Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that posttranscriptionally regulate gene expression by binding to 3′-untranslated regions (3′UTRs) of target mRNAs. Kaposi's sarcoma-associated herpesvirus (KSHV), a virus linked to malignancies including primary effusion lymphoma (PEL), encodes 12 miRNA genes, but only a few regulatory targets are known. We found that KSHV-miR-K12-11 shares 100% seed sequence homology with hsa-miR-155, an miRNA frequently found to be up-regulated in lymphomas and critically important for B-cell development. Based on this seed sequence homology, we hypothesized that both miRNAs regulate a common set of target genes and, as a result, could have similar biological activities. Examination of five PEL lines showed that PELs do not express miR-155 but do express high levels of miR-K12-11. Bioinformatic tools predicted the transcriptional repressor BACH-1 to be targeted by both miRNAs, and ectopic expression of either miR-155 or miR-K12-11 inhibited a BACH-1 3′UTR-containing reporter. Furthermore, BACH-1 protein levels are low in cells expressing either miRNA. Gene expression profiling of miRNA-expressing stable cell lines revealed 66 genes that were commonly down-regulated. For select genes, miRNA targeting was confirmed by reporter assays. Thus, based on our in silico predictions, reporter assays, and expression profiling data, miR-K12-11 and miR-155 regulate a common set of cellular targets. Given the role of miR-155 during B-cell maturation, we speculate that miR-K12-11 may contribute to the distinct developmental phenotype of PEL cells, which are blocked in a late stage of B-cell development. Together, these findings indicate that KSHV miR-K12-11 is an ortholog of miR-155.
MicroRNAs (miRNAs) are 19- to 23-nucleotide (nt) noncoding RNAs that posttranscriptionally regulate gene expression through translational inhibition and/or mRNA degradation. In the nucleus, precursor miRNAs are processed by Drosha and DGCR8, exported into the cytoplasm via exportin 5, and subsequently processed by Dicer (3, 5). One strand of the cytoplasmic miRNA duplex is incorporated into the RNA-induced silencing complex, which guides the binding of mature miRNAs to 3′-untranslated regions (3′UTRs) of target mRNAs (3, 5). Originally identified in Caenorhabditis elegans as being important regulators of development, it is now known that all metazoan organisms encode miRNAs (3, 5, 36). Recently, miRNAs have been identified in several DNA viruses including polyomaviruses and herpesviruses (for reviews, see references 16, 58, and 59). Epstein-Barr virus (EBV) encodes at least 17 miRNAs (9, 21, 52), while 12 miRNA genes have been reported for Kaposi's sarcoma-associated herpesvirus (KSHV) (8, 51, 56), a virus linked to Kaposi's sarcoma and lymphoproliferative disorders (11, 12). Unlike metazoan miRNAs, which are often highly conserved between species (5), viral miRNAs are much less conserved between related viruses.
Investigating miRNA targets is complicated by the fact that miRNAs require only limited complementarity for 3′UTR binding (5, 6, 39). Consequently, a single miRNA can modulate the expression of multiple genes. Analysis of the entire human transcriptome in silico in combination with experimental verification has revealed that the binding of an miRNA to a specific 3′UTR is critically dependent on 5′ nucleotides (nt) 2 to 7 of the mature miRNA. This sequence, termed the “seed,” classifies miRNAs into families (38).
We hypothesized that virally encoded miRNAs, like virokines, may have been captured from the host genome and/or coevolved within herpesvirus genomes. Since the seed represents the most important determinant for biological activity, we aligned seed sequences of all human herpesvirus-encoded miRNAs with the human miRNA database. Here, we report on a KSHV-encoded miRNA, which has an identical seed sequence to that of hsa-miR-155, and provide evidence that both miRNAs can regulate a common set of cellular genes.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
miRNA sensor plasmids were constructed using pGL3-Promoter (Promega). Oligonucleotides were annealed and inserted at FseI and XbaI. Constructs were verified by restriction enzyme digestion and were as follows: 5′-CTAGACCCCTATCACGATTAGCATTAACTCGAGCCCCTATCACGATTAGCATTACGGCCGG-3′ (miR-155 fwd), 5′-CCTTAATGCTAATCGTGATAGGGCTCGAGTTAATGCTAATCGTGATAGGGGT-3′ (miR-155 rev), 5′-CTAGAATCGGACACAGGCTAAGCATTAATTATTATCGGACACAGGCTAAGCATTAAGGCCGG-3′ (miR-K12-11 fwd), and 5′-CCTTAATGCTTAGCCTGTGTCCGATAATAATTAATGCTTAGCCTGTGTCCGATT-3′ (miR-K12-11 rev).
3′UTR sequences were obtained from the Ensembl database (www.ensembl.org) or the NCBI and were PCR amplified (TripleMaster; Eppendorf) from either BCBL-1 or 293 genomic DNA (DNAzol; Molecular Research Center, Inc., OH). Primers were designed using Vector NTI (Invitrogen) and are indicated in Table S2 in the supplemental material. PCR products were cloned into pCRII-TOPO (Invitrogen), excised, and inserted into the 3′UTR of pGL3-Promoter. Constructs were confirmed by restriction enzyme digestion and/or sequencing.
For the construction of pmiR-K12-11 and pmiR-155, ∼180 nt encompassing the stem-loop pre-miRNA were PCR amplified from BCBL-1 genomic DNA using primers indicated in Table S2 in the supplemental material. PCR products were TOPO cloned, excised with HindIII and XhoI, and inserted into pcDNA3.1V5/HisA (Invitrogen) at corresponding sites. KSHV miRNA expression vectors were described previously by Samols et al. (57).
2′ O-methylated (2′OMe) RNA oligonucleotides were synthesized by Dharmacon, Inc., and are antisense to the mature miRNA sequence. All bases were modified at the 2′ position.
QuikChange site-directed mutagenesis was performed using the primers indicated in Table S3 in the supplemental material according to the manufacturer's protocols (Stratagene). Primer design was done using PrimerX (www.bioinformatics.com). Each 6-mer seed match site was changed to an XhoI restriction enzyme site, and mutants were analyzed by restriction enzyme digestion.
Cell lines and transfections.
BCP-1 (a gift from Denise Whitbey at the NCI), BC-1, VG-1, JSC-1 (gifts from Dirk Dittmer at the University of North Carolina), RAJI (a gift from Sankar Swaminathan at the University of Florida), BCBL-1, and BJAB cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum and 5% penicillin-streptomycin (Gibco). 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 5% penicillin-streptomycin. For stable cell lines, cells were transfected with 10 μg plasmid DNA using Lipofectamine 2000 (Invitrogen) and selected with 500 μg/ml G418 (Mediatech, Inc.) for 4 weeks. Transfections for luciferase assays were performed in 6-well or 24-well plates with Lipofectamine 2000 according to the manufacturer's protocols. Cells were cultured 48 to 72 h prior to harvest. Transfections for BJAB cells were done by nucleofection using cell line solution T, program O-17, according to the manufacturer's instructions (Amaxa, Inc.).
Immunoblotting.
Whole-cell lysates were harvested 72 h posttransfection. Lysates corresponding to 1 × 105 cells were separated on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto Immobilon-P membranes (Millipore). Blots were probed for BACH-1 or actin (Santa-Cruz Biotechnology, Inc.) and developed with peroxidase-conjugated antibodies and chemiluminescent substrate (Pierce).
Affymetrix array-based gene expression profiling.
RNA isolation was performed using the RNeasy kit according to the manufacturer's instructions (QIAGEN). RNA labeling was done using the GeneChip eukaryotic one-cycle target labeling assay as directed by Affymetrix and used to probe Affymetrix U133 2.0 Plus Human GeneChips. Hybridization was performed at 45°C for 16 h, and the chips were then washed and stained using Affymetrix protocol EukGE-WS2v5_450. Array scanning and analysis were performed as described previously (57).
Luciferase assays.
Luciferase activity was quantified using the Luciferase assay system (Promega) according to the manufacturer's protocols. Briefly, transfected 293 cells were lysed in cell culture lysis reagent (Promega), and 20% of each cell lysate was assayed for firefly luciferase activity. Light units were normalized to total protein, determined using the BCA protein assay kit (Pierce) according to the manufacturer's instructions, or Renilla luciferase, using a dual luciferase reporter kit (Promega).
RNA extraction for RT-PCR.
Total RNA was prepared using RNA-Bee (Tel-Test, Inc., TX) according to the manufacturer's protocols. For detection of BIC expression, 1 μg of DNase-treated RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and random hexamers in the presence of RNase-OUT recombinant RNase inhibitor (Invitrogen) to create a cDNA pool. Ten percent of each reverse transcription (RT) reaction was used for PCR amplification. BIC and β-actin primers are listed in Table S2 in the supplemental material.
Northern blot analysis.
Thirty micrograms of total RNA was loaded onto 15% 8 M urea polyacrylamide gel and transferred onto Genescreen Plus (Perkin-Elmer) following electrophoresis. Probe labeling was performed using T4 polynucleotide kinase (New England Biolabs) in the presence of [α-32P-γ]ATP.
Microarray data accession number.
The Gene Expression Omnibus accession number for the microarray data is GSE9264.
RESULTS
DNA tumor virus-encoded miRNAs share seed sequence homology with human oncogenic miRNAs.
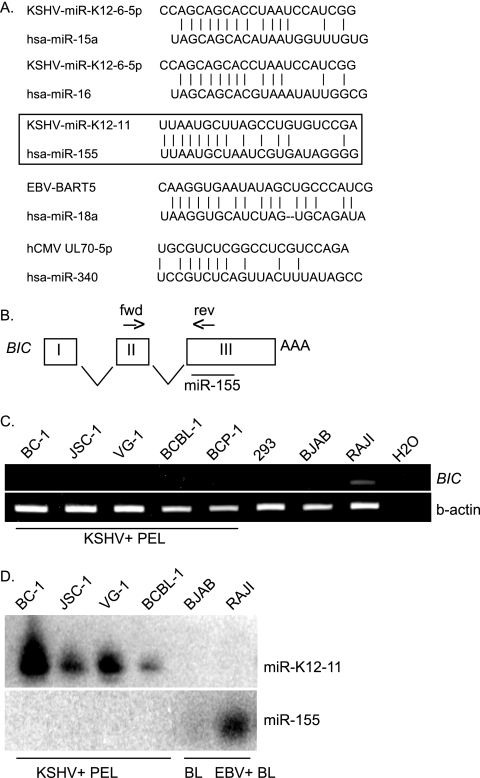
To determine whether seed sequences, known to be critically important for mRNA target recognition, exhibit conservation, we performed alignments of all known KSHV, EBV, and human cytomegalovirus miRNAs against an miRNA registry containing 474 human miRNA sequences (http://microrna.sanger.ac.uk). 5′ nt 2 to 7 of both cloned (8, 9, 22, 51, 52, 56) and predicted (21) miRNAs were analyzed, which revealed that at least four viral miRNAs harbor seed sequences homologous to those of human miRNAs (Fig. 1A). 5′ nt 1 to 8 of KSHV-miR-K12-11 are 100% identical to hsa-miR-155, while 5′ nt 3 to 10 of miR-K12-6-5p are homologous to hsa-miR-15a and hsa-miR-16. Additionally, 5′ nt 2 to 7 of EBV-BART5 are identical to hsa-miR-18a/b (Fig. 1A). Interestingly, both miR-15a and miR-16 are thought to have tumor suppressor activity and induce apoptosis by silencing BCL2 (13). In contrast, miR-155 and miR-18 are aberrantly expressed in many human malignancies. Human miR-18 is expressed from the miR-17-92 cluster on chromosome 13q31, a region often amplified in B-cell lymphomas, and has been proposed to be oncogenic since overexpression accelerates c-myc-induced lymphomagenesis in transgenic mice (24, 26).
FIG. 1.
Herpesvirus miRNAs share seed sequence homology with human miRNAs. (A) miRNA sequences from KSHV, EBV, and human cytomegalovirus were aligned with sequences in the human miRNA database. Shown are those viral miRNAs exhibiting the greatest sequence homology to human miRNAs. (B) miR-155 is expressed from exon 3 of the non-protein-coding BIC. Primers (forward [fwd] and reverse [rev]) for RT-PCR analysis of BIC expression are indicated. (C) RT-PCR analysis of BIC expression in KSHV-infected PEL cell lines. The RAJI cell line, an EBV-infected BL cell line, was used as a positive control. H2O indicates the no-template control. (D) Northern blot analysis of KSHV-miR-K12-11 and hsa-miR-155 expression in PEL and BL cells. Twenty-five micrograms of total RNA was loaded per lane and hybridized to probes for either miR-K12-11 or miR-155.
miR-155 is an evolutionarily conserved miRNA processed in humans from exon 3 of the non-protein-coding BIC (B-cell integration cluster) RNA (37, 62) (Fig. 1B). BIC was originally identified in chicken B-cell lymphomas, which develop following infection with avian leukosis virus (60). Tam et al. were the first to show that a region of the BIC RNA, now known to encode the miR-155 hairpin precursor, accelerated c-myc-associated lymphomagenesis in chickens (62). More recently, both BIC and miR-155 have been found to be highly expressed in several B-cell lymphomas as well as breast, lung, and colon cancers (28, 34, 64, 65). Additionally, transgenic mice overexpressing mmu-miR-155 from a B-cell-specific promoter exhibit splenomegaly and lymphopenia at early ages and develop high-grade B-cell neoplasms by 6 months of age (15). Together, these observations strongly suggest that miR-155 is an oncogenic miRNA. In humans, miR-155 expression can be detected in activated B and T cells (10, 23). Most recently, it was shown that a precise transient miR-155 expression burst is critically important for germinal-center reactions and B-cell development (54, 63). Thus, the dysregulation of miR-155 can have important implications in B-cell differentiation processes and may also be involved lymphomagenesis.
miR-155 is not expressed in primary effusion lymphomas, which express high levels of miR-K12-11.
Based on the identical seed sequences of miR-155 and miR-K12-11 and the association of KSHV with primary effusion lymphoma (PEL) and multicentric Castleman's disease (11), we hypothesized that miR-K12-11 and miR-155 might regulate a common set of cellular genes, thereby contributing to pathogenesis. First, we analyzed BIC expression in latently KSHV-infected PEL-derived cell lines by RT-PCR. EBV-positive Burkitt's lymphoma (BL) cells (RAJI) expressed readily detectable levels of BIC (Fig. 1C). In contrast, all KSHV-positive PEL lines tested did not express BIC (Fig. 1C) and did not express miR-155, as determined by Northern blot analysis (Fig. 1D). PEL cells do, however, highly express miR-K12-11, as shown in Fig. 1D and as previously reported (8, 51, 56). Hence, we hypothesized that miR-K12-11 could mimic miR-155-dependent regulation in PELs.
Generation of miR-155 and miR-K11 expression and sensor vectors.
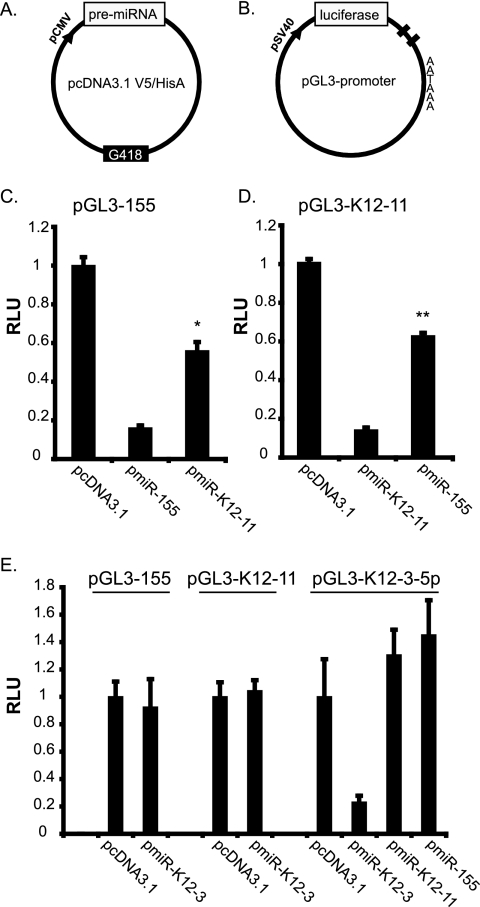
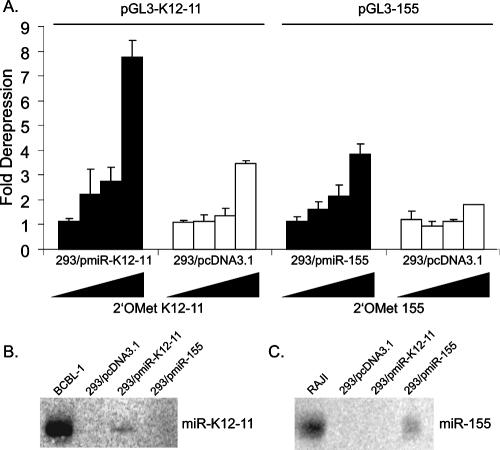
To test this idea, we generated pmiR-155 and pmiR-K12-11 expression vectors (Fig. 2A). To monitor miRNA expression from these constructs, miRNA sensor vectors (pGL3-155 and pGL3-K12-11) containing two antisense complementary binding sites within the 3′UTR of a luciferase reporter (Fig. 2B) were generated. Cotransfection of each miRNA expression vector with the respective sensor showed a 70% to 80% knockdown of luciferase expression (Fig. 2C and D, middle bars). Note that this inhibition reflects a small interfering RNA-like transcript-mediated effect since the miRNA and target 3′UTR are 100% complementary.
FIG. 2.
Importance of the seed sequence for miRNA targeting. (A) Schematic of miRNA expression vector. A region encompassing the pre-miRNA for either miR-155 or miR-K12-11 was PCR amplified and cloned downstream of the cytomegalovirus promoter in pcDNA3.1. (B) Schematic of miRNA sensor vector. Two antisense complementary binding sites for each miRNA were inserted into the 3′UTR of pGL3-Promoter (Promega) downstream of the luciferase gene. (C and D) Luciferase expression from miRNA sensors (pGL3-155 or pGL3-K12-11) is down-regulated in response to ectopic miRNA expression. 293 cells were transfected with 40 ng of the indicated sensor and 800 ng miRNA expression vector using Lipofectamine 2000 (Invitrogen). Lysates were analyzed at 72 h posttransfection (Promega). Relative light units (RLU) are normalized to total protein determined by BCA (Pierce). * indicates a P value of <0.01, and ** indicates a P value of <0.01 by Student's t test. (E) Luciferase assays to control for ectopic miRNA effects. miRNA expression and sensor vectors for KSHV-miR-K12-3-5p were cotransfected with plasmids as indicated in C and D.
Next, we tested whether miR-155 could target the miR-K12-11 sensor and vice versa. As shown in Fig. 2C and D (right bars), both miRNAs inhibited luciferase expression from both reporters approximately 50%, as expected for miRNA-dependent inhibition. Importantly, neither miR-155 nor miR-K12-11 had any effect on an unrelated sensor (pGL3-K12-3-5p), and an unrelated miRNA, miR-K12-3, did not affect either the miR-K12-11 or miR-155 sensors (Fig. 2E), validating the specificity of these assays. Hence, the importance of the miRNA seed for 3′UTR targeting is confirmed by the fact that both miR-155 and miR-K12-11 can inhibit luciferase expression from their opposite sensor vectors.
miR-155 and miR-K12-11 can target the BACH-1 3′UTR.
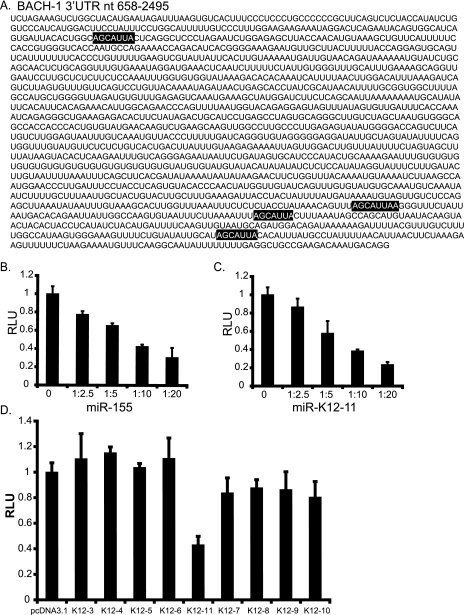
For miR-155, target genes associated with hematopoietic stem cell differentiation (19) and the angiotensin II type I receptor have been reported (42). No targets have been identified for KSHV miR-K12-11. Using three target prediction algorithms (miRanda, PicTar, and TargetScan) (17, 30, 35, 38), we identified BACH-1 (Btb and CNC homolog 1) as a candidate target for both miR-155 and miR-K12-11, as it contains four perfect seed match sites within its 3,315-nt-long 3′UTR (Fig. 3A). BACH-1 is a transcriptional regulator that binds NF-E2 sites and coordinates transcription with small Maf (musculoaponeurotic fibrosarcoma oncogene homolog) proteins (26a).
FIG. 3.
BACH-1 is targeted by miR-K12-11 and miR-155. (A) The BACH-1 3′UTR contains four seed match sites (highlighted) based on three target prediction programs (miRanda, PicTar, and TargetScan). (B and C) A region of the BACH-1 3′UTR (nt 658 to 2495) encompassing the predicted miRNA binding sites was cloned downstream of luciferase (pGL3-BACH1). 293 cells were cotransfected with 40 ng pGL3-BACH-1 and increasing amounts (100, 200, 400, and 800 ng) of pmiR-155 or pmiR-K12-11. Ratios on the x axis indicate the amount of miRNA vector to reporter. pcDNA3.1 was used as filler. Relative light units (RLU) are normalized to total protein determined by BCA (Pierce). (D) Additional KSHV miRNAs do not target the BACH-1 3′UTR. 293 cells were cotransfected with 40 ng pGL3-BACH-1, 10 ng pEF-RL (Renilla), and 800 ng of either pcDNA3.1 or KSHV miRNA expression vector. Light units are normalized to Renilla luciferase. For B to D, average data from three independent transfections are shown.
To examine whether BACH-1 can be regulated by miR-155 and/or miR-K12-11, we cotransfected pGL3-BACH-1, containing nt 658 to 2495 of the BACH-1 3′UTR inserted downstream of the luciferase gene, and increasing amounts of miR-155 or miR-K12-11 expression vectors. Luciferase expression was inhibited in a dose-dependent fashion, demonstrating that the BACH-1 3′UTR can be targeted by both miRNAs (Fig. 3B and C).
We next asked whether the additional KSHV miRNAs could target the BACH-1 3′UTR. Cotransfection of pGL3-BACH-1 with individual KSHV miRNA expression vectors (described in reference 57) revealed that of the nine different KSHV miRNAs tested, only miR-K12-11 targeted the BACH-1 3′UTR (Fig. 3D). To further confirm miRNA specificity, derepression assays utilizing miRNA antagomirs were performed. 2′OMe oligoribonucleotides antisense to miRNAs are potent inhibitors of RNA-induced silencing complex-associated miRNA activity (25, 45). Cotransfection of miR-155 or miR-K12-11 expression vectors with the BACH-1 reporter, in addition to an miRNA-specific antagomir, released the inhibitory miRNA effect on the reporter (data not shown). Together, these data validate the observed expression differences from the BACH-1 reporter as being miR155/miR-K12-11 dependent.
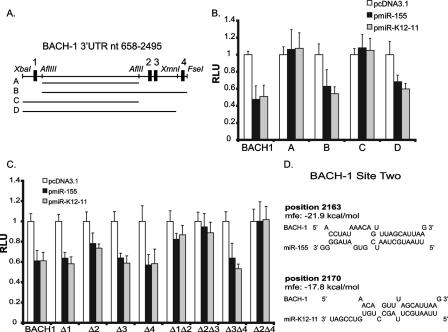
Seed match site 2 is essential for combinatorial regulation of BACH-1 by miR-155 and miR-K12-11.
To confirm that the observed BACH-1 inhibition depends on the presence of miR-155 and miR-K12-11 seed match sites, a mutational analysis of the BACH-1 3′UTR was performed. First, 3′UTR truncation mutants were generated by deleting one or more of the seed match sites (Fig. 4A). Deletion of all four sites or sites 2, 3, and 4 abrogated the miRNA-dependent inhibition, while deletion of site 1 only had no effect on luciferase expression (Fig. 4B), demonstrating that seed match sites 2, 3, and 4 within the BACH-1 3′UTR are likely targeted.
FIG. 4.
Seed match site 2 is essential for combinatorial regulation of BACH-1 by miR-155 and miR-K12-11. (A) Schematic of the BACH-1 3′UTR with potential miRNA seed match binding sites (sites 1 to 4) and restriction enzyme sites. “A” to “D” are truncation mutants generated by restriction enzyme digestion. (B) Mapping of miRNA binding sites. BACH-1 fragments A to D were inserted into pGL3-Promoter, and 40 ng of each reporter was cotransfected with 10 ng pEF-RL and 800 ng either pcDNA3.1, pmiR-155, or pmiR-K12-11 into 293 cells. (C) Site-directed mutagenesis to validate mapping data. Individual or double seed match sites within pGL3-BACH-1 were mutated to an XhoI site. Forty nanograms of each reporter was cotransfected with 10 ng pEF-RL and 800 ng miRNA expression vectors as described above (B). For B and C, lysates were harvested at 72 h, and relative light units (RLU) were normalized to Renilla luciferase. Shown are average data for five to eight independent transfections. (D) Potential miR-155 and miR-K12-11 binding to site 2 within the BACH-1 3′UTR (RNA-hybrid) (53). The binding position within the 3′UTR and the minimum free hybridization energy (mfe) for each miRNA are shown.
To confirm this, mutations of individual or multiple seed match sites were introduced into the BACH-1 3′UTR by site-directed mutagenesis. Mutagenesis of binding site 1, 3, or 4 alone had no effect on the miRNA-dependent inhibition of the reporter; however, a modest but significant increase in luciferase expression was observed after mutating binding site 2 alone (Fig. 4C). Analysis of double mutants showed that the deletion of site 2 in combination with any other site completely abrogated the effect of both miR-155 and miR-K12-11 on the BACH-1 reporter (Fig. 4C). In contrast, deletion of sites 3 and 4 together had no effect on luciferase expression (Fig. 4C). Together, these data demonstrate that site 2 in combination with either site 1, 3, or 4 is critically important for BACH-1 targeting.
Site 2 represents an 8-mer site for both miRNAs containing the seed match (nt 2 to 7) plus a match at position 8 and an A at position 1 (Fig. 4D) (38). Very recently, Grimson et al. reported a set of criteria that further contribute to the affinity by which an miRNA binds to target mRNAs. We note that the binding of miR-155/miR-K12-11 to site 2 within the BACH-1 3′UTR fulfills several of these criteria, which include an AU-rich environment surrounding the miRNA binding site (>70%), a seed sequence preceded and followed by an A (Fig. 4D), and the overall positioning within the periphery of the 3′UTR (Fig. 3A) (20).
Analysis of BACH-1 expression in PEL and BL cell lines.
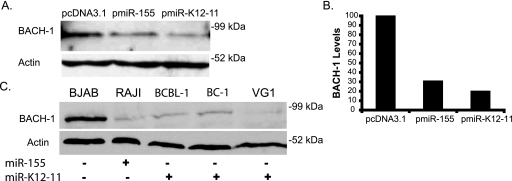
Following the identification of BACH-1 as a potential target using bioinformatic tools and the confirmation of its regulation by luciferase reporter assays, we asked whether both miRNAs can regulate endogenous BACH-1 protein levels in BJAB cells, an EBV- and KSHV-negative BL-cell line which does not express either miRNA (Fig. 1D). Western blot analysis revealed that the transient transfection of either miR-155 or miR-K12-11 expression vectors leads to 70% and 80% decreases in BACH-1 protein levels, respectively, compared to a control vector, further confirming BACH-1 as a target (Fig. 5A and B).
FIG. 5.
BACH-1 is down-regulated in miR-155- and miR-K12-11-expressing cells. (A) Transient expression of miR-155 or miR-K12-11 affects BACH-1 protein levels. BJAB cells were transfected with 2 μg miRNA expression vector or pcDNA3.1 control (nucleofection) (Amaxa, Inc.). Lysates were harvested at 72 h and probed for endogenous BACH-1 or actin. (B) Graph of BACH-1 levels normalized to actin (analyzed by NIH ImageJ) for transfected BJAB cells shown in A. (C) PEL cells expressing miR-K12-11 and BL cells expressing miR-155 express low amounts of BACH-1 protein. Lysates from 1 × 105 BJAB, RAJI, BCBL-1, BC-1, or VG-1 cells were probed for endogenous BACH-1 and actin.
Next, we asked whether the expression levels of BACH-1 coincide with miRNA expression in BL or PEL lines. miR-155 is expressed in EBV-positive BL RAJI cells, while miR-K12-11 is highly expressed in PEL cells (Fig. 1D). Western blot analysis revealed that both miR-155 (Raji)- and miR-K12-11 (BCBL-1, BC-1, and VG-1)-expressing cells expressed very low levels of BACH-1, while BJAB cells expressed high levels of BACH-1 (Fig. 5C). Together, these findings and the above-mentioned mutagenesis data demonstrate that both miRNAs target BACH-1 in lymphoid cells, which is relevant for the biology of KSHV.
Gene expression profiling reveals a common set of down-regulated genes in response to miRNA expression.
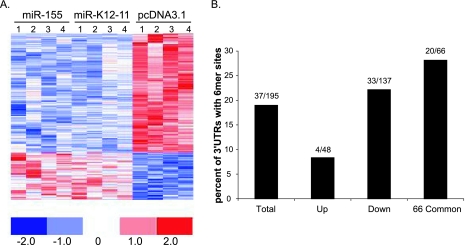
To investigate the effect of miR-155/miR-K12-11 expression on the cellular transcriptome, we generated 293 cells stably expressing miR-155, miR-K12-11, or the vector control (pcDNA3.1) and performed Affymetrix-based gene expression profiling. miRNA expression in 293/pmiR-155 and 293/pmiR-K12-11 cells was confirmed both by luciferase derepression assays using specific miRNA sensor vectors and antagomirs (Fig. 6A) and Northern blot analysis (Fig. 6B and C). To avoid the risk of analyzing integration events, we examined cell pools rather than individual cell clones. RNA was harvested from two independent cultures of each cell line at two time points. A total of 12 samples were used to synthesize cRNA and hybridized to Affymetrix U133 2.0 Plus Human GeneChips containing over 47,000 human probe sets. RNA extraction, quantification, cRNA synthesis, hybridization, and washing steps were performed as recommended by the manufacturer and as previ ously described (57). Figure 7A shows a heat map representing the expression profiles of genes significantly altered in the presence of miR-K12-11 or miR-155. In both 293/pmiR-155 and 293/pmiR-K12-11 cells, a total of 195 annotated genes, represented by 208 probe sets, were altered compared to the vector control. Of those genes altered, we found 137 genes (70%) to be down-regulated. A cross-validation analysis showed that of these 137 genes, 78 exhibited a cross-validation of 75% or greater. This analysis, based on t values of signal intensities, gives statistically robust data while including probe sets with relatively small changes (n-fold). A common set of 66 genes was significantly down-regulated between −1.11- and −11.05-fold in response to both miR-155 and miR-K12-11 (see Table S1 in the supplemental material).
FIG. 6.
miRNA expression in stable cell lines. (A) Luciferase derepression assays were performed using 293/pmiR-K12-11, 293/pmiR-155, or 293/pcDNA3.1 cells using 40 ng of the indicated sensor vector and increasing amounts (20 to 80 pmol) of a 2′OMe antagomir specific to the miRNA of interest. 2′OMe K12-10 and 100 ng of pcDNA3.1 were used as filler. Light units are normalized to total protein by BCA (Pierce). (B and C) Northern blot analysis of miRNA expression in stable cell lines. Thirty micrograms of total RNA was loaded per lane and hybridized to a probe for either miR-K12-11 (B) or miR-155 (C). RNA from BCBL-1 and RAJI cells was used as a positive control.
FIG. 7.
Gene expression profiling reveals a common set of target genes. (A) Genes are down-regulated in response to miRNA expression. Colors represent changes in variance-normalized gene expression differences for individual genes represented by the probe sets as indicated on the color scale. Four samples were used for each cell line. One hundred ninety-five genes were changed in response to miRNA expression: 137 genes were down-regulated, and 48 were up-regulated. Sixty-four genes showed >75% cross-validation. (B) The 3′UTRs of down-regulated genes are enriched for seed match sites. 3′UTRs of altered genes were scanned for seed match sites. Shown is the percentage of 3′UTRs with seed match sites for either the total number of altered genes (total), up-regulated (up) or down-regulated (down) genes, or the 66 commonly down-regulated genes (66 common). Thirty-three out of 137 down-regulated genes contained 6-mer seed match sites within their 3′UTRs, while 20 out of the 66 commonly down-regulated genes contained seed match sites.
Down-regulated genes contain potential seed match binding sites.
Using a previously published ad hoc miRNA seed match scanning algorithm (57), we examined the 3′UTRs of those genes exhibiting altered expression patterns in response to miR-155 or miR-K12-11 for potential miRNA binding sites. Of the 137 genes found to be down-regulated in both 293/pmiR-155 and 293/pmiR-K12-11 cells, 33 3′UTRs (24%) contained 6-mer (nt 2 to 7) sites, including 13 7-mer (nt 2 to 8) sites, for both miRNAs. Within the set of 66 genes commonly down-regulated by both miRNAs, 20 3′UTRs (30%) contained seed match sites (Fig. 7B). Importantly, within the 48 genes that were up-regulated in the presence of miRNA expression, only 4 (8%) contained seed match sites, further suggesting that the list of down-regulated genes is significantly enriched for miRNA binding sites (Fig. 7B).
Following the identification of these new potential miRNA targets, we examined pathways affected by miRNA expression using Biocarta. The majority of pathways included cell signaling, cell division, and T-cell activation. Additionally, we identified several genes with known roles in apoptosis, such as LDOC1, PHF17, BCL6B, BCLAF1, and BCL2L11 (27, 46). The significance and potential roles of these genes in KSHV biology will be discussed below.
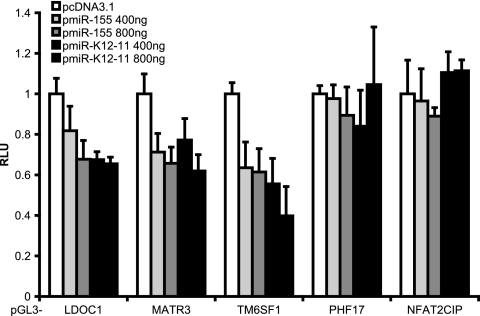
Luciferase reporter assays confirm miRNA targeting of LDOC1, MATR3, and TM6SF1.
Based on the bioinformatic data, we chose five genes for further examination and cloned their 3′UTRs into pGL3-Promoter downstream of the luciferase gene. Reporter constructs were cotransfected into 293 cells with pmiR-155, pmiR-K12-11, or pcDNA3.1 as a control. We observed a significant inhibition of luciferase from reporters containing the 3′UTRs of LDOC1, MATR3, and TM6SF1 in the presence of miR-155 or miR-K12-11 (Fig. 8). In contrast, neither miRNA had any effect on the PHF17 or NFAT2CIP 3′UTRs (Fig. 8). Thus, these assays confirmed LDOC1, MATR3, and TM6SF1 as being targets of both miR-155 and miR-K12-11.
FIG. 8.
Validation of candidate target genes. The 3′UTRs of LDOC1, MATR3, TM6SF1, PHF17, and NFAT2CIP contained at least one seed match site and were cloned into pGL3-Promoter downstream of luciferase. 3′UTR reporters were cotransfected into 293 cells with 10 ng pEF-RL and either 800 ng pcDNA3.1 or the indicated amounts of miRNA expression vector to determine miRNA targeting. Four hundred nanograms of pcDNA3.1 was used as filler for the transfection of 400 ng miRNA expression vector. Lysates were harvested at 72 h, and relative light units (RLU) were normalized to Renilla luciferase. For pGL3-LDOC1, pGL3-MATR3, and pGL3-TM6SF1, a 1.5- to 2.5-fold decrease in luciferase was observed with miR-155 and miR-K12-11 expression. Shown are average data from three to six independent transfections.
DISCUSSION
The goal of this study was to determine whether the known oncogenic miRNA hsa-miR-155 and the tumor virus-encoded miRNA miR-K12-11, which both contain identical seed match sequences, can regulate a common set of target genes and, therefore, might target similar regulatory pathways. Using a combination of bioinformatic tools and gene expression profiling, we identified 66 genes that are potentially targeted by both miRNAs. A total of 30% of these common targets contained seed match sites within their 3′UTRs, and reporter assays verified that three genes, LDOC1, MATR3, and TM6SF1, were regulated by both miRNAs (Fig. 8). Additional targets are currently under study. miR-K12-11 is the first example of a DNA tumor virus-encoded miRNA that regulates targets similar to those of its host counterpart.
Interestingly, this is not an anomaly, since alignments revealed additional herpesvirus-encoded miRNAs that share seed sequence homology with human miRNAs (Fig. 1A). KSHV miR-K12-6-5p has homology to miR-15a and miR-16, both associated with tumor suppressor activity (13), while EBV BART5 has homology to miR-18, a member of the oncogenic miR-17-92 cluster (24). This suggests that a subset of herpesvirus miRNAs represent cellular homologues that have arisen either by capturing host miRNA genes or through coevolution with their hosts. The fact that the pre-miRNAs of miR-155 and miR-K12-11 are vastly different would suggest coevolution similar to precursors of some let-7 miRNA family members, who share a common seed sequence and can target similar 3′UTRs (1, 31) (www.microrna.sanger.ac.uk).
By utilizing reporter assays and detailed mutational analyses, we found that both miR-155 and miR-K12-11 target seed match sites within the BACH-1 3′UTR (Fig. 3 and 4). In contrast to these reporter assays, the RNA levels of BACH-1 were not significantly affected in miRNA-expressing 293 cells as determined by expression profiling. Importantly, BACH-1 protein levels were down-regulated in BJAB cells transiently transfected with miRNA expression vectors (Fig. 5A). Furthermore, we observed low levels of BACH-1 in PEL or BL lines expressing both miRNAs compared to BJAB cells (Fig. 5C). Additionally, BACH-1 mRNA levels are down-regulated in latently KSHV-infected endothelial cells (4). BACH-1 is a broadly expressed transcriptional repressor, which regulates genes involved in the hypoxia response, such as heme-oxygenase 1 (HMOX1) (26a). Increased levels of HMOX1, which enhance cell survival and proliferation, have been reported following de novo KSHV infection of endothelial cells (44). While the increase in HMOX1 has been attributed in part to the KSHV G-protein-coupled receptor (41), the observed down-regulation of BACH-1 may be partially due to miR-K12-11 expression during latency.
The seed sequence homology of these two naturally occurring miRNA orthologs also provides an opportunity to analyze the contribution of sequences outside the seed match to mRNA targeting and target site selection. A detailed analysis of the BACH-1 3′UTR demonstrated that both miR-155 and miR-K12-11 preferentially utilized seed match site 2 for targeting; however, sites 3 and 4 also contributed (Fig. 4B and C). Further analysis of the 66 down-regulated genes using miRanda showed that >30% of 3′UTRs are predicted to contain miR-155 or miR-K12-11 binding sites, a majority of which (>50%) contain exact seed match sites (not shown) (17). Very recently, the Bartel laboratory published a new model outlining additional determinants of miRNA target site selection for more accurate target prediction (20). Analysis of the four seed match sites within the BACH-1 3′UTR revealed that target site 2, which conveyed miRNA-dependent inhibition (Fig. 4C), fulfilled several of those new criteria. Hence, a detailed analysis of additional targets will also aid a better understanding of how sequences outside the seed sequence contribute to miRNA targeting.
The biological consequences of these newly identified miRNA targets are currently under investigation. Using Biocarta, we found that the majority of pathways affected by genes that changed in response to miRNA expression included cell signaling, cell division, apoptosis, and T-cell activation. Additionally, we identified several miR-155- and miR-K12-11-responsive genes known to be aberrantly expressed in human malignancies. These include LDOC1, a regulator of apoptosis (27, 46), as well as several Bcl-2 and Bcl-6 family members (BCL2L11, BCL6B, and BCLAF1).
BCL2L11, known as Bim, inactivates the Bcl-2-like proteins, thereby inducing apoptosis (2, 14). BCLAF1 (Bcl-2 associated transcription factor 1 [Btf]) has been shown to interact with the antiapoptotic Bcl-2 and Bcl-xL family members and, when overexpressed, to induce apoptosis (32). BCL6B, a transcriptional repressor also known as BAZF, is an NF-κB regulator and homolog of the Bcl-6 oncogene; however, unlike Bcl-6, which is ubiquitously expressed, BAZF is expressed predominantly in the heart, lung, and, importantly, stimulated lymphocytes (49). Both Bim and BAZF have roles in cellular immunity. Bim is required for the apoptosis of activated T cells following viral infection, which is critical for limiting antiviral immune responses (50). BAZF is required for secondary responses of memory CD8+ T cells (40) and was recently linked to the homeostasis of hematopoietic progenitors (7). Thus, the observed down-regulation of these genes by miR-155 or miR-K12-11 suggests important roles in the regulation of immune responses to infection as well as in targeting apoptosis to regulate cell survival during lymphocyte differentiation. It is interesting that, to date, of the only ∼50 experimentally determined human miRNA targets, a significant proportion are regulators of apoptosis, such as targets of miR-15a and miR-16 (13). Furthermore, KSHV miRNAs have recently been reported to target apoptosis regulators (22, 57).
While there are only a few experimentally confirmed targets for miR-155, it was the first human miRNA suggested to be oncogenic (61) and was recently shown to be important for lymphocyte differentiation and immunity. miR-155 is expressed in activated macrophages following treatment with Toll-like receptor ligands, suggesting that miR-155 has a role in innate immunity (48). Recently, it was shown that BIC/miR-155 plays a significant role in B- and T-lymphocyte maturation (54, 63). BIC is absent in resting and progenitor B cells but induced upon activation; miR-155 is expressed shortly thereafter and regulates the germinal center reaction during B-cell maturation (63). The late stages of B-cell development, particularly the affinity maturation in conjunction with helper T-cell signaling, are critically dependent upon a timely miR-155 expression whereby both the induction and down-regulation of miR-155 are important (63).
Intriguingly, KSHV-associated PELs are of B-cell lineage and have a distinct developmental phenotype. PELs have rearranged immunoglobulin genes as well as somatic point mutations within rearranged immunoglobulin variable genes (18, 43), indicating that B-cell activation during the germinal center reaction has occurred. Accordingly, the PEL developmental phenotype was classified as plasmablastic (33), and expression profiling studies suggest that PEL represent B cells blocked late in their differentiation pathway towards an antibody-secreting plasma cell (29). We showed that PEL cells do not express miR-155 but do express high levels of miR-K12-11 (Fig. 1D). Exactly some of these late steps seem to be dependent upon a short miR-155 expression burst during normal B-cell development (63).
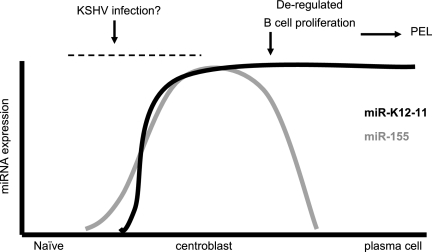
Based on our data demonstrating common targets for both miRNAs, we propose a model in which sustained miR-K12-11 expression continues to regulate a set of miR-155 targets at a stage when miR-155 is not expressed and thereby directly contributes to dysregulated nonmature B-cell proliferation and, potentially, lymphomagenesis (Fig. 9). Clearly, this hypothesis will have to be tested in appropriate in vivo models, such as NOD/SCID mice, that allow the study of human B-cell maturation.
FIG. 9.
Model of a potential role for miR-K12-11 in lymphomagenesis. Germinal-center-dependent B-cell maturation is dependent on a precise miR-155 expression burst whereby both induction and shutoff are equally important (54, 63). In contrast, after naïve B cells are infected with KSHV, sustained miR-K12-11 expression could contribute to proliferation in the absence of terminal differentiation into a mature antibody-producing B cell, a phenotype which is congruent with that characterized for PEL cells (29).
In conclusion, our data show that the oncogenic human miRNA miR-155 and the virally encoded KSHV miRNA miR-K12-11 can regulate a common set of target genes and that miR-K12-11 is an ortholog of miR-155. The recent link of miR-155 to B-cell maturation suggests a similar role for miR-K12-11, which may contribute directly to KSHV pathogenesis.
Finally, we found a total of four viral miRNAs (Fig. 1A) harboring seed match sequence identity to cellular miRNAs. Most importantly, three of these are potential orthologs of human miRNAs known to have either oncogenic or tumor suppressor activity. Hence, these observations strongly suggest that DNA tumor viruses have utilized molecular mimicry of miRNA genes to regulate host cellular environments in the same fashion as the long list of homologous cytokines, growth factors, and immunomodulatory genes commonly found in DNA tumor virus genomes (47, 55).
Supplementary Material
Acknowledgments
This work was supported by University of Florida Shands Cancer Center start-up funds and by a Bankhead-Coley Florida Biomedical Research grant to R.R. R.L.S. was supported by a National Cancer Institute Training Grant in Cancer Biology (5T32CA009126-30), and M.A.S. was supported by National Institutes of Health T32 training grant GM07250 at Case Western Reserve University.
Footnotes
Published ahead of print on 19 September 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abbott, A. L., E. Alvarez-Saavedra, E. A. Miska, N. C. Lau, D. P. Bartel, H. R. Horvitz, and V. Ambros. 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9:403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, J. M., and S. Cory. 2007. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26:1324-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 4.An, F. Q., H. M. Folarin, N. Compitello, J. Roth, S. L. Gerson, K. R. McCrae, F. D. Fakhari, D. P. Dittmer, and R. Renne. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 80:4833-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke, J., A. Stark, R. B. Russell, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PLoS Biol. 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broxmeyer, H. E., S. Sehra, S. Cooper, L. M. Toney, S. Kusam, J. J. Aloor, C. C. Marchal, M. C. Dinauer, and A. L. Dent. 2007. Aberrant regulation of hematopoiesis by T cells in BAZF-deficient mice. Mol. Cell. Biol. 27:5275-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, T. B., M. Borok, I. E. White, I. Gudza, B. Ndemera, A. Taziwa, A. Weinberg, and L. Gwanzura. 2003. Relationship of Kaposi sarcoma (KS)-associated herpesvirus viremia and KS disease in Zimbabwe. Clin. Infect. Dis. 36:1144-1151. [DOI] [PubMed] [Google Scholar]
- 11.Cesarman, E. 2002. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Recent Results Cancer Res. 159:27-37. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 13.Cimmino, A., G. A. Calin, M. Fabbri, M. V. Iorio, M. Ferracin, M. Shimizu, S. E. Wojcik, R. I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 102:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cory, S., D. C. Huang, and J. M. Adams. 2003. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22:8590-8607. [DOI] [PubMed] [Google Scholar]
- 15.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C. M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 103:7024-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38:S25-S30. [DOI] [PubMed] [Google Scholar]
- 17.Enright, A. J., B. John, U. Gaul, T. Tuschl, C. Sander, and D. S. Marks. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fais, F., G. Gaidano, D. Capello, A. Gloghini, F. Ghiotto, S. Roncella, A. Carbone, N. Chiorazzi, and M. Ferrarini. 1999. Immunoglobulin V region gene use and structure suggest antigen selection in AIDS-related primary effusion lymphomas. Leukemia 13:1093-1099. [DOI] [PubMed] [Google Scholar]
- 19.Georgantas, R. W., III, R. Hildreth, S. Morisot, J. Alder, C. G. Liu, S. Heimfeld, G. A. Calin, C. M. Croce, and C. I. Civin. 2007. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA 104:2750-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimson, A., K. K. Farh, W. K. Johnston, P. Garrett-Engele, L. P. Lim, and D. P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, A., J. J. Gartner, P. Sethupathy, A. G. Hatzigeorgiou, and N. W. Fraser. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 442:82-85. [DOI] [PubMed] [Google Scholar]
- 23.Haasch, D., Y. W. Chen, R. M. Reilly, X. G. Chiou, S. Koterski, M. L. Smith, P. Kroeger, K. McWeeny, D. N. Halbert, K. W. Mollison, S. W. Djuric, and J. M. Trevillyan. 2002. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell. Immunol. 217:78-86. [DOI] [PubMed] [Google Scholar]
- 24.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutvagner, G., M. J. Simard, C. C. Mello, and P. D. Zamore. 2004. Sequence-specific inhibition of small RNA function. PLoS Biol. 2:E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang, H. W., and J. T. Mendell. 2006. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94:776-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Igarashi, K., and J. Sun. The heme-BACH1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 8:107-118. [DOI] [PubMed]
- 27.Inoue, M., K. Takahashi, O. Niide, M. Shibata, M. Fukuzawa, and C. Ra. 2005. LDOC1, a novel MZF-1-interacting protein, induces apoptosis. FEBS Lett. 579:604-608. [DOI] [PubMed] [Google Scholar]
- 28.Iorio, M. V., M. Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65:7065-7070. [DOI] [PubMed] [Google Scholar]
- 29.Jenner, R. G., K. Maillard, N. Cattini, R. A. Weiss, C. Boshoff, R. Wooster, and P. Kellam. 2003. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 100:10399-10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human MicroRNA targets. PLoS Biol. 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120:635-647. [DOI] [PubMed] [Google Scholar]
- 32.Kasof, G. M., L. Goyal, and E. White. 1999. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 19:4390-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein, U., A. Gloghini, G. Gaidano, A. Chadburn, E. Cesarman, R. Dalla-Favera, and A. Carbone. 2003. Gene expression profile analysis of AIDS-related primary effusion lymphoma (PEL) suggests a plasmablastic derivation and identifies PEL-specific transcripts. Blood 101:4115-4121. [DOI] [PubMed] [Google Scholar]
- 34.Kluiver, J., S. Poppema, D. de Jong, T. Blokzijl, G. Harms, S. Jacobs, B. J. Kroesen, and A. van den Berg. 2005. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 207:243-249. [DOI] [PubMed] [Google Scholar]
- 35.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495-500. [DOI] [PubMed] [Google Scholar]
- 36.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 37.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735-739. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15-20. [DOI] [PubMed] [Google Scholar]
- 39.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115:787-798. [DOI] [PubMed] [Google Scholar]
- 40.Manders, P. M., P. J. Hunter, A. I. Telaranta, J. M. Carr, J. L. Marshall, M. Carrasco, Y. Murakami, M. J. Palmowski, V. Cerundolo, S. M. Kaech, R. Ahmed, and D. T. Fearon. 2005. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc. Natl. Acad. Sci. USA 102:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marinissen, M. J., T. Tanos, M. Bolos, M. R. de Sagarra, O. A. Coso, and A. Cuadrado. 2006. Inhibition of heme oxygenase-1 interferes with the transforming activity of the Kaposi sarcoma herpesvirus-encoded G protein-coupled receptor. J. Biol. Chem. 281:11332-11346. [DOI] [PubMed] [Google Scholar]
- 42.Martin, M. M., E. J. Lee, J. A. Buckenberger, T. D. Schmittgen, and T. S. Elton. 2006. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J. Biol. Chem. 281:18277-18284. [DOI] [PubMed] [Google Scholar]
- 43.Matolcsy, A., R. G. Nador, E. Cesarman, and D. M. Knowles. 1998. Immunoglobulin VH gene mutational analysis suggests that primary effusion lymphomas derive from different stages of B cell maturation. Am. J. Pathol. 153:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McAllister, S. C., S. G. Hansen, R. A. Ruhl, C. M. Raggo, V. R. DeFilippis, D. Greenspan, K. Fruh, and A. V. Moses. 2004. Kaposi sarcoma-associated herpesvirus (KSHV) induces heme oxygenase-1 expression and activity in KSHV-infected endothelial cells. Blood 103:3465-3473. [DOI] [PubMed] [Google Scholar]
- 45.Meister, G., M. Landthaler, Y. Dorsett, and T. Tuschl. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizutani, K., D. Koike, S. Suetsugu, and T. Takenawa. 2005. WAVE3 functions as a negative regulator of LDOC1. J. Biochem. (Tokyo) 138:639-646. [DOI] [PubMed] [Google Scholar]
- 47.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell, R. M., K. D. Taganov, M. P. Boldin, G. Cheng, and D. Baltimore. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okabe, S., T. Fukuda, K. Ishibashi, S. Kojima, S. Okada, M. Hatano, M. Ebara, H. Saisho, and T. Tokuhisa. 1998. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol. Cell. Biol. 18:4235-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pellegrini, M., P. Bouillet, M. Robati, G. T. Belz, G. M. Davey, and A. Strasser. 2004. Loss of Bim increases T cell production and function in interleukin 7 receptor-deficient mice. J. Exp. Med. 200:1189-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 52.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 53.Rehmsmeier, M., P. Steffen, M. Hochsmann, and R. Giegerich. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez, A., E. Vigorito, S. Clare, M. V. Warren, P. Couttet, D. R. Soond, S. van Dongen, R. J. Grocock, P. P. Das, E. A. Miska, D. Vetrie, K. Okkenhaug, A. J. Enright, G. Dougan, M. Turner, and A. Bradley. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samols, M. A., J. Hu, R. L. Skalsky, A. M. Maldonado, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samols, M. A., and R. Renne. 2006. Virus-encoded microRNAs: a new chapter in virus-host cell interaction. Future Virol. 1:233-242. [Google Scholar]
- 59.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 20:3-7. [DOI] [PubMed] [Google Scholar]
- 60.Tam, W., D. Ben-Yehuda, and W. S. Hayward. 1997. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol. Cell. Biol. 17:1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tam, W., and J. E. Dahlberg. 2006. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer 45:211-212. [DOI] [PubMed] [Google Scholar]
- 62.Tam, W., S. H. Hughes, W. S. Hayward, and P. Besmer. 2002. Avian bic, a gene isolated from a common retroviral site in avian leukosis virus-induced lymphomas that encodes a noncoding RNA, cooperates with c-myc in lymphomagenesis and erythroleukemogenesis. J. Virol. 76:4275-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thai, T. H., D. P. Calado, S. Casola, K. M. Ansel, C. Xiao, Y. Xue, A. Murphy, D. Frendewey, D. Valenzuela, J. L. Kutok, M. Schmidt-Supprian, N. Rajewsky, G. Yancopoulos, A. Rao, and K. Rajewsky. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604-608. [DOI] [PubMed] [Google Scholar]
- 64.van den Berg, A., B. J. Kroesen, K. Kooistra, D. de Jong, J. Briggs, T. Blokzijl, S. Jacobs, J. Kluiver, A. Diepstra, E. Maggio, and S. Poppema. 2003. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer 37:20-28. [DOI] [PubMed] [Google Scholar]
- 65.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.