poxvirus decapping enzymes.
It will be interesting to determine whether D9 exhibits any RNA sequence specificity, since microarray analyses suggest some selectivity in cellular mRNA degradation (2, 5). In this respect, we did not detect any gross changes in either host cell or viral protein synthesis after infection with a VACV D9 deletion mutant (14). In contrast, host and early viral mRNAs were detected for longer times than usual after infection with a D10 deletion mutant. Eventually, however, the RNAs were degraded, possibly with the assistance of D9 since prolonged expression of early proteins, including D9, would be expected to occur in the absence of D10. Partial complementation of D10 by D9 would explain our current inability to delete both genes in the same virus.
Acknowledgments
We thank Wolfgang Resch for preparing the sequence alignments and Wolfgang Resch, George Katsafanas, and Teri Shors for helpful discussions.
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Baldick, C. J., Jr., and B. Moss. 1993. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J. Virol. 67:3515-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brum, L. M., M. C. Lopez, J. C. Varela, H. V. Baker, and R. W. Moyer. 2003. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 315:322-334. [DOI] [PubMed] [Google Scholar]
- 3.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouet, P., X. Robert, and E. Courcelle. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerra, S., L. A. Lopez-Fernandez, A. Pascual-Montano, M. Munoz, K. Harshman, and M. Esteban. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 77:6493-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer, L. A., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 7.Laidlaw, S. M., and M. A. Skinner. 2004. Comparison of the genome sequence of FP9, an attenuated, tissue culture-adapted European strain of fowlpox virus, with those of virulent American and European viruses. J. Gen. Virol. 85:305-322. [DOI] [PubMed] [Google Scholar]
- 8.Lee-Chen, G. J., and E. G. Niles. 1988. Transcription and translation mapping of the 13 genes in the vaccinia virus HindIII D fragment. Virology 163:52-63. [DOI] [PubMed] [Google Scholar]
- 9.Martin, S. A., and B. Moss. 1975. Modification of RNA by mRNA guanylytransferase and mRNA (guanine-7-)methyl-transferase from vaccinia virions. J. Biol. Chem. 250:9330-9335. [PubMed] [Google Scholar]
- 10.McLennan, A. G. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63:123-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milligan, J. F., D. R. Groebe, G. W. Witherell, and O. C. Uhlenbeck. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15:8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss, B. 2007. Poxviridae: the viruses and their replicaton, p. 2905-2946. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, and M. A. Martin (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Moss, B., and N. P. Salzman. 1968. Sequential protein synthesis following vaccinia virus infection. J. Virol. 2:1016-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish, S., and B. Moss. 2006. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J. Virol. 80:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parrish, S., W. Resch, and B. Moss. 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc. Natl. Acad. Sci. USA 104:2139-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennington, T. H. 1974. Vaccinia virus polypeptide synthesis: sequential appearance and stability of pre- and post-replicative polypeptides. J. Gen. Virol. 25:433-444. [DOI] [PubMed] [Google Scholar]
- 17.Piccirillo, C., R. Khanna, and M. Kiledjian. 2003. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shors, T., J. G. Keck, and B. Moss. 1999. Down regulation of gene expression by the vaccinia virus D10 protein. J. Virol. 73:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steiger, M., A. Carr-Schmid, D. C. Schwartz, M. Kiledjian, and R. Parker. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA 99:12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Z., and M. Kiledjian. 2001. Functional link between the mammalina exosome and mRNA decapping. Cell 107:751-762. [DOI] [PubMed] [Google Scholar]