Abstract
Host translation shutoff induced in picornavirus-infected cells is a well-known phenomenon. The mechanisms by which separate genera of the picornavirus family achieve this shutoff differ. This study examined alterations in the cellular translational components in HeLa cells infected with encephalomyocarditis virus (EMCV), a cardiovirus. In agreement with previous reports, EMCV induced a marked decrease in host mRNA translation. The inhibition correlated with the appearance of a significantly enhanced 80S peak in cells and a concomitant decrease in polysome abundance. Characterization of the 80S material revealed that these ribosomes were virtually devoid of mRNA. Viral protein 2A was tightly associated with some of the free 40S ribosome subunits, but it was not present in the 80S pool which accumulated after infection. Expression of 2A protein in cells in the absence infection was able to modulate the cellular translational environment to increase the ratio of internal ribosome entry site-dependent translation to cap-dependent translation of a reporter construct. The results provide further evidence for a role of 2A protein in the mechanism of cardiovirus-induced host translational shutoff.
Picornavirus genomes are single-stranded, polyadenylated mRNAs which lack 5′ cap structures. Translation of the large open reading frame is dependent on an internal ribosome entry site (IRES) and involves direct recruitment of ribosomes onto the 5′ untranslated region. The product of the open reading frame is a polyprotein (Fig. 1A), de facto a 220-kDa zymogen, encoding a distinctive cohort of self-activating proteolytic functions that catalyze subsequent release of the requisite components for genome replication and progeny assembly (23). Picornavirus translation, processing, replication, and assembly are predominantly cytoplasmic, localizing to foci on the rough and smooth endoplasmic reticulum. Indeed, by using recombinant transcript RNAs, it is possible to recapitulate the entire virus life cycle in cell extracts in the complete absence of nuclei (16, 27).
FIG. 1.
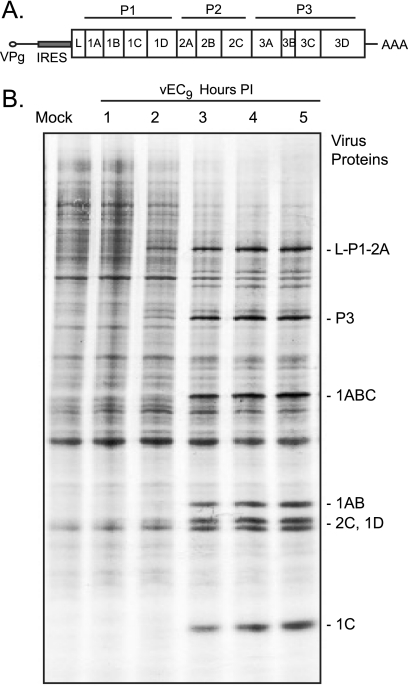
Protein synthesis in EMCV-infected HeLa cells. (A) Gene order within the EMCV polyprotein shows 2A at the border of the P1 and P2 regions (not to scale). (B) Autoradiogram after SDS-PAGE of samples from pulse-labeled HeLa cells infected with vEC9 (MOI of 100). At the indicated time postinfection (PI), the cells were treated with [35S]Met and then harvested 30 min later, as described in Materials and Methods.
During infection, however, picornaviruses are adept at modulating the host cell environment, and there are dramatic changes in host gene expression, transcription, translation, and nucleocytoplasmic trafficking (1, 3, 19, 21). As a consequence, innate antiviral immune responses and cell-cell signaling pathways are severely compromised. Some of the shutoff mechanisms are common to all members of the family. Protease 3C or its precursors, for example, cleave a variety of cytoplasmic and nuclear factors that normally contribute to host transcription (29). Protein and RNA trafficking through nuclear pores is also compromised, either by proteolytic cleavage of essential pore components (3) or by interference with the Ran GTPase system (21). Cap-dependent translational shutoff mechanisms are more variable and differ according to the genus of virus. Enteroviruses, such as poliovirus and rhinovirus, encode a second protease, 2Apro, that, among other activities, cleaves eukaryotic translation initiation factor 4G (eIF4G) (9, 24), effectively preventing recruitment of eIF4E-bound cap structures into 43S preinitiation complexes. Foot-and-mouth disease virus, an aphthovirus, achieves similar translational shutoff with another protease, Lpro, an enzyme that also targets eIF4G at a site near that of the enterovirus 2Apro (6).
Host translational shutoff by cardioviruses is less well understood. Encephalomyocarditis virus (EMCV) and Mengo virus do not induce cleavage of eIF4G (17). Viral proteins 2A and L are not proteases (24), and the shutoff of host protein synthesis, while clearly evident, is not as rapid or extensive as that caused by poliovirus (13). Instead, the initial descriptions of cardiovirus shutoff suggested that EMCV RNA, with its especially effective IRES, might simply out-compete capped mRNAs for translation initiation factors (13). Other studies took note of an unusual buildup of 80S ribosome complexes in Mengo virus-infected L cells and proposed the activation or synthesis of a putative inhibitor, presumably a viral protein, which might have the effect of trapping host mRNAs in defective 80S initiation complexes (19). More recently, experiments with BHK cells reported EMCV-dependent changes in 4E-BP1 phosphorylation patterns (26), a finding relevant to translational shutoff because 4E-BP1 is a regulator of eIF4E availability and direct competitor of eIF4G-eIF4E interactions (7, 26). This particular study also reported a link between the observation of 4E-BP1 phosphorylation changes and the presence of viral protein 2A. Although EMCV 2A is neither an analog nor homolog to the enterovirus 2Apro, deletions in EMCV 2A abrogated host-protein shutoff and, additionally, were partially defective in viral polyprotein processing. The tested deletions were not necessarily lethal to the virus, but they caused small-plaque phenotypes when mutant virus was plated on HeLa cells (26).
The EMCV 2A protein is 143 amino acids long, with a pI of 10.3. The carboxyl terminus harbors a 19-amino-acid autocatalytic cleavage cassette required for cotranslational polyprotein scission (12). Several amino acids upstream of this site is a segment with a high degree of conservation among all cardioviruses and a consensus sequence [(G/P)-(K/R)3-X1-4-(G/P)] identical to that which confers nuclear localization to several yeast ribosomal proteins (25). The remainder of 2A is still undefined in terms of structure or function. Cell fractionation studies carried out more than 20 years ago identified 2A (protein “G”) in crude preparations of infected-cell ribosomes (15). It was further noted that under appropriate conditions, 2A could bind nonspecifically to RNAs of various sequences (8). More recently, antibodies and confocal imaging characterized a strong 2A signal in infected HeLa nucleoli at the site of ribosome biogenesis. Despite this 2A signal, rRNA synthesis seemed to continue unabated in these nucleoli until cell lysis, and additional cytoplasmic pools of 2A were detected in the same cells, building concomitantly with the nucleolar pools throughout the infection (1). These collective observations suggested a novel 2A-dependent host-protein shutoff mechanism whereby cardioviruses might deliberately trigger continued rRNA synthesis so that nucleolar 2A could be built into new ribosomal subunits. When exported to the cytoplasm, these modified subunits might be presumed to have a strong 2A-directed preference for viral over host mRNA, effectively preventing host translation. Put simply, it was proposed that the virus might induce the cell to make toxic ribosomes that work only on the viral IRES (1). The current study was undertaken to characterize the nature of ribosomes during EMCV infection and determine whether they contain 2A or whether the presence of 2A in the absence of infection influences ribosome selection of capped or IRES mRNAs.
MATERIALS AND METHODS
Virus and cells.
H1-HeLa cells (ATCC CRL1958) were grown in suspension cultures with modified Eagle's medium (MEM) supplemented with 10% calf serum and 1% fetal bovine serum. Virus vEC9, a recombinant EMCV derivative containing a shortened poly(C) tract (11), was used for all infections. For virus amplification, HeLa cells (1 × 108) were washed with phosphate-buffered saline (PBS), resuspended (2 × 107 cells/ml) in MEM, and then treated (at a multiplicity of infection [MOI] of 10 PFU) with vEC9. After attachment (30 min at 20°C) with gentle agitation, cultures were diluted (5 × 105 cells/ml) with MEM (at 37°C) supplemented with 10% calf serum. Incubation (at 37°C) was in a shaking water bath (1).
Metabolic labeling.
Confluent HeLa monolayers (on 30-mm plates) were infected with pEC9 (at an MOI of 100 PFU). The virus was allowed to attach (30 min at 20°C) before the plates were washed (two times in PBS) and overlaid with MEM lacking methionine (Sigma). Thirty minutes prior to harvest (1 to 5 h postinfection [p.i.]), [35S]Met was added (100 μCi/plate; Amersham). Cells were washed (two times in PBS) and lysed (30 mM Tris-HCl, pH 7.4, 140 mM NaCl, 0.5% NP-40), and the incorporation of label into acid-insoluble material was quantified by scintillation counting. The protein content was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli) and subsequent autoradiography.
Ribosomes.
Ribosome profiles were characterized as described previously (22). Briefly, 15 min prior to harvest, cycloheximide (100 μg/ml) was added to infected HeLa cells. The cells were collected, washed (three times in PBS), resuspended in buffer A (20 mM Tris, pH 7.4, 150 mM KCl, 30 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml cycloheximide, 0.5% NP-40), and then lysed (5 min at 0°C). After clarification (10,000 × g for 15 min at 4°C) the S10 supernatants (typically, 43 optical density at 260 nm [OD260] units) were layered onto linear sucrose gradients (15 to 45%) and subjected to centrifugation (34,000 rpm for 210 min in an SW-41 rotor). The gradients were fractionated from the bottom (OD254; Amersham AktaPrime UV monitor), and acid-insoluble material (trichloroacetic acid [TCA]) was collected from each sample before fractionation by SDS-PAGE and Western analysis. Samples for measurements with dissociated subunits were prepared similarly, except that the cells were lysed with buffer A containing EDTA (30 mM) instead of MgCl2, and fractionation (38,000 rpm for 5 h in an SW-41 rotor) used 10 to 30% sucrose gradients.
Ribosome pelleting and factor removal by salt washes have been described previously (5). Infected cell samples (three to four times more than for gradient samples) were collected, lysed, clarified (15,000 × g for 15 min at 4°C), and then matched for total protein content (Bradford assay; Bio-Rad) before pelleting (55,000 rpm for 45 min at 4°C in a TLS-55 rotor). The supernatant (S-200a) was retained, and the new pellet (P-200a) was resuspended with agitation (for 1 h at 4°C) in buffer 1 (30 mM HEPES, pH 7.4, 2.5 mM MgAc2, 2 mM dithiothreitol, 0.3 mM phenylmethylsulfonyl fluoride, 0.01% NP-40, 10% glycerol) containing 150 mM KCl. Sequential extraction procedures using 500 mM KCl and then 750 mM KCl produced successively more stringent salt-washed ribosomes (P-200b and P-200c) and released auxiliary protein factors (S-200b and S-200c). All materials were collected by acid precipitation (with TCA) before gel fractionation and Western analysis.
Western analysis.
Protein samples were fractionated by SDS-PAGE and then transferred by electroporation (12 V for 80 min; Idea Scientific Co.) onto polyvinylidene difluoride membranes (Immobilon P; Millipore) in transfer buffer (25 mM Tris, pH 8.0, 0.19 M glycine, 20% methanol). After treatment with blocking solution (5% [wt/vol] nonfat dry milk, 0.05% [wt/vol] Tween-20 in TBS; 1 h at 20°C), the membranes were rinsed with TBS (20 mM Tris-HCl, pH 7.6, 140 mM NaCl) and reacted overnight with primary antibody (in MT buffer containing 2.5% [wt/vol] nonfat dry milk-0.05% [wt/vol] Tween-20 in TBS) at 4°C with agitation. The blots were rinsed (three times in TBS), treated with horseradish peroxidase-conjugated secondary antibody (in MT buffer), and rinsed (three times in TBS) before the bands were visualized by chemiluminescence (ECL kit; Amersham Bioscience) after exposures sufficient to detect the requisite signals. Murine monoclonal antibody 5A12 (1:2,000 dilution) against Mengo virus 2A has been described previously (1). Polyclonal antibodies against S6 (1:1,000; Cell Signaling Inc.), eIF4E (1:1,000; Cell Signaling Inc.), and L28 (1:500; Santa Cruz Biotech) were purchased, as were the appropriate secondary (Sigma) antibodies (1:8,000).
RNA detection.
Gradient samples from ribosome profile analyses were reacted with phenol-chloroform, and the total RNA was collected after ethanol precipitation. For a relative quantitation of polyadenylated RNA, fraction-equivalent samples were reacted with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and an oligo(dT) primer in the presence of [α-32P]dCTP (0.25 μCi/ml) according to the manufacturer's instructions. Incorporation of label into acid-insoluble material (TCA) from duplicate samples was monitored by scintillation counting.
Comparative viral RNA levels were determined from similar gradient samples by real-time PCR. Reverse transcription (using Moloney murine leukemia virus) used primer 823 (Table 1). Amplification reactions (30 to 40 cycles) used primers 997 and 998 in the presence of SYBR green PCR master mix, using an Applied Biosystems 7900HT Fast Real-Time PCR system. DNA from pEC9 was used to calibrate a standard curve, according to the manufacturer's instructions (Applied Biosystems) using SDS, version 2.2.1, software.
TABLE 1.
Primer sequences
| Primer name | Sequence |
|---|---|
| A1 | TTCGGCTAGCATGAGTCCAAATGCCCTAGACATT |
| A2 | CATGAAGGGCCCTGGATTTGTCTCAAT |
| 823 | AAGAACCCTGTCTGCAGTATG |
| 997 | TCTCTTTCCTGAGCAACGGGAACT |
| 998 | ACCGCCTGGTAGGTATTCTTCCAA |
Reporter cDNAs.
Bicistronic reporter plasmid pF/R-wt has been described previously (4). To introduce 2A, the relevant segment from pEC9 was amplified by PCR (primers A1 and A2) and then ligated with the largest fragment resulting from the ApaI digest of plasmid pME-NPGP-luCM (a generous gift from Cheryl Dvorak). The product fused the 2A gene in frame with the firefly luciferase (FLuc) sequence and therefore encoded a functional EMCV primary cleavage cassette (10) between the two genes. After additional PCR amplification, the product was exchanged with the analogous NheI-to-MluI segment in pF/R-wt, to create p2A-F/R-wt. Translation releases 2A from FLuc, extending its amino terminus by eight amino acids (PFMFKPTS-FLuc) relative to pF/R-wt. The activities of reporter plasmids were assayed after transfection into HeLa cells (0.8 μg of cDNA; 1.8 × 105 cells/well; 80 to 90% confluence) using a liposome technique (Lipofectamine 2000; Invitrogen) and serum-free medium (Opti-MEM; Gibco). Expressed enzyme (Renilla luciferase [RLuc]or FLuc) activity was monitored in triplicate from harvested cell lysates using a Dual-Luciferase Reporter assay system (Promega) according the manufacturer's instruction.
RESULTS
Protein synthesis.
The effect of EMCV infection on protein synthesis profiles was assessed by pulse incorporation of [35S]Met (Fig. 1B). Beginning at 2 h p.i. viral precursors L-P1-2A and P3 labeled strongly enough to become visible. From 3 h onward, new protein synthesis was dominated by these precursors and their cleavage products (e.g., 1ABC, 1AB, 1C, and 1D). The 2-h and 3 h-time points also marked a clear decrease in overall cellular protein synthesis, with the missing bands most apparent near the top of the gel. In repeated experiments the conversion from cellular to viral synthesis was never absolute at any time point or for any MOI (10 to 100 PFU/cell) (all data not shown), and multiple nonviral (cellular) bands labeled consistently throughout the infection.
Ribosome profiles.
Mengo virus infection is reported to have a profound effect on the format of the ribosome pools in L cells (19). EMCV infection of HeLa cells did likewise (Fig. 2). Relative to mock-infected samples, gradient profiles captured at 2 h p.i. showed a loss of polysome material from the bottom of the gradients and concordant increase in the 80S peak. The trend became more pronounced as the infection progressed in that the 3-h profile had more 80S than the 2-h profile, and the 5-h profile had more than the 4-h profile. By summing the curve areas for these matched samples (equal cells per gradient), the polysome-to-monosome ratio was measured at about six times higher for mock-infected cells than for samples taken 5 h p.i. The progressive conversion from polysome to monosome did not seem to affect the standing pools of 40S and 60S subunits, which were maintained at similar levels, approximately 1:1, in all samples before and after infection.
FIG. 2.
Ribosome profiles. HeLa cells in suspension culture were infected with vEC9 (MOI of 10) or left uninfected (mock). Cycloheximide was added 15 min prior to harvest (each panel indicates time postinfection). Equivalent cytoplasmic lysates derived from matched numbers of cells were fractionated in parallel on sucrose gradients. The OD254 distribution was monitored continuously while collecting the gradients drop-wise from the bottom of the tubes (left to right). The 80S, 60S, and 40S locations are indicated. The areas under the curves for polysomes (P) and monosomes (M, 80S) were summed (ImageQuant software), and results are presented as relative ratios.
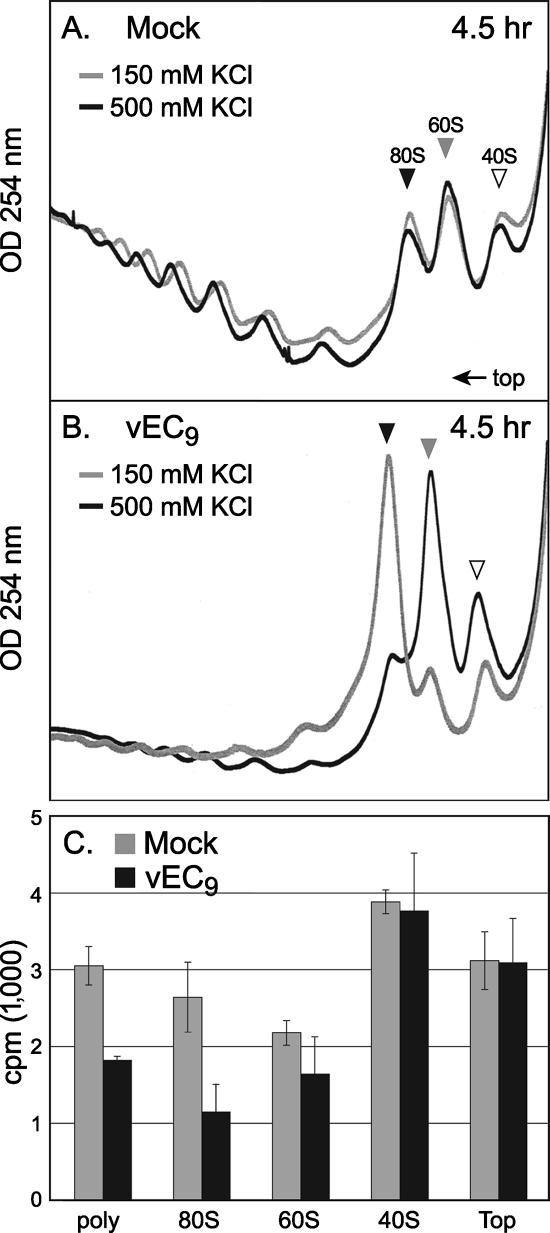
Experiments based on poliovirus infections have reported that picornavirus RNA is translated predominantly on “heavy” polysomes (18). However, heavy polysomes are not characteristic of Mengo virus infections (19), and polysome disaggregation profiles differ considerably when disaggregation is induced by poliovirus or EMCV (20). Therefore, it was necessary to test whether the increase in 80S subunits during EMCV infection could be due, in part, to augmented viral translation on monosomes or, alternatively, to long-lived cellular mRNAs preferentially captured from the depleted polysomes. Translating ribosomes bound to mRNA are stable under conditions of high salt (28), and, indeed, when samples from mock-infected cells were fractionated in the presence of 150 mM or 500 mM KCl, there was little change in the polysome or monosome (80S) profiles (Fig. 3A). Parallel samples from infected cells reacted differently. Almost all of the infection-induced 80S material dissociated into 60S and 40S at 500 mM KCl (Fig. 3B), suggesting that the majority of this peak was not bound to mRNA or actively translating.
FIG. 3.
Ribosome profiles with salt. (A) Ribosome isolation conditions were similar to those described in the legend of Fig. 2 (mock), except that half of the harvested cells were lysed in buffer containing 150 mM KCl and the other half in buffer with 500 mM KCl. The respective sucrose gradients maintained those salt conditions. (B) vEC9-infected HeLa cells (MOI of 10) were treated similar to the method described for panel A. (C) Aliquots from the polysome (poly), 80S, 60S, 40S, and top of the gradients (top), treated with 150 mM KCl, were extracted with phenol-chloroform and then analyzed for poly(A) RNA as described in Materials and Methods. The plot records the average [α-32P]dCTP incorporation from duplicate samples.
If the above assumption is true, the distribution of mRNA in these gradients should not shift in parallel to the 80S increase during infection. Total RNA was isolated from equivalent gradient fractions (150 mM KCl) and reverse transcribed in oligo(dT)-dependent reactions with [32P]dCTP. Samples from the 80S peak of mock-infected cells (Fig. 3C, gray bars) contained more than twice the amount of poly(A) template as analogous samples from infected cells (black bars). The mock-infected cells also had more poly(A) RNA in the polysome and 60S fractions, although the 40S and “top” pools were nearly equivalent. Therefore, in concordance with previous observations with Mengo virus infection (19), EMCV infection decreased the total mRNA content, on a per cell basis, for both the polysome and 80S fractions. When the experiment was repeated using reverse transcription-PCR to detect viral RNA, the weakest EMCV signal was found consistently in the 80S region, regardless of whether the samples were harvested at 4 h, 5 h (not shown), or 6 h p.i. (Fig. 4B). In these samples, packaged (or packaging) viral RNA localized most strongly as infectious PFU in the 135S region and as replication complexes (156S) near the bottom of the gradient. The combined RNA detection data suggest that only a minority of the 80S subunits, or perhaps a discrete pool, was associated with any mRNA during infection. Presumably, these templates alone were responsible for the steady-state synthesis of viral proteins and of those cellular proteins whose translation was not affected by infection. The salt-sensitive 80S subunits induced during infection were devoid of mRNA and presumably composed of “vacant couples” or empty pairings of 40S with 60S (28).
FIG. 4.
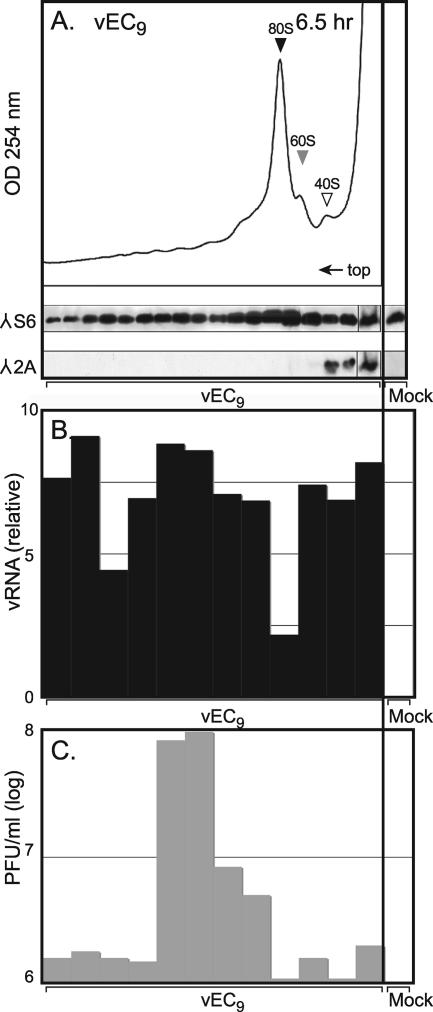
2A and viral RNA distribution. (A) Cytoplasmic lysates (43 OD260 units/gradient) from vEC9-infected HeLa cells (MOI of 10 at 6.5 h p.i.) were fractionated on sucrose gradients as in Fig. 2. Equivalent samples from each fraction were probed by Western analysis using a polyclonal antibody against ribosomal protein S6 or a monoclonal antibody against Mengo virus 2A. (B) Samples from a parallel gradient were extracted with phenol-chloroform and then probed by real time-PCR for the relative concentration of EMCV RNA. (C) Titers of infectious virus in samples from the same gradient (without phenol-chloroform extraction) were determined by plating onto HeLa cell monolayers. Data from the right-most samples are from control, mock-infected cells, treated in parallel. The top fraction represents a pool. λ, antibody target.
Location of 2A.
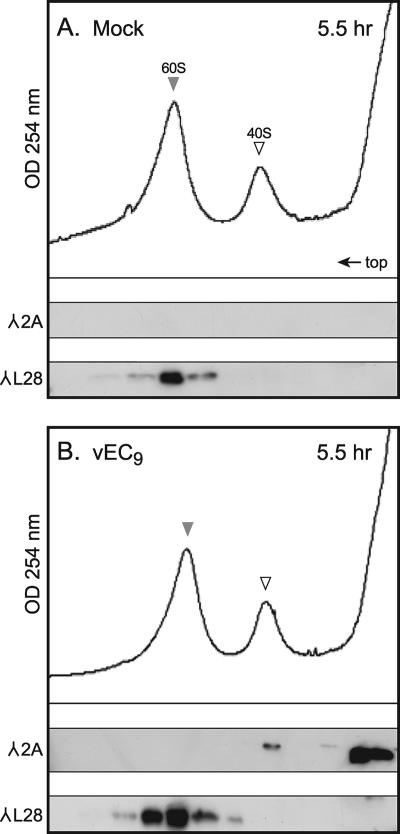
To determine if 2A was localizing to, or perhaps catalyzing, the 80S couplings, ribosome gradients from infected cells were probed for 2A using Western analysis (2). Surprisingly, the 80S peak was not immunoreactive (Fig. 4A). Instead, the majority of 2A localized to the top of the gradient, with only a small portion extending into the 40S region. Marker antibodies against S6, an integral 40S protein, showed the expected distribution throughout the gradient for a natural cohort of polysomes, 80S, 60S, and 40S subunits. To verify the 2A location, infected S10 lysates were treated with EDTA to dissociate the ribosomes into subunits of 40S plus 60S and then were refractionated (Fig. 5). Again, the majority of 2A localized near the top of the gradient with a weak but reproducible signal in the 40S peak. This signal did not overlap with L28, an integral 60S protein. In repeated experiments, this small portion of 2A clearly cofractionated with 40S in a manner well delineated from the majority of 2A at the top of the gradient.
FIG. 5.
2A in fractionated ribosomes. Cytoplasmic lysates from mock-infected (A) or vEC9-infected (B) cells (MOI of 10) were harvested at 5.5 h p.i. into a buffer containing 30 mM EDTA. After gradient fractionation (10 to 30% sucrose), Western analysis probed for the presence of L28 ribosomal protein and 2A, as described in Materials Methods and in the legend of Fig. 4. λ, antibody target.
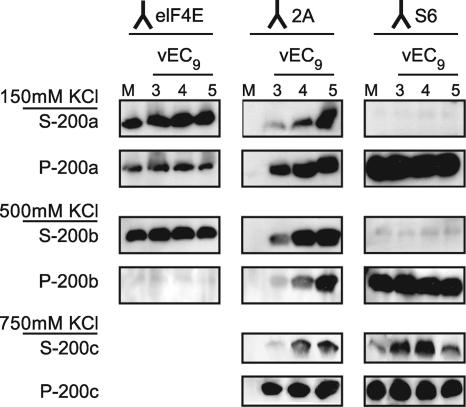
Protein 2A is reported to have a low-level binding affinity for viral RNA and for certain RNA homopolymers (8). To test whether the 2A migration with 40S was incidental, mock-infected and infected cytoplasmic extracts were harvested at 3, 4, or 5 h p.i. Sequential centrifugation steps were combined with increasing KCl concentrations to provide progressively more stringent washing conditions (Fig. 6). Consistent with the ribosome profiles, at 150 mM KCl, 2A was detected in the pellets (P-200a) and supernatants (S-200a) from all infected samples. Subsequent extraction of the pellets with 500 mM salt released eIF4E, a translation initiation factor, into the supernatant (S-200b). A portion of the previous 2A signal was also released, although some remained in the pellet (P-200b), as did all of the S6 signal. At 750 mM salt, the subunits began to disintegrate (5) and consequently, some (∼10%) of S6 and some (∼10%) of the remaining 2A were released to the supernatant (S-200c). Yet, the 3-, 4-, and 5-h-infected sample pellets (P-200c) still had detectable 2A signals. When mixed with naive ribosomes or cytoplasmic extracts, recombinant 2A does not sediment in the 40S region of gradients or pellet into the P-200 fractions (data not shown). Therefore, whatever the nature of the interaction that allowed a minor portion of viral 2A to pellet in the same fractions as ribosomes, it was held very tightly in this context and was probably not incidental.
FIG. 6.
2A interaction. HeLa cells infected with vEC9 (MOI of 10) cells were harvested at 3, 4, or 5 h p.i. into buffer containing 150 mM KCl. After a clarification step to remove nuclei, the extracts were pelleted at 200,000 × g and then washed with increasingly higher salt concentrations, as described in Materials and Methods. Equivalent samples from each subsequent supernatant or pellet were retained for fractionation by SDS-PAGE and Western analysis using antibodies against eIF4E (translation factor), 2A, or S6 (integral ribosome protein). Only the portions of the blots relative to these proteins are shown. λ, antibody target.
2A translation effects.
Host translational shutoff is usually monitored under conditions of infection or after transfection with viral replicons (2, 26). But it is difficult to assign culpability when multiple viral proteins or pathways can influence the observations. Preliminary experiments which expressed EMCV 2A at low levels from a bicistronic plasmid showed the full-length protein, but not a deleted derivative, localized predominantly to nucleoli, and this was somewhat toxic to expression of the upstream cap-dependent cistron (1). The effects of 2A on wild-type IRES versus cap translation could not be monitored from that experimental design.
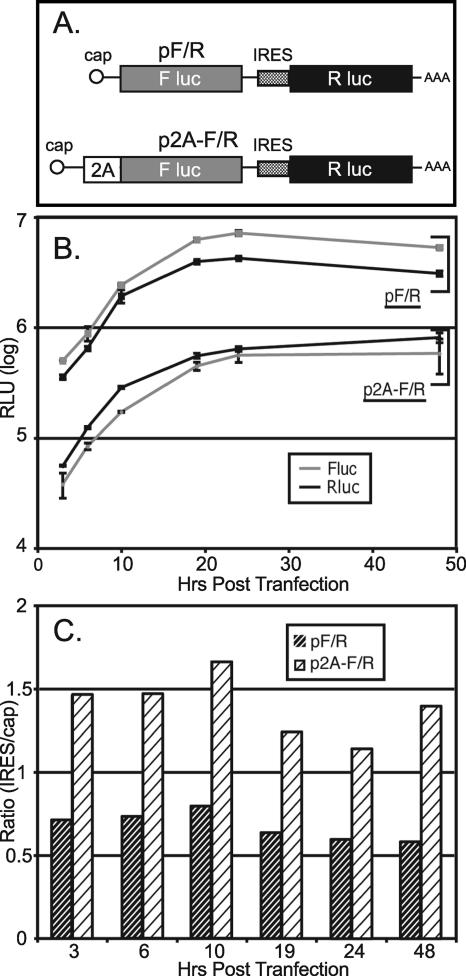
Plasmid pF/R-wt encodes an active FLuc gene under the transcriptional control of a cytomegalovirus promoter (Fig. 7A). Translation of FLuc, after transfection, is cap dependent. Transcripts also encode an active RLuc gene, whose translation is controlled by a wild-type EMCV IRES. In RNA-programmed reticulocyte extracts or cDNA-transfected cells, this configuration reproducibly synthesizes 8 to 10 times more RLuc than FLuc, reflecting the high level of IRES activity. But since these enzymes have inverse specific activities (FLuc activity is ∼10× that of RLuc), the measured luciferin turnover (in relative light units [RLU]) is roughly equivalent (4). In repeated trials with controlled doses of pF/R, the RLuc/FLuc activity (IRES/cap) gave a steady 0.6 to 0.8 RLU ratio over the course of 48 h (Fig. 7B and C). The 2A protein was introduced by linking it in frame to FLuc. During translation, these proteins are released from each other by the efficient activity (>95%) (data not shown) of the primary cleavage cassette at the carboxy terminus of 2A. Transfection into HeLa cells with matched samples of cDNA showed that both cistrons produced an average of six- to eightfold lower protein activity when directed by p2A-F/R-wt than with pF/R. The mRNA levels, as measured by reverse transcription-PCR, were not affected by 2A when this protein was delivered from a bicistronic construct in the absence of infection (Y. Bochkov, personal communication). Therefore, the observed downregulation of reporter expression must have been translational in origin. Characteristically with 2A, there was a one- to twofold larger reduction in cap-dependent expression than in IRES-dependent expression, with an RLU activity ratio of about 1.2 to 1.6 over the course of 48 h.
FIG. 7.
2A and bicistronic plasmids. (A) Reporter construct pF/R has been described previously (4). p2A-F/R is a tri-cistronic derivative plasmid, which additionally encodes EMCV 2A, as described in Materials and Methods. (B) After transfection of HeLa cells, expressed luciferase activities (RLuc or FLuc) were monitored in cell lysates by standard protocols. The number of RLU reported for each sample is for triplicates at each time point. Sample variance, when larger than the individual data symbol, is shown. (C) The RLuc/FLuc ratio (RLU) for each plasmid time point shown in panel B is represented graphically.
DISCUSSION
The presence of protein 2A during EMCV or Mengo virus infections is linked to the ability of the virus to shut off cap-dependent host translation. But the mechanism by which 2A affects this response remains elusive. Unlike the enterovirus or rhinovirus protein with the same name, the cardiovirus 2A is not a protease. It has no identifiable catalytic motifs. In BHK cells infected with EMCV, the mammalian target of rapamycin protein kinase pathway is inhibited, leading to dephosphorylation of 4E-BP1, a known suppressor of cap-dependent translation (26). Correlation of this activity as a cause or consequence of cap-dependent shutoff is difficult to establish, however. Viral mutants with deletions in the 2A protein are less effective at triggering 4E-BP1 dephosphorylation, but these same strains are also partially defective in the 2A-dependent primary cleavage of their viral polyproteins. The lower fecundity and smaller plaques might well result from lower processing efficiencies rather than from a competitive disadvantage with host translation (10). Moreover, HeLa and L cells infected with EMCV do not have altered 4E-BP1 phosphorylation patterns (our unpublished data).
In attempts to explore other phenotypes associated with 2A, we undertook an examination of ribosomes within infected cells. HeLa nucleoli resonate brightly with antibodies specific to this protein (1). Microarray data have suggested that the nuclear presence of 2A during infection might be linked to virus-induced polymerase II (Pol II) shutoff pathways. Although Pol I and Pol III continue synthesis throughout infection, direct measurements of rRNA also document a heightened rate of turnover, dependent on 2A, relative to uninfected cells. The results led to a model predicting during infection a rapid, virus-induced ribosome turnover into a 2A-modified subunit pool, with a consequent shift in ribosomal preference for viral over cap-dependent translation (2).
Our gradient profiles found, instead, a significant conversion of existing ribosome subunits into salt-sensitive vacant 80S couplings, mostly devoid of mRNA, be it viral or cellular. To be sure, viral translation and a reduced level of cellular translation continued throughout infection, but starting at about 2 h p.i., the shift from polysomes into the enhanced 80S peak was dramatic and characteristic of a productive infection. Given the recent discovery that EMCV protein L, another small nonstructural component, binds to Ran GTPase and inhibits all active nucleocytoplasmic transport, including ribosome egress (21), it now seems unlikely that any nucleolar forms of 2A traverse back into the cytoplasm as permanent ribosome modifications. Indeed, our 2A antibodies could not detect this protein in any cytoplasmic 80S material (Fig. 4) even though viral RNA was being translated there preferentially over that of the host (Fig. 1). Of the cytoplasmic 2A which was detected, only a small portion was associated with 40S subunits. None was associated with 60S or 80S. Most of the 2A which pelleted with ribosomes could be removed with low-salt washes, but the remainder required practical disruption of the integral protein-rRNA connections with 750 mM salt. We do not know the nature of the tight 2A-40S interaction or whether it is a protein-protein or protein-rRNA interaction. The normal cohort of translation factors (e.g., eIF4E) could be removed from similar ribosome fractions with much lower salt (i.e., 500 mM salt). To date, we do not have any recombinant 2A preparations which can mimic this tight association with 40S subunits when mixed in vitro, so the practical functionality of this observation in the cap-dependent shutoff pathway remains unclear. Perhaps we are observing a tight but transient association which affects many ribosomes or, alternatively, a permanent modification of a small cohort of subunits.
We do know that delivery of 2A into cells via bicistronic vectors was toxic to the translation of both cap and IRES reporter activity (Fig. 7), albeit cap-linked expression was reproducibly inhibited about twice as effectively as IRES-linked expression when the reporters and 2A were present in the same cells. Ribosome profiles from such samples are impractical because of the background of untransfected cells and because the induced 2A is at a substantially lower overall concentration than found in infected samples. This vector system is useful, however, in that it demonstrates at least one assayable activity of 2A in the absence of infection, and it may allow 2A mapping experiments for those portions of the protein responsible for reduced reporter expression. The fact that cap and IRES expression were both affected in these assays suggests that cardiovirus translational shutoff during infection may actually involve multiple viral components. Perhaps 2A binds and sequesters one or more specific ribosome-associated factors, either depleting the existing cytoplasmic pools or triggering relocalization from nucleolar fractions. Ribosomes depleted of eIF3 or eIF5, for example, are known to manifest as 80S vacant couples (14). But transient modification of 40S by 2A might be difficult to measure, and, indeed, the general cellular pools of these factors are reported to be unaltered by EMCV (13). Our preliminary (unpublished) detection experiments using antibodies against eIF4 and the major eIF3 subunits have also failed to detect overt virus-induced changes in these populations, but the data do not preclude subunit remodeling or phosphorylation events, for example. In the environment of an infection, the strategic advantage to the IRES could be restored, selectively or competitively, by other viral proteins (L, perhaps) or if concomitant transcriptional and nucleocytoplasmic transport shutoff pathways were also activated. Given the assays described here, it should now be possible to reconstruct the viral protein conditions which lead to salt-sensitive couplings of 40S with 60S and identify the required factors.
Acknowledgments
This work was supported by NIH grant AI-17331 to A.C.P., by an NSF Graduate Research Fellowship to R.P.G., and by NIH National Research Service Award T32 GM07215 to R.P.G.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Aminev, A. G., S. P. Amineva, and A. C. Palmenberg. 2003. Encephalomyocarditis viral protein 2A localizes to nucleoli and inhibits cap-dependent mRNA translation. Virus Res. 95:45-57. [DOI] [PubMed] [Google Scholar]
- 2.Aminev, A. G., S. P. Amineva, and A. C. Palmenberg. 2003. Encephalomyocarditis virus (EMCV) proteins 2A and 3BCD localize to nuclei and inhibit cellular mRNA transcription but not rRNA transcription. Virus Res. 95:59-73. [DOI] [PubMed] [Google Scholar]
- 3.Belov, G. A., P. V. Lidsky, O. V. Mitkitas, D. Egger, K. A. Lukyanov, K. Bienz, and V. Agol. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78:10166-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkov, Y., and A. C. Palmenberg. 2006. Translational efficiency of an EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. BioTechniques 41:238-290. [DOI] [PubMed] [Google Scholar]
- 5.DeStefano, J., E. Olmsted, R. Panniers, and J. Lucas-Lenard. 1990. The alpha subunit of eucaryotic initiation factor 2 is phosphorylated in mengovirus-infected mouse L cells. J. Virol. 64:4445-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devaney, M. A., V. N. Vakharia, R. E. Lloyd, E. Ehrenfeld, and M. J. Grubman. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gingras, A. C., Y. Svitkin, G. J. Belsham, A. Pause, and N. Sonenberg. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and polio. Proc. Natl. Acad. Sci. USA 93:5578-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorbalenya, A. E., K. M. Chumakov, and V. I. Agol. 1978. RNA-binding properties of nonstructural polypeptide G of encephalomyocarditis virus. Virology 88:183-185. [DOI] [PubMed] [Google Scholar]
- 9.Gradi, A., Y. V. Svitkin, W. Sommergruber, H. Imataka, S. Morino, T. Skern, and N. Sonenberg. 2003. Human rhinovirus 2A proteinase cleavage sites in eukaryotic initiation factors (eIF) 4GI and eIF4GII are different. J. Virol. 77:5026-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, H., and A. C. Palmenberg. 2001. Deletion mapping of the encephalomyocarditis virus 2A protein and the adjacent primary cleavage site. J. Virol. 75:7215-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, H., and A. C. Palmenberg. 1995. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their Mengo virus counterparts. J. Virol. 69:2697-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn, H., and A. C. Palmenberg. 1996. Mutational analysis of the encephalomyocarditis virus primary cleavage. J. Virol. 70:6870-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jen, G., B. M. Detjen, and R. E. Thach. 1980. Shutoff of HeLa cell protein synthesis by encephalomyocarditis virus and poliovirus: a comparative study. J. Virol. 35:150-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolupaeva, V. G., A. Unbehaun, I. B. Lomakin, C. T. T. Hellen, and T. V. Pestova. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11:470-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medvedkina, O. A., I. V. Scarlat, N. O. Kalinina, and V. I. Agol. 1974. Virus-specific proteins associated with ribosomes of Krebs-II cells infected with encephalomyocarditis virus. FEBS Lett. 39:4-9. [DOI] [PubMed] [Google Scholar]
- 16.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 17.Mosenkis, J., S. Daniels-McQueen, S. Janovec, R. Duncan, J. W. B. Hershey, J. A. Grifo, W. C. Merrick, and R. E. Thach. 1985. Shutoff of host translation by encephalomyocarditis virus infection does not involve cleavage of the eucaryotic initiation factor 4F polypeptide that accompanies poliovirus infection. J. Virol. 54:643-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penman, S., K. Scherrer, Y. Becker, and J. Darnell. 1963. Polyribosomes in normal and poliovirus-infected HeLa cells and their relationship to messenger-RNA. Proc. Natl. Acad. Sci. USA 49:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pensiero, M. N., and J. Lucas-Lenard. 1985. Evidence for the presence of an inhibitor on ribosomes in mouse L cells infected with mengovirus. J. Virol. 56:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plagemann, P. 1968. Mengovirus replication in Novikoff rat hepatoma and mouse L cells: effects on synthesis of host-cell macromolecules and virus-specific synthesis of ribonucleic-acid. J. Virol. 2:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter, F. W., Y. Bochkov, A. J. Albee, C. Wiese, and A. C. Palmenberg. 2006. A picornavirus protein disrupts nucleocytoplasmic transport by targeting Ran GTPase. Proc. Natl. Acad. Sci. USA 103:12417-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu, X., Z. Yang, S. Zhang, L. Shen, A. W. Dangel, J. H. Hughes, K. L. Redman, L. Wu, and C. Y. Yu. 1998. The human DEVH-box protein Ski2W from the HLA is localized in nucleoli and ribosomes. Nucleic Acids Res. 26:4068-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 24.Skern, T., B. Hampolz, A. Guarne, A. Fita, E. M. Bergmann, J. Petersen, and M. N. James. 2002. Structure and function of picornavirus proteases. ASM Press, Washington, DC.
- 25.Stuger, R., A. C. J. Timmers, H. A. Raue, and J. van't Riet. 2000. Nuclear import of ribosomal proteins: evidence for a novel type of nucleolar localization signal, p. 205-217. In R. A. Garrett, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, DC.
- 26.Svitkin, Y. V., H. Hahn, A. C. Gingras, A. C. Palmenberg, and N. Sonenberg. 1998. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J. Virol. 72:5811-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taub, F., and T. C. Johnson. 1975. The mechanism of polyribosome disaggregation in brain tissue by phenylalanine. Biochem. J. 151:173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]