Abstract
Recent studies have demonstrated that influenza A virus infection activates the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway by binding of influenza NS1 protein to the p85 regulatory subunit of PI3K. Our previous study proposed that two polyproline motifs in NS1 (amino acids 164 to 167 [PXXP], SH3 binding motif 1, and amino acids 213 to 216 [PPXXP], SH3 binding motif 2) may mediate binding to the p85 subunit of PI3K. Here we performed individual mutational analyses on these two motifs and demonstrated that SH3 binding motif 1 contributes to the interactions of NS1 with p85β, whereas SH3 binding motif 2 is not required for this process. Mutant viruses carrying NS1 with mutations in SH3 binding motif 1 failed to interact with p85β and induce the subsequent activation of PI3K/Akt pathway. Mutant virus bearing mutations in SH3 binding motif 2 exhibited similar phenotype as the wild-type (WT) virus. Furthermore, viruses with mutations in SH3 binding motif 1 induced more severe apoptosis than did the WT virus. Our data suggest that SH3 binding motif 1 in NS1 protein is required for NS1-p85β interaction and PI3K/Akt activation. Activation of PI3K/Akt pathway is beneficial for virus replication by inhibiting virus induced apoptosis through phosphorylation of caspase-9.
Influenza A viruses are important pathogens that are contagious and cause acute respiratory disease in humans and different animal species. The RNA segment 8 of influenza A virus encodes two proteins: NS1 and NS2/NEP (19). NS1 is a multifunctional protein, which is translated from unspliced mRNA (20). Two functional domains have been identified in the NS1 protein: the RNA-binding domain near the N terminus and the effector domain in the C terminus (28). The RNA-binding activity of NS1 protein correlates with its ability to inhibit cellular pre-mRNA splicing (23, 29). RNA binding by NS1 is also required to efficiently counteract cellular alpha/beta interferon (IFN-α/β) functions (10) by inhibiting the activation of protein kinase R (PKR) (13, 24) and transcription factors NF-κB, IRF-3, and IRF-7 (36, 40). Within the effector domain of NS1, two binding sites for cellular proteins were identified. The binding site for the cleavage and polyadenylation specificity factor is positioned around amino acid 186 and the poly(A)-binding protein II binding site is located between amino acids 223 and 237 (22). These binding sites are required for the inhibition of 3′-end processing of cellular pre-mRNAs. New studies showed that NS1 protein interacts with various cellular proteins and thus is involved in regulating different functions. NS1 of influenza A virus was found to bind to RIG-I and inhibit downstream activation of IRF-3, thus preventing the transcriptional induction of IFN-β (26). NS1 was also found to bind directly to PKR (21) via the amino acid sequence from amino acids 123 to 127, leading to inhibition of PKR activation (27).
Phosphatidylinositol 3-kinases (PI3Ks) are a family of cellular heterodimeric enzymes that consist of a regulatory subunit (p85) and a catalytic subunit (p110). PI3K is activated by binding of the Src homology 2 (SH2) domains in the p85 subunit to autophosphorylated receptor or nonreceptor tyrosine kinases or some viral proteins in the cytoplasm (4, 14, 32, 34, 35). After activation, the p110 subunit of PI3K phosphorylates the lipid substrate phosphatidylinositol-4,5-bisphosphate (PIP2) to produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) (38). This molecule serves as a lipid second messenger and is able to regulate phosphorylation of a number of kinases, including Akt. Akt is activated via phosphorylation at Thr-308 and Ser-473 (1). Phosphorylated Akt plays a central role in modulating diverse downstream signaling pathways associated with cell proliferation, migration, differentiation, and the prevention of apoptosis (5, 41).
Recently, increasing amounts of information have demonstrated that influenza A virus infection leads to PI3K/Akt pathway activation by interaction of the viral NS1 protein with the p85 subunit of PI3K (8, 11, 31). Previously, we proposed three motifs in the NS1 protein, namely, the SH2 binding motif (amino acids 89 to 93 [YXXXM]), the SH3 binding motif 1 (amino acids 164 to 167 [PXXP]), and the SH3 binding motif 2 (amino acids 212 to 216 [PPXXP]) may be involved in NS1-p85 interaction and PI3K/Akt pathway activation. Furthermore, we showed that mutant virus PR8-SH2/SH3mt, which encodes NS1 with a total of six mutations, including Y89F and five substitutions for proline, failed to bind to p85 and thus did not activate PI3K/Akt pathway (31). Consistent with our data, mutational studies by Hale et al. revealed that both Tyr-89 and Met-93 are essential for the interaction of NS1 with p85β (11).
The present study was initiated to determine the role of the two SH3 binding motifs in NS1 protein in p85β interaction and PI3K/Akt pathway activation. We performed mutational analysis and report here that SH3 binding motif 1 is essential for the NS1-p85β interaction and PI3K/Akt pathway activation, whereas SH3 binding motif 2 is not required for this process. Consistent with no PI3K/Akt activation, mutant viruses containing mutations of SH3 binding motif 1 are attenuated for growth and are more proapoptotic than wild-type (WT) virus. Our data suggest that SH3 binding motif 1 in NS1 mediates influenza A virus-induced activation of the PI3K/Akt pathway, which is beneficial for virus replication by inhibiting virus-induced apoptosis.
MATERIALS AND METHODS
Cells and virus.
A549 cells (human lung carcinoma cells) and 293T (human embryonic kidney) cells were maintained in Dulbecco modified Eagle medium (Invitrogen) containing 10% fetal bovine serum. Madin-Darby canine kidney (MDCK) cells were cultivated in minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum. Influenza A/PR/8/34 (H1N1) (PR8) was propagated at 37°C in 11-day-old embryonated chicken eggs. Virus titers were determined on MDCK cells by plaque assay.
Plasmids.
Mutations in the NS1 protein coding sequence were introduced into the plasmid pHW198-NS (15). Plasmid pHW-NS-SH3-mf-1 encodes mutant NS1 with prolines at 164 and 167 replaced by alanines (P164-167A). It was generated by replacing the 48 nucleotides between MfeI and BbvCI with an oligonucleotide pair (Fw [5′-AAT TGT TGG CGA AAT TTC TGC ATT GGC TTC TCT TGC AGG ACA TAC TGC-3′] and Bw [5′-TCA GCA GTA TGT CCT GCA AGA GAA GCC AAT GCA GAA ATT TCG CCA AC-3′]). Plasmid pHW-NS-SH3-mf-2 encoding mutant NS1 with a proline at 212 replaced by serine and prolines at 213 and 216 replaced by alanines (P212S/P213-216A) was generated by site-directed mutagenesis using the primers NS-P212S/P213-216A Fw (5′-ATG AGA ATG GGA GAT CTG CAC TCA CTG CAA AAC AGA AAC G-3′) and NS-P212S/P213-216A Bw (5′-CGT TTC TGT TTT GCA GTG AGT GCA GAT CTC CCA TTC TCA T-3′). Plasmid pHWNS-SH3-mf-1/2mt encodes NS1 protein with mutations at P164-167A and P212S/P213-216A. It was generated by replacing the 48 nucleotides of pHW-NS-SH3-mf-2 with the oligonucleotide pair as described above.
WT NS1 and its respective mutants were PCR amplified by using the plasmids described above as a template and cloned into pcDNA3.1(−) (Invitrogen) at the NheI and EcoRI sites, resulting in plasmids pcDNA-NS1, pcDNA-NS1-SH3-mf-1, pcDNA-NS1-SH3-mf-2, and pcDNA-NS1-SH3-mf-1/2, respectively. All of the mutations were confirmed by restriction enzyme digestions, followed by DNA sequencing.
Plasmid pGEX-4T3-p85β was purchased from Addgene (Addgene plasmid 1406). This plasmid encodes mouse p85β and was used in a glutathione S-transferase (GST) pull-down assay. Mouse p85β gene was isolated by digesting pGEX-4T3-p85β with EcoRI/XhoI and then ligated into pcDNA4HisMax-B at EcoRI/XhoI sites (Invitrogen), generating plasmid pcDNA4-HisMax-mp85β. This plasmid was used in an Ni-Sepharose bead pull-down assay.
Generation of NS1 mutant viruses.
NS1 mutant viruses were generated by using an eight-plasmid reverse genetics system described by Hoffmann et al. (16). Plasmids pHW191-PB2, pHW192-PB1, pHW193-PA, pHW194-HA, pHW195-NP, pHW196-NA, pHW197-M, and pHW198-NS (15) were kindly obtained from E. Hoffmann and R. G. Webster (St. Jude Children's Research Hospital, Memphis, TN). The cocultured MDCK and 293T cells (3 × 105 cells per well of each six-well plate) were transfected with eight plasmids (pHW191-PB2, pHW192-PB1, pHW193-PA, pHW194-HA, pHW195-NP, pHW196-NA, and pHW197-M and each mutant NS1-encoding plasmid) by using TransIT LT-1 (Mirus) according to the manufacturer's instructions. Briefly, 2 μl of TransIT LT-1 per 1 μg of DNA was mixed, incubated at room temperature for 45 min, and added to the cells. After 6 h, the DNA-transfection mixture was replaced by 1 ml of Opti-MEM (Invitrogen). Twenty-four hours later, 1 ml of Opti-MEM containing TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (1 μg/ml) was added to the cells. Seventy-two hours later, the supernatant was harvested and passaged once on MDCK cells. Mutant viruses were rescued and named PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2. The viruses were propagated in 9- to 10-day-old embryonated chicken eggs and characterized by sequencing of the reverse transcription-PCR product derived from the NS segment.
Antibodies.
Phospho-Akt (Ser-473; 193H12) rabbit monoclonal antibody (MAb), phospho-Akt (Thr-308; 244F9) rabbit MAb, rabbit polyclonal PARP antibody, rabbit polyclonal Akt antibody, horseradish peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG), and HRP-conjugated anti-mouse IgG were purchased from Cell Signaling Technology. Rabbit polyclonal p-caspase-9 (Ser-196), monoclonal PI3K p85β (T15; non-cross-reactive with p85α, sc56934) antibody, rabbit IgG, and mouse IgG were purchased from Santa Cruz Biotechnology. Monoclonal mouse anti-His6 antibody was purchased from BD Biosciences. Monoclonal M1 antibody was purchased from Serotec. Alkaline phosphatase-conjugated anti-rabbit IgG and Cy2-conjugated anti-mouse IgG were purchased from Jackson Immunoresearch. Rabbit polyclonal NS1 was generated in our laboratory (30).
Western blot analysis.
Western blotting was performed as described previously (30) with minor modifications. Briefly, A549 cells (1 × 106) or MDCK cells (7 × 105) were plated into 35-mm dishes and were mock infected or infected with influenza viruses at a determined multiplicity of infection (MOI). At the indicated times, cell monolayers were washed with 0.01 M phosphate-buffered saline (PBS; 0.138 M NaCl, 0.0027 M KCl [pH 7.4]) and lysed with cell lysis buffer (Cell Signaling Technology) supplemented with 1 mM phenylmethanesulfonyl fluoride (Sigma). The lysates were collected and incubated on ice for 10 min. Lysates were cleared by centrifugation for 5 min at 12,000 × g at 4°C. The supernatant was analyzed for total protein content by using a Bradford assay (Bio-Rad). A portion (30 μg) of total protein was resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked for nonspecific binding with Tris-buffered saline (0.1 M Tris [pH 7.6], 0.9% NaCl) containing 0.1% Tween 20 and 10% skim milk for 1 h at room temperature. For examination of NS1 and His-tagged protein, cell lysates were probed with NS1 (1:2,000) or His6 (1:3,000) antibody, followed by incubation with alkaline phosphatase-conjugated anti-rabbit IgG (1:10,000). The immunoblots were then visualized by incubating with BCIB/NBT premix solution (Sigma). For examination of phosphorylated Akt, total Akt, p85β, p-caspase-9, or PARP, a primary antibody was diluted according to the manufacturer's suggestion and applied overnight at 4°C. A secondary antibody of HRP-conjugated anti-rabbit IgG or HRP-conjugated anti-mouse IgG was then added at room temperature for 1 h. The immunoblots were visualized with an enhanced chemiluminescence reagent (ECL Advance Western Blotting Detection Kit; GE Healthcare).
Coimmunoprecipitation analysis.
A549 cells were mock infected or infected by the viruses at an MOI of 1. At 6 h postinfection (p.i.), cell lysates were prepared as described above. Cell lysates (500 μg) were precleared by 5 μg of rabbit IgG and protein A-Sepharose beads (GE Healthcare). Then, 5 μg of NS1 antibody or rabbit IgG was immobilized to protein A-Sepharose beads, followed by incubation with precleared cell lysates for 2 h at 4°C. After extensive washes, the precipitated proteins were subjected to SDS-PAGE, followed by Western blotting with NS1 or p85β antibody.
Transfection.
293T cells were seeded in a six-well plate at a density of 106/well. A total of 1 μg of pcDNA3.1(−), pcDNA-NS1, pcDNA-NS1-SH3-mf-1, pcDNA-NS1-SH3-mf-2, or pcDNA-NS1-SH3-mf-1/2 was transfected by using CaCl2 according to a protocol described previously (17). After 24 h, cell lysates were prepared as described above and subjected to a GST pull-down assay.
293T cells seeded in a six-well plate were cotransfected by 1 μg of pcDNA-NS1 or its derivatives and 1 μg of pcDNA4HisMax-mp85β using the CaCl2 method. Twenty-four hours later, cell lysates were prepared and subjected to a Ni-Sepharose bead pull-down assay.
GST and Ni-Sepharose bead pull-down assay.
Escherichia coli BL21 cultures expressing GST-p85β fusion protein or GST alone were grown to mid-log phase and induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 22°C for overnight. The bacterial pellets were resuspended in PBS and sonicated to lyse the cells. Triton X-100 was added to the lysates to a final concentration of 1%, and the mixture was incubated for 30 min to aid in solubilization of the fusion protein. Pellets were removed by centrifugation at 12,000 × g for 10 min at 4°C. The protein concentration was measured by Bradford assay. Aliquots of the supernatant were stored at −20°C.
A total of 250 μg of each GST fusion protein lysates was bound to 25 μl of 50% pre-equilibrated glutathione-agarose beads slurry (GE Healthcare) for 1 h at 4°C; the beads were then washed three times with radioimmunoprecipitation assay buffer (0.5 M Tris [pH 8.0], 0.15 M NaCl, 0.1% SDS, 1% NP-40, 1% deoxycholate) containing 1× Complete protease inhibitor cocktail (Roche). Then, 100 μl of lysates (1 μg/μl) from infected A549 cells or transfected 293T cells was incubated with glutathione-agarose bead GST fusion protein complexes in PBS. After 2 h of incubation at 4°C, the beads were washed five times in radioimmunoprecipitation assay buffer. Bound proteins were resolved by SDS-PAGE, followed by Western blotting with NS1 antibody.
Next, 200 μl of cotransfected cell lysates (0.5 μg/μl) was incubated with 50 μl of 50% Ni-Sepharose 6 Fast Flow (GE Healthcare) slurry. The reaction volume was made up to 500 μl with PBS and incubated for 2 h at 4°C. After five washes, bound proteins were resolved by SDS-PAGE, followed by Western blotting with NS1 or His6 antibody or Coomassie blue staining.
Immunofluorescence staining.
MDCK cells (2 × 104/well) were plated on an eight-well chamber slide and infected by WT or mutant viruses at an MOI of 1. At predetermined times postinfection, cells were fixed in a mixture of acetone-methanol (1:1) for 15 min at −20°C. After rehydration with PBS, cells were incubated with polyserum NS1 or monoclonal M1 antibody for 1 h at room temperature. Cells were rinsed three times with PBS and incubated with Cy2-conjugated goat anti-rabbit or anti-mouse IgG for 45 min at room temperature. Finally, the cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Roche) for 5 min. Images were obtained on a Carl Zeiss Axiovert 200M inverted fluorescence microscope.
RESULTS
Generation and characterization of mutant viruses carrying mutations in NS1 SH3 motifs.
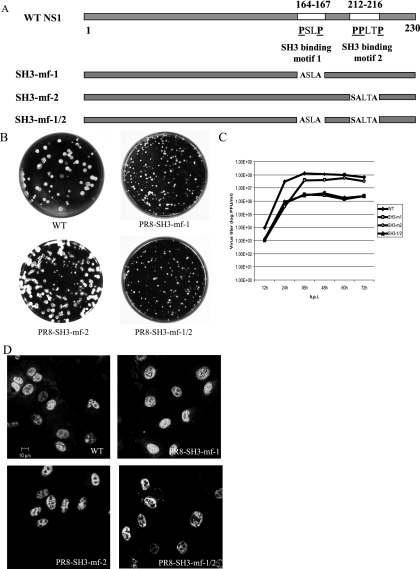
In order to investigate the contribution of two SH3 binding motifs on NS1 to the binding of p85 and the subsequent activation of the PI3K/Akt pathway, we generated three mutant viruses by reverse genetics. PR8-SH3-mf-1 carries mutations in NS1 SH3 binding motif 1, where prolines at amino acids 164 and 167 were replaced by alanines. PR8-SH3-mf-2 bears mutations in NS1 SH3 binding motif 2, where prolines at amino acids 212, 213, and 216 were mutated. To avoid introducing any mutations in NS2 protein, the proline at amino acid 212 was replaced by serine and the prolines at amino acids 213 and 216 were replaced by alanines. PR8-SH3-mf-1/2 harbors mutations in NS1 SH3 binding motifs 1 and 2 (Fig. 1A). These mutant viruses allowed us to identify the role of each individual motifs, as well as whether the two SH3 binding motifs work synergistically in the PI3K/Akt signaling pathway. The genotype of the mutant viruses were characterized and confirmed by DNA sequencing of the reverse transcription-PCR product derived from the NS gene of mutant viruses. Mutant viruses PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2 grown in 9- to 10-day-old embryonated eggs had titers of 8.79 × 107 PFU/ml, 2.13 × 108 PFU/ml, and 8.9 × 107 PFU/ml, respectively. We examined the replication potential of the mutant viruses in MDCK cells by monitoring the plaque size and multiple cycle growth kinetics. As shown in Fig. 1B, PR8-SH3-mf-1 and PR8-SH3-mf-1/2 formed small plaques, whereas PR8-SH3-mf-2 formed plaques similar in size to those of the WT virus, indicating that PR8-SH3-mf-1 and PR8-SH3-mf-1/2 are attenuated for growth. To assess the degree of attenuation for the mutant viruses, we compared the multiple cycle growth kinetics of the mutant viruses to that of the WT PR8 in MDCK cells. MDCK cells were infected at an MOI of 0.001, supernatant was harvested at 12-h intervals until 72 h p.i., and virus titers were determined by plaque assay. All of the viruses reached a plateau at 36 h p.i. PR8-SH3-mf-1 and PR8-SH3-mf-1/2 grew to titers approximately 1 to 1.5 log lower than the WT virus. PR8-SH3-mf-2 grew slightly slower than WT virus after 60 h p.i. (Fig. 1C). Finally, to determine whether the mutant NS1 proteins show a different subcellular localization in comparison with the WT NS1, MDCK cells were infected with viruses at an MOI of 1; at 7 h p.i. the cells were fixed, permeabilized, and stained with NS1 antibody. NS1 proteins encoded by PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2, like the WT NS1 protein, are localized in the nuclei of infected cells (Fig. 1D).
FIG. 1.
Characterization of mutant viruses. (A) Schematic diagram showing the locations of two SH3 binding motifs on WT NS1 and the changes in amino acid sequence in NS1 encoded by mutant viruses. (B) Plaques formed by WT, PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2 viruses on MDCK cells. (C) Multiple cycle growth curves of WT, PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2 on MDCK cells. Cells were infected in triplicate with each virus at an MOI of 0.001. Media were collected at 12-h intervals until 72 h p.i., and titers were determined by plaque assay on MDCK cells. The mean titer values at each time point were plotted with the associated standard deviation displayed as error bars. (D) Intracellular localization of NS1 protein encoded by WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 mutant viruses. Cells were infected by the viruses at an MOI of 1; at 7 h p.i. the cells were fixed, permeabilized, and stained with NS1 antibody, followed by Cy2-conjugated anti-rabbit IgG.
SH3 binding motif 1 in the NS1 protein is required for PI3K/Akt pathway activation.
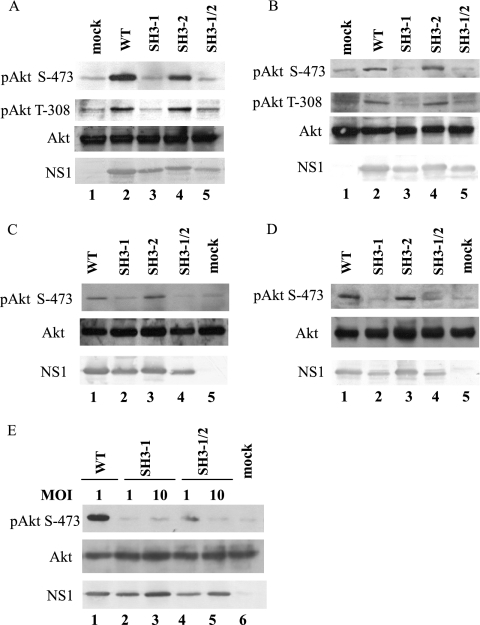
We have shown that influenza A virus infection leads to Akt phosphorylation at Ser-473 at 6 h p.i. and is sustained for the remainder of the infection (30). In order to examine the ability of PR8-SH3-mf-1, PR8-SH3-mf-2, and PR8-SH3-mf-1/2 to activate the PI3K/Akt pathway, A549 and MDCK cells were serum starved overnight and infected by the mutant viruses at an MOI of 1. Phosphorylation of Akt at Ser-473 and Thr-308, as well as the levels of Akt and NS1, were assessed by Western blotting at 8 and 16 h p.i. (Fig. 2). At 8 and 16 h p.i., a striking amount of phosphorylation of Akt at both Ser-473 and Thr-308 was detected in A549 cells that were infected by WT and PR8-SH3-mf-2 (Fig. 2A, lanes 2 and 4; Fig. 2B, lanes 2 and 4), but neither pAkt (Ser-473) nor pAkt (Thr-308) was detected in mock, PR8-SH3-mf-1 and PR8-SH3-mf-1/2 infected cells (Fig. 2A, lanes 1, 3 and 5; Fig. 2B, lanes 1, 3 and 5). Similarly, phosphorylation of Akt at Ser-473 could be detected in MDCK cells infected by WT and PR8-SH3-mf-2 at both time points (Fig. 2C, lanes 1 and 3; Fig. 2D, lanes 1 and 3) but not in PR8-SH3-mf-1-, PR8-SH3-mf-1/2-, and mock-infected MDCK cells (Fig. 2C, lanes 2, 4, and 5; Fig. 2D, lanes 2, 4, and 5). We did not detect phosphorylation of Akt at Thr-308 in MDCK cells. This may be the result of insufficient antibody sensitivity coupled with less PI3K/Akt pathway activation in this cell type. Viral infection of the MDCK cells produced less pAkt (Ser-473) phosphorylation compared to that observed in A549 cells. This, coupled with the reduced immunoreactivity of the pAkt (Thr-308) antibody compared to the pAkt (Ser-473) antibody, may have made phosphorylation on Thr-308 difficult to detect in the MDCK cells. Western blotting with total Akt antibody showed an equal amount of Akt in all samples, indicating that the changes in Akt phosphorylation by infection with different viruses were not due to an altered total level of Akt (Fig. 2A to D). NS1 expression levels in A549- and MDCK-infected cells were also assessed. In both types of cells, PR8-SH3-mf-2 produced similar amount of NS1 compared to WT virus, whereas PR8-SH3-mf-1 and PR8-SH3-mf-1/2 produced slightly less NS1 protein than WT. This might be due to a destabilizing effect of the mutations or to the reduced viral replication rate noted in Fig. 1C. Nonetheless, the inability of PR8-SH3-mf-1 and PR8-SH3-mf-1/2 to phosphorylate Akt was significantly more marked than the slight reduction in NS1 levels. Further, phosphorylation of Akt was still not observed when higher MOIs (MOI of 10) were used to infect cells with mutant viruses PR8-SH3-mf-1 (Fig. 2E, lane 3) and PR8-SH3-mf-1/2 (Fig. 2E, lane 5), where NS1 expression was increased to levels comparable to that of WT infection at an MOI of 1 (Fig. 2E, lanes 3 and 5 versus lane 1).
FIG. 2.
SH3 binding motif 1 in NS1 is required for PI3K/Akt activation. A549 cells (A, B, and E) or MDCK cells (C and D) were mock, WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 infected at an MOI of 1 (A, B, C, and D) or at an MOI as indicated (E). Cell lysates prepared at 8 (A, C, and E) or 16 h (B and D) p.i. were subjected to Western blotting with phospho-Akt (Ser-473), phopho-Akt (Thr-308), Akt, or NS1 antibody.
SH3 binding motif 1 in the NS1 protein is required for the NS1-p85β interaction in vitro.
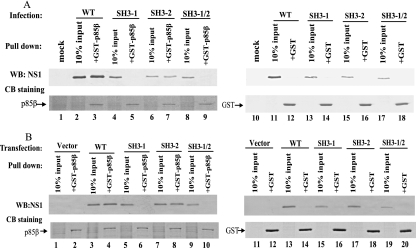
Previous studies showed that influenza A virus activates PI3K/Akt pathway by binding of NS1 to the PI3K regulatory subunit p85 (8, 11, 31). Three distinct genes encoding class IA PI3K regulatory subunits exist in mammals: Pik3r1 (p85α), Pik3r2 (p85β), and Pik3r3 (p55PIK) (9, 37). p85α is ubiquitously expressed and is thought to be the major response pathway for most stimuli, whereas p85β also is widely expressed but at a lower level than p85α (39). Hale et al. demonstrated that NS1 binds to p85β but not p85α (11). Our previous results have shown that viruses containing NS1 mutations in SH3 binding motif 1 were not able to activate PI3K/Akt. To ascertain the biological relevance of NS1-p85β interaction and PI3K/Akt pathway activation, we examined whether the NS1 protein encoded by the mutant viruses would interact with p85β using a GST pull-down assay. A549 cells were mock, WT, or mutant virus infected at an MOI of 1. Cell lysates were prepared at 6 h p.i. and were incubated with GST-p85β fusion protein or GST protein immobilized on beads. Pulled-down proteins were analyzed by Western blotting with polyclonal NS1 antibody (Fig. 3A). WT NS1 and NS1 encoded by PR8-SH3-mf-2 interacted with p85β efficiently (Fig. 3A, lanes 3 and 7). However, NS1 encoded by PR8-SH3-mf-1 and PR8-SH3-mf-1/2 could not bind to p85β at all (Fig. 3A, lanes 5 and 9). Neither NS1 protein nor its derivatives can interact with GST protein (Fig. 3A, lanes 12, 14, 16, and 18). Ten percent input was loaded as a control (Fig. 3A, lanes 2, 4, 6, 8, 11, 13, 15, and 17). An equal amount and the integrity of the GST and GST-p85β fusion proteins were demonstrated by Coomassie blue staining of the bound protein resolved by SDS-PAGE.
FIG. 3.
SH3 binding motif 1 in NS1 is essential for NS1-p85β interaction in vitro. A549 cells were mock, WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 infected at an MOI of 1. (A) Cell lysates were prepared at 6 h p.i. 293T cells were transfected with pcDNA-3.1(−), pcDNA-NS1, pcDNA-NS1-SH3-mf-1, pcDNA-NS1-SH3-mf-2, or pcDNA-NS1-SH3-mf-1/2. (B) Cell lysates were prepared at 24 h posttransfection. GST-p85β or GST was immobilized to beads and incubated with the infected or transfected cell lysates. Precipitated proteins were subjected to either Western blotting with NS1 antibody or SDS-PAGE, followed by Coomassie blue staining.
To examine whether NS1 and its respective mutants, unaccompanied by other viral components, would interact with p85β, WT NS1 gene and its respective mutants in SH3 binding motifs were cloned into the pcDNA3.1(−) vector. 293T cells were transfected with empty vector, pcDNA-NS1, pcDNA-NS1-SH3mf-1, pcDNA-NS1-SH3mf-2, or pcDNA-NS1-SH3mf-1/2. Cell lysates were prepared at 24 h posttransfection and were subjected to a GST pull-down assay. Figure 3B shows that WT NS1 and NS1 with mutations in SH3 binding motif 2 specifically interacted with p85β (Fig. 3B, lanes 4 and 8). However, NS1 encoded by pcDNA-NS1-SH3mf-1 and pcDNA-NS1-SH3mf-1/2 failed to bind p85β (Fig. 3B, lanes 6 and 10). No interactions with p85β were seen in vector pcDNA3.1(−)-transfected cells (Fig. 3B, lane 2). GST did not interact with NS1 (lanes 12, 14, 16, 18, and 20). An equal amount and the integrity of GST and GST-p85β were verified by Coomassie blue staining. These data suggest that SH3 binding motif 1 in NS1 mediates in vitro NS1-p85β interaction, regardless whether the NS1 protein is derived from virus infection or DNA transfection.
NS1 SH3 binding motif 1 is required for NS1-p85β binding in tissue culture.
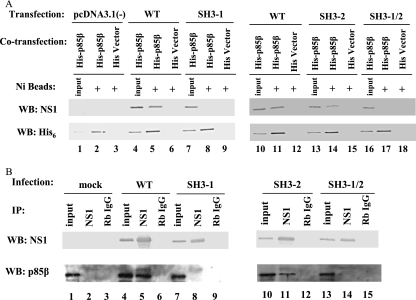
To verify whether the observed interactions take place in mammalian cells, we constructed plasmid pcDNA4HisMax-mp85β, where His-tagged mouse p85β is under the control of the cytomegalovirus promoter. Thus, 293T cells were cotransfected with pcDNA4HisMax-mp85β and either pcDNA3.1(−), pcDNA-NS1, pcDNA-NS1-SH3mf-1, pcDNA-NS1-SH3mf-2, pcDNA-NS1-SH3mf-1/2, or empty vector pcDNA4HisMax-B with NS1 expressing plasmids. At 24 h posttransfection, cell lysates were prepared and incubated with Ni-Sepharose beads. Precipitated proteins were analyzed by Western blotting with NS1 or His6 antibody. As seen in Fig. 4A, 10% of input was loaded as a control (Fig. 4A, lanes 1, 4, 7, 10, 13, and 16). Western blotting with His antibody shows that p85β could be detected in the precipitated complexes (Fig. 4A, lanes 2, 5, 8, 11, 14, and 17). In agreement with the in vitro GST-pull-down assay results using infected and transfected cells, while WT NS1 and NS1-SH3-mf-2 could be coprecipitated with p85β (Fig. 4A, lanes 5, 11, and 14), NS1 protein could not be coprecipitated with p85β in empty vector pcDNA3.1(−)-, pcDNA-NS1-SH3mf-1-, and pcDNA-NS1-SH3mf-1/2-transfected cells (Fig. 4A, lanes 2, 8, and 17). None of the NS1 proteins was precipitated with vector pcDNA4HisMaxB (Fig. 4A, lanes 6, 9, 12, 15, and 18).
FIG. 4.
SH3 binding motif 1 in NS1 is essential for NS1-p85β interaction in tissue culture. (A) 293T cells were cotransfected with either pcDNA3.1(−), pcDNA-NS1, pcDNA-NS1-SH3-mf-1, pcDNA-NS1-SH3-mf-2, or pcDNA-NS1-SH3-mf-1/2 each, together with pcDNA4HisMax-mp85β. As a negative control, pcDNA3.1(−), pcDNA-NS1, or its respective mutant plasmids were cotransfected with empty vector pcDNA4HisMaxB. At 24 h posttransfection, cell lysates were prepared and incubated with Ni-Sepharose beads. Precipitated proteins were subjected to Western blotting with NS1 or His6 antibody. Ten percent input was loaded as a control. (B) A549 cells were mock, WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 infected at an MOI of 1. Cell lysates were prepared at 6 h p.i., precleared by rabbit IgG-protein A, and incubated with either NS1 antibody-protein A or rabbit IgG-protein A. Precipitated proteins were subjected to Western blotting with NS1 or p85β antibody.
To verify that the results were not due to overexpression of His-p85β, we performed a coimmunoprecipitation assay to investigate the interactions of NS1 and its respective mutants with the endogenous p85β. A549 cells were mock infected or infected with viruses at an MOI of 1. At 6 h p.i., cells lysates were prepared, precleared with rabbit IgG, and incubated with NS1 antibody- or normal rabbit IgG-protein A complexes. Immunoprecipitated proteins were detected by p85β or NS1 antibody. As shown in Fig. 4B, endogenous p85β was present in the input samples in either mock-infected or virus-infected cells (Fig. 4B, lanes 1, 4, 7, 10, and 13). As expected, p85β was coimmunoprecipitated in complex with NS1 using an NS1 antibody in WT virus and PR8-SH3-mf-2 virus-infected samples (Fig. 4B, lanes 5 and 11). In contrast, p85β was not coimmunoprecipitated from PR8-SH3-mf-1- and PR8-SH3-mf-1/2-infected cells (Fig. 4B, lanes 8 and 14). No p85β was detected in samples that were immunoprecipitated using a control rabbit IgG (lanes 3, 6, 9, 12, and 15). Western blotting with NS1 antibody showed that WT NS1 protein and its respective mutants were immunoprecipitated by NS1 antibody (lanes 5, 8, 11, and 14). Thus, an intact SH3 binding motif 1 is required for NS1 to associate with p85β in cells.
Mutant virus PR8-SH3-mf-1 and PR8-SH3mf-1/2 are more proapoptotic.
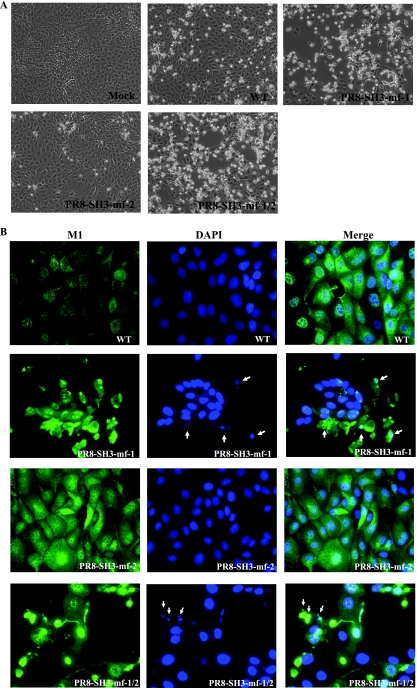
Phosphorylated Akt plays a central role in modulating diverse downstream signaling pathways associated with cell proliferation, migration, differentiation, and the prevention of apoptosis (5, 41). In particular, Akt has been shown to promote the survival of a wide range of cell types (6, 18). Several proapoptotic proteins have been identified as substrates for Akt phosphorylation, including the Bcl2 family member BAD, caspase-9, and GSK-3β (18). To examine the effect of PI3K/Akt pathway activation in influenza A virus-infected cells, we examined virus induced cytopathic effect in MDCK cells. As shown in Fig. 5A, a remarkable morphological difference was observed between cells infected with PR8-SH3-mf-1 and PR8-SH3-mf-1/2 compared to WT PR8 at 16 h p.i. There were more PR8-SH3-mf-1, PR8-SH3-mf-1/2 than WT-infected cells detached from the culture dish; the cells that remained attached displayed enhanced rounding and shrinkage. Severe cell death was not observed in PR8-SH3-mf-2-infected cells. To confirm that PR8-SH3-mf-1 and PR8-SH3-mf-1/2 are more proapoptotic than WT virus, MDCK cells were infected by WT and mutant viruses at an MOI of 1. At 16 h p.i., the cells were fixed, permeabilized, and stained with anti-M1 antibody, followed by DAPI staining. M1 staining allowed us to differentiate infected from uninfected cells and therefore visualize the morphology of infected cells in greater detail. As shown in Fig. 5B, WT- and PR8-SH3-mf-2-infected cells exhibited similar morphology, with no sign of severe cell death. DAPI staining showed the cells infected with WT or PR8-SH3-mf-2 virus exhibited uniform intact nuclei. In contrast, PR8-SH3-mf-1- and PR8-SH3-mf-1/2-infected cells underwent apoptosis with morphological changes, including cell shrinkage, membrane blebbing, and chromatin condensation (Fig. 5B).
FIG. 5.
Cells infected by mutant viruses containing SH3 motif 1 mutation exhibit more severe cytopathic effect and morphological changes characteristic of apoptosis. MDCK cells were mock, WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 infected at an MOI of 1. At 16 h p.i., the morphology of the infected cells was either documented by light microscopy (A) or by immunofluorescent staining with M1 antibody and DAPI, followed by fluorescence microscopy (B). Arrows indicate the fragmented chromatin.
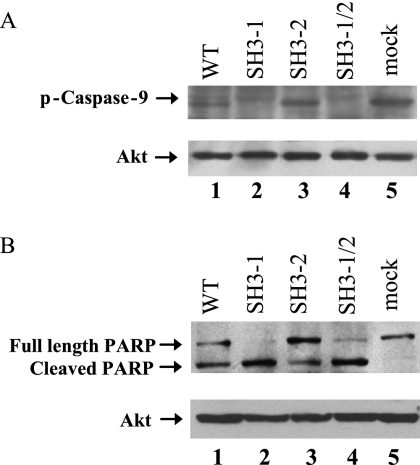
Apoptosis is mediated by a family of cysteine proteases termed caspases. It has been reported that activated Akt can phosphorylate caspase-9 at Ser-196 and thus inhibit caspase activity (3). Therefore, we examined the phosphorylation of caspase-9 in WT- and mutant virus-infected cells. MDCK cells were mock infected or infected by viruses at an MOI of 1. At 16 h p.i. cell lysates were prepared and were subjected to Western blotting with p-caspase-9 (Ser-196) antibody (Fig. 6A). Elevated p-caspase-9 was detected in WT- and PR8-SH3-mf-2-infected cells, as well as in mock-infected cells (Fig. 6A, lanes 1, 3, and 5). However, no phosphorylation of caspase-9 at Ser-196 occurred in cells infected by PR8-SH3-mf-1 and PR8-SH3-mf-1/2 viruses (Fig. 6A, lanes 2 and 4). PARP is a major downstream substrate for activated caspase-9. Activation of caspase-9 results in cleavage of full-length PARP (116 kDa) into two fragments (89 and 24 kDa). We then examined whether phosphorylation of caspase-9 would lead to inhibition of PARP cleavage in virus-infected cells. MDCK cells were mock infected or infected with viruses at an MOI of 1. At 24 h p.i. cell lysates were prepared and PARP cleavage was assessed by Western blotting with PARP antibody. As shown in Fig. 6B, full-length PARP was cleaved completely in PR8-SH3-mf-1- and PR8-SH3-mf-1/2-infected cells (Fig. 6B, lanes 2 and 4), where caspase-9 was not phosphorylated (Fig. 6A, lanes 2 and 4). In WT-, PR8-SH3-mf-2-, and mock-infected cells, where caspase-9 was phosphorylated, full-length PARP was detected (Fig. 6B, lanes 1, 3, and 5), indicating that PARP cleavage was inhibited to some extent. In both sets of samples, equal loading of the cellular protein was monitored by determining the total Akt level.
FIG. 6.
Influenza A virus induced pAkt phosphorylates caspase-9, leading to inhibition of apoptosis. MDCK cells were mock, WT, PR8-SH3-mf-1, PR8-SH3-mf-2, or PR8-SH3-mf-1/2 infected at an MOI of 1. At 16 (A) and 24 h (B) p.i., cell lysates were prepared and subjected to Western blotting with phospho-caspase-9 (Ser196) (A) or PARP (B) antibody, respectively. The total Akt level was monitored to verify equal loading of the samples.
DISCUSSION
Recent studies have demonstrated that influenza A virus infection activates PI3K/Akt pathway in the late phase of infection. The PI3K/Akt pathway activation is attributed to the viral NS1 protein, which can specifically interact with PI3K regulatory subunit p85 (7, 8, 11, 30, 31). In an effort to define the domains in NS1 that are responsible for p85 interaction and PI3K/Akt pathway activation, we previously constructed a mutant virus that encodes NS1 protein with mutations in three SH-binding domains, namely, a potential SH2 binding motif (Y89XXXM), SH3 binding motif 1 (amino acids 164 to 167 [PXXP]), and SH3 binding motif 2 (amino acids 212 to 216 [PPXXP]). Infection of this virus failed to activate the PI3K/Akt pathway (31), which prompted us to investigate the contribution of each individual motif in interacting with p85 and the subsequent activation of PI3K/Akt pathway. Mutational studies on the SH2 binding site were conducted by Hale et al. (11). In the present study, we focused on the two SH3 binding motifs. By using a genetic approach, we investigated the contribution of each individual SH3 binding motif to the PI3K/Akt pathway activation. We constructed three mutant viruses. To examine the role of the SH3 binding motif 1, we mutated prolines at amino acids 164 and 167 into alanines. To investigate the role of the SH3 binding motif 2, we replaced the proline at amino acid 212 by serine and the prolines at amino acids 213 and 216 with alanines. To examine whether both motifs work synergistically in PI3K/Akt activation, we mutated both motifs simultaneously (Fig. 1A). We examined the ability of these mutant viruses to activate the PI3K/Akt pathway at late times of infection. In agreement with previous results, WT virus infection leads to phosphorylation of Akt at Ser-437. In addition, we demonstrated that Akt was also phosphorylated at Thr-308 in A549 cells (Fig. 2A and B), which implies that influenza A virus-induced pAkt could be fully functional in regulating downstream effectors. Mutant virus PR8-SH3-mf-1 infection did not lead to Akt phosphorylation at either site, whereas mutant virus PR8-SH3-mf-2 readily activated the PI3K/Akt pathway, as did the WT virus (Fig. 2). These results suggest the SH3 binding motif 1 on the NS1 protein is required for PI3K/Akt pathway activation, whereas SH3 binding motif 2 is not necessary for this activation. Mutant virus carrying double motif mutations was not able to activate the pathway, further confirming that the SH3 binding motif 1 within NS1 is essential for PI3K/Akt pathway activation.
Next, we examined whether NS1 containing mutations in the SH3 binding motifs could interact with the p85 subunit of PI3K. We used GST-p85α and GST-p85β fusion proteins in an in vitro GST pull-down assay and found that NS1-p85β interaction was much stronger and more specific than NS1-p85α interactions (data not shown). Previously, we immunoprecipitated NS1 with a p85pan antibody which recognizes both isoforms of p85α and p85β (31). We speculated that NS1 precipitated by p85pan antibody is attributed to p85β. To confirm this, we performed immunoprecipitations with p85pan antibody, followed by Western blotting with p85α (25) or p85β antibody. Our results showed that both p85α and p85β were immunoprecipitated by p85pan antibody. Reciprocally, immunoprecipitation with NS1 antibody showed that p85β was present in the immunoprecipitated complexes in WT virus-infected cells (Fig. 4B), whereas p85α was absent (data not shown). These data further confirmed the findings of Hale et al. that NS1 is able to bind directly and efficiently to the p85β subunit of PI3K but not to the related p85α isoform (11). Hence, our subsequent investigations on the NS1-p85 interaction were focused on the p85β isoform.
In the GST pull-down assay, WT NS1 and NS1 with the SH3 binding motif 2 mutation, which derived either from virus infection or from plasmid transfection, could efficiently interact with p85β. In contrast, neither NS1 with SH3 binding motif 1 mutation nor NS1 with double motif mutations could interact with p85β (Fig. 3). In the Ni-Sepharose bead pull-down assay and the immunoprecipitation assay, both exogenous and endogenous p85β interacted with WT and SH3 binding motif 2 mutated NS1 but not with SH3 binding motif 1 or SH3 binding motif 1/2 mutated NS1 (Fig. 4). Notably, this pattern of interactions was also observed in NS1-transfected cells, indicating that SH3 binding motif 1-dependent NS1-p85β interaction does not need any other viral components to mediate complex formation. The GST-, Ni-Sepharose bead pull-down assay and coimmunoprecipitation data suggest that SH3 binding motif 1 is required for the NS1-p85β interaction, whereas SH3 binding motif 2 is not essential. These results are consistent with the phenotype of these viruses in terms of their ability to activate the PI3K/Akt pathway. Therefore, we conclude that NS1 SH3 binding motif 1 is critical for the NS1-p85β interaction and the subsequent PI3K/Akt pathway activation. The recently reported three-dimensional structure of NS1 (2) shows that the SH3-binding motif 1 lies exposed in the turn between the 6th β-strand and the large α-helix. The location of this motif is accessible to the p85β subunit, which provides the rationale that the SH3 binding motif 1 mediates the NS1-p85β interaction. It is noted that mutated NS1 proteins have a different mobility compared to WT NS1. Interestingly, SH3 binding motif 1 and motif 1/2 mutants migrate slightly faster than the WT or the motif 2 mutant. Whether this is due to the lack of posttranslational modification on SH3 motif 1 or SH3 motif 1/2 NS1 protein as a result of PI3K/Akt pathway deficient still needs to be determined. As to the other potential binding site YXXXM (SH2 binding motif) in NS1, mutational analysis by Hale et al. revealed that both Tyr-89 and Met-93 are essential for the interaction of NS1-p85β. However, SH2 binding motifs typically require phosphorylation on Tyr (33). Thus far, there is no evidence to suggest that Tyr-89 is phosphorylated during virus infection. Thus, it is not clear whether the SH2 binding motif on NS1 mediates the NS1-p85β interaction since mutation of Met-93 could potentially destabilize the NS1 dimer (12). Mutation of the p85β SH2 domain may help to resolve this question.
PI3K/Akt pathway has been implicated in cell survival and antiapoptosis control. Phosphorylated Akt can phosphorylate diverse substrates involved in the regulation of apoptosis. One of the mechanisms used by Akt to prevent apoptosis is to phosphorylate pro-caspase-9 and inhibit its protease activity, thus inhibiting apoptosis (3). Our results showed that WT and PR8-SH3-mf-2 virus infections, which activate the PI3K/Akt pathway, resulting in phosphorylation of caspase-9, do not induce severe apoptosis. In contrast, mutant viruses PR8-SH3-mf-1 and PR8-SH3-mf-1/2, which lack the ability to activate PI3K/Akt pathway, do not phosphorylate caspase-9 and are therefore more proapoptotic. Recently, a study using the PI3K specific inhibitor showed that influenza A virus activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses (8). Our data confirm and expand that observation using a genetic approach. Moreover, we demonstrate that the mutant viruses which are unable to activate the PI3K/Akt pathway are attenuated in growth, suggesting a role for activated PI3K/Akt in efficient influenza A virus replication. This finding is in agreement with our previous observation obtained using the PI3K inhibitor LY294002 (30).
Taken together, our study characterizes the role of two SH3 binding motifs in NS1 protein in the NS1-p85β interaction and PI3K/Akt pathway activation. We also reveal the biological function of the activated PI3K/Akt pathway in influenza A virus infection: activated PI3K/Akt mediates phosphorylation of caspase-9 and thereby blocks apoptosis of infected cells, leading to efficient virus replication.
Acknowledgments
We thank E. Hoffmann and R. G. Webster (St. Jude Children's Research Hospital) for providing PR8 plasmids and L. Cantley (Harvard Medical School, Boston, MA) for sharing the plasmid pGEX-4T3-p85β via Addgene. We are grateful to N. Berube for technical assistance.
L.A.B. holds the Canadian Research Chair in Vaccinology. Y.Z. holds a Canadian Institutes of Health Research (CIHR) New Investigator Award. This study was supported by grants from the CIHR, Natural Sciences and Engineering Research Council of Canada, to Y.Z.
This study is published as VIDO manuscript series no. 479.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Alessi, D. R., M. Andjelkovic, B. Caudwell, P. Cron, N. Morrice, P. Cohen, and B. A. Hemmings. 1996. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15:6541-6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. [DOI] [PubMed] [Google Scholar]
- 3.Cardone, M. H., N. Roy, H. R. Stennicke, G. S. Salvesen, T. F. Franke, E. Stanbridge, S. Frisch, and J. C. Reed. 1998. Regulation of cell death protease caspase-9 by phosphorylation. Science 282:1318-1321. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter, C. L., K. R. Auger, M. Chanudhuri, M. Yoakim, B. Schaffhausen, S. Shoelson, and L. C. Cantley. 1993. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 268:9478-9483. [PubMed] [Google Scholar]
- 5.Datta, S. R., A. Brunet, and M. E. Greenberg. 1999. Cellular survival: a play in three Akts. Genes Dev. 13:2905-2927. [DOI] [PubMed] [Google Scholar]
- 6.Dudek, H., S. R. Datta, T. F. Franke, M. J. Birnbaum, R. Yao, G. M. Cooper, R. A. Segal, D. R. Kaplan, and M. E. Greenberg. 1997. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275:661-665. [DOI] [PubMed] [Google Scholar]
- 7.Ehrhardt, C., H. Marjuki, T. Wolff, B. Nurnberg, O. Planz, S. Pleschka, and S. Ludwig. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defense. Cellular Microbiology. [DOI] [PubMed]
- 8.Ehrhardt, C., T. Wolff, S. Pleschka, O. Planz, W. Beermann, J. G. Bode, M. Schmolke, and S. Ludwig. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fruman, D. A., S. B. Snapper, C. M. Yballe, L. Davidson, J. Y. Yu, F. W. Alt, and L. C. Cantley. 1999. Impaired B-cell development and proliferation in absence of phosphoinositide 3-kinase p85α. Science 283:393-397. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 11.Hale, B. G., D. Jackson, Y. H. Chen, R. A. Lamb, and R. E. Randall. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. USA 103:14194-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale, B. G., and R. E. Randall. 2007. PI3K signalling during influenza A virus infections. Biochem. Soc. Trans. 35:186-187. [DOI] [PubMed] [Google Scholar]
- 13.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiles, I. D., M. Otsu, S. Volinia, M. J. Fry, I. Gout, R. Dhand, G. Panayotou, F. Ruiz-Larrea, A. Thompson, and N. F. Totty. 1992. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell 70:419-429. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, E., S. Krauss, D. Perez, R. Webby, and R. G. Webster. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165-3170. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackel-Cram, C., L. A. Babiuk, and Q. Liu. 2007. Upregulation of fatty acid synthase promoter by hepatitis C virus core protein: genotype-3a core has a stronger effect than genotype-1b core. J. Hepatol. 46:999-1008. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy, S. G., A. J. Wagner, S. D. Conzen, J. Jordan, A. Bellacosa, P. N. Tsichlis, and N. Hay. 1997. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 11:701-713. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A. 1989. The genes and proteins of influenza viruses, p. 1-87. In R. M. Krug (ed.), The influenza viruses. Plenum Press, Inc., New York, NY.
- 20.Lamb, R. A., and P. W. Choppin. 1979. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. USA 76:4908-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, Y., X. Y. Qian, and R. M. Krug. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817-1828. [DOI] [PubMed] [Google Scholar]
- 24.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 25.Mahon, E. S., A. D. Hawrysh, R. B. Chagpar, L. M. Johnson, and D. H. Anderson. 2005. A-Raf associates with and regulates platelet-derived growth factor receptor signaling. Cell Signal. 17:857-868. [DOI] [PubMed] [Google Scholar]
- 26.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236-243. [DOI] [PubMed] [Google Scholar]
- 28.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 30.Shin, Y. K., Q. Liu, S. K. Tikoo, L. A. Babiuk, and Y. Zhou. 2007. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J. Gen. Virol. 88:942-950. [DOI] [PubMed] [Google Scholar]
- 31.Shin, Y. K., Q. Liu, S. K. Tikoo, L. A. Babiuk, and Y. Zhou. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 88:13-18. [DOI] [PubMed] [Google Scholar]
- 32.Skolnik, E. Y., B. Margolis, M. Mohammadi, E. Lowenstein, R. Fischer, A. Drepps, A. Ullrich, and J. Schlessinger. 1991. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell 65:83-90. [DOI] [PubMed] [Google Scholar]
- 33.Songyang, Z., S. E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W. G. Haser, F. King, T. Roberts, S. Ratnofsky, and R. J. Lechleider. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767-778. [DOI] [PubMed] [Google Scholar]
- 34.Stoyanov, B., S. Volinia, T. Hanck, I. Rubio, M. Loubtchenkov, D. Malek, S. Stoyanova, B. Vanhaesebroeck, R. Dhand, and B. Nurnberg. 1995. Cloning and characterization of a G protein-activated human phosphoinositide-3 kinase. Science 269:690-693. [DOI] [PubMed] [Google Scholar]
- 35.Street, A., A. Macdonald, K. Crowder, and M. Harris. 2004. The Hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 279:12232-12241. [DOI] [PubMed] [Google Scholar]
- 36.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terauchi, Y., Y. Tsuji, S. Satoh, H. Minoura, K. Murakami, A. Okuno, K. Inukai, T. Asano, Y. Kaburagi, K. Ueki, H. Nakajima, T. Hanafusa, Y. Matsuzawa, H. Sekihara, Y. Yin, J. C. Barrett, H. Oda, T. Ishikawa, Y. Akanuma, I. Komuro, M. Suzuki, K. Yamamura, T. Kodama, H. Suzuki, K. Yamamura, T. Kodama, H. Suzuki, S. Koyasu, S. Aizawa, K. Tobe, Y. Fukui, Y. Yazaki, and T. Kadowaki. 1999. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat. Genet. 21:230-235. [DOI] [PubMed] [Google Scholar]
- 38.Toker, A., and L. C. Cantley. 1997. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387:673-676. [DOI] [PubMed] [Google Scholar]
- 39.Ueki, K., D. A. Fruman, C. M. Yballe, M. Fasshauer, J. Klein, T. Asano, L. C. Cantley, and C. R. Kahn. 2003. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J. Biol. Chem. 278:48453-48466. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao, R., and G. M. Cooper. 1995. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science 267:2003-2006. [DOI] [PubMed] [Google Scholar]