Abstract
In order to facilitate the generation of mutant viruses of varicella-zoster virus (VZV), the agent causing varicella (chicken pox) and herpes zoster (shingles), we generated a full-length infectious bacterial artificial chromosome (BAC) clone of the P-Oka strain. First, mini-F sequences were inserted into a preexisting VZV cosmid, and the SuperCos replicon was removed. Subsequently, mini-F-containing recombinant virus was generated from overlapping cosmid clones, and full-length VZV DNA recovered from the recombinant virus was established in Escherichia coli as an infectious BAC. An inverted duplication of VZV genomic sequences within the mini-F replicon resulted in markerless excision of vector sequences upon virus reconstitution in eukaryotic cells. Using the novel tool, the role in VZV replication of the major tegument protein encoded by ORF9 was investigated. A markerless point mutation introduced in the start codon by two-step en passant Red mutagenesis abrogated ORF9 expression and resulted in a dramatic growth defect that was not observed in a revertant virus. The essential nature of ORF9 for VZV replication was ultimately confirmed by restoration of the growth of the ORF9-deficient mutant virus using trans-complementation via baculovirus-mediated gene transfer.
Varicella-zoster virus (VZV), the causative agent of chicken pox (varicella) and shingles (herpes zoster), is a highly cell-associated herpesvirus both in vitro and in vivo (7, 23). Similar to the situation with a closely related alphaherpesvirus, Marek's disease virus (MDV), VZV spreads directly from cell to cell, presumably utilizing the machinery involved in adherens junction modeling and architecture. Infectious virus is released into the environment only after lytic VZV replication in the skin and respiratory mucous membranes of infected individuals (2, 43). Efficient and timely coordinated interaction between the tegument, a proteinaceous structure surrounding the icosahedral nucleocapsid, and envelope membrane (glyco) proteins plays a crucial role in the secondary envelopment and spread of herpesviruses in general and VZV in particular (35). It has been shown for related viruses, such as herpes simplex virus, that the inner layer of tegument is tightly associated with the nucleocapsid, whereas the outer layer of tegument provides the link to envelope proteins (34, 36). The current model of herpesviral tegumentation predicts that the inner layer of tegument is added to the nucleocapsid in the nucleus and de-enveloped particles in the cytoplasm, while proteins representing the outer layer are thought to accumulate at cytoplasmic membranes where they make contact with their respective partners, either other tegument proteins or membrane proteins or both (34-36). Through these intricately regulated interactions, final secondary envelopment of particles is facilitated and, ultimately, infectious virus is produced and released.
Determination of the role of individual tegument or membrane (glyco)proteins in the replication cycle of various herpesviruses is not trivial, because in vitro, many of the functions appear redundant (34, 35). Analyses of viral mutants have shown, however, that similar sets of structural proteins are required for most alphaherpesviruses. VZV and MDV seem to be the exceptions to the rule. Not only do they share important biological properties with respect to growth in vitro and in vivo but they also exhibit striking similarities with respect to the membrane (glyco)proteins, as well as tegument proteins, that they absolutely require for replication. For example, both glycoprotein E (gE) and gI, which form a noncovalently linked complex in infected cells and virions, are essential for the growth of both viruses, at least in certain cell types, while they are dispensable in all other alphaherpesviruses tested (3, 8, 25, 37, 42, 50, 63, 65). In contrast, the major viral transactivator encoded by VZV ORF10 (MDV UL48) is dispensable for the growth of either virus, but is essential for most related viruses (17, 19, 20, 58, 61). In this context, it must be noted that the MDV UL49 product, the ORF9 homologue of VZV, was shown to be absolutely required for the growth of the chicken virus. The facts that this tegument protein was shown to physically interact with gE in the case of related herpesviruses and that gE is essential for both VZV and MDV (4, 21, 29, 30, 38, 50) prompted us to investigate the role of the ORF9 product in the VZV life cycle.
Since mutagenesis of herpesviruses using standard techniques that are based on homologous recombination in cultured cells by default results in a steady positive selection for viral replication, manipulation of genes that are crucial for viral replication in noncomplementing cells is virtually impossible. In addition, propagation in cultured cells increases the risk of introducing compensatory mutations within the viral genome. The generation of stable trans-complementing cell culture systems for virus mutagenesis is not always possible, because herpesviral genes are expressed at a defined stage of viral replication, at a certain level, and can even be toxic if expressed in the absence of other viral proteins. These drawbacks of conventional mutagenesis are even more dominant in the case of slowly replicating, highly cell-associated viruses, making it virtually impossible to transfer virus from trans-complementing to noncomplementing cells. As the first step out of this predicament, herpesviral genomes were cloned as overlapping cosmid clones in Escherichia coli, which makes all the bacterial methods for sequence modification available but is also prone to selective pressure, at least at the sites of recombination of the individual cosmids during virus reconstitution (15, 48, 57, 59). In the case of a replication-deficient virus reconstituted from a cosmid clone, it is virtually impossible to discriminate between failure of cosmid recombination, a lack of trans-complementation, the essential nature of the manipulated sequence, or any unwanted fatal second-site mutations within viral sequences. As an improvement over cosmid cloning and a logical next step, a number of herpesvirus genomes, including the VZV genome, were previously cloned as bacterial artificial chromosomes (BAC). Full-length viral DNA sequences can be maintained in E. coli and manipulated with targeted or random techniques, and the final mutant viruses are derived immediately after transfection of permissive cells (1, 5, 13, 32, 33, 40, 47, 49, 52, 53).
Herpesvirus genomes range from approximately 100 to 300 kb in size and are densely packed with open reading frames totaling ∼70 to ∼220 genes. They also contain direct and indirect repeat sequences and sequence duplications (10, 31, 60). Having such optimized genomes also means that overlapping regulatory sequences are found within open reading frames, along with polycistronic transcription units, expression of multiple splice variants, antisense RNAs, and microRNAs (11, 12, 22, 26, 45, 46). To avoid collateral and spurious effects of herpesvirus mutagenesis, the need for minimal sequence modifications emerges. Such techniques, in addition to the desirable removal of previously introduced mini-F sequences that are necessary for maintenance in bacteria, should result in markerless modification or at least in modifications that will result in the introduction of only very small sequences, such as frt or loxP sites (59).
We here report on the generation of an infectious clone of the intensively characterized and fully sequenced VZV strain P-Oka, which served as the starting point for the generation of the widely used V-Oka vaccine strain (54). The infectious BAC was generated by utilizing previously generated cosmid clones that were modified using the Red recombination technique (39, 41, 64). A recently developed method that allows markerless modification of cloned DNA in E. coli was used to introduce a sequence duplication into the mini-F vector, which results in the removal of all vector sequences upon virus reconstitution in eukaryotic cells (55). Finally, mutants that were unable to express the major tegument protein encoded by ORF9 were generated, which were unable to replicate unless the protein was provided in trans, indicating that this major regulatory and structural tegument protein is essential for VZV replication.
MATERIALS AND METHODS
Viruses and cells.
Wild-type P-Oka and mutant viruses were propagated and passaged in human melanoma cells (MeWo) grown in Dulbecco's minimal essential medium or in minimal essential medium (MEM) containing 10% fetal bovine serum (FBS) and a mixture of penicillin and streptomycin (Invitrogen). Virus was passaged and amplified by cocultivation of infected with uninfected cells at ratios of 1/2 to 1/5. Virus stocks were frozen down in growth media supplemented with 8% dimethyl sulfoxide and stored in liquid nitrogen.
Construction of transfer vectors.
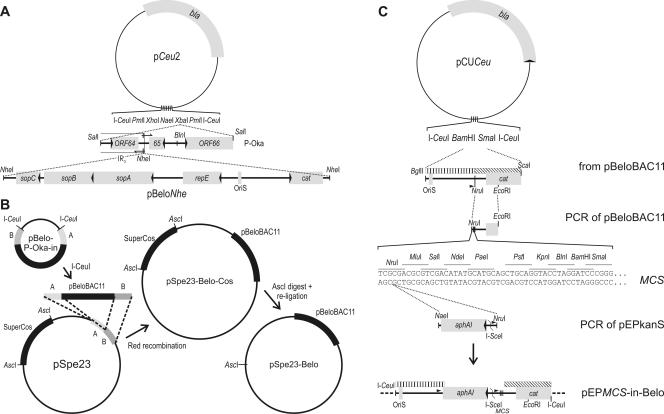
For the construction of the mini-F transfer vector, plasmid pUC18 was PCR amplified with primers 5′-CTTCTA GAC ACG TGT TCG CTA CCT TAG GAC CGT TAT AGT TAC GAG TAA TCA TGG TCA TAG CTG TTT C-3′ and 5′-TGTCTA GAA GCC GGC ACT CGA GCA CGT GTT CGC TAC CTT AGG ACC GTT ATA GTT ACG TCA CTG GCC GTC GTT TTA C-3′, cleaved with XbaI, and religated to derive pCeu2. The 2.6-kb SalI fragment of the rightmost cosmid, pSpe23 (41), was ligated into the XbaI site of pCeu2. The entire construct was amplified via inverse PCR using primers 5′-AGAGCT AGC GGA TAA AAG CGT ACT GTT TTT TAT T-3′ and 5′-AGAGCT AGC CGG TGT ATG TTT TAA ATT TAT T-3′, inserting an NheI restriction site between the poly(A) sites of ORF64 and ORF65. The resulting plasmid was termed pCeu2.6Nhe (Fig. 1A).
FIG. 1.
Schematic of vector construction. (A) pBelo-P-Oka-in. In a first step, recombinant plasmid pCeu2 was constructed by inverse PCR and self-ligation of pUC18, modifying the MCS. The 2.6-kb SalI fragment of P-Oka was cloned into the engineered XbaI site. Another inverse PCR/ligation step was used to insert an NheI site close to the borders of internal repeat short (IRS) and unique short sequences. The mini-F replicon, pBeloNhe, was linearized and cloned into the newly inserted NheI site. (B) pSpe23-Belo. The mini-F replicon with adjoining flanks A and B was released from plasmid pBelo-P-Oka-in by cleavage with I-CeuI and inserted into pSpe23 via Red recombination (55). Subsequently, SuperCos sequences were removed by AscI digestion and self-ligation. (C) pEPMCS-in-Belo. The 1.7-kb BglII-ScaI fragment of pBeloBAC11 was cloned into pCUCeu (56). A small portion of the pBelo sequence was replaced by a PCR product to introduce an MCS. A PCR was performed to add NaeI and NruI sites and a small sequence duplication (arrowheads) to the aphAI-I-SceI cassette. Cloning of that fragment into the NruI site of the MCS resulted in the en passant-ready transfer vector pEPMCS-in-Belo. The pBelo flanks used for Red recombination with pBelo-derived BACs are indicated with striped bars.
To introduce an NheI site into the mini-F vector pBeloBAC11 (New England Biolabs) and to remove its cos and loxP sequences, the mini-F plasmid was cleaved with SalI and the 6.4-kb fragment was ligated with the XhoI-treated oligo-homodimer of primer 5′-AGCCTC GAG CTA GCT CGA GGC TA-3′. The derived plasmid, pBeloNhe, was cleaved with NheI and cloned into the NheI site of pCeu2.6Nhe, resulting in recombinant plasmid pBelo-P-Oka-in (Fig. 1A).
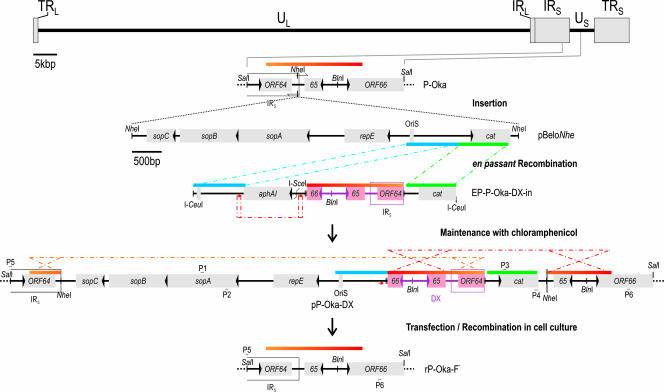
The pEP-P-Oka-DX-in transfer vector for insertion of duplicated sequences into the mini-F plasmid was generated as follows. First, a 1.7-kb BglII-ScaI-fragment was cloned via BamHI/SmaI into pCUCeu (56). The segment flanked by NruI and EcoRI was replaced by a PCR product (5′-TACGGA ATT CCG GAT GAG CAT TC-3′ and 5′-CCCGGG ATC CTG CAG TCG ACA CGT GGC GCG CCT AGG TAT AAA TAC CTG TGA CGG AAG ATC AC-3′) of the equivalent sequence extended with a multiple cloning site (MCS). Then, an aphAI-I-SceI PCR product derived from pEPkan-S using primers 5′-GGC GCG CCG GCC AGT GTT ACA ACC AAT TAA CC-3′ and 5′-TCGCGA TAA GCT CAT GGA GCG GCG TAA CCG TCG CAC AGG AAG GAC AGA GAT AGG GAT AAC AGG GTA ATC GAT TT-3′, which included a short duplication of pBeloBAC11 sequences, was inserted into a singular NruI site (55), ultimately resulting in the universal transfer vector pEPMCS-in-Belo (Fig. 1C). A 1.6-kb PCR fragment of P-Oka (primers 5′-ACTAGT GCT TCT GCG CAC AAT GCC ACA G-3′ and 5′-ACTAGT GAA CGG TAC TTC GCC GCC GCG C-3′) was cloned into BlnI of the MCS to derive pEP-P-Oka-DX-in (Fig. 2).
FIG. 2.
Insertion and self-excision of mini-F replicon. Schematic of the insertion of pBelo sequences between ORF64 and ORF65. A 1.6-kb sequence of P-Oka was inserted between the bacterial origin of replication (OriS) and cat to create an inverse duplication of VZV sequences adjoining the mini-F replicon. The orange-red gradient bar indicates such duplicate sequences and their orientation. Dot-and-dash lines indicate recombination events of identical sequences (same-colored bars) for a first Red recombination and of short duplications (red arrow) for the removal of the kanamycin resistance gene aphAI. The BAC with duplicated sequences was termed pP-Oka-DX and maintained in E. coli under steady chloramphenicol selection. Upon transfection of MeWo cells, two anti-parallel recombination events (orange and red dot-and-dash lines) released mini-F sequences and resulted in rP-Oka-F−. Primers P1 (5′-TTAACT CAG TTT CAA TAC GGT GCA G-3′) and P2 (5′-TGGGGT TTC TTC TCA GGC TAT C-3′) were used to screen for the presence of mini-F sequences, and P3 (5′-AGGCAT TTC AGT CAG TTG CTC-3′) and P4 (5′-TGCCAC TCA TCG CAG TAC TG-3′) were used to screen for the presence of cat. The loss of the pBelo insert was detected by utilizing primers P5 (5′-ATTGGA AGC GAC GTC GAC AC-3′) and P6 (5′-TGTAAC GCG CGT AAT ACA GAT CG-3′). TRL, terminal repeat long; UL, unique long; IRL, internal repeat long; IRS, internal repeat short; US, unique short; TRS, terminal repeat short.
Red recombination.
Red recombination was performed exactly as described above, using E. coli strain EL250 harboring the respective cosmid or BAC DNA (27, 55). All primers used for mutagenesis were polyacrylamide gel electrophoresis purified (Integrated DNA Technologies [IDT]). To perform en passant mutagenesis, a linear DNA fragment (PCR products using pEPkan-S as a template or a linearized plasmid) was used to introduce a cassette of aphAI, an I-SceI site, and a short sequence duplication into the target sequence in a first step. The resulting intermediates were selected with 30 μg/ml kanamycin and 30 μg/ml chloramphenicol on agar plates. An I-SceI-expressing plasmid (pBAD-I-sceI) was then introduced into EL250 harboring desired cointegrates, the expression of the homing endonuclease was induced with 0.5% arabinose, and a second Red recombination was performed. The loss of the aphAI-I-SceI cassette was detected via replica plating and restriction fragment length polymorphism (RFLP) analysis, and the identities of the introduced mutations were confirmed by nucleotide sequencing (55).
Reconstitution of recombinant VZV from cosmid DNA.
Recombinant pP-Oka-derived VZV was reconstituted by transfecting human melanoma (MeWo) cells with pSpe23-Belo and the three other cosmid clones, pFsp73, pSpe14, and pPme2, using a CalPhos mammalian transfection kit (Clontech) exactly as previously described (13, 41).
Generation of ORF9 mutants by en passant mutagenesis.
Primers 5′-TTTGGA TAT TTC ACG ACC CTA TCG TTT ATT TAC GTG CCG GCA TCT TCC GAC GGT GAT AGG GAT AAC AGG GTA ATC GAT TT-3′ and 5′-TTAGAG CGA CAA AGT CTG TCA CCG TCG GAA GAT GCC GGC ACG TAA ATA AAC GAT AGG CCA GTG TTA CAA CCA ATT AAC C-3′ were used to amplify the aphAI-I-SceI cassette present in pEPkan-S. The pP-Oka9ΔM mutants resulting from the en passant procedure were analyzed by RFLP and sequencing (data not shown). Using primers 5′-TGGATA TTT CAC GAC CCT ATC GTT TAT TTA CGT AAT GGC TAG CTC CGA CGG TGA CAT AGG GAT AAC AGG GTA ATC GAT TT-3′ and 5′-TGCATT AGA GCG ACA AAG TCT GTC ACC GTC GGA GCT AGC CAT TAC GTA AAT AAA CGG CCA GTG TTA CAC CAA TTA ACC-3′, a revertant clone from pP-OkaΔ9M, termed pP-Oka9Mr, was generated by en passant mutagenesis as well. The repaired ORF9 coded for the wild-type amino acid sequence but harbored an NheI site close to its start codon, which allowed unequivocal identification via RFLP.
Reconstitution of VZV from BAC DNA.
BAC DNA for transfections was prepared using affinity chromatography following the manufacturer's instructions (DNA Maxiprep system; QIAGEN). Subsequently, MeWo cells were transfected with Lipofectamine 2000 (Invitrogen). Briefly, 1 to 4 μg BAC DNA was cotransfected with 200 ng pCMV62, a plasmid which contains the VZV immediate early (IE) gene ORF62 under the control of a cytomegalovirus (CMV) promoter (44). The DNA-Lipofectamine solution was added into the cells in 1 ml of MEM and incubated for 4 h. The transfection medium was then replaced with MEM-10% FBS.
Immunofluorescence.
Cells were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.1% saponin, and blocked with 10% neonatal calf serum in PBS. Primary antibodies, monoclonal anti-ORF62 antibody (Chemicon), monoclonal anti-gB antibody (US Biological), and ORF-9 polyclonal antibody (6) (a kind gift of William Ruyechan, SUNY Buffalo, NY), were each diluted 1:500 in PBS containing 10% neonatal calf serum and 0.02% sodium azide. Secondary antibodies, including Alexa 568-conjugated goat anti-rabbit and Alexa 488-conjugated goat anti-mouse antibodies, were diluted 1:1,000 in PBS. Samples were examined under a fluorescence microscope and photographed with a digital camera (Zeiss Axiovert 25 and Axiocam).
Multistep growth kinetics.
The multistep growth kinetics of P-Oka-, rP-Oka-, rP-Oka9ΔM-, and rP-Oka9Mr-derived viruses were determined by inoculating 1 × 106 MeWo cells with 100 PFU of cell-associated virus. Infected cells were trypsinized on days 1, 2, 3, 4, and 5 and titrated by cocultivation of 10-fold dilutions of trypsinized cells with fresh MeWo cells. Five days after coseeding, viral plaques were stained by indirect immunofluorescence using anti-gB antibodies (US Biological) and counted, exactly as described elsewhere (56). All titer determinations were done in triplicate.
Generation of recombinant baculoviruses expressing ORF9.
A shuttle plasmid for the generation of recombinant baculoviruses by Tn7 transposition was generated as described previously (9). Briefly, the polyhedron promoter of the shuttle plasmid pFastBac1 (Invitrogen) was removed by digestion with SnaBI and HpaI and replaced with a 3.1-kb NruI-Bst1107I fragment from pcDNA3, which contains the human CMV (HCMV)-IE promoter/enhancer, the MCS, and the polyadenylation signal followed by the simian virus 40 promoter-neomycin phosphotransferase II expression cassette. The resulting plasmid was named pFastM. ORF9 was inserted into the MCS of pFastM using BamHI and XhoI restriction sites, resulting in the recombinant plasmid pFastMORF9.
The expression cassette of pFastMORF9 was incorporated into the baculovirus genome by using Tn7 site-specific transposition according to the manufacturer's instructions with the Bac-to-Bac expression system (Invitrogen). The resulting bacmid DNA was purified and transfected into Sf9 cells using Cellfectin (Invitrogen). After incubation at 27°C for 4 days, the resulting recombinant baculovirus, bMORF9, was propagated in Sf9 cells and viral titers were determined in a conventional plaque assay using neutral red agarose overlays (9). One milliliter of bMORF9-containing supernatant was used for infection of MeWo cells seeded in six-well-plates (25 PFU/cell). Incubation was allowed for 30 min at room temperature, the cells were centrifuged for 5 min at 200 × g, and finally, 10 mM butyrate was added to the cells to facilitate transgene expression (9). After 4 h of incubation, cells were washed and 2 ml of Dulbecco's minimal essential medium-10% FBS was applied.
RESULTS
Generation of an infectious P-Oka BAC clone.
The generation of the P-Oka BAC was based on overlapping cosmid clones that had been established earlier (41). Mini-F vector sequences were inserted into one of the P-Oka cosmids via Red recombination. Briefly, the pSpe23 cosmid was electroporated into E. coli strain EL250 (27) and the recombinant plasmid pBelo-P-Oka-in (Fig. 1B) was linearized with I-CeuI. The 9.4-kbp fragment containing linear mini-F sequence and sequences flanking the junction between the VZV internal repeat short and unique short was electroporated into recombination-competent EL250 harboring pSpe23 (27, 55). Chloramphenicol-resistant bacteria were selected and screened by RFLP analyses for correct insertion of the pBeloBAC sequences into the rightmost P-Oka cosmid (data not shown). The derived construct, pSpe23-Belo-Cos, was cleaved with AscI to remove the SuperCos sequences and religated, resulting in pSpe23-Belo (Fig. 1B). The generated mini-F clone, which now was located within the rightmost P-Oka sequences, was transfected with the other overlapping cosmid clones to reconstitute a P-Oka virus mutant with the mini-F insertion. Total cellular and viral DNA isolated from MeWo cells infected with the P-Oka-BAC virus was cleaved with the enzymes AscI, which is unable to cleave P-Oka-BAC sequences, and λ exonuclease to digest cellular DNA. The remaining P-Oka-BAC DNA (pP-Oka) was precipitated and electroporated into EL250 cells. Bacteria containing the full-length VZV genome were selected with chloramphenicol and screened by RFLP analysis (Fig. 3A).
FIG. 3.
(A) Southern blot of P-Oka, pP-Oka, pP-Oka-DX, and rP-Oka-F−. BAC DNA was prepared from E. coli harboring pP-Oka or pP-Oka-DX. MeWo cells were infected with the parental virus P-Oka or with the recombinant rP-Oka-F− and harvested when >80% infected, and total DNA was prepared. BAC DNA (10 μg) or viral DNA (20 μg) was digested with BamHI or NcoI, separated on a 1% agarose gel, and transferred to a nylon membrane. VZV BAC-specific bands were detected using random digoxigenin-labeled (DIG DNA labeling kit; Roche), sonicated pP-Oka DNA. (B) rP-Oka has growth kinetics that are very similar to those of wild-type P-Oka. Multistep growth kinetics of P-Oka and rP-Oka were determined using MeWo cells. Means and standard deviations of the results of three independent experiments are given.
Reconstitution and characterization of recombinant pP-Oka-derived VZV.
In order to demonstrate that the insertion of the mini-F sequence in the VZV genome did not have a detrimental effect on viral replication in vitro, recombinant rP-Oka was reconstituted from the full-length infectious clone (pP-Oka) and growth kinetics were determined. Human melanoma cells (MeWo) were transfected with pP-Oka clones by lipofection. The efficiency was increased by cotransfection with the pCMV62 plasmid (44). Transfected cells were analyzed by immunofluorescence microscopy to detect plaque formation as early as 4 days posttransfection. The determination of the multistep growth kinetics revealed that BAC-derived rP-Oka exhibited growth properties that were virtually identical to those of the parental, wild-type P-Oka virus (Fig. 3B). Therefore, neither the increased size of the viral genome nor the insertion of the mini-F sequences into the chosen genomic locus had measurable effects on viral replication.
Self-excision of mini-F sequences upon rP-Oka reconstitution.
Mini-F sequences of pP-Oka were removed by employing a stabilized inverse sequence duplication. The self-excision is based on the duplication within the mini-F of a VZV sequence that is present adjacent to the mini-F insertion site. The duplication was introduced between the origin of replication and the antibiotic resistance gene (Fig. 2) by first constructing an en passant-ready transfer construct, pEP-P-Oka-DX-in from pEPMCS-in-Belo (43, 55) (Fig. 1C). A 4.5-kb I-CeuI fragment was used to insert the 1.6-kb sequence duplication into mini-F sequences of pP-Oka via en passant mutagenesis in inverse orientation. The resulting BAC, pP-Oka-DX, was maintained in E. coli by standard chloramphenicol selection. The loss of vector sequences in E. coli through intramolecular recombination is prevented by the inverse orientation of the internal sequence duplication and its placement between the chloramphenicol resistance gene cat and the mini-F origin OriS (data not shown). Also, recombination of the duplication and identical sequences present in the terminal repeat short would separate the resistance gene and the mini-F replicon, which would be lethal to the bacteria. Consequently, pP-Oka-DX was stable in E. coli due to the constant selective pressure for plasmid replication and chloramphenicol resistance.
After stable maintenance of pP-Oka-DX was established in E. coli, MeWo cells were transfected with pP-Oka-DX. Since there is no positive selection for maintenance of the mini-F replicon or chloramphenicol resistance in eukaryotic cells, intra- and intermolecular recombination leading to the removal of the chloramphenicol resistance gene, the mini-F replicon, and the duplicated sequences can take place. Reconstituted rP-Oka-F− virus reconstituted from the pP-Oka-DX BAC was passaged and harvested for DNA preparation, and the loss of the mini-F sequences was confirmed via PCR and sequencing (Fig. 2 and 4). After the first passage of rP-Oka-F−, the absence of the pBeloBAC cassette was observed for the majority of genomes, but a small fraction of viral genomes still harbored cat or the mini-F replicon until passage four. The rP-Oka-F− genome was also analyzed via Southern blot analysis, and its restriction enzyme pattern was indistinguishable from that of P-Oka DNA (Fig. 3A).
FIG. 4.
PCR analysis of mini-F self-excision upon transfection. MeWo cells were transfected with pP-Oka-DX. Reconstituted virus was passaged by mixing infected and uninfected cells at a ratio of 1 to 5. DNA from virus passages was used to perform a PCR with primers P1 and P2 (top panel) or P3 and P4 (middle panel) to detect remaining pBelo sequences in viruses reconstituted from pP-Oka-DX. Primers P5 and P6 were used to show the restoration of the P-Oka wild-type genomic organization in rP-Oka-F− (see also Fig. 2). pP-Oka-DX DNA isolated from E. coli, as well as total DNA of MeWo cells infected with P-Oka or uninfected MeWo cells, were used as controls. Mini-F sequences are not detectable after five passages, and in a significant portion of reconstituted virus, the pBelo insert is lost immediately upon transfection. Primer sequences are given in the Fig. 2 legend. Numbers to the left indicate molecular size in base pairs.
Characterization of an ORF9-deficient VZV mutant.
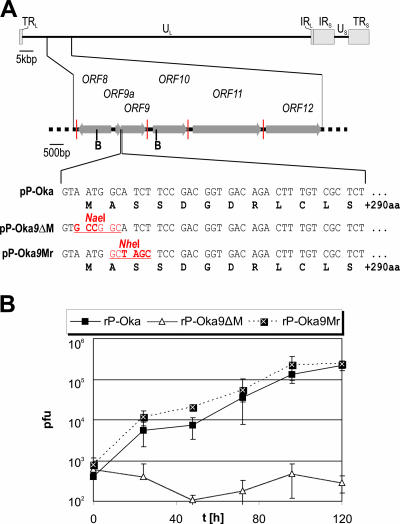
To create a P-Oka mutant unable to express the ORF9 protein, the start codon of the corresponding open reading frame (ORF9) was inactivated by markerless en passant mutagenesis (55). A revertant was also generated in which the start codon was repaired and an NheI site was introduced to allow unequivocal identification of the revertant (Fig. 5A). MeWo cells were transfected with pP-Oka DNA, DNA of the start codon mutant pP-Oka9ΔM, or the revertant pP-Oka9Mr to reconstitute the respective recombinant viruses (rP-Oka, rP-Oka9ΔM, and rP-Oka9Mr). Upon reconstitution, rP-Oka9ΔM exhibited drastically decreased plaque sizes in comparison to those of parental rP-Oka and the revertant virus pP-Oka9Mr (Fig. 6A). Immunofluorescence analysis using ORF9-specific polyclonal antibodies revealed that, as expected, rP-Oka9ΔM does not express ORF9 (Fig. 6A, upper panel) (6). On the other hand, ORF62 could be readily detected in the parental, the ORF9 deletion, and the revertant virus (Fig. 6A, lower panel). In order to confirm the dramatic growth defect of rP-Oka9ΔM, the multistep growth kinetics were determined. In contrast to rP-Oka9ΔM, whose growth curve stayed flat, the rP-Oka9Mr revertant virus, in which the start codon of ORF9 had been repaired, grew with kinetics that were virtually indistinguishable from those of wild-type P-Oka and the parental rP-Oka virus (Fig. 5B). Since rP-Oka9ΔM showed no replication that would be sufficient for virus propagation leading to self- excision of mini-F sequences, the characterization of ORF9 was performed using recombinant viruses still harboring pBeloBAC sequences.
FIG. 5.
Growth properties of an ORF9-negative VZV. (A) Genomic region of VZV ORF9 with a focus on the amino-terminal sequence of ORF9 is shown. The original nucleotide (light font) and amino acid sequences (bold black font) are given, as are the mutations introduced in pP-Oka9ΔM and its revertant (bold red font). Grey arrows, open reading frames; vertical red lines, polyadenylation signals; B, BamHI restriction sites; red underlined font, resulting restriction sites. (B) Multistep growth kinetics of rP-Oka, rP-Oka9ΔM, and rP-Oka9ΔMr. Kinetics determinations were performed in triplicate, and means and standard deviations (error bars) of the results are given. TRL, terminal repeat long; UL, unique long; IRL, internal repeat long; IRS, internal repeat short; US, unique short; TRS, terminal repeat short; aa, amino acids.
FIG. 6.
Immunofluorescence analysis and trans-complementation via baculovirus-mediated gene transfer. (A) Plaque analysis of rP-Oka, rP-Oka9ΔM, and rP-Oka9Mr. Plaques were analyzed by indirect immunofluorescence using anti-ORF9 polyclonal (6) and anti-ORF62 monoclonal (Chemicon, United States) antibodies at day 7 posttransfection. (B) Plaque analysis of rP-Oka, rP-Oka9ΔM, and rP-Oka9ΔM rescued with baculovirus gene delivery (right panel) at day 2 posttransfection. Plaques were analyzed via indirect immunofluorescence using anti-gB monoclonal antibody (US Scientific) at day 7 posttransfection. Scale bars are given in individual panels.
To confirm that the impaired growth was due only to the lack of ORF9 expression, we used baculovirus-mediated gene delivery to complement the growth of rP-Oka9ΔM. Baculoviruses have been shown to be an effective vehicle for the introduction of DNA sequences into mammalian cells since they neither replicate nor cause cytopathic effects (24). MeWo cells were transfected with pP-Oka9ΔM DNA and infected at 1, 2, 3, 4, or 5 days posttransfection with a recombinant baculovirus expressing ORF9 under the control of the HCMV-IE promoter. After BAC transfection and baculovirus infection, the cells were analyzed via immunofluorescence using anti-gB and anti-ORF9 antibodies at day 7 posttransfection (6). The trans-complementation via baculoviruses at days 1, 2, and 3 posttransfection resulted in replication and, consequently, significantly increased plaque sizes of rP-Oka9ΔM (Fig. 6B).
DISCUSSION
In this study, we demonstrated that the generation of a full-length BAC directly from overlapping cosmid clones is possible. We showed that Red mutagenesis could prove an efficient means to transfer a second bacterial origin of replication, here the mini-F replicon, into a preexisting cosmid clone. Having the BAC cassette within viral sequences ahead of virus reconstitution from the cosmids produces VZV BAC genomes without the need for (mostly inefficient) selection in cell culture. Furthermore, the removal of contaminating cellular chromosomal DNA from BAC DNA using restriction enzyme cleavage and λ exonuclease digestion before electroporation of the infectious VZV genome into E. coli was found to be vastly more efficient than standard methods of DNA preparation (33, 49). The strategy as described here, therefore, has the potential to be widely applicable for the cloning of herpesvirus genomes from already established cosmid clone systems, particularly with regard to highly cell-associated viruses.
Whenever bacterial sequences are used to manipulate viral genomes, the overall effect of introduced, foreign sequence scars, like the mini-F cassette itself, for viral replication is not predictable. The newly developed system applying an inversely oriented sequence duplication between the selection marker (antibiotic resistance gene) and the bacterial replicon allows stable maintenance of the herpesviral genome in E. coli. It also presents an in-cis tool to remove the mini-F cassette upon transfection. In the particular case described here, four passages in cultured cells were required to completely remove mini-F sequences from recombinant genomes as assayed by PCR. The highly cell-associated nature and slow replication kinetics of VZV, which could account for polyclonal growth of VZV within the same infected cell, could be one possible explanation for the maintenance of a few mini-F-containing genomes in the first few cell culture passages. In addition, the lack of a growth advantage of recombinant virus devoid of mini-F sequences (rP-Oka-F−) over viruses still carrying vector (rP-Oka-DX) could also contribute to the detection of mini-F sequences until passage four. Placing mini-F sequences within a gene that is essential for virus replication or using the developed methodology in faster-replicating viruses producing cell-free infectivity would increase the efficiency of self-excision of vector sequences (F. Wussow and B. K. Tischer, unpublished data). The combination of the VZV-BAC clone at hand, the established techniques to perform markerless mutagenesis in E. coli, and the construction of a self-excisable BAC allows the creation of clonal virus progeny with minimal sequence modification.
Similar to the situation with MDV, the generation of the full-length P-Oka BAC clone from the already-established system of overlapping cosmid clones allowed us to address genes that likely were crucial for VZV growth in cell culture. We were able to characterize the role of ORF9 in VZV replication. Immunofluorescence analyses of rP-Oka9ΔM with an ORF9-specific monoclonal antibody showed the lack of ORF9 production in infected cells, which were, however, reactive with an anti-ORF62 antibody. The repair of the ORF9 start codon restored ORF9 expression, as well as virus growth in cell culture, to levels observed with the parental virus (Fig. 6A). Complementation via baculovirus-mediated gene delivery was also able to rescue productive viral replication. These results clearly demonstrated that ORF9 is essential for the productive replication of VZV in cell culture, as is the case for MDV but unlike the situation with related human or animal alphaherpesviruses (14, 17, 18, 28). That supports the theory that highly cell-associated alphaherpesviruses mainly utilize the UL49-gE/gI pathway to spread from cell to cell, whereas the loss of the UL49 homologue in herpesviruses that produce cell-free virions is less crucial for virus propagation. The characterization of UL49-negative mutants of less cell-associated viruses allows the analysis of other, nonstructural functions of that tegument protein as well (51, 62), while with VZV and MDV, such a detailed characterization would required a very detailed dissection of respective functional domains. In addition, the highly cell-associated nature of VZV makes it virtually impossible to work with phenotypically rescued mutant viruses because such viruses cannot be purified from, e.g., trans-complementing cells.
Complementation of rP-Oka9ΔM with the baculovirus expression system, however, was able to restore virus growth and to significantly increase plaque sizes, albeit not to the levels of wild-type or revertant viruses. One explanation for the failure to completely restore VZV growth is that baculovirus-mediated gene expression decreases with time; therefore, the amounts of ORF9 produced may not be sufficient at the end of the experiment. Another reason might be that relatively low doses of recombinant baculovirus (multiplicity of infection, 25) were used, which resulted in the absence of ORF9 expression in a subset of cells and could limit the availability of ORF9 for replication of rP-Oka9ΔM. A low multiplicity of infection was chosen to prevent the overexpression of ORF9, which has been shown to lead to cytotoxic effects and likely is one of the reasons for the failure to generate permanent cell lines (B. B. Kaufer, B. K. Tischer, and N. Osterrieder, unpublished observations). Another explanation might be that ORF9 expression is controlled by the strong HCMV-IE promoter. This promoter is constitutively active, and transgene expression, therefore, cannot be modulated during the course of infection by virus-encoded transcription factors as is the case during regular VZV infection. Constant, not synchronized, high-level expression of ORF9 may be detrimental to virus replication, as has been suggested by the MDV system (16, 17).
In summary, we have generated an infectious herpesvirus (VZV) BAC clone from overlapping pieces of cloned viral DNA. Advances in focused and minimal sequence modification in bacteria allowed the characterization of the essential nature of ORF9 of the highly cell-associated herpesvirus VZV for in vitro replication. The established methodology is likely applicable to many other (herpes)viruses and has the advantages that the initial cloning step is independent of virus replication and that no selective pressure is applied because cosmid clones are usually derived from viral DNA packaged in nucleocapsids. Employing an inverse sequence duplication in the infectious BAC for cis recombination allowed scarless self-excision of the mini-F replicon, which not only is important for the generation of viruses and mutants free of foreign sequences but also represents an important step forward with respect to the recombineering of viral sequences in eukaryotic cells. In addition, since cosmid and mini-F plasmid origins of replication can be maintained in one and the same bacterial cell, as demonstrated here, the successive assembly of a full-length viral mini-F clone from overlapping cosmid clones in bacteria by applying Red recombination seems feasible and is currently being attempted. Viral genome assembly would avoid a eukaryotic stage in the generation of an infectious clone obtained without applying any positive or negative selection on the viral genomes.
Acknowledgments
The expert technical assistance of Neil G. Margulis is greatly appreciated. We thank Paul Kinchington, University of Pittsburgh, for providing plasmid pCMV62 and William Ruyechan, SUNY Buffalo, for the anti-ORF9 antibodies. We thank Helmut Fickenscher for inspiring discussions and careful perusal of the manuscript.
The study was supported by PHS grant 1R21AI061412 to N.O. F.W.'s work was funded by a grant from the medical faculty of Christian Albrecht University of Kiel (Research Center Integrative Neurobiology).
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin, A. M. 2006. Investigations of the pathogenesis of varicella zoster virus infection in the SCIDhu mouse model. Herpes 13:75-80. [PubMed] [Google Scholar]
- 3.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245-1258. [DOI] [PubMed] [Google Scholar]
- 4.Berarducci, B., M. Ikoma, S. Stamatis, M. Sommer, C. Grose, and A. M. Arvin. 2006. Essential functions of the unique N-terminal region of the varicella-zoster virus glycoprotein E ectodomain in viral replication and in the pathogenesis of skin infection. J. Virol. 80:9481-9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cilloniz, C., W. Jackson, C. Grose, D. Czechowski, J. Hay, and W. T. Ruyechan. 2007. The varicella-zoster virus (VZV) ORF9 protein interacts with the IE62 major VZV transactivator. J. Virol. 81:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, J. I. 1996. Varicella-zoster virus. The virus. Infect. Dis. Clin. N. Am. 10:457-468. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J. I., and H. Nguyen. 1997. Varicella-zoster virus glycoprotein I is essential for growth of virus in Vero cells. J. Virol. 71:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. J., D. J. Dargan, and N. D. Stow. 2002. Fundamental and accessory systems in herpesviruses. Antiviral Res. 56:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 13.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca, N. A., and P. A. Schaffer. 1985. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol. Cell. Biol. 5:1997-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219-2230. [DOI] [PubMed] [Google Scholar]
- 17.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grose, C., and T. I. Ng. 1992. Intracellular synthesis of varicella-zoster virus. J. Infect. Dis. 166(Suppl. 1):S7-S12. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Y. C., C. T. Tsai, Y. J. Chang, and J. H. Huang. 2003. Enhancement and prolongation of baculovirus-mediated expression in mammalian cells: focuses on strategic infection and feeding. Biotechnol. Prog. 19:373-379. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, H., S. E. Straus, and R. K. Williams. 1997. Varicella-zoster virus glycoproteins E and I expressed in insect cells form a heterodimer that requires the N-terminal domain of glycoprotein I. Virology 233:382-391. [DOI] [PubMed] [Google Scholar]
- 26.Krause, P. R., K. D. Croen, S. E. Straus, and J. M. Ostrove. 1988. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J. Virol. 62:4819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 28.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallory, S., M. Sommer, and A. M. Arvin. 1997. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J. Virol. 71:8279-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallory, S., M. Sommer, and A. M. Arvin. 1998. Analysis of the glycoproteins I and E of varicella-zoster virus (VZV) using deletional mutations of VZV cosmids. J. Infect. Dis. 178(Suppl. 1):S22-S26. [DOI] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 32.McGregor, A., and M. R. Schleiss. 2001. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol. Genet. Metab. 72:15-26. [DOI] [PubMed] [Google Scholar]
- 33.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9:423-429. [DOI] [PubMed] [Google Scholar]
- 37.Mijnes, J. D., L. M. van der Horst, E. van Anken, M. C. Horzinek, P. J. Rottier, and R. J. de Groot. 1996. Biosynthesis of glycoproteins E and I of feline herpesvirus: gE-gI interaction is required for intracellular transport. J. Virol. 70:5466-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffat, J., C. Mo, J. J. Cheng, M. Sommer, L. Zerboni, S. Stamatis, and A. M. Arvin. 2004. Functions of the C-terminal domain of varicella-zoster virus glycoprotein E in viral replication in vitro and skin and T-cell tropism in vivo. J. Virol. 78:12406-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagaike, K., Y. Mori, Y. Gomi, H. Yoshii, M. Takahashi, M. Wagner, U. Koszinowski, and K. Yamanishi. 2004. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 22:4069-4074. [DOI] [PubMed] [Google Scholar]
- 41.Niizuma, T., L. Zerboni, M. H. Sommer, H. Ito, S. Hinchliffe, and A. M. Arvin. 2003. Construction of varicella-zoster virus recombinants from parent Oka cosmids and demonstration that ORF65 protein is dispensable for infection of human skin and T cells in the SCID-hu mouse model. J. Virol. 77:6062-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olson, J. K., and C. Grose. 1998. Complex formation facilitates endocytosis of the varicella-zoster virus gE:gI Fc receptor. J. Virol. 72:1542-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osterrieder, N., J. P. Kamil, D. Schumacher, B. K. Tischer, and S. Trapp. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4:283-294. [DOI] [PubMed] [Google Scholar]
- 44.Perera, L. P., J. D. Mosca, W. T. Ruyechan, and J. Hay. 1992. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J. Virol. 66:5298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 46.Roizman, B., T. Kristie, J. L. McKnight, N. Michael, P. Mavromara-Nazos, and D. Spector. 1988. The trans-activation of herpes simplex virus gene expression: comparison of two factors and their cis sites. Biochimie 70:1031-1043. [DOI] [PubMed] [Google Scholar]
- 47.Rudolph, J., and N. Osterrieder. 2002. Equine herpesvirus type 1 devoid of gM and gp2 is severely impaired in virus egress but not direct cell-to-cell spread. Virology 293:356-367. [DOI] [PubMed] [Google Scholar]
- 48.Schaffer, P. A., S. K. Weller, B. A. Pancake, and D. M. Coen. 1984. Genetics of herpes simplex virus. J. Investig. Dermatol. 83:42s-47s. [DOI] [PubMed] [Google Scholar]
- 49.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schumacher, D., B. K. Tischer, S. M. Reddy, and N. Osterrieder. 2001. Glycoproteins E and I of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Virol. 75:11307-11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sciortino, M. T., B. Taddeo, M. Giuffre-Cuculletto, M. A. Medici, A. Mastino, and B. Roizman. 2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking solely the UL49 gene vary with respect to the defect in the UL41 gene encoding the host shutoff RNase. J. Virol. 81:10924-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suter, M., A. M. Lew, P. Grob, G. J. Adema, M. Ackermann, K. Shortman, and C. Fraefel. 1999. BAC-VAC, a novel generation of (DNA) vaccines: a bacterial artificial chromosome (BAC) containing a replication-competent, packaging-defective virus genome induces protective immunity against herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 96:12697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi, M., T. Otsuka, Y. Okuno, Y. Asano, and T. Yazaki. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet 2:1288-1290. [DOI] [PubMed] [Google Scholar]
- 55.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 56.Trapp, S., M. S. Parcells, J. P. Kamil, D. Schumacher, B. K. Tischer, P. M. Kumar, V. K. Nair, and N. Osterrieder. 2006. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 203:1307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Zijl, M., W. Quint, J. Briaire, T. de Rover, A. Gielkens, and A. Berns. 1988. Regeneration of herpesviruses from molecularly cloned subgenomic fragments. J. Virol. 62:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Einem, J., D. Schumacher, D. J. O'Callaghan, and N. Osterrieder. 2006. The alpha-TIF (VP16) homologue (ETIF) of equine herpesvirus 1 is essential for secondary envelopment and virus egress. J. Virol. 80:2609-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 60.Wang, N., P. F. Baldi, and B. S. Gaut. 2007. Phylogenetic analysis, genome evolution and the rate of gene gain in the Herpesviridae. Mol. Phylogenet. Evol. 43:1066-1075. [DOI] [PubMed] [Google Scholar]
- 61.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yedowitz, J. C., A. Kotsakis, E. F. Schlegel, and J. A. Blaho. 2005. Nuclear localizations of the herpes simplex virus type 1 tegument proteins VP13/14, vhs, and VP16 precede VP22-dependent microtubule reorganization and VP22 nuclear import. J. Virol. 79:4730-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshitake, N., X. Xuan, and H. Otsuka. 1997. Identification and characterization of bovine herpesvirus-1 glycoproteins E and I. J. Gen. Virol. 78:1399-1403. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 65.Zuckermann, F. A., T. C. Mettenleiter, C. Schreurs, N. Sugg, and T. Ben-Porat. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]