Abstract
The role of the high potential [3Fe-4S]1+,0 cluster of [NiFe] hydrogenase from Desulfovibrio species located halfway between the proximal and distal low potential [4Fe-4S]2+,1+ clusters has been investigated by using site-directed mutagenesis. Proline 238 of Desulfovibrio fructosovorans [NiFe] hydrogenase, which occupies the position of a potential ligand of the lacking fourth Fe-site of the [3Fe-4S] cluster, was replaced by a cysteine residue. The properties of the mutant enzyme were investigated in terms of enzymatic activity, EPR, and redox properties of the iron-sulfur centers and crystallographic structure. We have shown on the basis of both spectroscopic and x-ray crystallographic studies that the [3Fe-4S] cluster of D. fructosovorans hydrogenase was converted into a [4Fe-4S] center in the P238 mutant. The [3Fe-4S] to [4Fe-4S] cluster conversion resulted in a lowering of approximately 300 mV of the midpoint potential of the modified cluster, whereas no significant alteration of the spectroscopic and redox properties of the two native [4Fe-4S] clusters and the NiFe center occurred. The significant decrease of the midpoint potential of the intermediate Fe-S cluster had only a slight effect on the catalytic activity of the P238C mutant as compared with the wild-type enzyme. The implications of the results for the role of the high-potential [3Fe-4S] cluster in the intramolecular electron transfer pathway are discussed.
Hydrogenases catalyze the reversible oxidation of the most simple of chemical compounds, molecular hydrogen, according to the reaction: H2 ↔ 2H+ + 2e−. They are widely distributed among microorganisms including Archae, Bacteria, and Eukaryotes and are involved in many relevant biological processes where hydrogen is oxidized or evolved (1). In many anaerobes that use H2 as a source of energy, hydrogenases couple H2 oxidation to the reduction of electron acceptors such as carbon dioxide, sulfate, nitrate, fumarate, and sulfur, whereas H2 is produced via hydrogenase as a means of disposal of excess electrons during fermentation. These enzymes play a central role in the energy-generating mechanisms of sulfate-reducing bacteria of the genus Desulfovibrio that exhibit a strictly anaerobic mode of respiration of sulfate. They are involved both in the oxidation of hydrogen associated with the generation of a proton gradient and in the production of hydrogen through fermentation processes, depending on the growth conditions of these microorganisms (2, 3).
Hydrogenases are metalloenzymes that have been classified on the basis of their metal-centers composition in two major classes: iron-only ([Fe]) hydrogenases (4) and nickel-iron ([NiFe]) hydrogenases (5, 6). Extensive biochemical and biophysical studies have been performed on Desulfovibrio gigas [NiFe] hydrogenase (7–11). This enzyme is a heterodimeric periplasmic protein with a molecular mass of 89 kDa (28- and 60-kDa subunits) that contains four metal centers: one NiFe, one [3Fe-4S], and two [4Fe-4S] clusters (12, 13). The spectroscopic signatures of the enzyme redox centers indicate that the NiFe center, coordinated by four cysteine residues, is the catalytic site of the enzyme responsible of the heterolytic cleavage of H2, whereas the iron-sulfur clusters are secondary electron carriers (14, 15). Its natural electron acceptor is the low-potential tetraheme cytochrome c3 (16).
The D. gigas enzyme isolated under aerobic conditions is inactive and its activation requires incubation under reducing conditions, such as an hydrogen atmosphere. It exhibits two types of EPR signals: one centered at g = 2.01 detected only at low temperatures, which is assigned to an oxidized [3Fe-4S] cluster, and two Ni-related superimposed signals termed Ni-A and Ni-B (15). The most intense Ni-A signal is associated with the so-called “unready” state of hydrogenase whose activation requires prolonged exposure to hydrogen under anaerobic conditions. The minor Ni-B signal is associated with the “ready” state of the enzyme, which can be rapidly activated. Upon reduction under hydrogen, the enzyme displays an additional rhombic EPR signal, termed Ni-C, associated with one of the active states of the enzyme (15). In addition, the iron-sulfur clusters have been characterized at various stages of the reductive activation process by EPR and Mössbauer studies (11, 15) and magnetic interactions between the Ni-C form and a reduced [4Fe-4S] cluster have been described (15, 17).
The three-dimensional x-ray structure of the [NiFe] hydrogenase from D. gigas determined by x-ray crystallography revealed that the three iron-sulfur clusters are distributed along an almost straight line in the small subunit, with the [3Fe-4S] cluster intercalated between the two [4Fe-4S] clusters (12, 13). This sequential arrangement provides a plausible electron transfer pathway from the active site buried in the protein through the proximal [4Fe-4S] and [3Fe-4S] clusters to the distal [4Fe-4S] center located close to the molecular surface. However, although the middle position of the [3Fe-4S] cluster suggests an active redox role for this center, its high midpoint potential (−70 mV) as compared with the two [4Fe-4S] clusters (−290 and −340 mV, respectively) (11), casts doubt about its involvement in electron transfer.
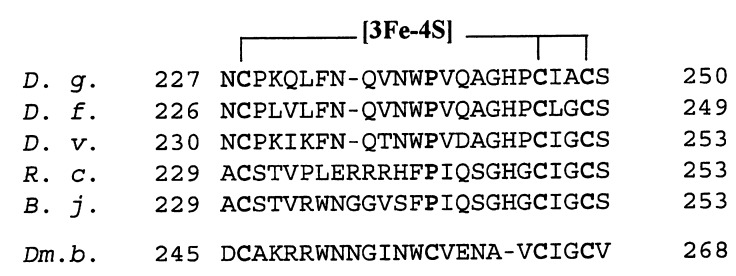
Recently, the [NiFe] hydrogenase from D. fructosovorans, which is highly homologous to D. gigas enzyme (18), has been cloned and expressed in a D. fructosovorans mutant lacking a wild-type copy of the enzyme (19, 20), thus opening the way to site-directed mutagenesis studies. To investigate the role of the [3Fe-4S] cluster in the intramolecular electron transfer, Pro-238 of D. fructosovorans hydrogenase, which is homologous to Pro-239 of D. gigas enzyme (Fig. 1) and occupies the position of a potential additional ligand to the intermediate Fe-S cluster (12) was replaced by a cysteine residue, as is the case of the native Desulfomicrobium baculatum [NiFeSe] hydrogenase (8) (Fig. 1).
Figure 1.
Alignment of amino acid sequences of various [NiFe] hydrogenase small subunits around the three cysteine ligands to the [3Fe-4S] center. The sequences were taken from refs. 8 (D. gigas [NiFe] hydrogenase, D. g.), 18 (D. fructosovorans, D. f.), 21 (D. vulgaris Miyazaki F, D. v.), 22 (Rhodobacter capsulatus, R. c.), 23 (Bradirhizobium japonicum, B. j.), and 8 (D. baculatum, Dm. b.). The cysteine cluster ligands and the conserved proline residue corresponding to position 239 in D. gigas enzyme are shown in bold.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions.
Escherichia coli strain DH5α, F−, endA1, hsdR17(rK− mK+), supE44, thi−1, λ−, recA1, gyrA96, relA1, Δ(argF− lacZYA)U169, φ80dlacZΔM15 was used as a host in the cloning of recombinant plasmids. The bacterium was routinely grown at 37°C in Luria–Bertani medium. Ampicillin at 100 μg/ml or chloramphenicol at 30 μg/ml were added when cell harbored pUC or pBCF4 derivatives, respectively. The method described by Inoue et al. (24) was used to generate high-efficiency E. coli competent cells. The cells were grown at 18°C in SOB medium. The recently constructed shuttle vector pBMC6 (25) was used in this study to carry the [NiFe] hydrogenase operon from D. fructosovorans. The 3.8-kb NsiI–DraI fragment from pHH7 that contains the hynABC genes from D. fructosovorans (20), was inserted in pBMC6 digested with PstI and SmaI. The resulting plasmid was named pBCF4. Plasmid pBCF4 was introduced into D. fructosovorans by electrotransformation (25).
D. Fructosovorans strain MR400 (hyn∷npt ΔhynABC) carrying a deletion in the [NiFe] hydrogenase operon (19) was grown anaerobically at 37°C in SOS medium (25). Kanamycine at 50 μg/ml was present routinely, and 30 μg/ml of thiamphenicol was added only when cells harbored the plasmid pBCF4.
Culture volumes were increased by a factor of 10 at each subculture from 10 ml to 10 liters. All transfers were performed by using strictly anaerobic procedures. Septum-stoppered serum bottles were used for 100-ml and 1-liter cultures for which the syringe-based anaerobic technique was used (26).
Site-Directed Mutagenesis.
The double PCR mutagenesis technique (27) was used to generate the point mutation in the hynA gene carried by the pBCF4 plasmid. The fragment HindIII–SacI from pBCF4 was subcloned into pUC18 in which the EcoRI site had been previously removed. This construct, called pUCmut, made unique the EcoRI site of the insert and allowed us to work with the 410-bp EcoRI–SacI cassette, which contains the Pro-238 target codon. Complementary mutagenic oligonucleotides B and C were designed to introduce the TGC Cys codon in place of the CCC Pro-238 codon. Oligonucleotide A was located upstream from the EcoRI site, and oligonucleotide D was located downstream from the SacI site. Vent DNA polymerase (New England Biolabs) was used in the PCR experiments. PCR1 performed in the presence of oligonucleotides A and C generated the mutated 5′ part of the cassette, and PCR2 performed in the presence of oligonucleotides B and D generated the mutated 3′ part of the cassette. In PCR3, the products from PCR1 and 2 were mixed to generate the whole mutated EcoRI–SacI fragment. After completion of PCR3, the mutated cassette was cloned in EcoRI–SacI-digested pUCmut. The sequences of four clones were verified. The HindIII–SacI fragment that carried the Cys-238 mutation then was cloned in the pBCF4 digested with HindIII and SacI. The mutated plasmid was introduced into D. fructosovorans MR400 by electrotransformation.
Protein Purification and Analysis.
Recombinant native D. fructosovorans [NiFe] and P238C mutant hydrogenases expressed in D. fructosovorans MR400 (pBCF4) strain were purified following the procedure previously reported (28) with minor modifications. Precautions were taken against oxygen by flushing buffers with purified argon, by washing each column with O2-free buffer and by collecting active fractions into sealed tubes under argon. In a typical experiment, 45-g wet weight cells stored at −80°C were thawed, and the soluble protein extract was prepared as described (29). The purification was achieved by using four chromatography steps (28), and the yield of pure enzyme was lower for the mutant (18 mg) than for recombinant native protein (30 mg).
Cytochrome c3 from D. fructosovorans was purified by using the procedure previously reported for the basic cytochrome c3 from D. africanus (30). The pure cytochrome c3 exhibited a purity index (A552red − A570red/A280ox) of 3.2.
Enzyme Assays.
Enzyme activity was measured at 30°C by using the spectrophotometric H2 uptake (31) and the manometric H2 evolution assay (29) with methyl viologen (1 mM) as mediator. H2 uptake activity also was determined by using D. fructosovorans tetraheme cytochrome c3 (25 μM) as electron acceptor (16). One unit of hydrogenase activity in the H2 uptake assay is defined as the amount of enzyme that catalyzes the reduction of 2 μmol methyl viologen or 0.5 μmol cytochrome c3 per min, corresponding to the consumption of 1 μmol of H2 per min. The D2/H+ exchange activity of the enzymes was measured as previously reported (32). Hydrogenase activity in the different assays was determined with the enzyme previously activated by prolonged incubation under H2 in a buffer containing 50 mM Tris⋅HCl and 1 mM methyl viologen (31) or in the same buffer with 20% of glycerol added as protecting agent and 10 μM cytochrome c3 (16).
Analytical Procedures.
Hydrogenase concentration of pure enzyme preparations were determined spectrophotometrically by using their extinction coefficient at 400 nm as described (29). Native and SDS/PAGE followed by Western blotting using a polyclonal antibody against D. fructosovorans [NiFe] hydrogenase (29) were performed as previously reported (33, 34). The metal content of the proteins was estimated by inductively coupled plasma emission spectroscopy using a Jobin-Yvon model JY 38 apparatus (Longjumeau, France).
EPR Spectroscopy and Redox Titrations.
Redox titrations were carried out at 23°C in an anaerobic cell containing a solution of purified hydrogenase (100 μM) in 50 mM Hepes (pH 8.0) buffer as previously described (35). At each desired potentials, a 150-μl sample was anaerobically transferred in an EPR tube and quickly frozen in liquid nitrogen.
EPR spectra were recorded on a Bruker ESP 300E spectrometer fitted with an Oxford Instrument ESR 900 helium-flow cryostat. For spin quantitation, the double integration of the signal recorded in nonsaturating conditions was compared with that given by a CuSO4 standard at the same temperature.
X-Ray Crystallography.
Crystals of the P238C mutant of D. fructosovorans hydrogenase were grown at room temperature under conditions [100 mM Mes buffer, pH 6.4, 100 mM NH4(CH3COO−), 25 mM octyl-β-d-glucopyranoside, and 24–26% polyethylene glycol (PEG) 6000 as precipitant], which are similar to those used for the native enzyme (36), except that the experiment was carried out in an anaerobic glove box (<1 ppm O2) because the mutant appeared more sensitive to O2 than the wild-type enzyme. Typically, crystals appeared after 3 days. A crystal of dimensions 0.3 × 0.2 × 0.05 mm3 was incubated a few minutes in a buffer containing 100 mM Mes, pH 6.4, 100 mM NH4(CH3COO−), 34% PEG 6000, and 20% of glycerol as cryoprotectant. The crystal then was flashed-cooled in liquid propane at −160°C and conserved in solid propane at −196°C.
Subsequently, a data set was collected at cryogenic temperature (−150°C) at the D2AM synchrotron beam line of the European Synchrotron Radiation Facility (Grenoble, France) by using a XRII-CCD detector (37). Data then were processed with xds (38), and further data reductions were made with the CCP4 package (39). The D. fructosovorans hydrogenase P238C mutant crystal unit cell is isomorphous with the corresponding native one at this same temperature. Data collection statistics are summarized in Table 1. The structure was solved by the molecular replacement method with the native D. fructosovorans hydrogenase structure as the starting model (code 1frf) using the amore program (40). The model of the native D. fructosovorans hydrogenase, including the [3Fe-4S] cluster and Pro-238, was refined against the 3-Å resolution data of the P238C mutant with the programs x-plor (41) and refmac (39). The model was adjusted first by a rigid-body rotation/translation and then by positional and individual B-factor refinements.
Table 1.
X-ray data statistics
| Resolution | 3 Å |
| Space group | P21 |
| Cell parameters | |
| a, b, c, Å | 68.35 99.73 182.61 |
| β, ° | 92.04 |
| λ, Å | 0.9798 |
| Observed reflections | 79,013 |
| Unique reflections | 33,794 |
| I/σ (I) | 8.9 |
| Completeness, % | 74 |
| Rsym,* % | 7.7 |
Rsym = 100 × (∑|I − 〈I〉|/∑I).
RESULTS
Expression, Characterization, and Catalytic Activity of [NiFe] Hydrogenase P238C Mutant.
The recombinant native D. fructosovorans [NiFe] hydrogenase and P238C mutant were expressed in D. fructosovorans MR400 pBCF4 strain as holoproteins and isolated as described. The purity of the proteins was confirmed by SDS/PAGE and Western blotting, which revealed specific bands at 28 and 60 kDa for both proteins. The final purity index (A400 nm/A280 nm) of the proteins was 0.28 and 0.27 for the native protein and the mutant, respectively (Table 2).
Table 2.
Physico-chemical and catalytic properties of native D. fructosovorans [NiFe] hydrogenase and P238/C mutant
| Enzyme | Wild type | P238/C mutant |
|---|---|---|
| Spectrophotometric properties | ||
| ɛ400 mM−1⋅cm−1 | 53 | 52 |
| A400/A280 | 0.28 | 0.27 |
| Metal content, atoms/mol | ||
| Ni | 1.05 ± 0.05 | 0.97 ± 0.06 |
| Fe | 11.95 ± 0.5 | 12.6 ± 0.8 |
| Catalytic activity | ||
| H2 uptake* | 330 | 205 |
| H2 production† | 65 | 104 |
| Cytochrome c3 reduction* | 1,320 | 812 |
| D2/H+ exchange activity‡ | 223 | 175 |
| Midpoint potentials, mV | ||
| Intermediate Fe-S cluster | +65 (3Fe) | −250 (4Fe) |
| [4Fe-4S]2+/1+ prox | −340 | −380 |
| [4Fe-4S]2+/1+ dist | −340 | −380 |
Specific activity expressed as reported in Materials and Methods.
Expressed as μmol H2 produced min−1⋅mg−1.
Expressed as μmol HD + H2 produced min−1⋅mg−1.
The iron content of the native hydrogenase was found to be close to 12 atoms per molecule (Table 2), in agreement with the presence of two [4Fe-4S] and one [3Fe-4S] clusters and one additional iron atom associated to nickel in the active site (12, 13). The absorption spectrum of the P238C mutant and its extinction coefficient at 400 nm (ɛ400 = 52 mM−1⋅cm−1) are similar to those of the native hydrogenase (Table 2). These properties are indicative of the presence of three Fe-S clusters in the mutant enzyme in agreement with the presence of at least 12 iron atoms per molecule (Table 2). In contrast to the wild-type enzyme, the as isolated P238C mutant Fe-S clusters exhibited oxygen sensitivity, which resulted in a decrease of its A400/A280 absorbance ratio and a loss of catalytic activity.
The P238C mutant is a functional enzyme that shows significant activity in the different reactions catalyzed by hydrogenase (Table 2). The H2 uptake activity of the mutant using methyl viologen as electron acceptor exhibited a decrease of 38% as compared with the wild-type values, whereas H2 production activity showed an increase of 60% with the same mediator as electron donor. In addition, a decrease of 38% of cytochrome c3 reduction activity was observed for the mutant, which is in good agreement with the H2 uptake activity obtained by using the viologen dye. These results indicate that the H2 uptake and H2 production activities of the P238C mutant were only slightly modified. In addition, the data suggest that the electron transfer pathway from the active site to the electron acceptor is similar when using either methyl viologen or cytochrome c3. The D2/H+ exchange activity of the P238C mutant exhibits a slight decrease (21%) as compared with the wild-type value (Table 2).
EPR Spectroscopy and Redox Properties.
Both the wild-type and the P238C mutant hydrogenases were titrated from about +300 mV to −480 mV. The EPR spectra were recorded at various temperature and microwave power conditions to analyze the redox behavior of the different metal centers.
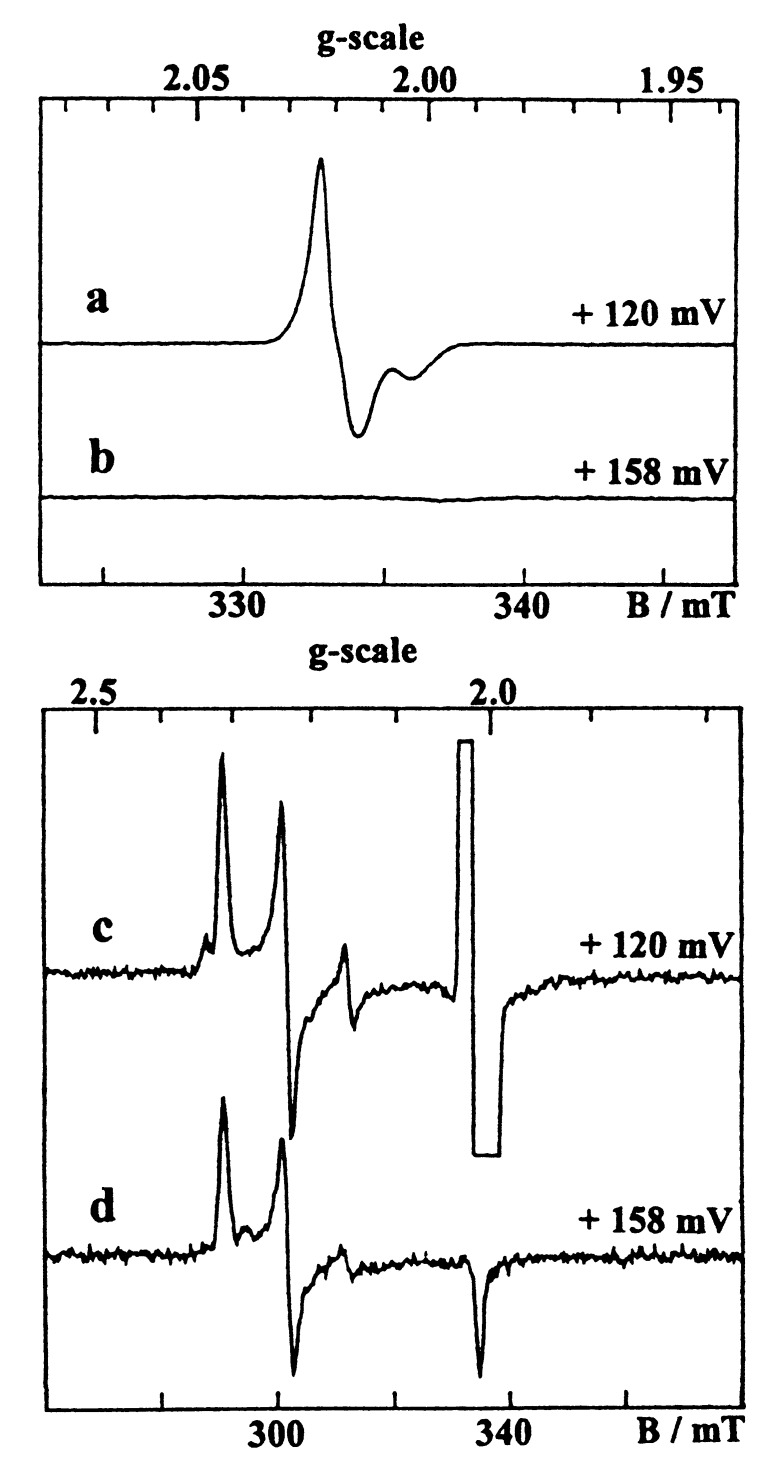
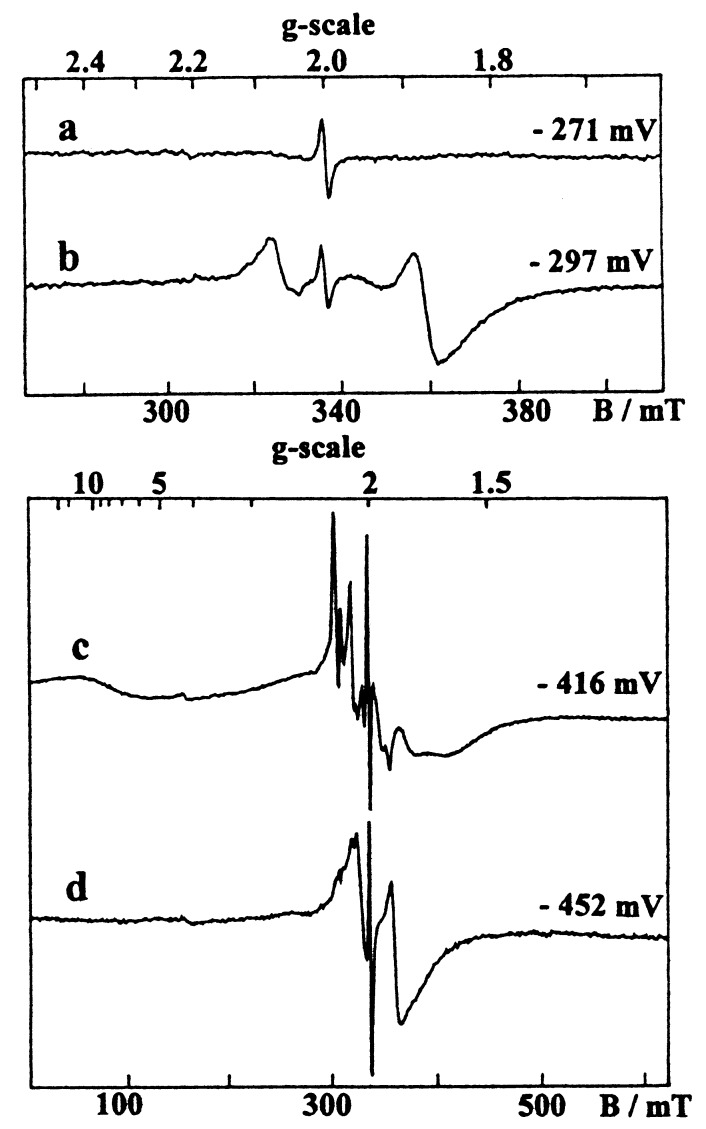
As previously described (28, 42), the oxidized wild-type enzyme exhibited a weakly anisotropic [3Fe-4S]1+ signal centered at g = 2.02 (Fig. 2a), which overlapped a Ni signal having a major Ni-A (g = 2.32, 2.23, 2.01) and a minor Ni-B (g = 2.34, 2.16, 2.01) components (Fig. 2c). The [3Fe-4S]1+ signal accounted for 0.85 spin/molecule, whereas the total Ni signal intensity corresponded to 0.7 spin/molecule, a substoichiometric quantitation usually found in [NiFe] hydrogenases (42, 43). Upon reduction, these signals disappeared progressively, leading to an EPR silent enzyme in the g = 2 region when poised at about −250 mV (Fig. 3a). The reduction of the [3Fe-4S] center is manifested by the increase of a broad line at g ≈ 12 arising from the S = 2 [3Fe-4S]0 clusters (Fig. 3c). At lower potentials (<−300 mV), the reduction of the two [4Fe-4S] centers of the wild- type hydrogenase gave, as in the D. gigas enzyme, an extremely broad spectrum (Fig. 3c), which results from their magnetic coupling with the [3Fe-4S]0 cluster (11). In addition, the appearance of the Ni-C species, which is associated with the active enzyme, gave at low temperature the split Ni-C signal arising from magnetic interactions with the proximal [4Fe-4S]1+ cluster (17) and characterized by major features at g = 2.21 and 2.10. This signal showed a bell-shaped variation as a function of the redox potential with a maximum at −360 mV.
Figure 2.
EPR spectra given by the hydrogenases from D. fructosovorans in the oxidized state. (a and c) Wild-type enzyme. (b and d) P238C mutant. Conditions: temperature, 12 K; microwave frequency, 9.422 GHz; microwave power, 0.04 mW; modulation amplitude, (a and b) 0.1 mT, (c and d) 1 mT.
Figure 3.
EPR spectra given by reduced hydrogenases from D. fructosovorans. (a and c) Wild-type enzyme. (b and d) P238C mutant. Conditions: temperature, 6 K; microwave frequency, 9.420 GHz; microwave power, (a and b) 0.1 mW, (c and d) 1 mW; modulation amplitude, 1 mT.
In the oxidized P238C mutant, a Ni signal similar to that of the wild-type enzyme though corresponding to a slightly different Ni-A/Ni-B ratio was observed, but interestingly, no [3Fe-4S] signal could be detected (Fig. 2 b and d). Upon reduction, the Ni signal disappeared, but below −200 mV, in contrast with the wild-type enzyme, a new-fast relaxing axial signal increased at g = 2.08 and 1.88 (Fig. 3b), which is typical of a reduced [4Fe-4S] center. At very low potentials (<−300 mV), this new signal broadened progressively, and the spin intensity of the whole spectrum increased from 1 to 3 spins/molecule, showing that the signal given by the fully reduced mutant (Fig. 3d) arises from three [4Fe-4S]1+ centers. In the P238/C mutant, the Ni-C signal behaved as in the wild-type enzyme.
Taken together, these results show clearly that the [3Fe-4S] cluster of the wild-type hydrogenase is no longer present in the P238C mutant and has been converted into a new [4Fe-4S] center. By fitting the amplitude variations of the various FeS signals as a function of the redox potential to Nernst curves, the midpoint potentials of the FeS clusters of the two enzymes were determined (Table 2). It then appears that the new [4Fe-4S] cluster lying between the proximal and distal [4Fe-4S] centers has a markedly more negative redox potential than the [3Fe-4S] cluster of the wild-type enzyme. In contrast, the potentials of the distal and proximal [4Fe-4S] centers are only slightly negatively shifted in the mutant. Moreover, it is worth noting that the properties of the Ni center were not significantly affected by the mutation, as shown by the similar redox behavior observed in both enzymes for the Ni-A, Ni-B, and Ni-C signals.
Structure of the P238C Mutant [NiFe] Hydrogenase.
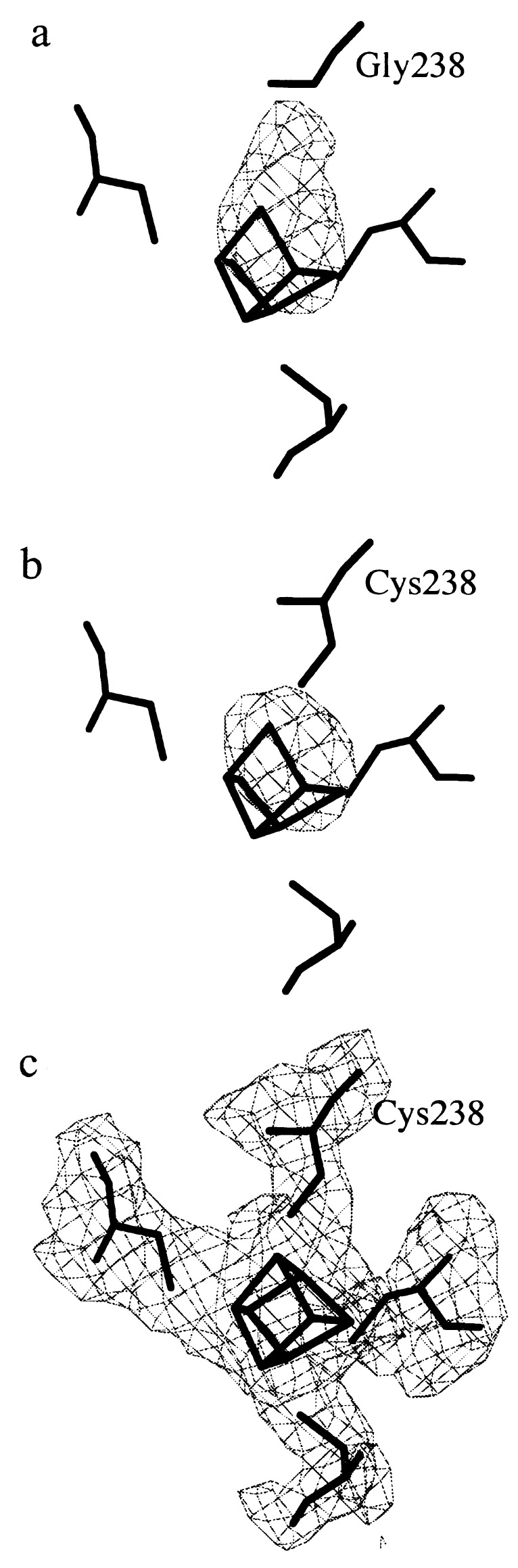
To model the mutation, the residue 238 was first included as a glycine with the program o (44). After one cycle of energy minimization (x-plor) (41), the resulting phases were used to calculate a difference Fourier map with (Fo-Fc) coefficients. This map showed residual density located between the [3Fe-4S] cluster and the polypeptide chain (Fig. 4a). Next, we modeled the residue 238 as a cysteine. In the difference map calculated with the new model phases, a strong peak (13.2 times the rms value of the map) remained and was at 2.5 Å from each of the Fe atoms of the [3Fe-4S] cluster (Fig. 4b). This peak could be explained by the presence of a fourth Fe atom at this position. The final model of the mutant enzyme containing three [4Fe-4S] clusters was refined and fitted to the electron density with (2Fo-Fc) and (Fo-Fc) Fourier maps (Fig. 4c). Besides the [FeS] clusters and a Ni-Fe pair in the active site, the final model contains 804 residues and 50 water molecules with temperature factor values <40 Å2. The standard and free R factors were 0.22 and 0.31, respectively in the range of 10 to 3 Å resolution. The model has rms deviations from ideality of 0.012 Å for bonds and 0.036 Å for angles.
Figure 4.
Three-fold averaged electron density maps (45) calculated with Fo-Fc coefficients in the 20–3 Å resolution range and contoured at the 3.2 σ level (a and b) or with 2Fo-Fc coefficients and contoured at the 1.1 σ level (c). (a) Fo-Fc map calculated with the native model in which Pro-238 has been substituted by a glycine. The map confirms the presence of a mutation close to the [3Fe-4S] cluster, as there is a significant electron density peak, which is not explained by the native model. (b) Fo-Fc map calculated with the native model in which Pro-238 has been substituted by a cysteine. (c) 2Fo-Fc map calculated with the final model of the mutant including both the Cys-238 and the third [4Fe-4S] cluster.
In our study, all the electron density is unambiguously explained by the final model including a Cys at position 238 in the small subunit and where a third [4Fe-4S] cluster is found instead of the original [3Fe-4S] center (Fig. 4c). The structures of D. fructosovorans native hydrogenase and of the P238C mutant superimpose with rms deviations of 0.288 Å for all 804 Cα atoms and 0.286 for the Cα of the residues within 5 Å from the [3Fe-4S] cluster. This finding shows that the structural rearrangement after mutation is minor, even around cysteine 238.
DISCUSSION
Understanding the role of the high-potential [3Fe-4S]1+,0 cluster of [NiFe] hydrogenase from Desulfovibrio species, which is located halfway between the proximal and distal low-potential [4Fe-4S]2+,1+ clusters, is important for elucidating the intramolecular electron transfer pathway involved in the catalytic cycle of the enzyme (12). In the present work, we have shown on the basis of spectroscopic and x-ray crystallographic data that the [3Fe-4S] cluster of D. fructosovorans [NiFe] hydrogenase was converted into a [4Fe-4S] center in the P238C mutant without significantly altering the spectroscopic and redox properties of the NiFe center and the two native [4Fe-4S] clusters. The mutation results in a functional enzyme that maintains its structural integrity and exhibits a decrease in midpoint potential of the modified center from +65 mV to −250 mV. This study provides a x-ray crystallographic structure of a mutant protein where a [3Fe-4S] to [4Fe-4S] conversion is observed. The structures of the inactive and activated aconitase already have provided direct crystallographic evidence for such a Fe-S cluster interconversion (46). However, in that case, the fourth ligand of the Fe atom inserted into the original [3Fe-4S] cluster was a water molecule, which occupied a similar site in the crystal structure of the inactive enzyme.
To our knowledge, the only other reported transformation of a [3Fe-4S] cluster into a [4Fe-4S] center by site-directed mutagenesis has been carried out in fumarate reductase from E. coli (47). Interestingly, the mutated cluster had a 285-mV drop in midpoint potential, a value that is similar to the one observed in the modified center of hydrogenase. In fumarate reductase, the [3Fe-4S] to [4Fe-4S] cluster conversion was accomplished by a Val-to-Cys substitution at the site normally occupied by the second cysteine residue in a typical ferredoxin arrangement (Cys-158, Cys-204, Val-207, and Cys-210). A structural description of this transformation is lacking because the three-dimensional structure of this enzyme has not yet been determined. In D. fructosovorans hydrogenase, the Cys arrangement coordinating the [3Fe-4S] cluster (Cys-227, Cys-245, and Cys-248) is not ferredoxin-like, but the residue Pro-238 that was mutated faces the empty potential fourth Fe site of the cluster (12). It is worth noting that the same position is occupied by Cys in the [NiFeSe] hydrogenase from D. baculatum, an enzyme that possesses three [4Fe-4S] clusters (Elsa Garcin, personal communication).
The spatial arrangement of the three Fe-S clusters in D. gigas hydrogenase has suggested a plausible electron transfer pathway from the active site buried in the protein to the exposed His ligand (His-185) of the distal [4Fe-4S] cluster. In this arrangement, the redox centers are separated by approximately 12 Å and a potential electron transfer pathway involving H bonds and covalent bonds has been identified (12). However, the midpoint potential of the [3Fe-4S] center (−70 mV), which is markedly higher than those of the [4Fe-4S] clusters (−290 and −340 mV) (11), appears to be thermodynamically unfavorable for electron transfer involving the [3Fe-4S] cluster.
The present work shows that the midpoint potential of the median [3Fe-4S] cluster of the wild-type D. fructosovorans hydrogenase is even higher (+65 mV) (Table 2). Therefore electron transfer from and to hydrogen (E′° = −414 mV) through the [4Fe-4S] (both Em = −340 mV) and the [3Fe-4S] clusters is likely to be much less favorable from a thermodynamic point of view. In this context, it is rather striking that the decrease by about 300 mV of the midpoint potential of the median cluster in the P238C mutant (−250 mV vs. +65 mV), which is kinetically more favorable, has such a small effect on the catalytic activity, as exemplified by the slight decrease of H2 uptake activity in the presence of methyl viologen (Em = −440 mV) or cytochrome c3 (average Em = −250 mV) (48) (Table 2) as electron acceptor. This finding indicates that if the median cluster is involved in electron transfer, electron transfer is not a rate-limiting step of the catalytic reaction. Another possible explanation could be that the median cluster (whether [4Fe-4S] or [3Fe-4S]) is not involved in electron transfer and merely plays a structural role in stabilizing the IIs domain of the small subunit (12). This explanation would imply a direct electron transfer between the two [4Fe-4S] clusters distant of about 20 Å that bypasses the [3Fe-4S] center. We also have to keep in mind that the [3Fe-4S] cluster could undergo a further two-electron reduction as it was reported for other [3Fe-4S]-cluster-containing proteins (49). However, the bielectronic character of the [3Fe-4S]0,2− redox transition and its very negative potential (−720 mV) seem to exclude this hypothesis. Another possibility is that the midpoint potential of the [3Fe-4S] center as measured by equilibrium redox titration is an apparent potential that is higher than the microscopic potential related to the redox states involved in the catalytic cycle of the enzyme as a result of redox cooperativity between the clusters.
Interestingly, the D2/H+ exchange activity, which reflects the intrinsic activity of the active site, including heterolytic splitting of hydrogen, proton transfer, and gas access to the active site, is also slightly decreased in the P238C mutant (Table 2). This decrease and the corresponding decrease of the H2 uptake activity in a similar extent could indicate that the catalytic activity is limited by proton transfer because electron transfer is not involved in the exchange activity and neither the properties of the active site nor the gas accessibility (36) are affected by this point mutation. In this respect, the pH dependence of both proximal and distal [4Fe-4S]2+,1+ clusters midpoint potentials (15) and the presence of the exposed His-185 ligand of the distal [4Fe-4S]2+,1+ cluster (12), likely to undergo redox-dependent protonation/deprotonation as in the Rieske protein (50), suggests the existence of a proton-coupled electron-transfer reaction during the catalytic cycle of hydrogenase as reported for photosystem II (51) and cytochrome oxidase (52). This hypothesis may imply that the electron and proton pathways use some common structural elements located in the “channel” connecting the active site with the distal [4Fe-4S] center. In this context, the behavior of the P238C mutant, which exhibits a slight decrease of the H2 uptake activity and a small increase in H2 production activity, could be explained if the [3Fe-4S] to [4Fe-4S] cluster conversion alters the proton transfer rate depending on the direction of the reaction. Further studies on other mutants will be needed for a rational explanation of electron and proton transfer pathways in [NiFe] hydrogenases.
Acknowledgments
We thank Dr. V. M. Fernandez for measurements of D2/H+ exchange activity. The help of M. Roth, P. Carpentier, and J.-L. Ferrer during x-ray data collection at the D2AM beamline of the European Synchrotron Radiation Facility is greatly appreciated. We are indebted to M. Charrel for precious assistance in site-directed mutagenesis experiments and to J. C. Germanique for the estimation of iron content of the proteins. We also thank Dr. M. Frey for many stimulating discussions and critical reading of this manuscript. This work was supported by Grant BI02-CT94-2041 from the European Commission Biotechnology Program.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The coordinate reported in this paper has been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1frf).
References
- 1. Adams M W W, Mortenson L E, Chen J-S. Biochim Biophys Acta. 1980;594:105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- 2.Odom J M, Peck H D., Jr Annu Rev Microbiol. 1984;38:551–592. doi: 10.1146/annurev.mi.38.100184.003003. [DOI] [PubMed] [Google Scholar]
- 3.Hatchikian E C, Fernandez V, Cammack R. In: Microbiology and Biochemistry of Strict Anaerobes Involved in Interspecies H2 Transfer. Bélaich J P, Bruschi M, Garcia J L, editors. New York: Plenum; 1990. pp. 53–73. [Google Scholar]
- 4.Adams M W W. Biochim Biophys Acta. 1990;1020:115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- 5.Przybyla A E, Robins J, Menon N, Peck H D. FEMS Microbiol Rev. 1992;88:109–136. doi: 10.1111/j.1574-6968.1992.tb04960.x. [DOI] [PubMed] [Google Scholar]
- 6.Albracht S P J. Biochim Biophys Acta. 1994;1188:167–204. doi: 10.1016/0005-2728(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 7.Hatchikian E C, Bruschi M, LeGall J. Biochem Biophys Res Commun. 1978;82:451–461. doi: 10.1016/0006-291x(78)90896-3. [DOI] [PubMed] [Google Scholar]
- 8.Voordouw G, Menon N K, LeGall J, Choi E S, Peck H D, Jr, Przybyla A E. J Bacteriol. 1989;171:2894–2899. doi: 10.1128/jb.171.5.2894-2899.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cammack R, Patil D, Aguirre R, Hatchikian E C. FEBS Lett. 1982;142:289–292. [Google Scholar]
- 10.Moura J J G, Moura I, Huynh B H, Krüger H-J, Teixeira M, Du Varney R C, DerVartanian D V, Xavier A V, Peck H D, Jr, LeGall J. Biochem Biophys Res Commun. 1982;108:1388–1393. doi: 10.1016/s0006-291x(82)80060-0. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira M, Moura I, Xavier A V, Moura J J G, LeGall J, DerVartanian D V, Peck H D, Jr, Huynh B H. J Biol Chem. 1989;264:16435–16450. [PubMed] [Google Scholar]
- 12.Volbeda A, Charon M H, Piras C, Hatchikian E C, Frey M, Fontecilla-Camps J C. Nature (London) 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 13.Volbeda A, Garcin E, Piras C, de Lacey A I, Fernandez V M, Hatchikian E C, Frey M, Fontecilla-Camps J C. J Am Chem Soc. 1996;118:12989–12996. [Google Scholar]
- 14.Fernandez V M, Hatchikian E C, Patil D S, Cammack R. Biochim Biophys Acta. 1986;883:145–154. [Google Scholar]
- 15.Cammack R, Patil D S, Hatchikian E C, Fernandez V M. Biochim Biophys Acta. 1987;912:98–109. [Google Scholar]
- 16.Niviere V, Hatchikian E C, Bianco P, Haladjian J. Biochim Biophys Acta. 1988;935:34–40. [Google Scholar]
- 17.Guigliarelli B, More C, Fournel A, Asso M, Hatchikian E C, Williams R, Cammack R, Bertrand P. Biochemistry. 1995;34:4781–4790. doi: 10.1021/bi00014a036. [DOI] [PubMed] [Google Scholar]
- 18.Rousset M, Dermoun Z, Hatchikian E C, Bélaich J P. Gene. 1990;94:95–101. doi: 10.1016/0378-1119(90)90473-5. [DOI] [PubMed] [Google Scholar]
- 19.Rousset M, Dermoun Z, Chippaux M, Bélaich J P. Mol Microbiol. 1991;5:1735–1740. doi: 10.1111/j.1365-2958.1991.tb01922.x. [DOI] [PubMed] [Google Scholar]
- 20.Rousset M, Dermoun Z, Wall J D, Bélaich J P. J Bacteriol. 1993;175:3388–3393. doi: 10.1128/jb.175.11.3388-3393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deckers H M, Wilson F R, Voordouw G. J Gen Microbiol. 1990;136:2021–2028. doi: 10.1099/00221287-136-10-2021. [DOI] [PubMed] [Google Scholar]
- 22.Leclerc M, Colbeau A, Cauvin B, Vignais P M. Mol Gen Genet. 1988;214:97–107. doi: 10.1007/BF00340186. [DOI] [PubMed] [Google Scholar]
- 23.Sayavedro-Soto L A, Powell G K, Evans H J, Morris R O. Proc Natl Acad Sci USA. 1988;85:8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue H, Nojima H, Okayama H. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 25.Rousset M, Casalot L, Rapp-Giles B J, Dermoun Z, de Philip P, Bélaich J P, Wall J D. Plasmid. 1998;39:114–122. doi: 10.1006/plas.1997.1321. [DOI] [PubMed] [Google Scholar]
- 26.Macy J M, Schroder I, Thauer R K, Kröger A. Arch Microbiol. 1986;144:147–151. [Google Scholar]
- 27.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatchikian E C, Traore A S, Fernandez V M, Cammack R. Eur J Biochem. 1990;187:635–643. doi: 10.1111/j.1432-1033.1990.tb15347.x. [DOI] [PubMed] [Google Scholar]
- 29.Pieulle L, Guigliarelli B, Asso M, Dole F, Bernadac A, Hatchikian E C. Biochim Biophys Acta. 1995;1250:49–59. doi: 10.1016/0167-4838(95)00029-t. [DOI] [PubMed] [Google Scholar]
- 30.Pieulle L, Haladjian J, Bonicel J, Hatchikian E C. Biochim Biophys Acta. 1996;1273:51–61. doi: 10.1016/0005-2728(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez V M, Hatchikian E C, Cammack R. Biochim Biophys Acta. 1985;832:69–79. [Google Scholar]
- 32.Hatchikian E C, Forget N, Fernandez V M, Williams R, Cammack R. Eur J Biochem. 1992;209:357–365. doi: 10.1111/j.1432-1033.1992.tb17297.x. [DOI] [PubMed] [Google Scholar]
- 33.Davis B J. Ann NY Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Guigliarelli B, Asso M, More C, Augier V, Blasco F, Pommier J, Giordano G, Bertrand P. Eur J Biochem. 1992;207:61–68. doi: 10.1111/j.1432-1033.1992.tb17020.x. [DOI] [PubMed] [Google Scholar]
- 36.Montet Y, Amara P, Volbeda A, Vernede X, Hatchikian E C, Field M J, Frey M, Fontecilla-Camps J C. Nat Struct Biol. 1997;4:523–527. doi: 10.1038/nsb0797-523. [DOI] [PubMed] [Google Scholar]
- 37.Moy J P. Nuclear Instr Method. 1990;A348:641–644. [Google Scholar]
- 38.Kabsch W. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 39.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 40.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 41.Brünger A. In: Crystallographic Computing 4: Techniques and New Technologies. Isaacs N W, Taylor M R, editors. Oxford: Clarendon; 1988. pp. 127–140. [Google Scholar]
- 42.Asso M, Guigliarelli B, Yagi T, Bertrand P. Biochim Biophys Acta. 1992;1122:50–56. doi: 10.1016/0167-4838(92)90126-x. [DOI] [PubMed] [Google Scholar]
- 43.Dole F, Fournel F, Magro V, Hatchikian E C, Bertrand F, Guigliarelli B. Biochemistry. 1997;36:7847–7854. doi: 10.1021/bi963171i. [DOI] [PubMed] [Google Scholar]
- 44.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1990;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Vellieux F M D A P, Hunt J F, Roy S, Read R J. J Appl Crystallogr. 1995;28:347–351. [Google Scholar]
- 46.Robbins A H, Stout C D. Proc Natl Acad Sci USA. 1989;86:3639–3643. doi: 10.1073/pnas.86.10.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manodori A, Cecchini G, Schröder, Gunsalus R P, Werth M T, Johnson M K. Biochemistry. 1992;31:2703–2712. doi: 10.1021/bi00125a010. [DOI] [PubMed] [Google Scholar]
- 48.Bianco P, Haladjian J. Biochimie (Paris) 1994;76:605–613. doi: 10.1016/0300-9084(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 49.Duff J L C, Breton J L J, Butt J N, Armstrong F A, Thomson A J. J Am Chem Soc. 1996;118:8593–8603. [Google Scholar]
- 50.Iwata S, Saynovits M, Link T A, Michel H. Structure. 1996;4:567–578. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 51.Lydakis-Simantiris N, Ghanotakis D F, Babcock G T. Biochim Biophys Acta. 1997;1322:129–140. [Google Scholar]
- 52.Wikström M. Nature (London) 1989;338:776–778. doi: 10.1038/338776a0. [DOI] [PubMed] [Google Scholar]