Abstract
Species B human adenoviruses (Ads) are often associated with fatal illnesses in immunocompromised individuals. Recently, species B Ads, most of which use the ubiquitously expressed complement regulatory protein CD46 as a primary attachment receptor, have gained interest for use as gene therapy vectors. In this study, we focused on species B Ad serotype 35 (Ad35), whose trimeric fiber knob domain binds to three CD46 molecules with a KD (equilibrium dissociation constant) of 15.5 nM. To study the Ad35 knob-CD46 interaction, we generated an expression library of Ad35 knobs with random mutations and screened it for CD46 binding. We identified four critical residues (Phe242, Arg279, Ser282, and Glu302) which, when mutated, ablated Ad35 knob binding to CD46 without affecting knob trimerization. The functional importance of the identified residues was validated in surface plasmon resonance and competition binding studies. To model the Ad35 knob-CD46 interaction, we resolved the Ad35 knob structure at 2-Å resolution by X-ray crystallography and overlaid it onto the existing structure for Ad11-CD46 interaction. According to our model, all identified Ad35 residues are in regions that interact with CD46, whereby one CD46 molecule binds between two knob monomers. This mode of interaction might have potential consequences for CD46 signaling and intracellular trafficking of Ad35. Our findings are also fundamental for better characterization of species B Ads and design of antiviral drugs, as well as for application of species B Ads as in vivo and in vitro gene transfer vectors.
Human adenoviruses (Ads) have been classified into six species (A to F), containing 51 serotypes. Species B Ads form two genetic clusters, B1 (Ad serotype 3 [Ad3], Ad7, Ad16, Ad21, and Ad50) and B2 (Ad11, Ad14, Ad34, and Ad35) (30). Most B1 serotypes are associated with acute respiratory disease and, unlike the species C Ads (e.g., Ad5), do not establish persistence (29). The B2 serotypes are mainly associated with infections of the kidney and urinary tract and are often fatal in immunocompromised individuals (11). Species B Ads can also be grouped based on their receptor usage (27). Group I Ads (Ad serotypes 16, 21, 35, and 50) nearly exclusively use CD46, a ubiquitously expressed membrane protein with complement regulatory functions that is upregulated on tumor and stem cells (6, 21, 25). Group II Ads (Ad serotypes 3, 7, and 14) utilize a still unidentified receptor X, but not CD46. Group III (Ad11) uses both CD46 and receptor X. Recently, gene transfer vectors based on species B Ads have shown promise for cell and gene therapy. Vectors derived from species B Ads or Ad5 vectors containing fibers from species B Ads efficiently transduce human cell types that are relatively refractory to infection with classical serotype Ad5 vectors, including tumor cells, hematopoietic stem cells, mesenchymal stem cells, dendritic cells, and lymphocytes (for a review, see reference 26). The most commonly used species B vectors contain Ad35 fibers (Ad35 or Ad5/F35), and therefore most studies on the interaction of species B Ads with CD46 have focused on Ad35. Ad35 binds through its trimeric fiber knob to CD46 with a high avidity (23, 27). While the interacting residues within CD46 have been localized to the two distal extracellular domains of CD46 (SCR1 and SCR2) (5-7, 20), the contact areas within the Ad35 knob have not been reported so far.
The structure of an Ad fiber knob domain was first determined in 1994, for the species C serotype Ad5, which uses the coxsackie and adenovirus receptor (CAR) for attachment (31). The homotrimeric knob structure is best described as a three-bladed propeller, whereby each blade contains multiple tightly packed beta-sheets (labeled A to J). The structure of a CD46-interacting Ad knob (Ad11), which is similar overall to other Ad knobs, was recently published (18). Crystallization of recombinant Ad11 knob bound to CD46 domains SCR1 and SCR2 revealed three critical contact regions within the FG, HI, and IJ loops of the fiber knob. This model is supported by studies demonstrating that binding of Ad11 virus to CD46 can be abolished by introduction of a single amino acid substitution (Arg279Gln) within the Ad11 HI loop (10). Although there is overlap in tropism between Ad11 and Ad35, Ad11 virus binds to CD46 with a higher avidity (27), which implies that the mechanism of Ad11-CD46 interaction cannot necessarily be translated to Ad35. In this study we show the affinity of the Ad35 knob to CD46, report the crystal structure for the Ad35 knob, identify four amino acids in the Ad35 knob that are critical for CD46 binding, and present a model for Ad35 knob interaction with CD46.
MATERIALS AND METHODS
Production of recombinant Ad35 knob proteins.
Recombinant Ad35 knobs (for cloning procedure, see reference 6) were produced using the QIAGEN protocol 7. Protein expression was induced for 5 h by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cells were harvested, and cell pellets were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), followed by incubation with 1 mg/ml lysozyme for 30 min on ice and sonication. Cellular debris was removed by centrifugation, and the supernatant was incubated with Ni-nitrilotriacetic acid (NTA) agarose at 4°C for 3 h. Beads were washed with 50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole, and 20% glycerol, and recombinant knob protein was eluted with 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, and 20% glycerol. For production of Ad35 knob without His tags, the Ad35 fiber knob sequence was cloned into pQE-30Xa (QIAGEN), and knob protein was purified and digested with factor Xa protease as suggested by the manufacturer. After digestion, factor Xa protease was removed using Xa Removal Resin, and cleaved His6-tagged peptides and undigested His6-tagged protein were captured by Ni-NTA affinity chromatography as described in the QIAGEN protocol.
Production of sCD46.
293 cells were transfected with 10 μg of a soluble CD46 (sCD46) expression plasmid (7), and stably expressing cell clones were selected with G418. Twenty clones were screened for sCD46 expression by a competition assay with 3H-labeled wild-type Ad35 virus (6). The clone with the highest sCD46 level was expanded, and supernatant was used for Western blotting and colony blot hybridization in a 1:1 dilution.
The CD46 peptide (sCD46-P), containing the knob-interacting CD46 domains SCR1 and SCR2, was produced and expressed in mutated CHO cells (CHO Lec 3.2.8.1) that have a limited glycosylation pattern (18). For surface plasmon resonance (SPR) analyses, the concentration of the sCD46-P was determined using the absorption signal at 280 nm and an extinction coefficient of 2.064 (absorption of 1 g/liter of solution of protein) calculated by the computer program ProtParam (8), based on the amino acid sequence of sCD46-P (EEPPTFEAMELIGKPKPYYEIGERVDYKCKKGYFYIPPLATHTICDRNHTWLPV SDDACYRETCPYIRDPLNGQAVPANGTYEFGYQMHFICNEGYYLIGE EILYCELKGSVAIWSGKPPICE).
Western blotting with purified Ad35 knobs.
Knob proteins were produced in Escherichia coli and purified by Ni-NTA agarose chromatography as described above. Recombinant knobs were separated by polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes. Protein samples were loaded in loading buffer (50 mM Tris-HCl, pH 6.8, 100 mM dithiothreitol, 2% sodium dodecyl sulfate, 10% glycerol, 0.2% bromophenol blue) with and without boiling. To detect whether recombinant Ad35 knobs bind to CD46, the blot was incubated with sCD46 in TBS (10 mM Tris-Cl, pH 7.5, 150 mM NaCl) and 3% milk for 1 h at room temperature (RT) and then washed three times for 10 min in TBS-0.05% Tween 20 (TBS-T) buffer. The blot was then incubated with anti-CD46 antibody (clone J4.48; Fitzgerald, Concord, MA) (1:50) in TBS and 3% milk for 1 h at RT and then washed three times for 10 min in TBS-T buffer. To visualize binding, the blot was incubated with goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) (BD Pharmingen, San Jose, CA) (1:1,000) in TBS and 3% milk for 1 h at RT, washed, and subjected to enhanced chemiluminescence substrate (Pierce, Rockford, IL). To assess Ad knob trimerization, the same blot was then washed three times for 10 min in TBS-T-0.1% Triton buffer and incubated with rabbit polyclonal anti-His6-HRP antibody (ab1187-100, lot 134173; Abcam, Cambridge, MA) in TBS and 3% milk for 1 h at RT. Binding was visualized by an enhanced chemiluminescence kit. Notably, this anti-His antibody recognizes the Ad35 knob only as a trimer. Knob trimers disintegrate into knob monomers if samples are boiled before loading.
Ad35 knob library.
The coding sequence of the Ad35 knob (amino acids 123 to 320) was obtained by PCR from Ad35 DNA using the primers P1 (5′-TTTAAGGATCCGGTGACATTTGTATAAAGGATAG-3′) and P2 (5′-ATGC TCTGCAGCTGCCGCTTCTGTAATGTAAGAAAAGAAAAAGGG-3′) and cloned into pQE100 (QIAGEN, Valencia, CA) for expression in E. coli with an N-terminal His6 tag. Random mutagenic PCR was performed based on a protocol published elsewhere (3, 4). Briefly, 20 fmoles of pQE-Ad35knob DNA template, 30 pmol (each) of PCR primer (Pmut1, 5′-CGTCAGCACGGATCCGGTGACATTTGTATAAAGGATAGTATTAACACCTTATGGACTGGA-3′; Pmut2, 5′-ctatAGTCTCTTCAAGCTTGGCTGCAGCTGCCGC-3′) 10× mutagenic buffer (2.5 μl, 3.5 μl, 5 μl, or 10 μl; 70 mM MgCl2, 500 mM KCl, 100 mM Tris [pH 8.3 at 25°C], and 0.1% [wt/vol] gelatin), 10 μl of 5 mM MnCl2, 10 μl of deoxynucleoside triphosphate mix (2 mM dGTP, 2 mM dATP, 10 mM dCTP, and 10 mM dTTP), and 5 units of Taq polymerase (Promega, Madison, WI) were mixed in a final volume of 100 μl. PCR conditions were 94 °C for 1 min, 45 °C for 1 min, and 72 °C for 1 min (30 cycles). The mutant PCR products (640 bp in length) were purified, digested with appropriate enzymes, and cloned into pQE100 vector (QIAGEN, Valencia, CA). For quality control of the random mutagenic library, the ligation product was transformed into E. coli M15 (QIAGEN, Valencia, CA) and plated on kanamycin and ampicillin plates, and 50 colonies were randomly picked for sequencing.
Colony assays.
The Ad35 knob mutant plasmid library was transformed into E. coli XL1-Blue or M15 host strains and plated on LB plates (master plate) with appropriate antibiotics. About 800 to ∼1,000 colonies were grown on one 15-cm petri dish the next day. After overnight growth, a 0.45-μm-pore-size Durapore filter membrane (Millipore, Billerica, MA) was placed on top of the colonies. The membrane was peeled off and placed carefully, with the colonies facing upwards, on two sheets of Whatman 3MM paper soaked in LB medium supplemented with antibiotics and 1 mM IPTG. Protein expression of the colonies was induced for 6 h at 30°C, after which the filter with the colonies was placed on top of a nitrocellulose filter and a Whatman 3MM paper soaked in native lysis buffer (20 mM Tris-Cl [pH 8], 300 mM NaCl, 50 mM MgCl2, 0.1 mg/ml lysozyme, 0.75 mg/ml DNase I, and one-half complete EDTA-free protease inhibitor cocktail tablet/10 ml [Roche, Palo Alto, CA]). The “filter sandwich” was incubated at room temperature for 30 min and then freeze-thawed four times for 10 min at −80 °C and 10 min at 30 °C. The nitrocellulose membrane was removed from the sandwich and blocked with 3% bovine serum albumin in TBS-T at 4°C overnight. CD46 binding and trimerization were analyzed as described for Western blotting. Colonies that showed binding to our anti-His antibody but not binding to CD46 were picked from the original master plates. DNA from these colonies was sequenced, and absence of binding was verified by Western blot analysis using purified knob protein.
Crystallization.
N-terminal His6-tagged Ad35 knob protein was expressed from pQE30-Ad35knob (6) and produced as described above. It was subsequently purified by Ni-NTA agarose affinity and gel filtration chromatography and dialyzed against 20 mM Tris-HCl (pH 8), 200 mM NaCl, and 5 mM dithiothreitol. The purified protein was crystallized using NeXtal DWBlock crystallization screening suites. Five different crystal forms appeared in 2 to 3 days. Four of them were much larger than the other but diffracted poorly. Crystals grew at 21°C in 30% polyethylene glycol 1000 and 0.2 M MgBr2 (pH 8.0). X-ray data were derived from frozen crystals, but due to the high concentration of polyethylene glycol 1000, no cryoprotectant was added. Native data sets were collected at National Synchrotron Radiation Research Center beam lines BL13B1 and BL13C1 in Taiwan, using an ADSC Quantum charged-coupled-device detector, and processed with HKL2000 software (17). The structure was solved by molecular replacement with the Open-EPMR program (15) using Ad3 head (Protein Data Bank [PDB] entry 1h7z) as a search model. The model was rebuilt using the O program (13) and refined using the CNS program with the maximum-likelihood target and automatic weight optimization (2).
Biacore biosensor analysis.
All analyses were carried out on a Biacore 3000 at 25°C. Research grade CM5 sensor chips, N-hydroxysuccinimide (NHS), N-ethyl-N′-(3-diethylaminopropyl)carbodiimide (EDC), ethanolaminehydrochloride, and HBSEP running buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, and 0.005% [vol/vol] surfactant P20) were purchased from the manufacturer (Biacore Inc., Piscataway, NJ). For amine coupling, carboxymethylated dextran surfaces on CM5 chips were activated using a standard NHS/EDC procedure. Ad35 knob and sCD46-P, containing the knob-interacting CD46 domains SCR1 and SCR2 (18), were diluted to no less than 20 nM in 20 mM sodium acetate (pH 4). Ad35 knob protein was immobilized on CM5 chips, and various concentrations of sCD46-P were injected over the activated surface until the desired surface densities were achieved. Activated, coupled surfaces were then quenched of reactive sites with 1 M ethanolamine (pH 8) for 3 to 5 min. All data were collected at 1 Hz using two replicate injections for each concentration of analyte. Flow rates during the experiment were maintained at 50 μl/min, and HBSEP running buffer was supplemented with 0.1 mg/ml bovine serum albumin to block the nonspecific binding. To completely remove remaining amounts of sCD46-P bound to the sensor chip surface, regenerations were performed by double 30-s injections of 20 mM sodium acetate solution (pH 4). Data processing and kinetic analysis were performed using Scrubber software (version 2.0; BioLogic Software, Campbell, Australia). Data processing included double referencing (16). The stoichiometry of the reaction was estimated from the equation:
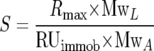 |
where S is the number of monomers of the analyte per monomer of the ligand in the surface protein complex, Rmax is maximum surface density determined from the kinetic model, MwL is the molecular weight of the immobilized ligand, RUimmob is the number of the response units of the ligand immobilized, and MwA is the molecular weight of the injected analyte.
Competition assays.
Wild-type Ad35 virus was propagated in 293 cells, labeled with [methyl-3H]thymidine, and purified as described before (24). HeLa cells were detached from culture dishes by incubation with Versene and washed with phosphate-buffered saline (PBS). A total of 105 cells per tube were resuspended in 50 μl of ice-cold adhesion buffer (Dulbecco's modified Eagle's medium supplemented with 2 mM MgCl2, 1% fetal calf serum, and 20 mM HEPES) containing different concentrations of Ad35 fiber knob protein and incubated on ice for 1 h. Then, 3H-labeled wild-type Ad35 virus was added in adhesion buffer at a multiplicity of infection of 8,000 viral particles (VP) per cell to a final volume of 100 μl. After 1 h of incubation on ice, cells were pelleted and washed twice with 0.5 ml of ice-cold PBS. After the last wash, the supernatant was removed, and the cell-associated radioactivity was determined by a scintillation counter. The number of VP bound per cell was calculated by using the virion-specific radioactivity and the number of cells.
Model analysis.
In order to generate a model for Ad35 bound to CD46, the crystal structure for the Ad35 fiber knob was superimposed onto the Ad11-CD46 structure (Research Collaboratory for Structural Bioinformatics [RCSB], PDB structure 2o39), using the modeling software What If (28). Structures for knobs or knob-CD46 interaction in ribbon or surface format were generated with the Deepview/Swiss-PdbViewer (9).
Protein structure accession numbers.
The structures of the Ad35 fiber knob (RCSB identifier, rcsb043750) has been deposited in the PDB under code 2QLK; the structure of the Ad35-CD46 interaction model has been deposited in the Protein Model DataBase under accession number PM0074963.
RESULTS AND DISCUSSION
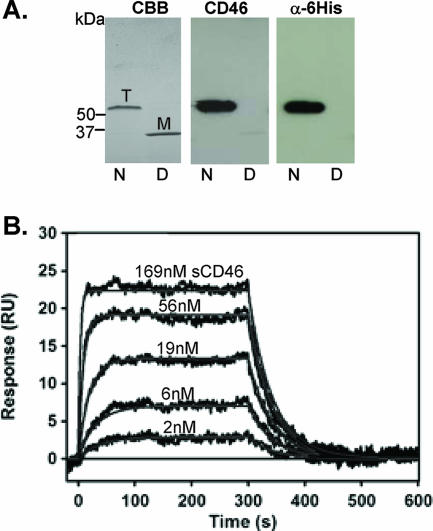
We first studied the interaction of recombinant Ad35 fiber knob and CD46 by Western blotting and SPR analyses. Recombinant Ad35 knob containing an N-terminal His6 tag was produced in E. coli and purified by affinity chromatography on Ni-agarose. Notably, the receptor-interacting knob domain is formed by the C terminus of the fiber, and an N-terminal His tag does not interfere with knob-receptor interactions (1, 18). In agreement with earlier studies (27), purified Ad35 knob formed trimers (Fig. 1A, left panel). For analysis of binding to CD46, we used sCD46, and binding was visualized with an anti-CD46 monoclonal antibody (MAb) that does not interfere with the Ad35 knob-CD46 interaction. Only the trimeric Ad35 form interacted with sCD46 (Fig. 1A, middle panel). Interestingly, we identified a rabbit polyclonal anti-His6-HRP antibody (ab1187-100, lot no. 134173; Abcam) that recognizes only the N-terminal His-tagged Ad35 knob in its trimeric form (Fig. 1A, right panel).
FIG. 1.
Analysis of recombinant Ad35 knob. (A) Western blot analysis. Purified Ad35 knob was run as native (N) or denatured protein (D) on a polyacrylamide gel. Coomassie brilliant blue staining (CBB) revealed the trimeric knob form (T), which is converted into monomers after boiling (M). Blots were analyzed for CD46 binding by subsequent incubation with sCD46, anti-CD46 MAb, and anti-mouse IgG-HRP (middle panel). After stripping, the same filters were incubated with an anti-His6-HRP antibody (right panel). (B) Kinetic response data for sCD46 binding to biosensor surface containing Ad35 knob (50 RU). Experimental data (black) represent the responses of duplicate injections of various concentrations of sCD46 (indicated above the corresponding curves). Global fits of these data according to a 1:1 interaction model are shown in red (see text for resulting kinetic constants).
We then used SPR to study the affinity and stoichiometry of the interaction between recombinant Ad35 knob and sCD46-P, which contains the knob-interacting CD46 domains SCR1 and SCR2 (Fig. 1B). Because preliminary experiments indicated that binding of Ad35 knob to the sensor surface containing immobilized sCD46-P was complex and could not be modeled by a simple 1:1 interaction scheme, we used surfaces with covalently attached Ad35 knob and injected various concentrations of sCD46-P. The values for the kinetic binding constants were determined by globally fitting the data as described in Materials and Methods. The association rate constant was measured to be (1.504 ± 0.004) × 106 M−1 s−1, and the dissociation rate constant was 0.0234 ± 0.0001 s−1, corresponding to a complex half-life of 29.6 s. The KD (equilibrium dissociation constant) as determined from the ratio of these constants was 15.55 ± 0.05 nM. As the Biacore instrument measures the mass of molecules bound to the sensor surface, the stoichiometry of the Ad35 knob-sCD46-P binding reaction can be determined (see Materials and Methods). The observed Rmax of 24.4 RU, corresponding to a stoichiometry of 0.86, is consistent with a 1:1 interaction model; i.e., the surface protein complex contains one molecule of soluble sCD46-P per monomer of Ad35 knob. We recently cloned the Ad35 knob sequence into pQE-30Xa, which allows for expression and purification of a His-tagged Ad35 knob containing a factor Xa protease-cleavable linker. In this protein, the His tag can be removed from the purified protein by factor Xa protease cleavage. We then compared the affinities of purified wild-type Ad35 knob preparations to CD46 and found KD values of 13.73 nM and 15.35 nM for the His-tagged Ad35 knob and the Ad35 knob without His tag, respectively. This indicates that the N-terminal His tag does not affect the Ad35 knob-CD46 interaction.
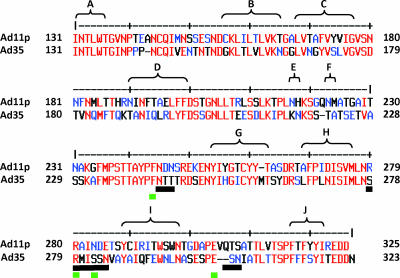
We next attempted to identify the amino acid residues within the knob that are critical for CD46 binding by using an approach based on a library of Ad35 knob mutants expressed in E. coli. To generate this library, we used mutagenic PCR (3, 4) in an approach that on average generated one to two amino acid mutations per knob. The Ad35 knob mutant library in E. coli XL1-Blue was plated on agar plates, and knob expression was induced by IPTG. We used a two-step colony blotting protocol to simultaneously screen the library for knob trimerization and binding to sCD46. Notably, fiber knob trimerization is required for Ad binding to receptors (12), and analysis of trimerization allowed us to exclude mutations that reduce CD46 binding by causing major conformational changes within the Ad35 knob. Knob trimerization was assessed with the anti-His6-HRP antibody that recognizes only trimeric knob forms. Binding of CD46 was visualized as described for Western blotting (Fig. 1A). More than 98% of colonies negative for CD46 binding were also negative for binding to the anti-His6-HRP antibody, indicating that the vast majority of mutations interfered with trimerization and/or protein solubility. DNA from colonies that were positive for trimerization but negative for CD46 binding was sequenced. A first screening round of ∼10,000 colonies revealed four amino acid residues (Phe242, Arg279, Ser282, and Glu302) that abolished Ad35 knob binding to CD46 without affecting knob trimerization in colony assays (Fig. 2). The identified residues were in areas that encompassed the three contact regions reported for the Ad11 knob. Further rounds of screening did not uncover other regions, indicating that all the detectable CD46-interacting areas had been found.
FIG. 2.
Library identification of amino acids involved in Ad35-CD46 binding. (A) Amino acid alignment of the Ad11 and Ad35 fiber knob sequences. Numbers represent the fiber amino acid sequences for Ad11p (GenBank accession AAN62521) and Ad35 (GenBank accession AP_000601). β-Sheet regions (A to J) are shown. Mutated amino acids from the Ad35 fiber knob that decrease CD46 binding are shown by green lines, and contact amino acids between the Ad11 fiber and CD46 (18) are shown by black lines.
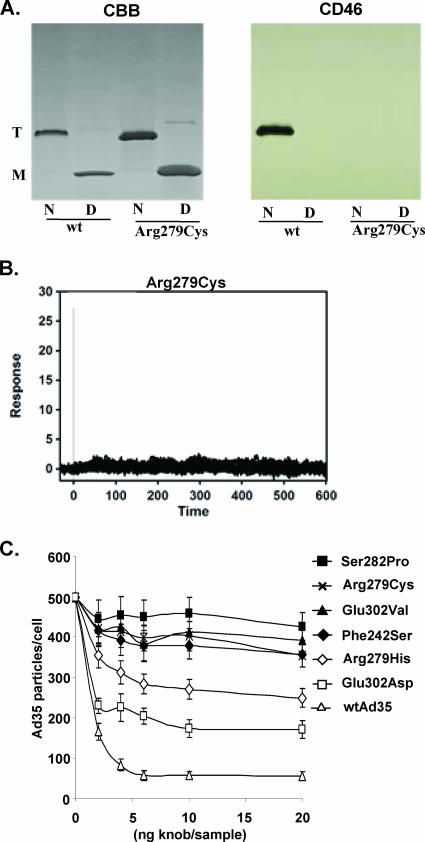
To assess the functional properties of mutants, knobs containing substitutions in positions 242, 279, 282, and 302 were purified. These mutations completely ablated binding to sCD46 as analyzed by Western blotting and SPR analysis. An example for mutant Arg279Cys is shown in Fig. 3A and B. As a more relevant functional assay, we used Ad35 knob mutants as competitors for attachment of Ad35 virus to HeLa cells, a cell line that expresses ∼50,000 CD46 molecules per cell (27). While 5 ng of wild-type Ad35 knob almost completely inhibited Ad35 binding, Ad35 knobs with the mutations Phe242Ser, Arg279Cys, Ser282Pro, and Glu302Val were not able to block Ad35 binding at the highest concentration tested (20 ng) (Fig. 3C). However, when the positively charged Arg279 residue was substituted for a similar histidine or the negatively charged Glu302 was substituted for negatively charged aspartate, the ability to block Ad35 binding was not completely ablated. In short, competition studies validated the functional importance of the Ad35 residues identified by library screening.
FIG. 3.
Analysis of functional properties of selected Ad35 knob mutants. (A) Binding of Arg279Cys mutant to sCD46 in Western blot analysis. Purified Ad35 wild-type and mutant knobs were run as native (N) or as denatured (D) protein on a polyacrylamide gel. Coomassie brilliant blue staining (CBB) revealed the trimeric knob form (T), which is converted into monomers after boiling (M). Blots were analyzed for CD46 binding by subsequent incubation with sCD46, anti-CD46 MAb, and anti-mouse IgG-HRP. (B) Biacore response data for sCD46 binding to biosensor surface containing Arg297Cys knob (127 RU). Experimental data represent the responses of duplicate injections of various concentrations of sCD46 (169 nM, 56 nM, 19 nM, 6 nM, and 2 nM). Experimental conditions were the same as for wild-type Ad35 knob (Fig. 1B). (C) Virus binding competition assay. A total of 1 × 105 HeLa cells were preincubated with different concentrations of recombinant Ad35 fiber knobs at 4°C for 1 h before incubation with [3H]thymidine-labeled wild-type Ad35 (wtAd35) at a multiplicity of infection of 8,000 VP/cell for another hour, and the number of VP bound per cell was determined after washing with PBS.
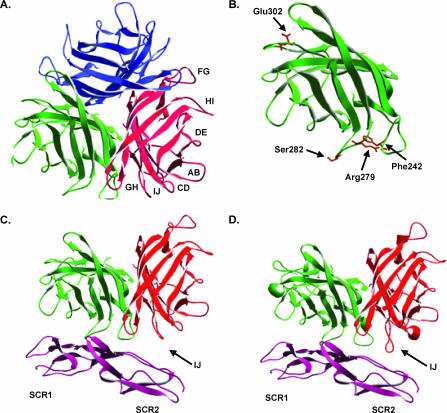
To properly evaluate the residues identified by library screening, the Ad35 crystal structure was resolved by X-ray crystallography. The best Ad35 knob crystals grew to a size of 0.01 by 0.02 by 0.02 mm, which is only 1/100 of the Ad2 crystal size reported by Xia (31). The crystals diffracted to a 2.0-Å resolution and belonged to space group R3 with the following cell dimensions: a = b = 87.8 Å; c = 53.9 Å; α = β = 90°; γ = 120°. These dimensions are very similar to those of Ad2. The generated model of the Ad35 structure has good stereochemistry with an R factor of 20% and an Rfree of 25.7%. Data collection and refinement statistics are shown in Table 1. The solvent content is only 27.3%, reflecting a tightly packed unit cell, which explains why the small crystal size could diffract to high resolution. The final model contains 183 residues (residues 130 to 219 and 229 to 321 of the Ad35 knob) as parts of the EF/FG loops are invisible in the electron density map. There are nine Ad35 knob monomers per unit cell, while Ad2 knob crystals contained only six monomers per unit cell. The superposition of Ad35 trimer with Ad11 trimer shows a root mean square deviation for the Ca atoms of 0.85 Å, which implies that the core structures between Ad35 and Ad11 are highly similar. The trimeric Ad35 structure is shown in Fig. 4A. All mutations were in exposed loop regions within the globular Ad knob structure, underscoring their role in receptor binding (Fig. 4B).
TABLE 1.
Data collection and refinement statistics
| Parametera | Value for Ad35b |
|---|---|
| Data collection | |
| Space group | R3 |
| Unit cell (Å) | |
| a | 87.38 |
| b | 87.38 |
| c | 53.89 |
| Rmerge | 8.2 (50.1) |
| Redundancy | 4.7 (2.3) |
| Resolution (Å) | 40-2.02 (2.09-2.02) |
| Completeness (%) | 92.8 (58.0) |
| I/σ(I) | 17.3 (1.9) |
| Refinement statistics | |
| Final Rfactor/Rfree(%)c | 20.0/25.7 |
| No. of non-hydrogen atoms | |
| Protein | 1,436 |
| Phosphate | 6 |
| Water | 62 |
| RMS deviation from ideality | |
| Bond length (Å) | 0.010 |
| Angle (°) | 1.6 |
| Avg B factor (Å2) | |
| Protein | 44.9 |
| Phosphate | 49.6 |
| Water | 42.5 |
| Ramachandran plot (percent of amino acids in region) | |
| Most favored | 79.6 |
| Allowed | 17.3 |
| Generally allowed | 1.2 |
| Disallowed | 1.2 |
RMS, root mean square.
Values in parentheses correspond to the highest resolution shell.
For I/σ > 0.
FIG. 4.
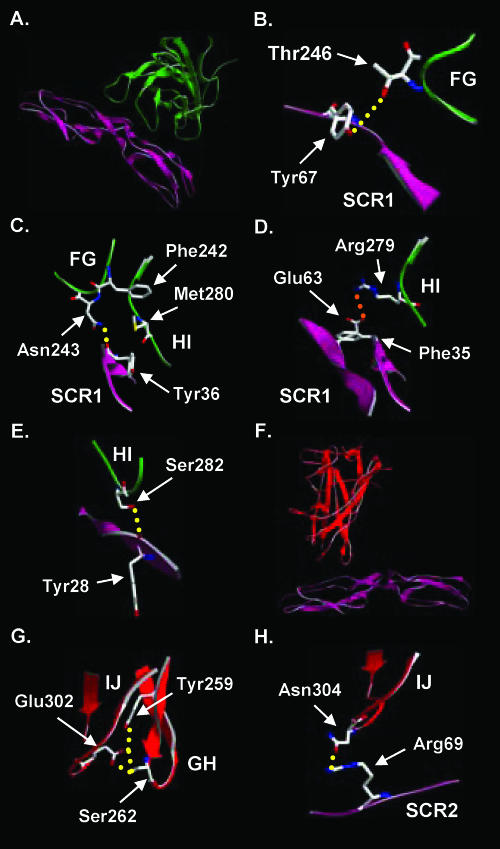
Structure of the Ad35 fiber knob. (A) Ribbon representation of the Ad35 trimeric fiber knob. Fiber monomers are represented in red, blue, and green, with loop regions between β-sheets indicated for the red monomer. (B) Locations of Ad35 fiber amino acids (brown), identified through library analysis, which, upon mutation, decrease binding to CD46 without affecting fiber trimer formation. (C) Representation of two Ad35 trimeric fiber subunits binding to CD46 domains SCR1 and SCR2, based on overlay of the Ad35 crystal structure onto PDB crystal structure 2o39. (D) Representation of two Ad11 trimeric fiber subunits binding to CD46 domains SCR1 and SCR2 from structure 2o39. Differences in proximity between SCR2 and the IJ loops of Ad35 and Ad11 are indicated.
In order to generate a model for Ad35 bound to CD46, the crystal structure for the Ad35 fiber knob was superimposed onto the Ad11-CD46 structure (18). Within the Ad11-CD46 structure, CD46 was shown to flex from its native bent conformation to a straightened rod-like conformation. For binding of Ad35 to CD46, it is also likely that CD46 adopts a straightened conformation; however, the degree of flexing is impossible to predict accurately. For modeling the Ad35-CD46 interaction, we assumed that CD46 had the same conformation as in the Ad11-CD46 structure. Overall analysis of the Ad35 and Ad11 structures revealed that the FG loop of Ad35 gets closer to CD46 domain SCR1 than the FG loop of Ad11 (data not shown), while the IJ loop of Ad11 is two amino acids longer than the IJ loop of Ad35, allowing it to get closer to CD46 domain SCR2 (Fig. 4C and D). Amino acids identified by library screening that ablate CD46 binding are discussed in the following paragraphs (Fig. 5A to H).
FIG. 5.
Location of amino acid residues in the Ad35 fiber knob that are involved in binding to CD46. (A to E) Amino acids in the HI and FG loops, identified through library or model structure analysis, that are involved in HI-FG loop interaction or binding to CD46 domain SCR1. (F to H) Amino acids in the IJ and GH loops, identified through library or structural analysis, that are involved in IJ-GH loop interaction or binding to CD46 domain SCR2. Hydrogen bonds are indicated by dashed yellow lines, and salt bridges are indicated by dashed orange lines.
Phe242.
Phe242 protrudes from the FG loop into the region between the FG and HI loops (Fig. 5C) but is not in close enough proximity to directly interact with CD46 domain SCR1. However, the neighboring amino acids Asn243 and Thr246 likely form hydrogen (H) bonds with the carbonyl group of amino acid Tyr36 and the side chain of Tyr67 in SCR1, respectively (Fig. 5B and C). Furthermore, Phe242 likely stabilizes the proximity of the FG and HI loops to SCR1 by creating a region of hydrophobicity between the FG loop, HI loop, and SCR1 (Fig. 5C). The aromatic ring of Phe242 and the aromatic ring of CD46 residue Tyr36 project toward the HI loop forming a “hydrophobic sandwich” around Met280 within the HI loop. As mutation of Phe242 to serine completely inhibited the ability of the corresponding knob to block Ad35 binding to HeLa cells (Fig. 3C), mutation of the large and hydrophobic Phe242 residue to serine likely changes the proximity of the FG and HI loops (toward each other and CD46) and eliminates the hydrophobic sandwich.
Arg279.
As with Arg280 of Ad11, Arg279 in the Ad35 structure forms a salt bridge with Glu63 of SCR1, which is stabilized by docking of the hydrophobic portion of Arg279 against the Phe35 side chain of CD46 (Fig. 5D). Mutation of Arg279 to Cys almost completely inhibited the ability of the Ad35 fiber knob to block Ad35 binding to HeLa cells (Fig. 3C), likely through ablation of the direct interaction with SCR1 residue Glu63. In contrast, mutation of Arg279 to histidine in the Ad35 fiber knob resulted in less severe but significant inhibition of Ad35 binding to HeLa cells, likely because histidine, although similar, forms a weaker salt bridge as the amino acid side chain is shorter.
Ser282.
Ser282 in the Ad35 knob, which corresponds to amino acid Asn283 of Ad11, forms an H bond with the carbonyl group of amino acid Tyr28 in SCR1 (Fig. 5E). Mutation of Ser282 to proline or alanine (data not shown) inhibited the ability of the Ad35 fiber knob to block Ad35 binding to HeLa cells more than any other identified mutation (Fig. 3C), and this is likely due to elimination of H-bond formation with Tyr28 due to the absence of the OH side chain. Considering the relatively small Gibbs free energy contribution of one H bond, this is not likely the key interaction in formation of the Ad35-CD46 complex, and an alternative explanation is that the H bond with Ser282 is critical for keeping the HI loop and Arg279 in the correct position.
Glu302.
Glu302 of the Ad35 knob projects from the IJ loop into the space between the IJ and GH loops. It appears to stabilize the spatial proximity of the IJ and GH loops within the monomer by directly forming two H bonds with Ser262 within the GH loop and one H bond with Tyr259 in the GH loop (Fig. 5G). In the Ad11 structure Glu303, which in turn forms an H bond with Tyr261, is further away from the analogous Ser264 in the GH loop. Therefore, the interaction between the IJ and GH loops is likely stronger for Ad35. In Ad35, Glu302 is close to Asn304, which forms an H bond with Arg69 in SCR2 (Fig. 5H), and reduction in GH/IJ loop stability likely affects this interaction. In Ad11, Glu305, which is analogous to Ad35 residue Asn304, forms an H bond with the amino group of Leu72 in CD46 (18). For Ad35, CD46 interaction might occur with either Arg69 or Leu72 since the level of CD46 flexing is predicted only for Ad35. Mutation of Glu302 to valine almost completely inhibited the ability of the Ad35 fiber knob to block Ad35 binding to HeLa cells. This is likely due to the fact that the interactions between Glu302 with amino acids Tyr259 and Ser262 within the GH loop are no longer present, which probably helps orient the IJ loop (in particular Asn304) in relation to CD46. Mutation of Glu302 to aspartate in the Ad35 fiber knob resulted in less severe inhibition of Ad35 binding to HeLa cells. As both glutamate and aspartate contain carboxyl side chains, it is possible that the same interactions occur with residues Tyr259 and Ser262 within the GH loop, but the orientation of the IJ loop may change somewhat due to the shorter side chain of aspartate. Notably, a targeted GH loop Ser262Ala substitution (as well as GH loop Cys257Ala, Thy258Ala, Met260Ala, Thr261Ala, Asp264Ala, Arg265Ala, and Ser266Ala) did not ablate binding to CD46. The GH loop substitution Tyr259Ala (as well as Tyr263Ala and Ley267Ala) could not be evaluated for CD46 binding because the corresponding knob formed inclusion bodies. It is possible that a dual Tyr259/Ser262 mutation would be needed to sufficiently disrupt the GH/IJ loop interaction mediated by Glu302 and subsequently inhibit binding to CD46. Further mutagenesis studies on the GH loop are required to delineate residues critical for CD46 binding.
Our model of Ad35-CD46 interaction differs from the model of Ad11-CD46 interaction in a number of ways. In our model the FG loop of the Ad35 knob is closer to CD46 than the Ad11 FG loop, and the IJ loop of Ad35 is shorter and does not get as close to CD46 as the Ad11 IJ loop. We have also suggested a role for the GH loop, through interaction with Glu302, in stabilizing the interaction of the Ad35 IJ loop with CD46, and we have delineated the role of Phe242 in forming a hydrophobic sandwich that stabilizes the FG and HI loops alongside SCR1 upon binding.
The existence of multiple contact residues and the fact that the contact areas in the FG and HI loops are on opposite sides of the Ad35 monomer to the contact area in the IJ loop imply that one CD46 unit binds between two Ad35 knob monomers (as described for the Ad11-CD46 interaction). This indicates that CD46-interacting species B Ads developed a different strategy to bind to their receptor than Ads that interact with CAR (19). For example, within the Ad5 knob the critical CAR binding residues cluster only in one area of the knob (14, 19). The specific configuration of Ad35 binding to CD46 could also explain why CD46-interacting species B Ads are released more slowly from endosomes upon uptake into cells than Ad5 (22).
Ad35-based vectors are attractive for vaccination, in part because of the low serum prevalence of neutralizing Ad35 antibodies in the human population. They are about to be used in clinical trials for vaccination against human immunodeficiency virus (http://clinicaltrials.gov/show/NCT00472719), malaria (http://clinicaltrials.gov/ct/show/NCT00371189), and tuberculosis (http://www.aeras.org/documents/Crucell-AerasclinicaltrialPR). Our findings provide a basis for the retargeting of these promising vectors to important targets for gene therapy. Furthermore, understanding the structural details of the Ad35-CD46 interaction might help in the design of small peptides or molecules to inhibit Ad35 infection in immunosuppressed patients.
Acknowledgments
This work was supported by NIH grants HLA078836 (A.L.) and AI065429 (D.S.); a grant from Academia Sinica, Taiwan (Y.-C.L.); and grants from the Deutsche Forschungsgemeinschaft (SFB685 and STE1163) (T.S.). Portions of this research were carried out at the National Synchrotron Radiation Research Center, a national user facility supported by the National Science Council of Taiwan, Republic of China. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine.
O.K. designed, performed, and analyzed the Biacore studies.
We thank Roland Strong (FHCRC, Seattle, WA) for providing access to Biacore analysis.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 2.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 3.Cadwell, R. C., and G. F. Joyce. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136—S140. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, R. C., and G. F. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28-33. [DOI] [PubMed] [Google Scholar]
- 5.Fleischli, C., S. Verhaagh, M. Havenga, D. Sirena, W. Schaffner, R. Cattaneo, U. F. Greber, and S. Hemmi. 2005. The distal short consensus repeats 1 and 2 of the membrane cofactor protein CD46 and their distance from the cell membrane determine productive entry of species B adenovirus serotype 35. J. Virol. 79:10013-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaggar, A., D. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 7.Gaggar, A., D. M. Shayakhmetov, M. K. Liszewski, J. P. Atkinson, and A. Lieber. 2005. Localization of regions in CD46 that interact with adenovirus. J. Virol. 79:7503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guex, N., and M. C. Peitsch. 1997. Swiss-Model and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson, D. J., A. Segerman, K. Lindman, Y. F. Mei, and G. Wadell. 2006. The Arg279Gln [corrected] substitution in the adenovirus type 11p (Ad11p) fiber knob abolishes EDTA-resistant binding to A549 and CHO-CD46 cells, converting the phenotype to that of Ad7p. J. Virol. 80:1897-1905. [Author's correction, 80:5101.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hierholzer, J. C. 1992. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 5:262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, J. S., and J. A. Engler. 1996. Domains required for assembly of adenovirus type 2 fiber trimers. J. Virol. 70:7071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 14.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 74:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kissinger, C. R., D. K. Gehlhaar, B. A. Smith, and D. Bouzida. 2001. Molecular replacement by evolutionary search. Acta Crystallogr. D 57:1474-1479. [DOI] [PubMed] [Google Scholar]
- 16.Myszka, D. G. 1999. Improving biosensor analysis. J. Mol. Recognit. 12:279-284. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. J. Carter and R. M. Sweet (ed.), Methods in enzymology: macromolecular crystallography. Academic press, San Diego, CA. [DOI] [PubMed]
- 18.Persson, B. D., D. M. Reiter, M. Marttila, Y. F. Mei, J. M. Casasnovas, N. Arnberg, and T. Stehle. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14:164-166. [DOI] [PubMed] [Google Scholar]
- 19.Roelvink, P. W., G. Mi Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai, F., S. Murakami, K. Kawabata, N. Okada, A. Yamamoto, T. Seya, T. Hayakawa, and H. Mizuguchi. 2006. The short consensus repeats 1 and 2, not the cytoplasmic domain, of human CD46 are crucial for infection of subgroup B adenovirus serotype 35. J. Control. Release 113:271-278. [DOI] [PubMed] [Google Scholar]
- 21.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 77:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sirena, D., B. Lilienfeld, M. Eisenhut, S. Kalin, K. Boucke, R. R. Beerli, L. Vogt, C. Ruedl, M. F. Bachmann, U. F. Greber, and S. Hemmi. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone, D., and A. Lieber. 2006. New serotypes of adenoviral vectors. Curr. Opin. Mol. Ther. 8:423-431. [PubMed] [Google Scholar]
- 27.Tuve, S., H. Wang, C. Ware, Y. Liu, A. Gaggar, K. Bernt, D. Shayakhmetov, Z. Li, R. Strauss, D. Stone, and A. Lieber. 2006. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J. Virol. 80:12109-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 29.Wadell, G. 1984. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110:191-220. [DOI] [PubMed] [Google Scholar]
- 30.Wadell, G., M. L. Hammarskjold, G. Winberg, T. M. Varsanyi, and G. Sundell. 1980. Genetic variability of adenoviruses. Ann. N. Y. Acad. Sci. 354:16-42. [DOI] [PubMed] [Google Scholar]
- 31.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]