Abstract
Viroids are small, circular, single-stranded RNA molecules that, while not coding for any protein, cause several plant diseases. Viroids rely for their infectious cycle on host proteins, most of which are likely to be involved in endogenous RNA-mediated phenomena. Therefore, characterization of host factors interacting with the viroid may contribute to the elucidation of RNA-related pathways of the hosts. Potato spindle tuber viroid (PSTVd) infects several members of the Solanaceae family. In an RNA ligand screening we have previously isolated the tomato protein Virp1 by its ability to specifically interact with PSTVd positive-strand RNA. Virp1 is a bromodomain-containing protein with an atypical RNA binding domain and a nuclear localization signal. Here we investigate the role of Virp1 in the viroid infection cycle by the use of transgenic lines of Nicotiana tabacum and Nicotiana benthamiana that either overexpress the tomato Virp1 RNA or suppress the orthologous Nicotiana genes through RNA silencing. Plants of the Virp1-suppressed lines were not infected by PSTVd or Citrus exocortis viroid through mechanical inoculation, indicating a major role of Virp1 in viroid infection. On the other hand, overexpression of tomato Virp1 in N. tabacum and N. benthamiana plants did not affect PSTVd KF 440-2 infectivity or symptomatology in these species. Transfection experiments with isolated protoplasts revealed that Virp1-suppressed cells were unable to sustain viroid replication, suggesting that resistance to viroid infection in Virp1-suppressed plants is likely the result of cell-autonomous events.
Viroids are small, circular, single-stranded RNA molecules that cause several plant diseases. Although the viroid genome does not code for any protein, it contains the genetic information required for its replication and movement, as well as for the induction of pathogenic effects (39). The almost self-complementary circular RNA results in secondary structures which provide several binding signals to host factors that directly or indirectly serve the biological cycle of the infectious agent, whereas in some viroids they fold into a catalytically active ribozyme (39).
Viroids follow the course of a typical viral infection: there is an initial entry into the cell and its compartments (nuclei or chloroplasts), followed by replication in infected cells and spread and replication in new tissues, eventually causing a pathogenic effect. Since viroids do not code for any protein, the pathogenic effect has to be the result of the interaction of viroid RNA with host factors and interference with their normal functions. The study of viroids, therefore, involves the analysis of both the viroid RNA molecule per se and its interaction with the host plant. The major questions being addressed at present in viroid biology are the identities of the host enzymes involved in viroid replication and of the host proteins that protect the circular viroid and facilitate its movement through the host and the relationship between the viroid and epigenetic modifications of the chromatin. The usefulness of viroids as research tools and viroid-host interactions have been recently extensively reviewed (6, 9).
Potato spindle tuber viroid (PSTVd) is the type species of the genus Pospiviroid and in general of the family Pospiviroidae. PSTVd contains a specific signal that directs it to the nucleus (15, 44, 46), where it replicates via a rolling-circle mechanism. This starts with transcription of the incoming, monomeric, circular positive-strand RNA into multimeric, linear negative strands. The latter then serve as the replication intermediates for enhanced production of multimeric, linear positive strands that are finally processed into unit-length, mature circular RNAs (1). While the negative strands are anchored in the nucleoplasm, the positive strands traffic into the nucleolus, presumably for processing (33, 36). The positive-strand PSTVd RNA moves from cell to cell via plasmodesmata (7), the plant organelles which provide cytoplasmic connections between cells. Systemic movement from organ to organ occurs through the phloem (31, 47).
Despite PSTVd positive-strand RNA being resistant to mammalian Dicer activity in vitro (2), PSTVd was shown to be a target of posttranscriptional gene silencing (PTGS) in infected plants (19, 32). Double-stranded intermediates are presumably present in the nucleus only during replication. However, PSTVd-derived short interfering RNAs (siRNAs) were shown to accumulate in the cytoplasm but not in the nucleus (3). Nevertheless, PTGS is unable to efficiently suppress viroid infectivity. PSTVd RNA resistance to silencing could be due to its subcellular localization, to yet-unknown silencing suppressor activities by the RNA itself, to protection conferred on the RNA by host proteins, or to its secondary structure. A very recent paper (20) reveals that PSTVd positive-strand RNA is a substrate for Dicer-like cleavage but is resistant to RNA-induced silencing complex-mediated degradation, probably by virtue of its secondary structure.
In our search for host factors that assist steps in the biological cycle of viroids, we have isolated a tomato protein by its ability to interact specifically with PSTVd positive-strand RNA in an RNA ligand screening (27). This protein, which has been named Virp1, was confirmed to interact with PSTVd positive-strand RNA in vivo by immunoprecipitation from infected tomato leaves (27). Virp1 contains a nuclear localization signal and a bromodomain, which is an acetyl-lysine binding domain (5, 45) that is found in a variety of proteins and is thought to play a role in chromatin dynamics (8, 18). The protein does not display a typical RNA binding domain, but the carboxy-terminal region has been proposed as an RNA binding domain per se (14, 26).
Virp1 is the first bromodomain-containing protein reported to bind RNA. Although Virp1 is expressed in different tissues of healthy plants (27), its function in the tomato plant is presently unknown. It has been suggested that during viroid infection the symptoms observed may be due, at least partly, to competition for Virp1 binding between the viroid and a still-unknown endogenous RNA. Moreover, several roles in the viroid infection cycle have been proposed for this protein (27). Since Virp1 exhibits a nuclear localization signal, it might transfer the viroid RNA to the nucleus and bring it into contact with chromatin through its chromatin-associated bromodomain. Hence, Virp1 might also have a function during viroid replication. It has also been suggested that Virp1 may be involved in the systemic spread of PSTVd in the host since a PSTVd mutant defective for Virp1 binding is unable to spread systemically (25).
Applying the yeast three-hybrid system, we localized the viroid RNA region that is responsible for specific interaction with Virp1 in the terminal right domain of PSTVd (25). Analysis of the specific binding of the tomato protein Virp1 to PSTVd RNA revealed that two asymmetric internal loops within the terminal right domain are responsible for the specific RNA-protein interaction, and we called this structural element an “RY motif.” Simultaneous sequence alterations in both RY motifs abolished the specific binding to Virp1 and infectivity (14). Although these findings provided a strong indication that Virp1 is a factor required for viroid infectivity, they were not conclusive with respect to the role of the protein in the viroid infection cycle and to its being essential and/or sufficient for PSTVd infection.
Overexpression and suppression of Virp1 in the plant could give valuable information regarding the role of the protein in the viroid infectious cycle. Although Virp1 has been isolated in tomato (Lycopersicon esculentum), we used Nicotiana benthamiana and Nicotiana tabacum plants for this type of experiment, since these species are better suited for transgenic manipulation. By providing experimental evidence that PSTVd is unable to infect host plants following mechanical inoculation in which Virp1 has been suppressed, we identify Virp1 as a host factor essential for the viroid infection cycle. On the other hand, since overexpression of tomato Virp1 in N. tabacum and N. benthamiana plants does not affect PSTVd infectivity or symptomatology in these species, the disparities between Virp1 orthologs are not sufficient to explain differences in viroid infectivity, cellular distribution, or symptomatology between different species. Based on transfection experiments in isolated Virp1-suppressed protoplasts, we suggest for Virp1 a role at the cellular level.
MATERIALS AND METHODS
Cloning and sequence analysis of Virp1 homologs.
Oligonucleotides used for reverse transcription-PCR (RT-PCR) of the Virp1 homologs were purchased from MWG-Biotech AG (Ebersberg, Germany) and were as follows: X1BclI, 5′-GCGTGATCAATGGCATCCGCCGTCTTA-3′; X1revXbaI, 5′-GGGGTCTAGACTCAAGAGTGTGCATCATCAGC-3′; X1BDA, 5′-CACGGCCGATGTACTTGCAGGGGAGTTCTTGGAA-3′; X1BDB, 5′-GTAGATCTGTCGACGGTGTCAAGGGCCTCAAGT-3′ (the regions of homology to the tomato Virp1 cDNA sequence are indicated in bold). Two-step RT-PCR amplifications were performed with the Thermoscript RT-PCR System kit (Invitrogen, Carlsbad, CA). Primers and products are depicted in Fig. 1A. The nature of the DNA products was verified by Southern blot analysis (not shown), and the DNA products were separated by agarose gel electrophoresis, eluted from the gel, and ligated into pGEMT-Easy vector (Promega, WI) with the pGEMT-Easy Kit II (Promega, WI) The resulting ligation mixture was used to transform competent Escherichia coli JM83. Several colonies from each ligation were selected and screened for the presence of the desired insert by EcoRI restriction. DNA sequencing of several clones was performed with SP6 and T7 primers by the Genome Lab at IMBB-FORTH (Heraklion, Greece). The sequences were assembled with the aid of computer software.
FIG. 1.
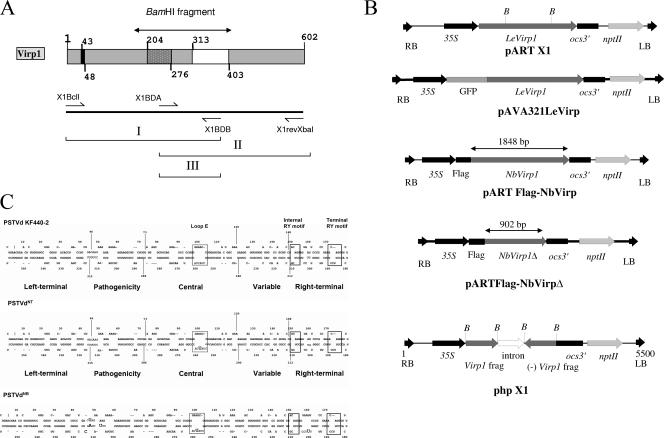
(A and B) Schematic representation of the tomato Virp1 protein (A, top); the location of the primers used for the RT-PCR (A, bottom); and the constructs used (B) for overexpression of Virp1, GFP-Virp1 fusion, FLAG-NbVirp1, FLAG-NbVirp1Δ, and suppression of Virp1 (from top to bottom, respectively) in N. benthamiana and N. tabacum transgenic plants. (A) (Top) Virp1 contains a nuclear localization signal (amino acids 43 to 48), a bromodomain (amino acids 204 to 276), and a potential RNA binding domain (amino acids 313 to 403). (B) nptII, selection gene; ocs3′, ocs terminator sequence; LB and RB, T-fragment left and right border sequences, respectively; 35S, cauliflower mosaic virus 35S promoter; B, BamHI sites delimiting the bromodomain. The truncated FLAG-NbVirp1Δ contains the first 902 bp of the NbVirp1 sequence. (C) Comparison of PSTVd strains KF 440-2, NT, and NB, showing the relative locations of the sequence differences between these three strains with respect to the loop E (dotted-line boxes) and RY motifs (solid-line boxes). In addition to a C259U substitution in the loop E motif of PSTVd-NT (shown by an oversized letter), strain PSTVd-NB also contains five additional mutations (shown by oversized letters).
Virp1 plasmid constructs. (i) GFP-Virp1 fusion construct.
For the generation of the fusion sequence the pAVA321 vector was used (41) and the whole LeVirp1 gene sequence was amplified by specific primers containing BglII (forward primer) and XbaI (reverse primer) restriction sites. The amplified product was then inserted at the respective pAVA321 plasmid vector sites, immediately downstream of the mGFP4 sequence (16). The expression cassette, 35S promoter-GFP-LeVirp1-35S terminator, was then subcloned in a pART27 binary vector (11). Control green fluorescent protein (GFP) agroinfiltration experiments were performed exactly as described before (22).
(ii) Sense-overexpressing construct.
A 2,203-bp RsaI fragment containing the whole open reading frame (ORF) from λVirp1 clone (AJ249592) (27) was introduced in the SmaI site of pART7 vector producing pART7/X1. The whole expression cassette of pART7/X1 containing the 35S promoter, the Virp1 gene sequence, and the ocs terminator was extracted from the plasmid by NotI digestion and subcloned in a NotI site of the binary vector pART27, producing transformation vector pART X1.
(iii) Production of Virp1 hairpin RNA.
To suppress Virp1 expression via RNA silencing, the BamHI 734-bp fragment containing the bromodomain sequence and the RNA binding domain sequence (Fig. 1A) was subcloned in sense and antisense orientation behind a 35S promoter of the pART7 vector, producing vector pART7/phX1. This fragment contains two regions of ≥21 bp with 100% identity with Nicotiana benthamiana and both Nicotiana tabacum orthologs. The two Virp1 fragments of the inverted repeat were separated by a 597-bp intron sequence (from the glutamate dehydrogenase [EC 1.4.1.2] gene of Vitis vinifera, the kind donation of K. Roubelakis-Angelakis). The expression cassette of pART7/phX1 including the 35S promoter, the Virp1 inverted repeat, and the ocs terminator was subcloned in pART27 as described for pART X1, generating construct phpX1. All constructs were sequenced before subsequent use.
Viroid strains and infections.
N. benthamiana or N. tabacum plants were mechanically inoculated using either a viroid transcript or infectious sap from tomato as described before (38). Specifically, 0.3 g of infected tomato leaf tissue was homogenized in 3 ml of buffer. One hundred microliters of the infectious homogenate was used to infect each individual N. benthamiana or tobacco leaf. Two leaves of each plant were inoculated. For N. benthamiana infections the PSTVd strain used was the severe isolate KF 440-2 transcribed with SP6 RNA polymerase from an EcoRI-linearized pHa106 plasmid. Strain PSTVdNB (34) was used for tobacco infections and was transcribed by T7 RNA polymerase from an EcoRI-linearized pTB110 plasmid (kindly provided by M. Wassenegger). Following inoculation, plants were kept in the greenhouse. Infection with PSTVd as seen by Northern hybridization could be detected 4 to 5 weeks postinoculation (p.i.). For the Citrus exocortis viroid (CEVd) inoculation, plasmid pCEV-MH1 (17) containing two head-to-tail-connected BamHI CEVd units was linearized with EcoRI for the transcription of the dimeric form of the CEVd positive strand. CEVd transcripts were used for the infection of tomato, and 4 weeks later infectious sap from one infected tomato plant was used for the N. benthamiana infections.
Virp1/PSTVd coimmunoprecipitation.
The FLAG sequence used for the immunoprecipitation experiments is MDYKDDDDK. The primers used for the PCR-based cloning were as follows: for tomato, FLAGVirp1-Fw, 5′-ATGGACTACAAAGACGATGACGACAAAGCATCCGCCGTCTTAGCTAG-3′, and Virp1-Rev, 5′-TCAAGAGTGTGCATCATCAG-3′. For N. benthamiana two overlapping PCRs were used and then the full FLAG-N. benthamiana sequence was obtained, taking advantage of the HindIII site that lies in the overlapping sequence of the two amplified parts of the gene. To generate the 5′ fragment, which was also then used as a truncated FLAG-Virp1 (FLAG-NbVirp1Δ), the primers were the following: FLAGVirp1, 5′-ATGGACTACAAAGACGATGACGACAAAGCA-3′, and NbVirp-Trunk-Rev, 5′-TCCTGTCGCTTCCGCCAT-3′. For the 3′ NbVirp1 fragment the primers used were NbVirp1-middle-Fw, 5′-GGCACGTTTTGAGGA-3′, and Virp1-Rev, 5′-TCAAGAGTGTGCATCATCAG-3′.
Virp1/PSTVd coimmunoprecipitation was performed by a modification of a published method (29). Briefly, agroinfiltrated tissue was collected 3 days p.i.; cross-linking of proteins and RNA was achieved by vacuum infiltration of 0.4% formaldehyde for 10 min. Cross-linking was quenched by 0.25 M glycine, and cross-linked tissue was frozen in liquid nitrogen. Proteins and RNA were extracted as indicated for Western hybridization (see the supplemental material) Following removal of cell debris, extract was incubated with anti-FLAG antibody (6 μl/ml; Sigma) and equilibrated G protein-Sepharose beads (4 Fast Flow; GE Healthcare Bio-sciences AB) overnight. Beads were then collected and washed five times with high-stringency buffer (50 mM Tris-Cl, pH 7.5, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA, 1 M NaCl, 3 M urea, 0.2 mM phenylmethylsulfonyl fluoride). Ribonucleoprotein complexes were then eluted and incubated in high-salt solution (200 mM NaCl) at 65°C for 4 h to reverse cross-links.
RNA was then extracted from the complexes with Tri reagent (Ambion, Austin, TX) according to the manufacturer's instructions and treated with DNase. RT-PCR was then performed using PSTVd-specific primers: PSTVdfw, ATCCCCGGGGAAACCTGGAGCGA, and PSTVdrev, CCCTGAAGCGCTCCTCCGAG. For the GFP and GFP-Virp1 coimmunoprecipitation experiments GFP antibody was used (Mobitec, Gottingen, Germany).
Plant transformations and plant growth conditions and agroinfiltration.
Transformation vectors pART27/X1 and pART27/phX1 were introduced into Agrobacterium tumefaciens strain LBA 4404 by triparental mating and were then used for the transformation of N. tabacum and N. benthamiana plants as described before (21). Explants and plants were grown at 25°C (day) and 18°C (night) in the growth chamber with a 16-h photoperiod provided by cool white fluorescent tube lights to give 90 μmol/m2/s photosynthetically active radiation. Plantlets were transferred to the greenhouse at a controlled temperature of 23°C. Agroinfiltration experiments were performed exactly as described previously (24).
Nucleic acid hybridizations.
Northern analyses for both mRNA and siRNA detection were performed according to previously published methods (21, 32). DNA probes were labeled by random priming (Invitrogen, Carlsbad, CA), and riboprobes were transcribed according to standard procedures (32). Hybridization to U1 RNA (a 156-base RNA of the spliceosomal snRNP complexes) using a potato U1 antisense probe was used as an internal standard to control RNA loading in short RNA Northern hybridizations as described before (4). The control U6 snRNA antisense probe was obtained as described in reference 3. Except where explicitly stated random prime DNA probes were prepared from a DNA fragment encompassing the whole tomato Virp1 ORF (RsaI-RsaI fragment). A 5′ probe was generated from an RsaI-BamHI fragment. The template used to transcribe the full-length PSTVd RNA was an EcoRI-linearized pHa106 plasmid (40) which was transcribed with SP6 RNA polymerase to generate the longer-than-unit-length PSTVd positive strand. HindIII-linearized pHa106 served as a template DNA to generate longer-than-unit-length PSTVd negative-strand transcripts with T7 RNA polymerase. The coat protein sequence of Potato virus Y (PVYN) was used to generate random-prime-labeled DNA probe as described previously (28) in order to detect PVY infectivity of transgenic protoplasts. 18S rRNA hybridization was used as a loading standard.
Protoplast preparation and transfection.
Protoplasts were prepared and inoculated as described before (10, 34) with minor modifications. Medium-size N. tabacum or N. benthamiana leaves were sterilized in 10% commercial bleach-Tween 20 and then washed three times with sterile water. Leaves were then transferred to petri dishes containing 10 ml substrate buffer (MS growth medium, pH 5.7, 0.4 M sucrose, 2 mM CaCl2, 25 mM morpholineethanesulfonic acid [MES]). An additional 10 ml of filter-sterilized substrate buffer plus 0.2 g cellulase and 0.1 g macerozyme (Onozuka R10; Apollo Scientific, Stockport, United Kingdom) was added to the petri dishes, which were then sealed with Parafilm and incubated at 25°C for 5 h in a dark chamber. The protoplast-containing solution was then transferred to a fresh tube, and 3 ml of culture medium (MS growth medium, pH 5.7, 0.4 M glucose, 2 mM CaCl2, 25 mM MES) was added on the top in order to create a gradient. Intact protoplasts were isolated from the interphase after centrifugation. Isolated protoplasts were pelleted at 40 × g and resuspended in culture medium to a final concentration of 0.5 × 106/ml. One milliliter of protoplast solution was used for electroporation in a Gene-Pulser (Bio-Rad, Hercules, CA) with 10 μg or 30 μg of total RNA extract from viroid-infected tomato leaves essentially as described before (10). Protoplasts were cultivated for 6 days prior to harvesting for transfection analysis. The protoplasts were then harvested by centrifugation and washed once with phosphate-buffered saline. RNA isolation from washed protoplasts was done as described for intact leaf tissue.
Staining and microscopy.
Leaf explants were stained with 4′,6′-diamino-2-phenylindole dihydrochloride (DAPI; Molecular Probes) in phosphate-buffered saline solution containing DAPI at a final concentration of 1 g/ml. Vacuum was applied for the first 10 min, and then explants were left in the staining solution overnight at 4°C. Protoplasts were stained for 16 h at 4°C in 1 μg/ml DAPI in culture medium (see above), washed and mounted in the same medium, and observed by fluorescence microscopy. Microscopic detection of GFP was performed using an inverted Nikon Eclipse E800 fluorescence microscope as described by Haseloff et al. (16) with a B-2E/C filter (excitation, 460 to 500 nm; diameter, 505 nm; baseline, 510 nm). DAPI-stained tissues and protoplasts were visualized with a UV-2E/C filter (excitation, 330 to 380 nm; diameter, 400 nm; baseline, 435 to 485 nm).
Nucleotide sequence accession numbers.
The sequences were deposited in the EBI database with accession numbers AJ504728 to AJ504732.
RESULTS
Virp1 homologs in species of the Solanaceae family.
The Solanaceae species Lycopersicon esculentum and Solanum tuberosum are known to be infected by the viroid PSTVd and dependent on the cultivar to show symptoms, while Nicotiana benthamiana is regarded as a symptomless host and Nicotiana tabacum as a nonhost for PSTVd KF440-2. Virp1 protein has been recently reported in tomato (Lycopersicon esculentum) to bind specifically to PSTVd and other related viroids (27). With the aim of assessing a correlation between the presence of the protein in a plant species and the plant's susceptibility to PSTVd, we looked for Virp1 orthologs in various Solanaceae species (L. esculentum, S. tuberosum, N. benthamiana, and N. tabacum). By Western blot assay, we could detect a protein of approximately 65 kDa in the four Solanaceae species (see Fig. S1A in the supplemental material). To investigate the transcription of the Virp1 genes, we performed Northern blot analysis on total RNA extracted from leaves (see Fig. S1B in the supplemental material). A signal of approximately 2.5 kb was detected in all four plant species examined. In N. tabacum the signal was fainter but was evident after longer exposure. With the purpose of cloning the Virp1 cDNAs from the different species in which Virp1 mRNAs were detected, we performed RT-PCRs with pairs of primers specific for different regions of tomato Virp1 sequence (Fig. 1A). The reactions yielded products of the expected sizes, the sequence identity of which was confirmed by Southern blot hybridization (data not shown). The fragments were cloned, and several individual clones were sequenced. The sequences of the fragments, which overlapped over a considerable length, were assembled with the aid of the appropriate software to yield the complete cDNA sequences, which were submitted to the EBI database (accession numbers AJ504728 to AJ504732). The sequence of the tomato cDNA is identical to the published Virp1 ORF sequence (27).
An analysis of the sequences revealed that two Virp1 genes are present in both N. tabacum and N. benthamiana, while a single copy of the gene was found in L. esculentum cv. “Rentita” and in S. tuberosum. The sequence similarity among the Virp1 orthologs, both at the nucleotide and at the amino acid level, was analyzed by employing the program CLUSTALW (see Alignment Fig. S1 and Alignment Fig. S2 in the supplemental material). An identity of 96% is shared at the nucleotide level between the potato and the tomato sequence, while there is an identity of 87 to 88% between the tomato sequence and any of the Nicotiana sequences and an identity of 88 to 89% between the potato sequence and any of the Nicotiana sequences. The Nicotiana sequences share an identity of 94 to 97% among them. The results of these comparisons are summarized in Table S1 in the supplemental material. Similar percentages of identities were observed at the amino acid level as summarized in Table S2 in the supplemental material. Since an overall high sequence conservation was observed in the Virp1 cDNAs among different Solanaceae species, differences observed between these species in relation to viroid infectivity are unlikely to be due to the lack of Virp1. However, it cannot be excluded in principle that disparities at the protein level between orthologs may affect plant-viroid interactions.
The infection of a plant by a viroid, and the emergence of symptoms, is a process that involves two actors, each of them with its own identity: the host plant and the pathogen viroid. PSTVd strain KF 440-2 (40) is capable of infecting tomato and N. benthamiana. A single-nucleotide C259U substitution in the central conserved region (shown by an oversized nucleotide letter in Fig. 1C) converts PSTVd isolate KF 440-2 from a noninfectious to an infectious, non-symptom-causing RNA for N. tabacum (variant PSTVdNT [42]). While PSTVdNT predominantly accumulated in the phloem, in N. tabacum PSTVNB (a strain derived in tobacco from PSTVNT) had higher accumulation levels and was present in the phloem, mesophyll, and epidermis (35). In addition to a C259U substitution in the loop E motif of PSTVdNT strain PSTVdNB also contains five additional mutations (shown by oversized nucleotide letters in Fig. 1C), only one of which is located in the terminal right domain where Virp1 interacts. As shown in Fig. 1C, there is no apparent correlation between the C259U substitution in PSTVdNT and the RY motif. Also, the only nucleotide mutated in PSTVdNB in the terminal right domain (G201U) does not appear to modify the secondary structure of the RY motifs.
Nicotiana Virp1 interacts in vivo with PSTVd.
We had previously shown that tomato Virp1 (LeVirp1) interacts with PSTVd in vivo (27). Here we performed an immunoprecipitation assay to verify the physical interaction between viroid and N. benthamiana Virp1 (NbVirp1) in vivo. NbVirp1, LeVirp1, and NbVirp1Δ (a truncated version encompassing amino acids 1 to 299 and thus predicted not to bind PSTVd positive-strand RNA, by similarity to a truncated version of LeVirp1), each carrying a FLAG epitope at the N terminus (Fig. 1B), were expressed in viroid-infected N. benthamiana leaves by agroinfiltration. Leaf extracts were subsequently subjected to anti-FLAG immunoprecipitation. PSTVd RNA coimmunoprecipitated with NbVirp1 and LeVirp1 but not with NbVirp1Δ or from healthy tissue (Fig. 2); this interaction withstood very stringent wash conditions (3 M urea). Therefore, we concluded that in Nicotiana as in tomato, Virp1 interacts strongly with the viroid in vivo.
FIG. 2.
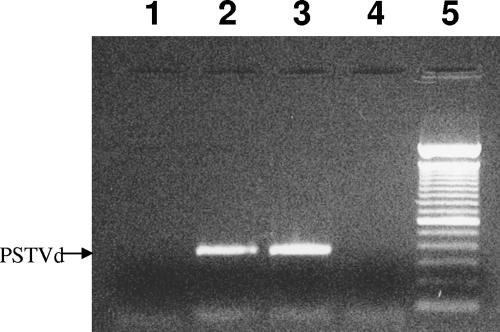
PSTVd coprecipitates with FLAG-NbVirp1. PSTVd-infected leaf tissue was infiltrated with FLAG-NbVirp1, FLAG-LeVirp1, and FLAG-NbVirp1Δ. An anti-FLAG antibody was then used to immunoprecipitate the tagged proteins, and RT-PCR using PSTVd-specific primers was then performed to detect coprecipitated viroid RNA. Lane 1, noninfiltrated tissue; lane 2, FLAG-LeVirp1; lane 3, FLAG-NbVirp1; lane 4, FLAG-NbVirp1Δ; lane 5, 100-bp DNA ladder.
Virp1 localizes in the nucleus of healthy and infected leaf cells.
To study the cellular and subcellular localization of Virp1, a 35S-GFP-LeVirp1 fusion construct was generated (pAVA321LeVirp, Fig. 1B). First, we performed again a coimmunoprecipitation experiment in PSTVd-infected N. benthamiana leaves agroinfiltrated with pAVA321LeVirp. This time, however, an anti-GFP antibody was used to immunoprecipitate the GFP-Virp1 fusion protein. The PSTVd sequence was then easily detected by RT-PCR in the immunoprecipitate (not shown), indicating that the fusion protein was still binding to the viroid. Viroid RNA could not be detected in the control GFP-infiltrated immunoprecipitate. The fusion protein, overexpressed by agroinfiltration in N. benthamiana leaves, was detected only in the nuclei (Fig. 3) of the infiltrated tissue, unlike the GFP control, which could be detected both in cytoplasm and in nucleoplasm (not shown). It has been previously suggested (25) that Virp1 moves from cell to cell, perhaps assisting the spreading of PSTVd in the host. Therefore, we reasoned that, if Virp1 moved from cell to cell, the fluorescence of GFP-LeVirp1 would be detected outside the agroinfiltrated area, while GFP alone would not spread outside the infiltrated region.
FIG. 3.
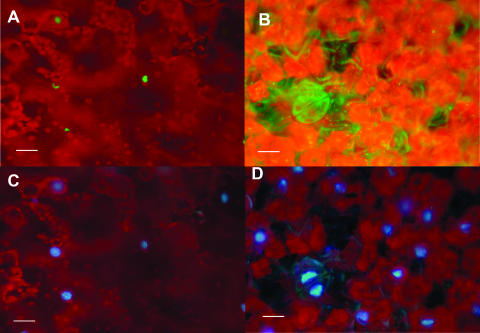
GFP-Virp1 is localized in the nucleus. Leaf area agroinfiltrated with GFP-Virp1 fusion construct. (A and B) GFP fluorescence reveals fusion protein localization in the nucleus (A), unlike unfused GFP, which can be found all over the cell (B). (C and D) Same areas as those in panels A and B, respectively, viewed for DAPI fluorescence. The red background is due to the natural fluorescence of chlorophyll. Bars, 25 μm.
The border zone of agroinfiltration was carefully marked and compared with the area in which GFP fluorescence could be detected p.i. GFP fluorescence was detected exclusively in the infiltrated zone, indicating that the GFP-Virp1 fusion does not spread, at least not in visible amounts (see Fig. S2 in the supplemental material). The same pattern of cellular and subcellular localization was recapitulated in infiltrations of viroid-infected N. benthamiana leaves as well as healthy and viroid-infected tomato leaves (not shown).
Overexpression of LeVirp1 in N. tabacum and N. benthamiana plants does not affect PSTVd infectivity or symptomatology in these species.
To analyze the function of Virp1 in viroid infection, we overexpressed Virp1 in N. tabacum and N. benthamiana plants. Ten transgenic N. tabacum and 11 transgenic N. benthamiana lines, estimated as single-locus transgenic lines for 35S-LeVirp1 (pARTX1, Fig. 1B) by their T1 segregation rates on kanamycin, were generated. Only these lines were used for further experimentation after homozygous plants for each line were obtained. In these lines the tomato Virp1 gene is present as a transgene in addition to the two already existing endogenous Virp1 homologs of both N. tabacum and N. benthamiana. During tissue culture for the generation of these lines we observed that over 20% of the regenerating N. tabacum shoots and over 50% of the regenerating N. benthamiana shoots eventually died, unlike the control GFP-transgenic regenerants. T1 transgenic lines demonstrated no obvious phenotypic abnormalities, except for mild epinasty at the cotyledon stage (data not shown). Virp1 mRNA expression levels, analyzed by Northern analysis for five N. tabacum and five N. benthamiana Virp1 transgenic lines, were generally higher in the transgenic lines than in the wild type (wt) (Fig. 4). Overexpression of Virp1 in the lines used for further experimentation was also verified at the protein level by Western blotting (see Fig. S6 in the supplemental material).
FIG. 4.
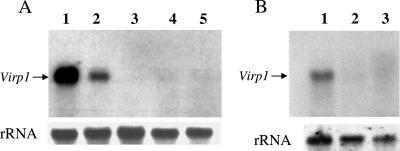
Overexpression of Virp1 in N. benthamiana plants. Lane 1, wt; lanes 2 to 5, transgenic lines BX1.2, BX1.7, BX1.10, and BX1.13, respectively. (Top) Virp1-specific DNA probe. (Bottom) Hybridization of the same membrane with an 18S rRNA probe as a loading control.
PSTVd strain KF 440-2, which induces severe symptoms in tomato, is infectious but asymptomatic in N. benthamiana and noninfectious in N. tabacum. In order to investigate whether the expression of the tomato Virp1 gene in N. benthamiana and N. tabacum is able to alter the infectivity and symptomatology of this viroid strain, we inoculated our Virp1-overexpressing lines with strain KF 440-2-infected tomato sap. Both the time course and the levels of viroid infection in the N. benthamiana transgenic lines were similar to those of the wt control (not shown). No symptoms were detected in Virp1-overexpressing N. benthamiana following PSTVd strain KF 440-2 infection. No infection was observed in N. tabacum Virp1-transgenic plants, indicating that expression of the tomato Virp1 did not alter the infectivity of strain KF 440-2 in N. tabacum plants (not shown).
Virp1-suppressed N. tabacum and N. benthamiana plants are resistant to mechanical inoculation by PSTVd and CEVd.
Further, we wanted to investigate the function of Virp1 in viroid infection by suppressing the Virp1 gene through RNA interference. Twelve N. tabacum and 10 N. benthamiana transgenic lines carrying the Virp1 hairpin-producing construct were generated (the transformation construct is schematically depicted in Fig. 1B). The majority of these plants (9/12 N. tabacum and 9/10 N. benthamiana plants) were found to carry one transgenic locus based on T1 germination rates on kanamycin-containing medium. To make additional investigations of transgene copy number, Southern blot hybridization was performed (not shown) using the marker gene (nptII) sequence as a probe in order to differentiate endogenous from transgenic Virp1 copies. Only lines that were confirmed to carry a single copy of the transgene were used for further experimentation. None of these lines demonstrated any obvious phenotypic abnormalities in the T1 or any subsequent generation.
Efficient suppression of Virp1 was confirmed for three out of four N. benthamiana and both N. tabacum lines analyzed (Fig. 5). N. benthamiana line ph20nb showed no suppression of the full-size Virp1 mRNA plus an additional more intense and much smaller transcript (see Fig. S3 in the supplemental material). This line was further used only as an additional control.
FIG. 5.
Generation of Virp1-suppressed lines with the use of hairpin constructs. Virp1 mRNA levels are strongly reduced in most N. benthamiana (A) and N. tabacum (B) transgenic lines. (A) (Top) Northern blot analysis of Virp1 mRNA in N. benthamiana. Lane 1, line BX1.13 overexpressing Virp1; lane 2, wt N. benthamiana; lanes 3 to 5, transgenic N. benthamiana in which expression of Virp1 is suppressed (lines ph5.2nb, ph10nb, and ph11nb, respectively). A 5′ region of the Virp1 sequence was used as a probe to avoid detection of the hairpin transcript. (Bottom) Hybridization of the same membrane with an 18S rRNA probe as a loading control. (B) (Top) Northern blot analysis of Virp1 expression levels in transgenic N. tabacum lines. Lane 1, wt N. tabacum; lanes 2 and 3, transgenic N. tabacum in which expression of Virp1 is suppressed (lines ph10nt and ph4nt, respectively). A 5′ region of the Virp1 sequence was used as a probe to avoid detection of the hairpin transcript. (Bottom) Loading control (18S rRNA).
To verify that the suppression of Virp1 was due to RNA silencing induced by the Virp1 hairpin, we analyzed the plants by Northern hybridization looking for Virp1-specific siRNAs. Virp1-specific siRNAs were found in all transgenic lines with reduced levels of Virp1 in both N. benthamiana (lines ph5.2nb, ph10nb, and ph11nb [see Fig. S4A in the supplemental material]) and N. tabacum (lines ph4nt and ph10nt [see Fig. S4B in the supplemental material]), confirming that suppression was due to RNA silencing. In line ph20nb, as expected, siRNAs could not be detected.
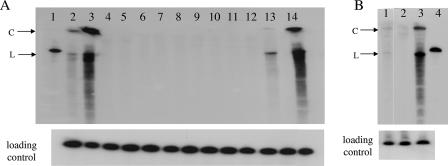
Next, we analyzed the ability of PSTVd to infect Virp1-suppressed transgenic plants. T1 N. benthamiana plants from lines ph5.2nb, ph10nb, ph11nb, and ph20nb and N. tabacum plants from lines ph4nt and ph10nt were inoculated with the appropriate viroid strain (KF 440-2 for N. benthamiana [40] and PSTVNB for N. tabacum [35]). Viroid inoculations were performed mechanically using infectious sap from tomato. Five weeks p.i. two medium-size leaves from the top of each plant were collected for RNA analysis. Northern hybridizations revealed PSTVd infection only in wt plants and plants of the ph20nb line and marginally in one transgenic line (ph4nt) (Fig. 6). The experiment was repeated at least twice for each of the above lines in the T2 generation, once with infectious sap and once with in vitro-synthesized viroid transcripts, with each experiment giving the same results as those for the T1 plants (not shown).
FIG. 6.
Transgenic N. benthamiana and N. tabacum lines suppressed for Virp1 cannot be infected by PSTVd. Detection of viroid infection by Northern blot analysis of RNA extracted from the upper leaves of transgenic lines 6 weeks p.i. Samples were separated by polyacrylamide gel electrophoresis on 6% gels. The probe was negative-strand RNA transcript of PSTVd KF 440-2 strain. (A) Lane 1, in vitro-transcribed PSTVd; lanes 2 and 3, wt N. benthamiana plants infected with 30 and 100 μl of inoculum, respectively; lanes 4 to 14, Virp1-suppressed transgenic lines, two or three plants each; lanes 4 to 6, line ph5.2nb; lanes 7 to 9, line ph10nb; lanes 10 to 12, line ph11nb; lanes 13 and 14, line ph20nb. (Bottom) The same membrane was hybridized with a U6 snRNA riboprobe as a loading control. (B) Lane 1, ph4nt; lane 2, ph10nt; lane 3, wt N. tabacum; lane 4, in vitro-transcribed PSTVd. (Bottom) Loading control (18S rRNA). C, size of circular form of viroid; L, size of linear form of viroid. The same source of inoculum was used for all experiments.
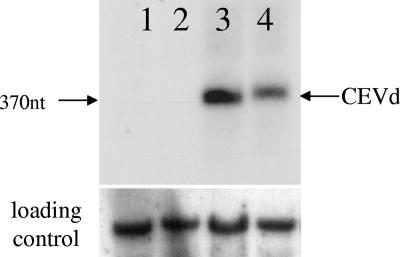
In a similar experiment CEVd transcripts (see Materials and Methods) were used to mechanically inoculate tomato seedlings. Five weeks later, sap from infected tomato plants was used to inoculate 10 wt and 10 Virp1-suppressed N. benthamiana plants. Six weeks p.i. newly developed leaves from two wt and two Virp1-suppressed plants, randomly selected, were isolated and analyzed for the presence of CEVd by Northern hybridization. Viroid could be detected in the wt but not in the Virp1-suppressed samples tested (Fig. 7).
FIG. 7.
CEVd is unable to infect Virp1-suppressed N. benthamiana. (Top) Northern blot analysis of Virp1-suppressed (ph11nb, lanes 1 and 2) and wt (lanes 3 and 4) plants inoculated with CEVd. Total RNA extracted from noninoculated leaves 6 weeks p.i. was separated on a 1.5% agarose gel; a CEVd-specific DNA probe was used. (Bottom) The same membrane was hybridized with a U1 snRNA riboprobe as a loading control. nt, nucleotide.
Virp1 has a major role in PSTVd infection.
To understand whether PSTVd is precluded from infecting Virp1-suppressed Nicotiana plants due to impairment of viroid replication in the cell, we analyzed the ability of the viroid to replicate in Virp1-suppressed N. tabacum protoplasts. Protoplasts from leaves of the Virp1-suppressed ph10nt line were isolated and were found to preserve the Virp1 suppression (see Fig. S5A in the supplemental material). These protoplasts were electroporated with RNA extracted from PSTVdNB-infected tomato and were then kept in culture for 6 days before total RNA was extracted. We could not detect PSTVd in Virp1-suppressed protoplasts by Northern hybridization analysis, indicating that PSTVd was unable to replicate in the Virp1-suppressed protoplasts. In contrast, PSTVd RNA quickly accumulated and could be readily detected in control wt protoplasts (Fig. 8). Similar results were obtained in N. benthamiana Virp1-suppressed protoplasts (not shown). To test for a more general compromised resistance to parasitic RNA, we infected Virp1-suppressed protoplasts with PVY. We found that PVY accumulated at similar levels in ph10nt protoplasts and in wt, indicating that, at least as far as this pathogen is concerned, Virp1-suppressed protoplasts do not show compromised resistance (see Fig. S5B in the supplemental material). Taken together these results indicate that N. tabacum plants with strongly reduced levels of Virp1 protein are unable to sustain significant PSTVd accumulation.
FIG. 8.
Virp1-suppressed N. tabacum protoplasts are unable to sustain viroid infection. Protoplasts from wt (lanes 1 and 2) and Virp1-suppressed (line ph10nt, lanes 3 and 4) N. tabacum were electroporated with total RNA extracted from the leaves of a tomato plant that had been infected with PSTVd strain PSTVdNb. Protoplasts were cultured for 6 days postelectroporation, and total RNA was analyzed by Northern blotting. The probe was negative-strand RNA transcript of PSTVd strain PSTVdNb. (Bottom) Loading control (18S rRNA).
DISCUSSION
Tomato Virp1 protein has been identified as specifically binding to PSTVd viroid RNA (27). Here, we show that this protein is absolutely required for PSTVd to infect Nicotiana benthamiana and Nicotiana tabacum by mechanical inoculation. We have identified and sequenced genes orthologous to tomato (Lycopersicon esculentum) Virp1, from N. benthamiana (two genes), Ν. tabacum (two genes), and potato (S. tuberosum, one gene). All the sequences analyzed are highly conserved both at the nucleotide and at the amino acid level. Although coding sequence conservation is generally high among these members of the Solanaceae family (37), it is likely that the very high sequence and domain conservation between the Virp1 orthologs and paralogs reflects functional conservation for the protein. Alignment between the Nicotiana Virp1 genes revealed that for each single N. benthamiana paralog there is a single N. tabacum ortholog with very high sequence identity and vice versa. As expected, conservation is even higher when sequences at specific domains are compared. The sequence identity is sufficiently high among all the orthologs that silencing of all Virp1 genes could be achieved by a single hairpin construct, using the tomato Virp1 clone.
Since PSTVd specifically binds Virp1, we deemed it interesting to study whether the differences in strain KF 440-2 infectivity between L. esculentum, N. benthamiana, and N. tabacum (infectious with symptoms, infectious without symptoms, and noninfectious, respectively) were related to the differences in Virp1 sequence in individual species. We therefore overexpressed the tomato Virp1 gene in the two Nicotiana species. Since we did not observe any alteration in PSTVd infectivity profile in Virp1-transgenic N. benthamiana and N. tabacum lines, we concluded that the differences in Virp1 sequences are not responsible, at least not solely, for infectivity and pathogenicity of PSTVd in the three Solanaceae species. While performing these experiments, we observed that only a relatively small fraction of the transgenic lines overexpressed Virp1. Moreover, in these plants LeVirp1 transcript levels were found to be much lower than those of a GFP transgene expressed from control transformation constructs (not shown). We, therefore, speculate that overexpression of Virp1 might be toxic for the plant. This toxicity was possibly manifested in the lower numbers of viable transformed lines which were obtained.
With the aim of elucidating the function of Virp1 in PSTVd infection, we silenced Virp1 mRNA by RNA interference. Northern blot analyses revealed that Virp1 accumulation was indeed knocked down in both N. benthamiana and N. tabacum transgenic plants carrying the LeVirp1 hairpin construct and that the suppression was the result of PTGS, as Virp1-specific siRNAs were detectable. Since Virp1 paralogs are highly conserved, our probes are expected to detect both sequences so that lack of mRNA hybridization signal indicates suppression of all endogenous Virp1 mRNAs. As is usually the case in PTGS (23, 43) the targeted sequences are not 100% suppressed and traces of the targeted mRNA can be still found in the steady-state Northern analysis. Although the high sequence conservation in all Solanaceae species studied suggests a conserved function for Virp1 in these plants, Virp1 knockdowns did not reveal any obvious phenotype under the conditions studied. It is possible that the low levels of Virp1 left are still sufficient to fulfill its physiological function. Alternatively, the role of Virp1 could be essential under different environmental or physiological conditions, in which Virp1-suppressed plants would then show a phenotype.
In this paper we describe Virp1 as the first host factor identified as being necessary for viroid infectivity without being essential for the plant. Mechanical inoculation of Virp1-suppressed plants (either N. benthamiana or N. tabacum) with the appropriate PSTVd strain did not result in systemic PSTVd infection in these lines. In addition, Virp1-suppressed N. benthamiana plants were not infected by another viroid of the Pospovirideae family, CEVd. CEVd has been shown before to bind Virp1 protein specifically in vitro (26). Nevertheless, it is possible that “resistance” to viroid infection in Virp1-suppressed plants could be broken by using other means of infection such as agroinfection or grafting onto infected wt rootstocks.
In contrast to wt N. tabacum protoplasts, where replication of the viroid takes place, following introduction of PSTVd RNA in the cells, PSTVd replication could not be established in protoplasts from Virp1-suppressed N. tabacum plants, indicating a role for Virp1 protein in the viroid replication cycle. It is possible that Virp1 is necessary for the replication of the viroid per se and/or necessary for the replication cycle of the viroid indirectly, for example by enabling the viroid to enter the nucleus, where it replicates. Since we show here that Virp1 is mainly localized in the nucleus and is strongly bound in vivo by PSTVd, it is possible that Virp1 may be hijacked by the viroid to assist the viroid's transport to the nucleus, where it would be largely protected from the activity of various components of the RNA interference machinery.
A phloem-specific protein (named CsPP2) has been isolated in cucumber and shown to bind to and move systemically with Hop stunt viroid (12, 13, 30). We have no evidence, either in silico or experimental, that Virp1 may have functional similarities with CsPP2. Nevertheless, a role for Virp1 in the uploading of PSTVd to the phloem cannot be excluded at present.
Based on the experimental evidence so far, we can only speculate about the function that Virp1 would serve for the host. We did not observe a strong phenotype upon suppression or overexpression of this protein in the Nicotiana species studied. The analysis of the domains found in the protein (a bromodomain, a putative RNA binding domain, and a nuclear localization signal) implies that Virp1 participation in a nuclear mechanism where RNA has an important role seems realistic. An obvious candidate would in this case be RNA-mediated DNA methylation, although such an involvement can only be speculative at present.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Martin Tabler.
We are grateful to M. Tzanopoulou, D. Yakoumakis, T. Saridaki, and K. Skreka for assistance in this work during their lab rotations; M. Gozmanova, K. Melzak, and N. Vrettos for critical reading of the manuscript and useful suggestions; and K. Gouskou for advice on immunoprecipitation experiments. We thank M. Wassenegger (AIPlanta, Neustadt, Germany) for plasmids.
This work was supported by the European Union (contracts QLG2-CT-2002-01673 VIS and 2004-005120 FOSRAK) and the national EPEAEK program.
Footnotes
Published ahead of print on 26 September 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 2.Chang, J., P. Provost, and J. M. Taylor. 2003. Resistance of human hepatitis delta virus RNAs to dicer activity. J. Virol. 77:11910-11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denti, M. A., A. Boutla, M. Tsagris, and M. Tabler. 2004. Short interfering RNAs specific for potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J. 37:762-769. [DOI] [PubMed] [Google Scholar]
- 4.Denti, M. A., A. E. Martinez de Alba, R. Sagesser, M. Tsagris, and M. Tabler. 2000. A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme. Nucleic Acids Res. 28:1045-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 6.Ding, B., and A. Itaya. 2007. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microbe Interact. 20:7-20. [DOI] [PubMed] [Google Scholar]
- 7.Ding, B., M. O. Kwon, R. Hammond, and R. Owens. 1997. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12:931-936. [DOI] [PubMed] [Google Scholar]
- 8.Filetici, P., P. Ornaghi, and P. Ballario. 2001. The bromodomain: a chromatin browser? Front. Biosci. 6:D866-D876. [DOI] [PubMed] [Google Scholar]
- 9.Flores, R., C. Hernandez, A. E. Martinez de Alba, J. A. Daros, and F. Di Serio. 2004. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 10.Gaire, F., C. Schmitt, C. Stussi-Garaud, L. Pinck, and C. Ritzenthaler. 1999. Protein 2A of grapevine fanleaf nepovirus is implicated in RNA2 replication and colocalizes to the replication site. Virology 264:25-36. [DOI] [PubMed] [Google Scholar]
- 11.Gleave, A. P. 1992. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20:1203-1207. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, G., and V. Pallas. 2001. Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant-Microbe Interact. 14:910-913. [DOI] [PubMed] [Google Scholar]
- 13.Gomez, G., and V. Pallas. 2004. A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with hop stunt viroid RNA. J. Virol. 78:10104-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gozmanova, M., M. A. Denti, I. N. Minkov, M. Tsagris, and M. Tabler. 2003. Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31:5534-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harders, J., N. Lukacs, M. Robert-Nicoud, T. M. Jovin, and D. Riesner. 1989. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haseloff, J., K. R. Siemering, D. C. Prasher, and S. Hodge. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillman, M. 1986. Molecularbiologische Analyse von Struktur-Funktionsbeziehungen des Citrus exocortis viroid. Ph.D. dissertation. University of Munich, Munich, Germany.
- 18.Horn, P. J., and C. L. Peterson. 2001. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 6:D1019-D1023. [DOI] [PubMed] [Google Scholar]
- 19.Itaya, A., A. Folimonov, Y. Matsuda, R. S. Nelson, and B. Ding. 2001. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant-Microbe Interact. 14:1332-1334. [DOI] [PubMed] [Google Scholar]
- 20.Itaya, A., X. Zhong, R. Bundschuh, Y. Qi, Y. Wang, R. Takeda, A. R. Harris, C. Molina, R. S. Nelson, and B. Ding. 2007. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81:2980-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalantidis, K., S. Psaradakis, M. Tabler, and M. Tsagris. 2002. The occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistance to the virus. Mol. Plant-Microbe Interact. 15:826-833. [DOI] [PubMed] [Google Scholar]
- 22.Kalantidis, K., M. Tsagris, and M. Tabler. 2006. Spontaneous short-range silencing of a GFP transgene in Nicotiana benthamiana is possibly mediated by small quantities of siRNA that do not trigger systemic silencing. Plant J. 45:1006-1016. [DOI] [PubMed] [Google Scholar]
- 23.Kerschen, A., C. A. Napoli, R. A. Jorgensen, and A. E. Muller. 2004. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566:223-228. [DOI] [PubMed] [Google Scholar]
- 24.Koscianska, E., K. Kalantidis, K. Wypijewski, J. Sadowski, and M. Tabler. 2005. Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum. Plant Mol. Biol. 59:647-661. [DOI] [PubMed] [Google Scholar]
- 25.Maniataki, E., A. E. Martinez de Alba, R. Sagesser, M. Tabler, and M. Tsagris. 2003. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9:346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez de Alba, A. E. 2000. Ph.D. dissertation. Universidad del Pais Vasco, Bilbao, Spain.
- 27.Martinez de Alba, A. E., R. Sagesser, M. Tabler, and M. Tsagris. 2003. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77:9685-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Missiou, A., K. Kalantidis, A. Boutla, S. Tzortzakaki, M. Tabler, and M. Tsagris. 2004. Generation of transgenic potato plants highly resistant to potato virus Y (PVY) through RNA silencing. Mol. Breed. 14:185-197. [Google Scholar]
- 29.Niranjanakumari, S., E. Lasda, R. Brazas, and M. A. Garcia-Blanco. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26:182-190. [DOI] [PubMed] [Google Scholar]
- 30.Owens, R. A., M. Blackburn, and B. Ding. 2001. Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant-Microbe Interact. 14:905-909. [DOI] [PubMed] [Google Scholar]
- 31.Palukaitis, P. 1987. Potato spindle tuber viroid: investigation of the long-distance, intra-plant transport route. Virology 158:239-241. [DOI] [PubMed] [Google Scholar]
- 32.Papaefthimiou, I., A. Hamilton, M. Denti, D. Baulcombe, M. Tsagris, and M. Tabler. 2001. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, Y., and B. Ding. 2003. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15:2566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi, Y., and B. Ding. 2002. Replication of potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Virology 302:445-456. [DOI] [PubMed] [Google Scholar]
- 35.Qi, Y., T. Pelissier, A. Itaya, E. Hunt, M. Wassenegger, and B. Ding. 2004. Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16:1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin, H., Y. Dong, and A. G. von Arnim. 2003. Epigenetic interactions between Arabidopsis transgenes: characterization in light of transgene integration sites. Plant Mol. Biol. 52:217-231. [DOI] [PubMed] [Google Scholar]
- 37.Rensink, W. A., Y. Lee, J. Liu, S. Iobst, S. Ouyang, and C. R. Buell. 2005. Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics 6:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabler, M., and H. L. Sanger. 1984. Cloned single- and double-stranded DNA copies of potato spindle tuber viroid (PSTV) RNA and co-inoculated subgenomic DNA fragments are infectious. EMBO J. 3:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]
- 40.Tsagris, M., M. Tabler, and H. L. Sanger. 1991. Ribonuclease T1 generates circular RNA molecules from viroid-specific RNA transcripts by cleavage and intramolecular ligation. Nucleic Acids Res. 19:1605-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Arnim, A. G., X. W. Deng, and M. G. Stacey. 1998. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221:35-43. [DOI] [PubMed] [Google Scholar]
- 42.Wassenegger, M., R. L. Spieker, S. Thalmeir, F. U. Gast, L. Riedel, and H. L. Sanger. 1996. A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226:191-197. [DOI] [PubMed] [Google Scholar]
- 43.Wesley, S. V., C. A. Helliwell, N. A. Smith, M. B. Wang, D. T. Rouse, Q. Liu, P. S. Gooding, S. P. Singh, D. Abbott, P. A. Stoutjesdijk, S. P. Robinson, A. P. Gleave, A. G. Green, and P. M. Waterhouse. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27:581-590. [DOI] [PubMed] [Google Scholar]
- 44.Woo, Y. M., A. Itaya, R. Owens, L. Tang, R. Hammond, H. Chou, M. Lai, and B. Ding. 1999. Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplasts. Plant J. 17:627-635. [Google Scholar]
- 45.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, Y., R. A. Owens, and R. W. Hammond. 2001. Use of a vector based on potato virus X in a whole plant assay to demonstrate nuclear targeting of potato spindle tuber viroid. J. Gen. Virol. 82:1491-1497. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, Y., L. Green, Y. M. Woo, R. Owens, and B. Ding. 2001. Cellular basis of potato spindle tuber viroid systemic movement. Virology 279:69-77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.