Abstract
The UL16 tegument protein of herpes simplex virus is conserved throughout the herpesvirus family. It has been reported to be capsid associated and may be involved in budding by providing an interaction with the membrane-bound UL11 protein. UL16 has been shown to be present in all the major locations that capsids are found (i.e., the nucleus, cytoplasm, and virions), but whether it is actually capsid associated in each of these has not been reported. Therefore, capsids were purified from each compartment, and it was found that UL16 was present on cytoplasmic but not nuclear capsids. In extracellular virions, the majority of UL16 (87%) was once again not capsid associated, which suggests that the interaction is transient during egress. Because herpes simplex virus (HSV) buds into the acidic compartment of the trans-Golgi network (TGN), the effect of pH on the interaction was examined. The amount of capsid-associated UL16 dramatically increased when extracellular virions were exposed to mildly acidic medium (pH 5.0 to 5.5), and this association was fully reversible. After budding into the TGN, capsid and tegument proteins also encounter an oxidizing environment, which is conducive to disulfide bond formation. UL16 contains 20 cysteines, including five that are conserved within a putative zinc finger. Any free cysteines that are involved in the capsid interaction or release mechanism of UL16 would be expected to be modified by N-ethylmaleimide, and, consistent with this, the amount of capsid-associated UL16 dramatically increased when virions were incubated with this compound. Taken together, these data suggest a transient interaction between UL16 and capsids, possibly modified in the acidic compartment of secretory vesicles and requiring a release mechanism that involves cysteines.
Herpesviruses are assembled with more than 40 different virally encoded proteins that comprise three morphologically distinct structures: the icosahedral nucleocapsid containing the viral DNA; the lipid envelope containing virus-encoded glycoproteins; and the tegument, the assortment of proteins between the nucleocapsid and the envelope. The specific mechanisms by which these various viral proteins come together to drive the assembly and budding processes are poorly understood.
The addition of tegument proteins to capsids during assembly is widely viewed as an ordered process of adding “layers” of protein (38). In this model, some tegument is added in the nucleus, more in the cytoplasm, and the rest at the trans-Golgi network (TGN) where maturation budding occurs (39). The data presented in this report challenge this view and suggest that the process is more dynamic than previously recognized. Specifically, the UL16 protein of herpes simplex virus (HSV) may be the first example of a tegument protein that associates with and releases from capsids during egress through low-pH compartments of the cell.
The UL16 protein is conserved throughout the herpesvirus family (41, 45, 69). As measured by immunofluorescence, this protein has been found to be primarily nuclear at early times postinfection, colocalizing with the major capsid protein VP5 and the scaffold protein VP22a, suggesting that UL16 may interact with capsids in the nucleus (41). The localization of UL16 changes to a mostly punctate perinuclear region at later times postinfection (41, 45). The actual function of this protein remains elusive, but it has been suggested that it participates in genome packaging and nucleocapsid maturation due to its weak association with DNA-containing type C capsids isolated from whole-cell lysates (45), its ability to bind single-stranded DNA in vitro (45), and its potential zinc finger domain (69).
More recently, UL16 has been identified as a binding partner of another conserved tegument protein, UL11 (30). This is a myristylated and palmitylated protein that targets to Golgi-derived vesicles in the absence of other viral proteins (29). Therefore, it is conceivable that the interaction between UL16 and UL11 might participate in targeting nucleocapsids to the site of maturation budding, where the virus obtains its final envelope. Consistent with roles in maturation, Vero cells infected with UL16 or UL11 deletion mutants have defects in virus production compared to wild type (3, 4, 6, 22, 23, 33, 34, 54, 59, 60).
The goal of the experiments described here was to ascertain at which cellular sites UL16 interacts with nucleocapsids. The data reveal that UL16 is associated with capsids isolated from the cytoplasm but not with those from the nucleus of infected cells. Moreover, very little UL16 remains capsid associated following envelope removal of extracellular virions. The amount of UL16 present on extracellular capsids is considerably increased when virions are exposed to mildly acidic pH, and this association is reversed when pH is returned to neutral. Since herpesviruses bud into TGN-derived vesicles, which are thought to have an acidic pH (16, 19), this may represent a possible mechanism for the association or release of UL16 from capsids. These findings raise the likely possibility that other changes within the tegument occur during passage through low-pH compartments during virus egress.
MATERIALS AND METHODS
Virus and cells.
Vero cells (ATCC CCL-81) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum, 5% bovine calf serum, penicillin, and streptomycin (Gibco 15140-148). Confluent monolayers of Vero cells were infected with the KOS strain of HSV type 1 (HSV-1) (61). Following infection, cells were incubated in Dulbecco's modified Eagle's medium supplemented with 2% fetal bovine serum, 25 mM HEPES buffer, glutamine (0.3 μg/ml), penicillin, and streptomycin. The HSV-1 UL31 deletion mutant and complementing cell line (clone 7) that expresses this protein were generously provided by Joel Baines (Cornell University) (28).
Antibodies.
UL16-, UL11-, VP22-, and VP5-specific antibodies were produced in rabbits and have been described previously (29, 30, 37). Rabbit polyclonal antibodies were generated against purified glutathione transferase-UL21 and glutathione transferase-UL46 antigens. Rabbit polyclonal VP16-specific antibodies were obtained from Clontech (product number 3844-1). Monoclonal VP5 antibodies, 8F5 and 5C, were generated against conformational epitopes on the capsid surface (kindly provided by Jay Brown, University of Virginia) (64). Monoclonal antibodies were used to detect the scaffold proteins VP21/VP22a (MCA406; Serotec, Washington, DC) (42). Rabbit polyclonal antibody raised against glycoprotein gE was a kind gift from Harvey Friedman (University of Pennsylvania).
UL16 chimeras.
Construction of plasmid pCMV.UL16-GFP (where CMV is cytomegalovirus and GFP is green fluorescent protein) used in this study has been described previously (30). To express UL16 lacking the GFP tag, pCMV.UL16 was generated by removal of the GFP sequence.
Confocal microscopy.
Vero cells were transfected with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). At 20 to 22 h posttransfection, cells were infected with HSV at a multiplicity of infection (MOI) of 5 PFU. At the various times postinfection, cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) and then viewed by using a Zeiss laser scanning microscope with a helium-argon laser (488-nm peak excitation).
Capsid analyses.
HSV capsids from the nucleus and cytoplasm were isolated as described by Sherman and Bachenheimer with some modifications (58). Two 850-cm2 roller bottles of confluent Vero cells were infected at an MOI of 10. At 16 to 20 h postinfection, cells were scraped into 20 ml of PBS, collected by centrifugation at 1,000 × g for 10 min, resuspended in 6 ml of NP-40 lysis buffer (0.5% NP-40, 150 mM NaCl, 1 M Tris-HCl, pH 8.0) containing protease inhibitors (P8340; Sigma), and incubated for 15 min on ice. The cytoplasmic fraction was separated from the nuclei by centrifugation at 1,000 × g for 10 min. The nuclei were washed three times and resuspended in 6 ml of NP-40 lysis buffer with protease inhibitors. The separated cytoplasmic and nuclear fractions were treated identically throughout the experiments. Capsids were released from the purified nuclei by freezing (−80°C) and then thawing three times (37°C), followed by cup sonication for 3 min at moderate power. Insoluble material from the nuclear and cytoplasmic fractions was cleared by centrifugation at 8,000 × g for 30 min. The capsids remaining in the soluble supernatant of the nuclear and cytoplasmic fractions were pelleted through a 1.7-ml 30% (wt/vol in TNE buffer [20 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM EDTA]) sucrose cushion in a SW41 rotor at 83,500 × g for 1 h. Pellets were then resuspended in 500 μl of TNE buffer, sonicated for 2 min at moderate power, and layered onto a 20 to 50% (wt/vol, sucrose in TNE buffer) continuous gradient. Gradients were then centrifuged at 74,000 × g for 1 h in an SW41 rotor. All centrifugation steps were carried out at 4°C. Fractions of 750 μl were collected from the top of the gradient with a piston gradient fractionator (Brandel). Trichloroacetic acid (TCA) was added to a final concentration of 13%, and the samples were incubated overnight at 4°C. The precipitated proteins were collected by centrifugation in a microcentrifuge at 18,000 × g for 30 min, washed with 100% ethanol, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (3.5% SDS, 8.5% β-mercaptoethanol, 130 mM dithiothreitol, 0.5 M urea, 290 mM Tris-HCl, pH 8.8), and boiled for 15 min at 95°C. Samples were separated in SDS-polyacrylamide (10% or 7%) gels and electrotransferred to nitrocellulose membranes. The enhanced chemiluminescence method of immunoblot analysis was performed according to the manufacturer's instructions (Amersham). Anti-UL16 and anti-VP5 were used as the primary antibodies at dilutions of 1:3,000 and 1:7,500 (in 5% nonfat milk in buffer containing 20 mM Tris, pH 7.6, 135 mM NaCl, and 0.1% Tween-20), respectively. The positions of B and C capsids in the gradient were confirmed by identification of the light-scattering bands as previously described (17, 58) and by immunoblotting for the scaffold proteins VP21 and VP22a (42).
Immunoprecipitation of capsids.
Cells were infected at an MOI of 10. At 16 to 20 h postinfection cells were washed with 5 ml of PBS, scraped into 1 ml of PBS, and collected by centrifugation at 1,000 × g for 10 min. The cells were resuspended into 1 ml of NP-40 lysis buffer with protease inhibitors and incubated for 15 min on ice. Whole-cell lysates were generated by sonication for 3 min. For nuclear and cytoplasmic separation, nuclei were first isolated by centrifugation at 1,000 × g for 10 min and then washed three times with NP-40 lysis buffer. Nuclei were lysed as described above. The lysates were clarified by centrifugation at 8,000 × g for 30 min, and supernatants were transferred to new tubes for immunoprecipitation analysis. Capsids and their associated proteins were immunoprecipitated with monoclonal antibodies 8F5 and C5, which recognize epitopes present on the mature capsid structure (9, 15, 36, 64). Immunoprecipitated proteins were separated by electrophoresis in SDS-10% polyacrylamide gels. The gels were then electrotransferred to nitrocellulose membranes and subjected to immunoblot analysis.
Analysis of UL16 in virions.
Purification of extracellular virions was performed as previously described with some modifications (31). Briefly, confluent monolayers of Vero cells were infected at an MOI of 10 for 22 h, the medium was collected, and cell debris was removed by centrifugation for 10 min at 1,000 × g. To isolate extracellular capsids, medium from infected plates was treated with NP-40 (final concentration, 0.5%) for 15 min at room temperature. Extracellular virions or capsids (in 10 ml) were pelleted from the medium by centrifugation for 1 h at 83,500 × g in an SW41 rotor through a 30% (wt/vol) sucrose cushion (1.7 ml). Virions or extracellular capsids were separated by SDS-PAGE in 7% polyacrylamide gels and analyzed by immunoblotting using VP5-, UL16-, and VP16-specific antibodies.
Trypsin treatment of virions.
Extracellular virions harvested as described above were resuspended in TNE buffer and separated into six equal fractions. Fractions were treated as previously described with minor modifications (see Fig. 5) (68). Following treatment, virions and capsids were repelleted through an additional sucrose cushion as described above. Pelleted virions and capsids were separated by SDS-PAGE in 10% gels and analyzed by immunoblotting using antibodies specific for VP5, VP16, and UL16.
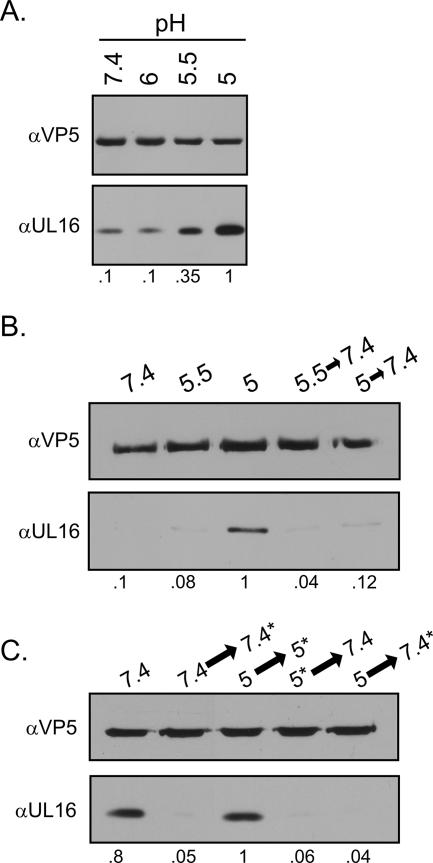
FIG. 5.
Effects of low pH on the association of UL16 with capsids. Extracellular virions were harvested 20 to 24 h postinfection, pelleted through a 30% sucrose cushion, and resuspended in medium buffered to the indicated pH values. (A) After an incubation of 1 h at 37°C, the viral envelopes were removed with NP-40 treatment for 10 min at room temperature. The released capsids were pelleted, dissolved in sample buffer, and analyzed by immunoblotting with anti-VP5 and anti-UL16 antibodies. (B) Virions were first incubated at the indicated pH values for 30 min at 37°C and then incubated for an additional 30 min, either at the same pH or at pH 7.4 (following titration with NaOH). The viral envelopes were removed with NP-40, and the released capsids were collected and analyzed by immunoblotting. (C) Virions were treated as described for the previous panel except that the timing of the NP-40 treatment was either before or after neutralization with NaOH, as indicated by the asterisks. Relative levels of UL16 quantitated by densitometry and normalized for VP5 are indicated bellow each figure. α, anti.
Low-pH treatment of extracellular virus.
HSV pelleted through a sucrose cushion was resuspended in cell culture medium buffered with 5 mM HEPES (Sigma), 5 mM 2-(N-morpholino)ethanesulfonic acid (Sigma), and 5 mM succinate (Sigma) to achieve a pH ranging from 7.4 to 5.0. Following a 1-h incubation at 37°C, virions were stripped of their membrane with NP-40 (0.5% final concentration) to release capsids. Virions or capsids were then pelleted by centrifugation, and the levels of UL16 remaining capsid associated were determined by immunoblotting. In reversibility experiments, the acid pH was titrated back to neutral pH with 1 M NaOH either before or after NP-40 treatment.
NEM treatment of extracellular virions.
Extracellular virions harvested at 18 to 24 h postinfection were treated with N-ethylmaleimide (NEM) for 30 min at 37°C, either before or after removal of the envelope with NP-40 (0.5% final concentration). A 10 μM final concentration of NEM was used to attain rapid modification of free cysteines (5). Following NEM treatment, virions and capsids were purified by centrifugation for 1 h at 83,500 × g, separated by SDS-PAGE in 7% or 10% gels, and analyzed by immunoblotting using antibodies specific for VP5, VP16, and UL16. Using densitometry, the percentages of UL16 and VP16 remaining bound to capsids following removal of the envelope were determined by dividing the amounts of protein present with capsids following envelope removal by the total amounts present in the virion, all normalized to VP5 levels.
RESULTS
The conserved UL16 protein is of interest because it may provide a physical link between the capsid and the membrane-bound UL11 protein during HSV budding. However, very little is known regarding the capsid association properties of UL16. This protein has been reported to be present at all locations where capsids are found (the nucleus, cytoplasm, and virion), but whether it is actually capsid associated in all these locations has never been shown for any herpesvirus. Therefore, the following experiments were pursued.
Subcellular localization of UL16-GFP in infected cells.
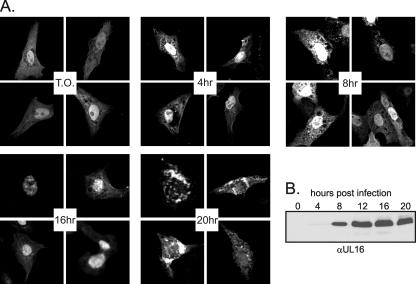
Previous immunofluorescence studies suggested that UL16 accumulates in the nuclei of infected cells up to 18 h postinfection but at later times is localized to punctate, cytoplasmic structures (41). Moreover, the early nuclear staining colocalized with that of capsid proteins VP5 and VP22a, raising the possibility that UL16 is added to capsids in the nucleus. In an attempt to confirm the localization properties of UL16 during the course of an infection, a transfection-infection assay was employed. Vero cells transfected with a plasmid that expresses UL16-GFP were subsequently infected with HSV and visualized by confocal microscopy at various times postinfection. The GFP chimera was found in both the cytoplasm and nucleus early during the infection, similar to results in noninfected (transfected only) cells (Fig. 1A). More intense nuclear fluorescence was observed until 18 h postinfection (Fig. 1A and data not shown) and preceded expression of the virus-encoded UL16, first detected at ∼8 h (Fig. 1B). Therefore, any viral factors responsible for nuclear retention must have been expressed earlier, which is consistent with the suggestion that UL16 might have a role in DNA replication or packaging (45). Localization of UL16-GFP changed dramatically at later times postinfection to cytoplasmic regions surrounding the nucleus of infected cells with very little nuclear fluorescence (Fig. 1A). Thus, an assay that does not require the use of antiserum confirmed the nuclear trafficking properties of UL16 and raised the possibility that this protein would be present on nuclear capsids.
FIG. 1.
Change in UL16-GFP localization during the course of infection. (A) Vero cells were transfected with pUL16-GFP. At 20 to 22 h posttransfection, cells were infected with HSV, fixed with paraformaldehyde at the indicated times postinfection, and viewed by confocal microscopy. T.O., transfected only. (B) Vero cells were infected with HSV, and at the indicated times postinfection, they were harvested and resuspended in sample buffer. Proteins were separated by SDS-PAGE in 10% gels, transferred to nitrocellulose, and detected by immunoblotting with a polyclonal antibody raised against UL16.
Association of UL16 with capsids.
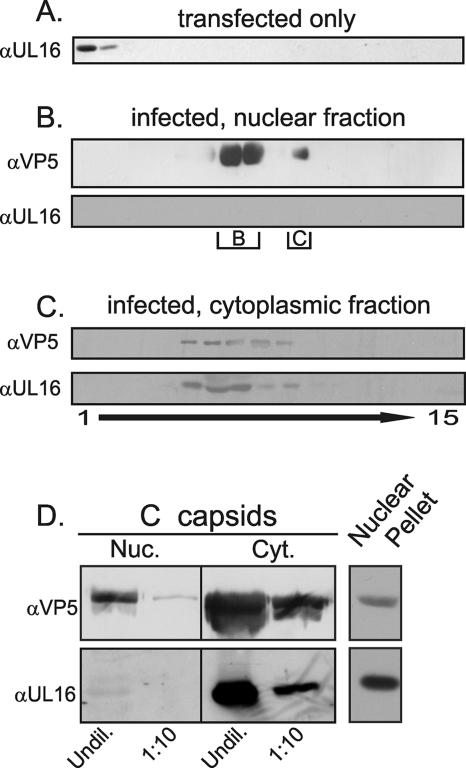
To test the prediction, capsids were released from the nuclei of detergent-disrupted, HSV-infected cells by freeze-thaw cycling and sonication. Capsids from this extract were first pelleted through a sucrose cushion and then sedimented in a sucrose gradient. Fractions were collected from the top, and proteins in each were precipitated with TCA prior to SDS-PAGE and immunoblotting with antibodies specific for UL16 and the VP5 capsid protein. Despite the large amount of UL16 present in the nucleus at 18 h postinfection, this protein was not found to be associated with the purified, intranuclear capsids (Fig. 2B). Indistinguishable results were obtained when intranuclear capsids were harvested at 16 h postinfection (data not shown). In contrast, UL16 was readily detected as a species cosedimenting with the heterogeneous population of capsids isolated from the cytoplasm using identical buffer and detergent conditions (Fig. 2C). Capsids isolated from the cytoplasm are thought to contain more tegument than those isolated from the nucleus, and this likely accounts for the sedimentation differences shown in panels B and C of Fig. 2. The additional tegument would increase the size of the particles, and therefore decrease the density, causing them to sediment at a slower rate in the gradient. When UL16 was expressed alone by transient transfection and subjected to sucrose gradient centrifugation, it was found in the top fractions (Fig. 2A), suggesting that it cannot form large aggregates on its own.
FIG. 2.
Cosedimentation of UL16 with cytoplasmic but not nuclear capsids. Vero cells were transfected with pUL16 (A) or infected with HSV at an MOI of 10 (B to D) for 16 to 22 h (representative 18-h time point shown). Detergent lysates of the transfected cells were analyzed directly in a 20 to 50% (wt/vol) sucrose gradient whereas nuclear and cytoplasmic capsids from the infected cells were first pelleted through a 30% sucrose cushion prior to sedimentation. Fractions were collected, and proteins were concentrated by TCA precipitation. The precipitates were dissolved in sample buffer and separated either in a 7% (for detection of VP5) or 10% (for UL16) polyacrylamide-SDS gel prior to immunoblot analyses with anti-VP5 or anti-UL16 rabbit polyclonal antibodies. The locations of B and C capsids in the gradients are indicated. (D) To compare the levels of UL16 present on equal amounts of nuclear (Nuc) and cytoplasmic (Cyt) C capsids, the relevant species were collected from the gradients, repelleted through a 30% sucrose cushion, and resuspended in sample buffer. Undiluted (Undil) or 1:10 dilutions of the samples were analyzed by immunoblotting with anti-VP5 and anti-UL16 sera. α, anti.
Most of the capsids in the nucleus are of the B rather than the mature, DNA-containing, export-competent C form (58), and if UL16 was associated only with the latter, then it might be difficult to detect. Therefore, C capsids from nuclei and cytoplasm were purified from a sucrose gradient and diluted so that equal amounts of VP5 from each compartment could be compared. Again, UL16 was readily detectable on the capsids from the cytoplasm but not those from the nucleus, even though it was present in the nuclear compartment (Fig. 2D). When quantitated by densitometry, the amount of UL16 normalized for VP5 levels was 40-fold higher on cytoplasmic capsids. Moreover, UL16 was not detected on intranuclear capsids produced by a mutant defective for UL31 expression (data not shown), which accumulates C capsids in the nucleus (8). Thus, UL16 copurifies with C capsids isolated from the cytoplasm of infected cells but not those from the nucleus.
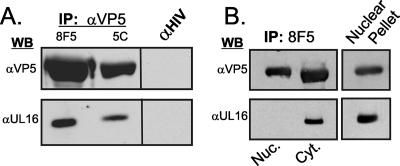
Although the sedimentation pattern of UL16 suggests that it is physically associated with capsids isolated from the cytoplasm, it is possible that the protein is part of a large cytoplasmic complex that just happens to purify with capsids. To address this, capsids from HSV-infected cell lysates were immunoprecipitated with two different monoclonal antibodies that recognize VP5 epitopes present only on the mature capsid surface (9, 15, 36, 64). As a negative control, an irrelevant mouse monoclonal (anti-human immunodeficiency virus) antibody was used. Subsequent immunoblot assays showed that UL16 is indeed associated with capsids immunoprecipitated from whole-cell lysates (Fig. 3A). As expected, envelope glycoprotein gE and “outer” tegument proteins UL11 and VP22 (all of which are released from virions in the presence of NP-40) did not coimmunoprecipitate with capsids in these experiments (data not shown). Moreover, the monoclonal antibodies did not immunoprecipitate UL16 from transfected cells (data not shown).
FIG. 3.
Coimmunoprecipitation of UL16 with cytoplasmic but not nuclear capsids. Vero cells were infected with HSV, and viral proteins were immunoprecipitated (IP) 18 h later from whole-cell lysates (A) or nuclear (Nuc) and cytoplasmic (Cyt) fractions (B) by using monoclonal antibodies 8F5 or 5C, which are specific for VP5 epitopes present only on mature capsids. A monoclonal antibody specific for human immunodeficiency virus (HIV) p24 was used as a negative control. The immunoprecipitated capsid and associated proteins were separated in an SDS-10% polyacrylamide gel and analyzed by immunoblotting with polyclonal rabbit serum specific for VP5 or UL16. WB, Western blotting; α, anti.
To further test the hypothesis that UL16 is associated only with cytoplasmic capsids, the immunoprecipitation analysis was repeated on capsids from nuclear and cytoplasmic fractions. UL16 was easily detected on capsids isolated from the cytoplasm but was not found associated with capsids from the nucleus, even though it was present in this compartment (Fig. 3B). While the experiments described here clearly demonstrate that UL16 is present on at least some of the capsids present in the cytoplasm, the results do not rule out the possibility that UL16 is weakly associated with nuclear capsids or is added just as the capsids exit the nucleus, events that would make detection very difficult.
Analysis of UL16 in virions.
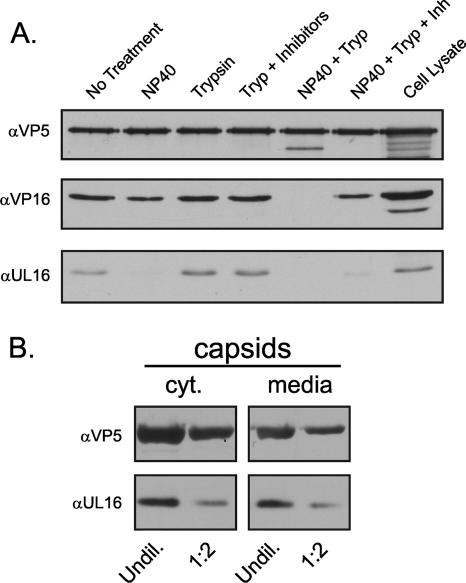
Previous studies suggested that UL16 is associated with HSV-1 (41) but not HSV-2 particles (45). To rigorously address whether UL16 is inside the virion and capsid associated, a trypsin sensitivity assay was used (Fig. 4A). UL16 is sensitive to trypsin (data not shown) but was found to be present in a form that was protected from trypsin cleavage unless the envelope was removed by NP-40 treatment, confirming that it was inside the virion. However, very little UL16 pelleted with extracellular capsids, even when trypsin was not present. Quantitation by densitometry of four independent experiments showed that only 13% ± 6% of UL16 remained associated with capsids after NP-40 treatment.
FIG. 4.
UL16-capsid interactions in virions. (A) Extracellular virions were harvested from HSV-infected Vero cells and pelleted through a 30% sucrose cushion. The samples were resuspended in TNE buffer, separated into six equal fractions, and treated as indicated for 15 min at room temperature. Following treatment, virions and capsids were pelleted through an additional sucrose cushion, resuspended in sample buffer, and separated by SDS-PAGE in 7% gels. Proteins were analyzed by immunoblotting using antibodies specific for VP5, VP16, and UL16. (B) Capsids were isolated from NP-40-treated cytoplasmic (cyt) lysates or extracellular medium by centrifugation through a 30% sucrose cushion. Capsid pellets were resuspended in sample buffer, and two different amounts were analyzed by immunoblotting using antibodies specific for VP5 and UL16. α, anti; Tryp, trypsin; Inh, inhibitors; Undil, undiluted.
If the UL16 interaction is transient during egress, then capsids isolated from the cytoplasm might contain more UL16 than those from extracellular virions. Comparisons of equal amounts of capsids purified from the cytoplasm and virions (normalized for VP5) revealed similar levels of UL16 (within a twofold difference) (Fig. 4B); however, the interpretation of this result is difficult because the cytoplasm contains capsids in all stages of assembly: as they exit the nucleus, traverse the cytoplasm, bud at internal membranes, and travel in vesicles to the cell surface for release. The low levels of capsid-associated UL16 present in extracellular virions (∼13%) may be an indication that the majority of this protein (∼87%) is packaged via another mechanism. Alternatively, these results may be indicative of a transient capsid association that is subsequently reversed to release the majority of UL16 into the “outer” virion space at some stage following budding (see Discussion).
Low-pH treatment of extracellular virions.
HSV is thought to bud into the TGN, which has a slightly acidic pH of 6.0 (16, 19). During transit to the cell surface, the pH of the secretory vesicles continues to drop to ∼5.5 (11). Since low pH is known to trigger conformational changes in many proteins (e.g., the hemagglutinin of influenza virus) (13, 53), it is possible that HSV might utilize this environment to trigger various maturation events, including release of UL16 from capsids. With this hypothesis in mind, extracellular virions were collected and incubated in medium buffered at various pH values. NP-40 was used to solubilize the virus envelope, and the capsids were then pelleted through a sucrose cushion. The amount of UL16 copelleting with capsids increased as the pH of the medium was made more acidic (Fig. 5A).
Because low pH might alter the structure of UL16—causing it to precipitate and pellet nonspecifically with capsids—the reversibility of the reaction was examined. Precipitated proteins often have difficulty resolubilizing, but this was not the case with UL16, which was completely released following neutralization (Fig. 5B). Moreover, the viral membrane was not needed for reversibility (Fig. 5C). Pelleting the pH 5.0-treated capsids through a sucrose cushion gave similar results whether the cushion had a pH of 5.0 or 7.4. Presumably, this is because the cushion is small, and the speed of sedimentation through that cushion is fast. These data provide evidence that dynamic changes may occur inside the virions as they travel through the secretory vesicles en route to the extracellular medium.
NEM treatment of virions.
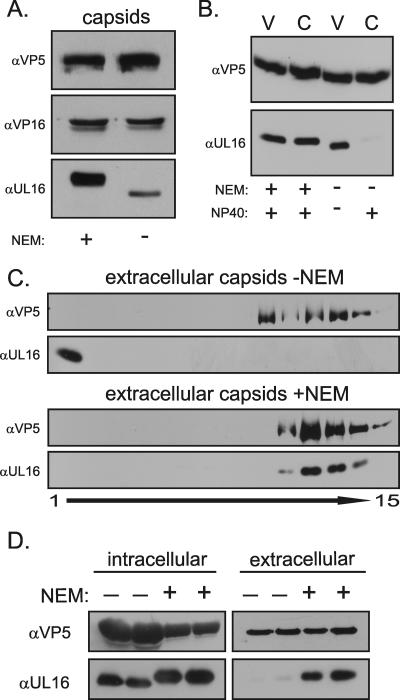
Upon completion of budding, the capsid and other internal components of the virion first encounter an oxidizing environment, which promotes disulfide bond formation (14, 65). Because UL16 contains 20 cysteines, 5 of which are conserved in a putative zinc finger (69), it seemed possible that one or more of these might be important for capsid association or release, perhaps as part of a disulfide-exchange reaction (18, 20, 27, 35, 40). Free cysteines are covalently modified when exposed to NEM, a group-specific chemical that has been used extensively in the study of protein structure and function (5, 7, 26, 56, 70). To examine the possible involvement of cysteines in the UL16-capsid interaction, extracellular virions were treated with NEM at 37°C. A dramatic increase in the amount of UL16 that copellets with capsids was observed (Fig. 6A). This was not observed with other tegument proteins tested: VP16, UL11, UL21, and UL46, which contain 6, 5, 5, and 11 cysteines, respectively (Fig. 6A and data not shown). The corresponding increase in apparent molecular mass of UL16 is indicative of multiple NEM modifications, which are not sensitive to reducing agents in sample buffer or to boiling prior to electrophoresis. NEM has a molecular mass of 125 Da, and the magnitude of the shift in mass of ∼1,500 Da suggests that approximately half of the 20 cysteines are modified. In several repeats of the experiment, the amount of capsid-associated UL16 within the virion (normalized to VP5) ranged between 68 to 85% following NEM treatment, whereas the amount was only 3 to 15% without NEM (a representative result is shown in Fig. 6B). At the concentration of NEM used, the titer of extracellular virions was reduced 4 orders of magnitude after 10 min of treatment at 37°C (data not shown), which unfortunately provides little insight because the drug can potentially act on many viral proteins.
FIG. 6.
Effects of NEM on the UL16-capsid interaction. (A) Extracellular virions were treated with or without NEM at 37°C. Viral membranes were removed with NP-40 treatment, and the capsids were pelleted through a 30% sucrose cushion prior to immunoblot analyses with antibodies specific for VP5, VP16, and UL16. (B) Extracellular virions were treated with or without NEM for 30 min and then treated with or without NP-40. The released capsids or intact virions (C or V, respectively) were pelleted through a 30% sucrose cushion prior to immunoblotting with the indicated antiserum. (C) Virions present in the medium were concentrated by centrifugation through a sucrose cushion and resuspended in a small volume of TNE buffer. The sample was divided into two equal portions, and these were treated with or without NEM for 30 min. Both samples then received NP-40 for 15 min at room temperature prior to sedimentation through parallel 20 to 50% sucrose gradients. Fractions were collected from the top and analyzed by immunoblotting with antibodies specific for VP5 and UL16. (D) Gradient-purified capsids treated with NEM or carrier prior to lysis for 15 min at 37°C from infected cytoplasm and medium were analyzed by immunoblotting for levels of VP5 and UL16. Gradients were run in duplicate. α, anti.
To determine whether NEM-modified UL16 is capsid associated rather than present in an aggregate or large complex of another sort, the samples were analyzed in a sucrose gradient following solubilization of the viral membrane with NP-40. In the absence of NEM treatment, all detectable UL16 remained at the top of the gradient while the capsids moved toward the bottom (Fig. 6C). In contrast, treatment with NEM resulted in all detectable UL16 cosedimenting with capsids. Attempts to demonstrate the UL16-capsid association in virions using monoclonal antibodies specific for VP5 (as described in the legend of Fig. 3) were not possible, presumably because the presence of tegument proteins that blocked epitope recognition, with the result that less than 10% of all extracellular capsids were immunoprecipitated from NP-40-treated virions (data not shown).
Because infectious virions can be obtained from the cytoplasm following disruption of infected cells at neutral pH, it seemed likely that the levels of UL16 associated with cytoplasmic capsids would increase if the cells were pretreated with NEM. This was found to be the case, as the quantity of UL16 present on cytoplasmic capsids (normalized to VP5) was fourfold higher if cells were treated with NEM prior to lysis (Fig. 6D). This is likely due to the action of NEM on virions present within secretory vesicles because NEM did not cause UL16 to associate with intranuclear capsids (data not shown).
DISCUSSION
The experiments described here provide a detailed analysis of the interaction of UL16 with capsids. The major finding is that the interaction is dynamic, with a binding and release mechanism that is pH regulated and likely involves cysteines. To our knowledge, this report provides the first evidence for a transient interaction of a tegument protein with a herpesvirus capsid during egress. However, the possibility that virus maturation events might take place as the virion travels through low-pH compartments was first suggested by the discovery that neutralization of cellular organelles blocks production of infectious HSV, even though budding still occurs (19). It is likely that other maturation events will be found to occur as the virion travels the egress pathway. The particular insights obtained regarding UL16 are discussed below.
Nuclear localization of UL16.
UL16-GFP was found to localize to the nucleus, even in the absence of the other HSV proteins. Because this 69-kDa chimera is predicted to be too large for passive entry through the nuclear pore complex (12), it presumably contains a nuclear localization signal or interacts with a host protein that enables its transport. In the context of an infection, there is an increase in the amount of UL16 (41) or UL16-GFP (this study) present in the nucleus; however, the interactions responsible for nuclear retention remain unknown.
What is the function of UL16 in the nucleus? This tegument protein contains a putative zinc finger (60) and has been shown to bind single-stranded DNA in vitro (45), suggesting that it may associate with the DNA packaging machinery along with UL6, UL15, UL17, UL28, UL32, and UL33 (1, 2, 10, 24, 25, 46, 47, 52, 57, 63, 67, 71). If so, then UL16 would be predicted to be associated with procapsids, which would not have been detected in the experiments described here because they are unstable, present in low numbers, and difficult to isolate (43, 50, 55). An association with procapsids would also be consistent with the observed colocalization of UL16 with capsid and scaffold proteins (41). To gain further insight, studies to identify the binding partners of UL16 in the nucleus are warranted.
Whatever its function in the nucleus, it is clear that UL16 is not stably present on the C capsids isolated from this compartment. Moreover, it was not detected on capsids made by a UL31-null virus (data not shown), which accumulate in the nucleus because of a defect in nuclear egress (8). It remains possible that UL16 associates with nuclear capsids, but this interaction cannot be detected under the lysis conditions used in this study because the association is weaker than that with cytoplasmic capsids. Also, UL16 might be added to capsids just as they bud into—or exit from—the perinuclear space, but the capsids present in that compartment at any given time are few in number and would not be easily detected in the experiments described here. In any case, it seems likely that the function of UL16 in the nucleus is not required for the packaging of this tegument protein into the mature virion. Therefore, it may be possible to find UL16 mutants that are packaged even though they fail to enter the nucleus.
Mechanism of UL16 packaging.
There are at least two potential mechanisms for the packaging of UL16 into virions. In the first, UL16 binds to C capsids prior to their arrival at the TGN for maturation budding, a model that is consistent with (but not proven by) the data presented here. From this point of view, it is particularly interesting that antibodies specific for mature capsids were unable to efficiently recognize their epitopes when extracellular virions were solubilized with NP-40 (data not shown) under conditions that readily enabled recognition of capsids from the cytoplasm (and nucleus). If antibody access is blocked following tegumentation, then the capsids that are immunoprecipitated from the cytoplasm with UL16 attached probably represent intermediates en route to the TGN. In this model, subsequent interaction of UL16 with membrane-bound UL11 would provide a bridging function to promote budding (30).
In the other mechanism of packaging, UL16 travels to the TGN independently of capsids, presumably by means of its interaction with UL11 (30). When capsids arrive at the membrane, their binding to the UL16-UL11 complex would then promote budding. If this model is correct, then the population of cytoplasmic capsids that has UL16 bound would be the capsids that have already entered the postbudding pathway. Because virions in this pathway have received their full complement of tegument proteins, they would not be expected to react with the antibodies, which argues against this model. However, most of the UL16 in extracellular virions (∼85%) was found to be free of capsids upon treatment with NP-40, and this raises the possibility that some is packaged without the need for that interaction. Alternatively, it is possible that all UL16 molecules are transiently associated with capsids during transport to the TGN, with most being released by the time the virion leaves the cell. The dramatic and reversible changes in UL16 binding that occur in response to pH clearly support the transient association model.
To gain further insight on the packaging mechanism, it will be important to identify the protein that links UL16 to the capsid. One candidate is UL21, a capsid-associated tegument protein that has recently been identified as a binding partner of UL16 in pseudorabies virus (21, 66). The HSV homolog of UL21 has been reported to interact (perhaps indirectly) with microtubules (62); hence, it is reasonable to speculate that UL16 and UL21 might be involved in transporting capsids to the TGN. Subsequent release of the complex would prevent capsids from reentering the egress pathway when the next cell is infected.
Dynamic interaction of UL16 with capsids.
Two entirely different methods were found for increasing the amount of UL16 that is associated with extracellular capsids (low pH and NEM), and the observations made with these provide evidence to suggest that the interaction is transient during capsid egress. Nevertheless, there is a fundamental but difficult question that remains unanswered: Are the UL16 molecules in an untreated virion actually bound to the capsid and merely released upon exposure to NP-40? If so, then it still must be the case that the structure of UL16 in extracellular virions is different from that on cytoplasmic capsids because the same NP-40 buffer conditions were used to isolate both forms.
It might be possible to ascertain the capsid association state of UL16 in virions through the use of chemical cross-linkers; however, such experiments are complicated by the many cysteines in this protein and their potential for creating abnormal disulfide bond linkages following virion disruption. NEM is often used to block any free cysteines that are present, and one interpretation of the data presented here is that modification of UL16 blocks a cysteine exchange reaction that is needed for release (and triggered by NP-40). However, it is possible that NEM changes the conformation of free UL16 so that it becomes capsid associated in the extracellular virion. If so, then the conformation of UL16 in extracellular virions must still be different from that in the cytoplasm, where capsid association was readily detected even in the absence of NEM. Moreover, NEM did not stimulate interaction with capsids in the nucleus.
The effects of pH on the binding of UL16 to capsids does not shed light on the native state of UL16 within the virion. That is, low pH might prevent an NP-40-induced conformational change that releases UL16, or it might cause UL16 to revert to a conformation that once again becomes competent for capsid binding. However, it seems more likely that treatments as different as pH and NEM would preserve an interaction rather than induce the particular conformation needed for capsid binding. Moreover, the UL16 homolog of human CMV (UL94) has been reported to be associated with extracellular capsids following NP-40 treatment (69), and thus that virus appears to lack the release mechanism of HSV. In any case, the reversibility of the pH effect does argue strongly that UL16 is not merely precipitated upon treatment. Moreover, UL16 was readily found to be associated with capsids from the cytoplasm at neutral pH, and this once again argues that the structure of this protein is very different by the time the virion is released from the cell.
Based on all available data, we hypothesize that UL16 is stably added to capsids prior to their arrival at the site of budding in the cytoplasm. Subsequently, something destabilizes this capsid interaction, but the mechanism is unknown. Treatments with NEM or low pH may provide clues as to what is involved (i.e., cysteine residues and transit through acidic compartments of the cell). Whatever the mechanism, it is clear that most of the UL16 in extracellular virions is not stably associated with capsids. Although the data presented here demonstrate a pH-dependent, reversible stabilization of UL16 with extracellular capsids, it is important to understand that in vitro treatment with low pH does not restore UL16 to the stable state found on cytoplasmic capsids. That is, when the pH is returned to pH 7.4, the stable interaction of UL16 with extracellular capsids is lost (unlike UL16 on cytoplasmic capsids, which are isolated at this pH). The inability to “reset” UL16 to its cytoplasmic state may be an indication that additional maturation events occur among tegument proteins during capsid egress.
There are many examples of viral maturation events that occur within the Golgi complex and secretory vesicles (32, 44, 49, 51). Most recently, structural changes in the West Nile virus capsid have been determined by cryo-electron microscopy as the virus particles travel through the low-pH environment of the TGN (72). Neutralization of organelle pH with ammonium chloride causes these immature particles to accumulate within the cell (48). The idea that herpesviruses could be utilizing the acidic pH of the secretory vesicles in a similar fashion to bring about changes in the tegument necessary for infectivity seems highly likely (19).
Acknowledgments
We extend special thanks to our colleagues Richard Courtney, Michael Murphy, Kevin O'Regan, and Nicholas Baird for helpful discussions and to Rebecca Craven for careful review of the manuscript. In addition, we thank Michelle Bucks and Jacob Marsh for technical help on the capsid isolation protocol and the NEM infectivity experiments, respectively.
This work was supported by NIH grants to J.W.W. (CA47482, AI071286) and in part by a training grant from the National Cancer Institute (CA60395).
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., C. Cunningham, D. Nalwanga, and A. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolen, J. B., and M. A. Israel. 1983. Inhibition of polyoma virus middle T antigen-associated tyrosyl kinase activity by N-ethylmaleimide. J. Biol. Chem. 258:15135-15140. [PubMed] [Google Scholar]
- 6.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooker, R. J., and C. W. Slayman. 1983. [14C]N-ethylmaleimide labeling of the plasma membrane [H+]-ATPase of Neurospora crassa. J. Biol. Chem. 258:222-226. [PubMed] [Google Scholar]
- 8.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham, C., A. J. Davison, A. Dolan, M. C. Frame, D. J. McGeoch, D. M. Meredith, H. W. Moss, and A. C. Orr. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303-311. [DOI] [PubMed] [Google Scholar]
- 11.Demaurex, N. 2002. pH homeostasis of cellular organelles. News Physiol. Sci. 17:1-5. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall, C., and R. A. Laskey. 1986. Protein import into the cell nucleus. Annu. Rev. Cell Biol. 2:367-390. [DOI] [PubMed] [Google Scholar]
- 13.Doms, R. W., A. Helenius, and J. White. 1985. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 260:2973-2981. [PubMed] [Google Scholar]
- 14.Frand, A. R., J. W. Cuozzo, and C. A. Kaiser. 2000. Pathways for protein disulphide bond formation. Trends Cell Biol. 10:203-210. [DOI] [PubMed] [Google Scholar]
- 15.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glomb-Reinmund, S., and M. Kielian. 1998. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology 248:372-381. [DOI] [PubMed] [Google Scholar]
- 19.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain, S., L. W. McGinnes, and T. G. Morrison. 2007. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J. Virol. 81:2328-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., S. Bottcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 25.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Quoc, K., D. Le Quoc, and Y. Gaudemer. 1981. Evidence for the existence of two classes of sulfhydryl groups essential for membrane-bound succinate dehydrogenase activity. Biochemistry 20:1705-1710. [DOI] [PubMed] [Google Scholar]
- 27.Li, P. P., A. Nakanishi, S. W. Clark, and H. Kasamatsu. 2002. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc. Natl. Acad. Sci. USA 99:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 29.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2006. Packaging determinants in the UL11 tegument protein. J. Virol. 80:10534-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 34.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 35.Markovic, I., T. S. Stantchev, K. H. Fields, L. J. Tiffany, M. Tomic, C. D. Weiss, C. C. Broder, K. Strebel, and K. A. Clouse. 2004. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 103:1586-1594. [DOI] [PubMed] [Google Scholar]
- 36.Matusick-Kumar, L., W. Hurlburt, S. P. Weinheimer, W. W. Newcomb, J. C. Brown, and M. Gao. 1994. Phenotype of the herpes simplex virus type 1 protease substrate ICP35 mutant virus. J. Virol. 68:5384-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 40.Mulvey, M., and D. T. Brown. 1994. Formation and rearrangement of disulfide bonds during maturation of the Sindbis virus E1 glycoprotein. J. Virol. 68:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nalwanga, D., S. Rempel, B. Roizman, and J. D. Baines. 1996. The UL 16 gene product of herpes simplex virus 1 is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236-242. [DOI] [PubMed] [Google Scholar]
- 42.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 43.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novoa, R. R., G. Calderita, P. Cabezas, R. M. Elliott, and C. Risco. 2005. Key Golgi factors for structural and functional maturation of bunyamwera virus. J. Virol. 79:10852-10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oshima, S., T. Daikoku, S. Shibata, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Characterization of the UL16 gene product of herpes simplex virus type 2. Arch. Virol. 143:863-880. [DOI] [PubMed] [Google Scholar]
- 46.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 47.Poon, A. P., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 49.Risco, C., J. L. Carrascosa, and T. K. Frey. 2003. Structural maturation of rubella virus in the Golgi complex. Virology 312:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 73:7952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato, S. B., K. Kawasaki, and S. Ohnishi. 1983. Hemolytic activity of influenza virus hemagglutinin glycoproteins activated in mildly acidic environments. Proc. Natl. Acad. Sci. USA 80:3153-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-Golgi network and is involved in cell-to-cell spread. Virology 308:23-36. [DOI] [PubMed] [Google Scholar]
- 55.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shenoy, S. K., and R. J. Lefkowitz. 2003. Trafficking patterns of beta-arrestin and G protein-coupled receptors determined by the kinetics of beta-arrestin deubiquitination. J. Biol. Chem. 278:14498-14506. [DOI] [PubMed] [Google Scholar]
- 57.Sherman, G., and S. L. Bachenheimer. 1987. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology 158:427-430. [DOI] [PubMed] [Google Scholar]
- 58.Sherman, G., and S. L. Bachenheimer. 1988. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology 163: 471-480. [DOI] [PubMed] [Google Scholar]
- 59.Silva, M. C., J. Schroer, and T. Shenk. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. USA 102:2081-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, K. O. 1964. Relationship between the envelope and infectivity of herpes simplex virus. Proc. Soc. Exp. Biol. Med. 115:814-816. [DOI] [PubMed] [Google Scholar]
- 62.Takakuwa, H., F. Goshima, T. Koshizuka, T. Murata, T. Daikoku, and Y. Nishiyama. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in over-expressing cells. Genes Cells 6:955-966. [DOI] [PubMed] [Google Scholar]
- 63.Taus, N. S., and J. D. Baines. 1998. Herpes simplex virus 1 DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology 252:443-449. [DOI] [PubMed] [Google Scholar]
- 64.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tu, B. P., S. C. Ho-Schleyer, K. J. Travers, and J. S. Weissman. 2000. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290:1571-1574. [DOI] [PubMed] [Google Scholar]
- 66.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weller, S. K., E. P. Carmichael, D. P. Aschman, D. J. Goldstein, and P. A. Schaffer. 1987. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology 161:198-210. [DOI] [PubMed] [Google Scholar]
- 68.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wing, B. A., G. C. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winslow, J. W. 1981. The reaction of sulfhydryl groups of sodium and potassium ion-activated adenosine triphosphatase with N-ethylmaleimide. The relationship between ligand-dependent alterations of nucleophilicity and enzymatic conformational states. J. Biol. Chem. 256:9522-9531. [PubMed] [Google Scholar]
- 71.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Y., B. Kaufmann, P. R. Chipman, R. J. Kuhn, and M. G. Rossmann. 2007. Structure of immature West Nile virus. J. Virol. 81:6141-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]